Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice
By combining proteomic and biochemical approaches with live cell imaging including FLIM-FRET and FRAP, Lamond and colleagues provide evidence for a direct involvement of hnRNP M in the spliceosome machinery through its interaction with the hCDC5L/hPLRG1 subcomplex. These results provide new insight into the mechanism whereby hnRNP M can modulate both 5′ and 3′ splice site choice.
Keywords: alternative pre-mRNA splicing, FLIM–FRET, CDC5L, hnRNP-M, spliceosome
Abstract
Heterogeneous nuclear ribonucleoprotein-M (hnRNP-M) is an abundant nuclear protein that binds to pre-mRNA and is a component of the spliceosome complex. A direct interaction was detected in vivo between hnRNP-M and the human spliceosome proteins cell division cycle 5-like (CDC5L) and pleiotropic regulator 1 (PLRG1) that was inhibited during the heat-shock stress response. A central region in hnRNP-M is required for interaction with CDC5L/PLRG1. hnRNP-M affects both 5′ and 3′ alternative splice site choices, and an hnRNP-M mutant lacking the CDC5L/PLRG1 interaction domain is unable to modulate alternative splicing of an adeno-E1A mini-gene substrate.
Introduction
The spliceosome comprises small nuclear ribonucleoprotein (snRNP) subunits, and additional protein splicing factors (Wahl et al, 2009), including the conserved cell division cycle 5-like (CDC5L) complex (called the nineteen complex NTC in yeast), which is essential for spliceosome assembly and catalysis. The human CDC5L and human pleiotropic regulator 1 (PLRG1) proteins are ‘core' components of this complex and are directly associated with and required for the first catalytic step of pre-mRNA splicing (Ajuh et al, 2000; Makarova et al, 2004).
Pre-mRNAs associate co-transcriptionally with heterogeneous nuclear ribonucleoproteins (hnRNPs; Dreyfuss et al, 1993). Many hnRNP proteins modulate alternative splicing of pre-mRNAs and affect the fate of heterogeneous nuclear RNAs by influencing their structure and/or by facilitating or hindering the interaction of their sequences with other pre-mRNA processing factors (Martinez-Contreras et al, 2007). hnRNP-M is an abundant component of human hnRNP complexes that can influence pre-mRNA splicing by regulating its own pre-mRNA splicing (Hase et al, 2006) or by affecting the regulation of alternative splicing of fibroblast growth factor receptor 2 (Hovhannisyan & Carstens, 2007). An antibody against hnRNP-M inhibited splicing in vitro, and heat-shock-induced inhibition of splicing was associated with a loss of hnRNP-M from spliceosomal complexes (Gattoni et al, 1996). Proteomic analyses of in vitro purified spliceosomes detected hnRNP-M in the pre-spliceosomal H-complex and throughout the spliceosome assembly (Rappsilber et al, 2002; Wahl et al, 2009). The interactions of hnRNP-M with other components of the spliceosome machinery remains unclear.
In this study, we combine a large-scale proteomic analysis with biochemical approaches and live-cell imaging, including Förster resonance energy transfer–fluorescence lifetime imaging microscopy (FLIM–FRET) and fluorescence recovery after photobleaching (FRAP), to analyse interactions between hnRNP-M and the pre-mRNA splicing machinery. We show that hnRNP-M interacts directly in vivo with CDC5L and PLRG1, and that hnRNP-M can modulate both 5′ and 3′ alternative splicing choices.
Results And Discussion
A comparative proteomic analysis of CDC5L and SKIP-containing complexes isolated from HeLa nuclear extracts was performed by immuno-affinity purification (IP) using CDC5L and SKIP antibodies, respectively (supplementary Fig S1A–E and supplementary Tables S2–S5 online). Mass spectrometric analysis showed that both complexes contained previously characterized CDC5L and SKIP interaction partners (Makarov et al, 2002) and additional proteins involved in RNA processing (supplementary Table S1A online), especially members of the hnRNP family. The hnRNP-M protein was abundant in both cases with 37 hnRNP-M peptides detected, that is, 47.1% of the final sequence coverage (supplementary Fig S1F–H online).
hnRNP-M–CDC5L complex interactions
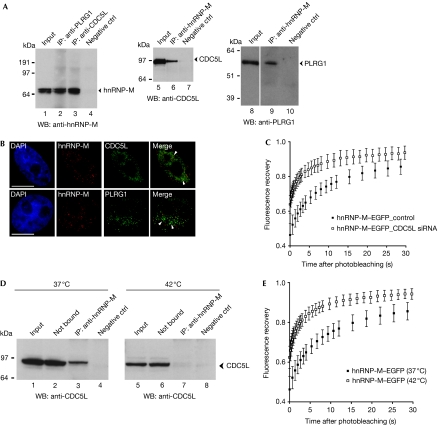
Antibodies directed against either CDC5L or PLRG1 proteins were used to immunoprecipitate proteins from whole-cell extracts. The isolated proteins were analysed by western blotting using an hnRNP-M antibody (Fig 1A). These results show specific co-IP of hnRNP-M, using both antibodies (Fig 1A, lanes 2 and 3). Reciprocal co-IP, using an hnRNP-M antibody, followed by detection with either CDC5L (Fig 1A, lane 6) or PLRG1 antibody (Fig 1A, lane 9) confirmed specific association of hnRNP-M with the CDC5L–PLRG1 complex. The IP protocols included treatment of extracts with both RNase-A and DNase-I, suggesting that association of hnRNP-M with these complexes might be specific. We infer that a subset of the pool of hnRNP-M proteins might be in complex with CDC5L–PLRG1 because less than 10% of the protein is recovered in the IP, relative to the input (Fig 1A, lanes 1–3).
Figure 1.
hnRNP-M interacts with components of the CDC5L complex. (A) Co-IP from total HeLa cell extracts using either anti-PLRG1 (lane 2) or anti-CDC5L (lane 3) and probed with anti-hnRNP-M (D2IF7). Co-IP with anti-hnRNP-M and blotted with anti-CDC5L (lane 6). Co-IP with anti-hnRNP-M and blotted with anti-PLRG1 (lane 9). (B) Co-immunofluorescence labelling was performed using anti-hnRNP-M with either anti-CDC5L (upper row) or anti-PLRG1 (lower row). Nuclei were stained with DAPI. Scale bar, 10 μm. (C) Mobility of hnRNP-M–GFP in control cells (scrambled short interfering RNA; black squares) and CDC5L-knockdown HeLa cells (CDC5L short interfering RNA; white squares) was monitored by FRAP. The relative fluorescence recovery kinetics are represented. Each data point shows the average±s.d. of more than 10 cells. (D) Heat shock. Co-IP using anti-hnRNP-M either at 37°C (left panel) or on heat shock for 1 h at 42°C (right panel) and probed with anti-CDC5L. hnRNP-M is associated with CDC5L under physiological conditions (lane 3) but not after heat shock (lane 7). (E) Mobility of hnRNP-M–GFP at 37°C (black squares) or on heat shock (white squares) was investigated by FRAP. The fluorescence recovery kinetics are shown. Each data point shows the average±s.d. of more than 10 cells. CDC5L, cell division cycle 5-like; DAPI, 4′,6-diamidino-2-phenylindole; FRAP, fluorescence recovery after photobleaching; GFP, green fluorescent protein; hnRNP, heterogeneous nuclear ribonucleoprotein; IP, immuno-affinity purification; PLRG1, pleiotropic regulator 1.
In vivo distribution of hnRNP-M was compared with both CDC5L and PLRG1 proteins by immunostaining experiments (Fig 1B, top and bottom rows, respectively). Both CDC5L and PLRG1 antibodies showed a speckled nuclear staining pattern as described previously (Ajuh et al, 2001). The hnRNP-M antibody instead showed a diffuse localization throughout the nucleoplasm, as seen previously (Carmo-Fonseca et al, 1991). Partial colocalization between hnRNP-M and both CDC5L and PLRG1 is observed in the diffuse area of the nucleoplasm, with little or no colocalization in the splicing speckles (Fig 1B, ‘Merge' arrowheads). The nuclear localization of hnRNP-M was compared with protein markers for various nuclear compartments (e.g. SC-35, Sm proteins and coilin; supplementary Fig S2A online) indicating that its association with CDC5L–PLRG1 is likely to occur within the diffuse nuclear compartment.
The diffuse nuclear compartment contains nascent transcripts that can be detected by transmission electron microscopy as perichromatin fibrils (Biggiogera & Fakan, 2008). Subnuclear distribution of both hnRNP-M and CDC5L proteins was analysed by immunoelectron microscopy by using CDC5L and hnRNP-M antibodies in thin sections of HeLa cell nuclei (supplementary Fig S2B online).
Immunolabelling of CDC5L was observed on the perichromatin fibrils, stained using the regressive EDTA technique (supplementary Fig S2B online, i, arrowheads), and also at the periphery of the interchromatin granule clusters (supplementary Fig S2B online, i, asterisk). Double labelling performed using hnRNP-M and CDC5L antibodies (black and white arrowheads, respectively) showed both proteins localized in tight proximity to a single RNA fibril, stained with either terbium citrate (supplementary Fig S2B online, ii) or by using the regressive EDTA technique (supplementary Fig S2B online, iii, iv). These data show the colocalization of hnRNP-M and CDC5L proteins on individual RNA fibrils. The mobility of green fluorescent protein (GFP)-tagged hnRNP-M wild type in control cells (scrambled short interfering RNA) and CDC5L-knockdown HeLa cells (CDC5L short interfering RNA) was monitored by FRAP. CDC5L knockdown caused an increase in the mobility of hnRNP-M (Fig 1C), suggesting it is regulated by interaction with the CDC5L protein.
The combination of fluorescence and electron microscopic data support the proteomic and co-IP data, indicating that a subset of hnRNP-M associates in vivo with complexes containing the CDC5L–PLRG1 proteins.
Heat shock disrupts hnRNP-M interactions
We tested whether hnRNP-M association with CDC5L and PLRG1 would be disrupted after heat shock, which inhibits transcription of most pre-mRNAs and blocks splicing (Bond, 1988; Biamonti, 2004). IPs were performed from both control (i.e. 37°C) and heat-shocked (1 h at 42°C) HeLa cell extracts using an hnRNP-M antibody. Isolated proteins were analysed with CDC5L antibody (Fig 1D). The CDC5L protein was specifically detected together with hnRNP-M at 37°C, but not at 42°C (Fig 1D, lanes 3 and 7). The levels of hnRNP-M show little or no change on heat shock (supplementary Fig S2C online). Loss of binding between hnRNP-M and the CDC5L complex after heat shock stress was addressed by FRAP experiments, showing increased mobility of hnRNP-M–EGFP after heat shock (Fig 1E). This suggests that the association of hnRNP-M with CDC5L is dependent on nascent pre-mRNA production.
In vivo hnRNP-M–CDC5L–PLRG1 interactions
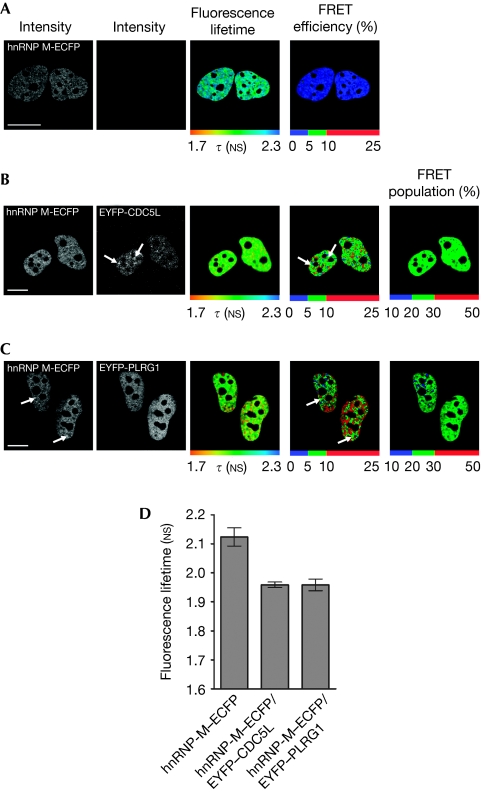
We investigated if hnRNP-M and CDC5L–PLRG1 proteins interact directly in vivo, by using FLIM–FRET. hnRNP-M–ECFP (enhanced cyan fluorescent protein) was transiently coexpressed in HeLa cells together with either enhanced yellow fluorescent protein (EYFP)-CDC5L or EYFP–PLRG1 proteins (Fig 2). As a positive control, FLIM–FRET assays were performed on HeLa cells coexpressing ECFP–PLRG1 and EYFP–CDC5L, which interact directly (supplementary Fig S3 online).
Figure 2.
hnRNP-M interacts directly with CDC5L and PLRG1 in vivo. HeLa cells were transfected with hnRNP-M–ECFP (A) alone or in combination with either (B) EYFP-CDC5L or (C) EYFP-PLRG1 and analysed using the FLIM–FRET technique. The fluorescence lifetime τ (NS) was detected using the TCSPC method, and its spatial distribution was analysed using the SPCimage software. Fluorescence lifetimes are depicted using a continuous pseudo-colour scale representing time values ranging from 1.7 to 2.3 NS. The FRET efficiency percentage is calculated and depicted using discrete colours representing FRET values ranging from 0 to 25%. The FRET species (%) was quantified for both hnRNP-M–CDC5L and hnRNP-M–PLRG1 interactions. Scale bars, 10 μm. (D) Statistical analysis from three independent experiments in at least 15 different cells for each condition is presented. CDC5L, cell division cycle 5-like; ECFP, enhanced cyan fluorescent protein; EYFP, enhanced yellow fluorescent protein; FLIM–FRET, Förster resonance energy transfer–fluorescence lifetime imaging microscopy; hnRNP, heterogeneous nuclear ribonucleoprotein; PLRG1, pleiotropic regulator 1; TCSPC, time-correlated single photon counting.
In contrast with cells expressing hnRNP-M–ECFP alone (Fig 2A), coexpression of either EYFP–CDC5L (Fig 2B) or EYFP–PLRG1 (Fig 2C) decreased the mean fluorescence lifetime of the donor ECFP, indicating a FRET interaction (Fig 2D).
As the FLIM–FRET assay measures the FRET efficiency for each pixel, inspection of the FRET efficiency distribution throughout the nucleus shows that the highest FRET values are predominantly in the diffuse nuclear domain with lower values in the speckles (Fig 2B,C, arrows indicate lower FRET). Further analysis of the fraction of the hnRNP-M–ECFP molecules showing a FRET interaction (Fig 2B,C, ‘FRET population %' panels) indicated that less than 30% of the total hnRNP-M protein pool was involved in this interaction.
As the CDC5L subcomplex is implicated in the activation of the second catalytic step of pre-mRNA splicing (Bessonov et al, 2008), the direct interactions between hnRNP-M and the CDC5L subcomplex detected by FLIM–FRET might correspond either to active sites of pre-mRNA splicing or to functional complexes that will be involved in splicing (House & Lynch, 2008). In combination with the electron microscopic and heat-shock data, this suggests that the interactions between hnRNP-M and both CDC5L and PLRG1 are either co-transcriptional or require nascent transcription.
Identification of interaction domain
The interaction between hnRNP-M and CDC5L/PLRG1 proteins was confirmed in vitro using a binding assay with in vitro translated hnRNP-M and Escherichia coli-expressed glutathione-S-transferase (GST)–CDC5L and GST–PLRG1 (supplementary Fig S4B,C online). No direct interaction was observed between GST–SPF30 and in vitro translated hnRNP-M (supplementary Fig S4B,C online, lane 2), consistent with previous data (Ajuh et al, 2001).
To map the region of hnRNP-M that binds directly to CDC5L and PLRG1 proteins, deletion mutants of hnRNP-M were made (supplementary Fig S4A online) and the size and level of expression were evaluated for each mutant (supplementary Fig S5A online). Full-length hnRNP-M and mutants 2 and 3 were pulled down by both GST–CDC5L (supplementary Fig S4B online, lanes 3, 5 and 6) and GST–PLRG1 (supplementary Fig S4C online, lanes 4, 6 and 7), respectively. By contrast, little or none of mutants 1, 4, 5 and 6 were pulled down with either GST–CDC5L (supplementary Fig S4B online, lanes 4 and 7; supplementary Fig S4D online, lanes 2 and 3) or with GST–PLRG1 (supplementary Fig S4C online, lanes 5 and 8; supplementary Fig S4D online, lanes 7 and 8). IP of cell extracts expressing YFP-mutant 1 and mutant 3 using an antibody against CDC5L showed that mutant 3, but not mutant 1, could be co-immunoprecipitated (supplementary Fig S5B online, lanes 6 and 7). Together, these results indicate that the central region of hnRNP-M, spanning amino acids 248–344, is required for binding to both CDC5L and PLRG1.
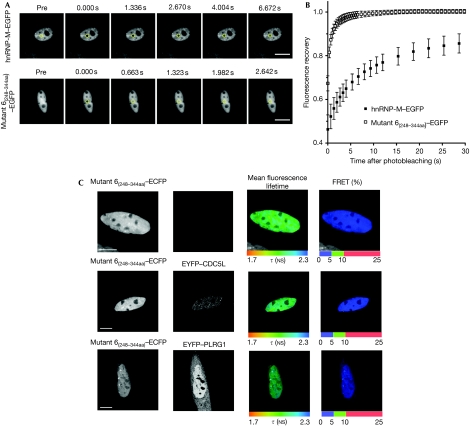
To test whether this binding region affects the properties of the hnRNP-M protein in vivo, both wild-type hnRNP-M and hnRNP-M mutant 6(248−344aa) protein (lacking the central CDC5L/PLRG1-binding region) were tagged with enhanced green fluorescent protein (EGFP), expressed in HeLa cells and analysed by FRAP (Fig 3A,B). These data show a difference in their rate of recovery from photobleaching, with mutant 6(248−344aa) showing more than 10-fold higher rate of fluorescence recovery as compared with wild-type hnRNP-M. Furthermore, mutant 6(248−344aa) shows a mobile fraction corresponding to essentially all the expressed protein, as compared with about 85% of the wild-type hnRNP-M protein.
Figure 3.
The nuclear dynamic of hnRNP-M–EGFP and hnRNP-M mutant 6(248−344aa)–EGFP as observed by FRAP. (A) A time-lapse series showing nucleoplasmic hnRNP-M–EGFP (top panels) and mutant 6–EGFP (bottom panels) FRAP. The photobleached area is outlined with yellow circles. (B) The kinetics of the relative fluorescence recovery of hnRNP-M–EGFP (black squares) and mutant 6–EGFP (white squares) are represented. The mobile fraction (%) and the half-life time recovery were calculated, respectively: hnRNP-M–EGFP, 84.4±5.3%, 6.40±0.04 s; mutant 6(248−344 aa)–EGFP, 100.0±3.2%, 0.47±0.02 s. Each data point shows the average±s.d. of more than 10 cells. Scale bars, 10 μm. (C) HeLa cells transfected with hnRNP-M mutant 6(248−344 aa)–ECFP alone (top row) or with either EYFP–CDC5L (middle row) or EYFP–PLRG1 (bottom row) were analysed by FLIM–FRET. Scale bars, 10 μm. CDC5L, cell division cycle 5-like; FRAP, fluorescence recovery after photobleaching; hnRNP, heterogeneous nuclear ribonucleoprotein; PLRG1, pleiotropic regulator 1.
The direct binding of hnRNP-M mutant 6(248−344aa) to both CDC5L and PLRG1 in vivo was analysed using the FLIM–FRET assay (Fig 3C). hnRNP-M mutant 6(248−344aa)–ECFP was transiently expressed in HeLa cells alone (Fig 3C, upper panels) or together with either EYFP–CDC5L (middle panels) or EYFP–PLRG1 (lower panels). Coexpression of the tagged CDC5L or PLRG1 proteins did not decrease the mean fluorescence lifetime of mutant 6(248−344aa)–ECFP, indicating no FRET and therefore no protein–protein interaction. In agreement, no difference in the mobility of the mutant 6(248−344aa)–EGFP was observed by FRAP after heat shock (supplementary Fig S6 online).
Together, the data are consistent with hnRNP-M wild-type but not mutant 6(248−344aa) protein, interacting directly with the CDC5L and PLRG1 proteins in vivo and thereby reducing its mobility. The FRAP data indicate roughly 10–20% of the tagged hnRNP-M engaged in the splicing machinery in vivo, consistent with the binding fraction of less than 30% estimated by FLIM–FRET.
hnRNP-M is involved in alternative splice site choice
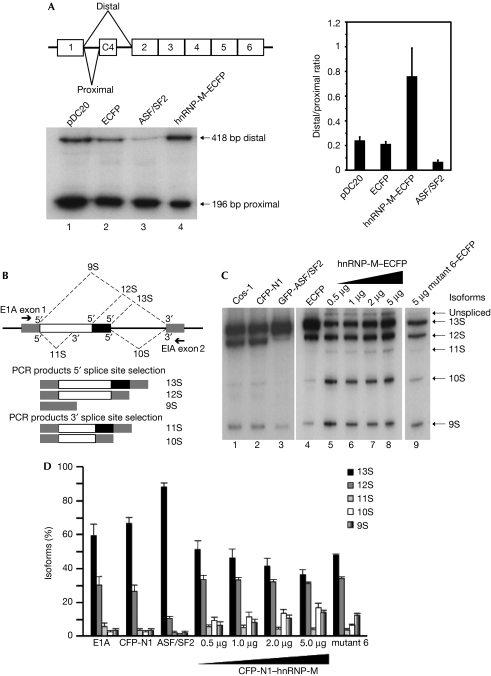
We investigated whether the direct interactions of hnRNP-M with CDC5L and PLRG1 were important for pre-mRNA splicing activity. As hnRNP-M was reported to promote exon skipping and exon inclusion (Hovhannisyan & Carstens, 2007), we analysed the ability of wild-type hnRNP-M to affect alternative splice site choice on two separate pre-mRNA substrates (Fig 4). By using expression of calcitonin/dhfr pre-mRNA from pDC20 reporter (Bai et al, 1999), the effect of increasing in vivo levels of hnRNP-M by transient coexpression in HeLa cells was analysed (Fig 4A) and compared with overexpression of the ASF/SF2 protein. Overexpression of exogenous hnRNP-M activated the distal 3′ splice site, increasing the level of the 418-bp band (Fig 4A, lane 4). By contrast, overexpression of ASF/SF2 protein promoted preferential use of the proximal 3′ splice site, increasing the ratio of the 196/418-bp bands (Fig 4A, compare lane 3 with lanes 1 and 4). These data show that increased expression of hnRNP-M has the opposite effect on alternative 3′ splice site choice to ASF/SF2 (Fig 4A, graph).
Figure 4.
hnRNP-M is involved in alternative splice site choice through its CDC5L/PLRG1-binding region. (A) Representation of pDC20 mini-gene containing the alternative 3′ splice sites. RT–PCR analysis of HeLa cells co-transfected with pDC20 and either the ECFP-N1 vector (lane 2) or hnRNP-M–ECFP (lane 4). Overexpression of hnRNP-M activates the distal 3′ splice site choice of the chimeric calcitonin/dhfr pre-mRNA. The relative distal-to-proximal 3′ splice site ratio was quantified (right panel). (B) Representation of E1A reporter mini-gene containing the alternative 5′ splice sites. The arrows indicate the positions of the exon primers used for RT–PCR. (C) Analysis of cells co-transfected with E1A and ECFP-N1 vector (lanes 2 and 4) or a gradient of hnRNP-M–ECFP (lanes 5–8) or the mutant 6(248−344aa)–ECFP (lane 9). (D) The relative amounts of the various isoforms were quantified. CDC5L, cell division cycle 5-like; ECFP, enhanced cerculean fluorescent protein; GFP, green fluorescent protein; hnRNP, heterogeneous nuclear ribonucleoprotein; PLRG1, pleiotropic regulator 1; RT–PCR, reverse transcription–PCR.
A more complex effect on alternative splice site choice is provided by the E1A mini-gene reporter, in which five distinct RNA isoforms can be generated by alternative splicing (Fig 4B). As previously observed (Caceres et al, 1994), overexpression of ASF/SF2 protein strongly activated the 13S 5′ splice site, enhancing the levels of the 13S RNA isoform and reducing the levels of 12S and 10S RNAs (Fig 4C, lane 3). By contrast, overexpression of hnRNP-M activated the most distal 5′ splice site, increasing the levels of the 9S RNA isoform and decreasing the levels of the 13S isoform, with little or no change in the 12S isoform (Fig 4C, lanes 5–8, and Fig 4D). The effect of hnRNP-M was concentration-dependent. This indicates that hnRNP-M facilitates preferential use of the distal over the proximal 5′ splice site in E1A pre-mRNA splicing. Our data are consistent with previous evidence that hnRNP-M can regulate splicing in metazoans. Furthermore, studies have shown that Hrp59, the homologue of hnRNP-M in Drosophila melanogaster, can regulate the splicing of its own transcript as well as that of another transcript by acting as a negative regulator of splicing (Hase et al, 2006).
Overexpression of hnRNP-M caused an enhancement in the levels of the 10S RNA isoform (Fig 4C,D, compare lanes 5–8 with lane 4), indicating stimulation of the distal 3′ splice site. Consistently, we observed by using FRET a direct in vivo interaction between hnRNP-M and both subunits of the splicing factor U2AF, which binds to the polypyrimidine tract adjacent to the 3′ intron–exon junction (supplementary Fig S7A,B online). This interaction was stronger with the U2AF65 subunit, which could be co-immunoprecipitated using anti-GFP antibodies (supplementary Fig S7C online).
Finally, we investigated whether the function of hnRNP-M in alternative splicing was affected by the central region mapped as essential for binding to the CDC5L/PLRG1 proteins. Overexpression of hnRNP-M mutant 6(248−344aa) was compared with full-length hnRNP-M using the E1A mini-gene assay (Fig 4C, lanes 8 and 9). The mutant 6(248−344aa) protein caused minimal change to the RNA isoform pattern, whereas overexpression of wild-type hnRNP-M enhanced the level of the 10S and reduced that of the 13S isoforms (Fig 4D). No direct interactions between mutant 6(248−344aa)–EGFP and both mCherry-tagged U2AF65 and U2AF35 were detected by FLIM–FRET in vivo (supplementary Fig S7D online). We conclude that loss of the CDC5L/PLRG1 interaction domain in hnRNP-M correlates with a loss of ability to modulate alternative splice site selection in this assay.
It is established from studies on ASF/SF2 and spliceosome proteins, that splicing factors can contribute to both constitutive and alternative splicing mechanisms and show pre-mRNA substrate-specific effects (Cazalla et al, 2005). We propose that hnRNP-M provides another example of this phenomenon and our data suggest that interactions with the core spliceosome factors in the CDC5L complex might influence the mechanism of splice site selection. In this regard, detection of additional interactions between hnRNP-M and U2AF65 by FRET and IP assays suggests that the effect of hnRNP-M on 3′ splice-site selection could result from titration of the 3′ splice-site-binding factor U2AF. A similar titration mechanism has been proposed for SRp53, which interacts with the U2AF65-related protein HCC1 and can activate weak 3′ splice sites in addition to its role at 5′ splice-sites (Cazalla et al, 2005). All three U2AF-type proteins, that is, U2AF65, PUF60 and HCC1, interact with SRp54, which has been implicated in early 3′ splice site recognition (Zhang & Wu, 1996). These factors, together with the U2AF35 subunit and SR proteins, determine the 3′ splice site ‘exon definition complex' and bend the pre-mRNA (Matlin et al, 2005).
In summary, by using many experimental approaches, we show a direct involvement of hnRNP-M in the spliceosome machinery through its interaction with the CDC5L/PLRG1 spliceosomal subcomplex. The data show that hnRNP-M can influence both constitutive and alternative splicing, and provide new insight into the mechanism whereby hnRNP-M can modulate both 5′ and 3′ splice site choice.
Methods
Antibodies and plasmids. The following antibodies were used: PLRG1 and CDC5L (Ajuh et al, 2001); PLRG1 (Everest Biotech, Oxford, UK); hnRNP-M 2A6 (sc-20001) and 1D8 (Datar et al, 1993); hnRNP D2IF7 (Gattoni et al, 1996); GFP (Roche Diagnostics, Indianapolis, IN, USA); SC35 (Sigma, St Louis, MO, USA); Sm (Abcam, Cambridge, UK); E1A reporter plasmid (Caceres et al, 1994); pDC20 reporter (Wang et al, 2002); and EYFP–CDC5L, EYFP–PLRG1, EYFP–U2AF35 and EYFP–U2AF65 (Chusainow et al, 2005).
Protein binding assays. We performed the assays as described by Ajuh et al (2001) using 0.2 nmol of the appropriate recombinant protein and 8–10 μl of the in vitro translation reaction.
Alternative splicing assays. We performed alternative splicing assays using E1A and pDC20 mini-gene reporters as described by Caceres et al (1994) and Wang et al (2002).
Heat shock. Heat-shock treatments were performed as described by Chiodi et al (2000).
Fluorescence lifetime imaging microscopy. We acquired and analysed FLIM–FRET measurements as described by Lleres et al (2007) and Ellis et al (2008).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Drs M.S. Swanson, J.P. Fuchs, J.F. Caceres, P. Ouyang, G. Biamonti and J. Tazi for reagents; Dr Bartosz Pilch from Prof Matthias Mann's lab for performing mass spectrometry analysis; the Lamond lab members for helpful suggestions; and Dr S. Swift of the CLS Light Microscopy Facility for advice and technical assistance. This work was supported by the Wellcome Trust and the European Alternative Splicing Network (Eurasnet). A.I.L. is a Wellcome Trust Principal Research Fellow.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI (2000) Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J 19: 6569–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajuh P, Sleeman J, Chusainow J, Lamond AI (2001) A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J Biol Chem 276: 42370–42381 [DOI] [PubMed] [Google Scholar]
- Bai Y, Lee D, Yu T, Chasin LA (1999) Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res 27: 1126–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R (2008) Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Biamonti G (2004) Nuclear stress bodies: a heterochromatin affair? Nat Rev Mol Cell Biol 5: 493–498 [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Fakan S (2008) Visualization of nuclear organization by ultrastructural cytochemistry. Methods Cell Biol 88: 431–449 [DOI] [PubMed] [Google Scholar]
- Bond U (1988) Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J 7: 3509–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Stamm S, Helfman DM, Krainer AR (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706–1709 [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond AI (1991) In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J 10: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Newton K, Caceres JF (2005) A novel SR-related protein is required for the second step of pre-mRNA splicing. Mol Cell Biol 25: 2969–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodi I, Biggiogera M, Denegri M, Corioni M, Weighardt F, Cobianchi F, Riva S, Biamonti G (2000) Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J Cell Sci 113: 4043–4053 [DOI] [PubMed] [Google Scholar]
- Chusainow J, Ajuh PM, Trinkle-Mulcahy L, Sleeman JE, Ellenberg J, Lamond AI (2005) FRET analyses of the U2AF complex localize the U2AF35/U2AF65 interaction in vivo and reveal a novel self-interaction of U2AF35. RNA 11: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar KV, Dreyfuss G, Swanson MS (1993) The human hnRNP M proteins: identification of a methionine/arginine-rich repeat motif in ribonucleoproteins. Nucleic Acids Res 21: 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG (1993) hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Ellis JD, Lleres D, Denegri M, Lamond AI, Caceres JF (2008) Spatial mapping of splicing factor complexes involved in exon and intron definition. J Cell Biol 181: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattoni R, Mahe D, Mahl P, Fischer N, Mattei MG, Stevenin J, Fuchs JP (1996) The human hnRNP-M proteins: structure and relation with early heat shock-induced splicing arrest and chromosome mapping. Nucleic Acids Res 24: 2535–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Yalamanchili P, Visa N (2006) The Drosophila heterogeneous nuclear ribonucleoprotein M protein, HRP59, regulates alternative splicing and controls the production of its own mRNA. J Biol Chem 281: 39135–39141 [DOI] [PubMed] [Google Scholar]
- House AE, Lynch KW (2008) Regulation of alternative splicing: more than just the ABCs. J Biol Chem 283: 1217–1221 [DOI] [PubMed] [Google Scholar]
- Hovhannisyan RH, Carstens RP (2007) Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem 282: 36265–36274 [DOI] [PubMed] [Google Scholar]
- Lleres D, Swift S, Lamond AI (2007) Detecting protein–protein interactions in vivo with FRET using multiphoton fluorescence lifetime imaging microscopy (FLIM). Curr Protoc Cytom 12: 10. [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Luhrmann R (2004) A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J 23: 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B (2007) hnRNP proteins and splicing control. Adv Exp Med Biol 623: 123–147 [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW (2005) Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang P, Lou PJ, Leu S, Ouyang P (2002) Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem Biophys Res Commun 294: 448–455 [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Wu JY (1996) Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol 16: 5400–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.