Recovery from a DNA-damage-induced G2 arrest requires Cdk-dependent activation of FoxM1
After DNA-damage checkpoint activation, Cdk activity is reduced in order to arrest the cell cycle. Alvarez-Fernández and colleagues demonstrate that during arrest, residual Cdk activity maintains FoxM1-dependent transcription and the expression of mitotic inducers, such as cyclin B1 and Plk1, in order to allow efficient re-entry into the cell cycle after checkpoint recovery.
Keywords: Cdk, checkpoint recovery, DNA damage, FoxM1, transcription
Abstract
Activation of the DNA-damage checkpoint culminates in the inhibition of cyclin-dependent kinase (Cdk) complexes to prevent cell-cycle progression. We have shown recently that Cdk activity is required for activation of the Forkhead transcription factor FoxM1, an important regulator of gene expression in the G2 phase of the cell cycle. Here, we show that FoxM1 is transcriptionally active during a DNA-damage-induced G2 arrest and is essential for checkpoint recovery. Paradoxically, Cdk activity, although reduced after checkpoint activation, is required to maintain FoxM1-dependent transcription during the arrest and for expression of pro-mitotic targets such as cyclin A, cyclin B and Plk1. Indeed, we find that cells need to retain sufficient levels of Cdk activity during the DNA-damage response to maintain cellular competence to recover from a DNA-damaging insult.
Introduction
DNA damage activates a checkpoint response that prevents cell-cycle progression until the lesion has been repaired. This checkpoint-dependent cell-cycle arrest is mediated by ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) kinases, which in turn activate the effector kinases Chk1 and Chk2, which can activate or inactivate several regulatory components of the cell-cycle machinery to enforce cell-cycle arrest (Bartek & Lukas, 2007). In the G2 phase of the cell cycle, activation of the DNA-damage checkpoint prevents the activation of cyclin-B–cyclin-dependent kinase 1 (Cdk1), the cyclin–Cdk complex that is essential for mitotic entry (Smits & Medema, 2001). Moreover, activation of the tumour suppressor p53 contributes to the maintenance of the arrest by direct repression of G2/M genes and induction of p21waf1, which acts as an inhibitor of several cyclin–Cdk complexes (Taylor & Stark, 2001).
One of the main regulators of the transcriptional programme in G2 is the Forkhead transcription factor FoxM1, which is essential for proper mitotic progression and chromosome stability (Laoukili et al, 2005; Wang et al, 2005). We have shown recently that FoxM1 activity in G2 depends strictly on phosphorylation by cyclin A–Cdk complexes (Laoukili et al, 2008a). In addition, others have shown recently that FoxM1 expression is negatively regulated by p53 (Barsotti & Prives, 2009; Pandit et al, 2009). These findings imply that FoxM1 activity is suppressed during DNA-damage response through combined inhibition of Cdk activity and suppression by p53, and it has been suggested that this could contribute to the cell-cycle delay imposed by the checkpoint (Barsotti & Prives, 2009). Contrary to this model, we report here that FoxM1 remains active during a DNA-damage-induced G2 arrest and is required for efficient recovery from the arrest.
Results And Discussion
FoxM1 is active during a DNA-damage-induced G2 arrest
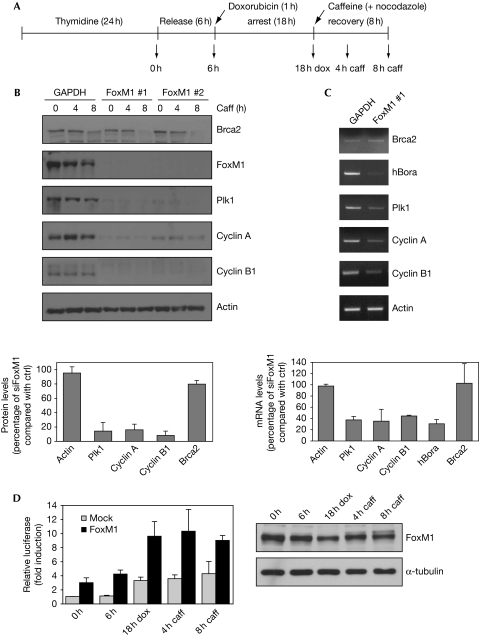
The idea that FoxM1 expression is negatively regulated by p53, and its activity requires Cdk-dependent phosphorylation, suggests that FoxM1 is inhibited during a DNA-damage response. However, in an unperturbed cell cycle, FoxM1 activity is essential for the expression of a large number of G2-specific genes, and this raises the question of how expression of these genes is regulated during a DNA-damage-induced arrest. To study this, we synchronized U2OS cells in G2 and applied the genotoxic drug doxorubicin to cause a DNA-damage-induced arrest. Subsequently, checkpoint silencing and recovery were induced by the addition of caffeine, an inhibitor of the checkpoint kinases ATM/ATR (Fig 1A). First, we analysed the effect of small inhibitory RNA (siRNA)-mediated FoxM1 depletion on the expression of several known FoxM1 target genes in DNA-damage-arrested G2 cells (supplementary Fig S1 online). Interestingly, expression of cyclin A/B and Plk1 was reduced dramatically in DNA-damage-arrested cells depleted of FoxM1 (Fig 1B), and this correlated with a marked reduction in the messenger RNA level of those genes (Fig 1C). However, expression of the repair protein BRCA2, another reported FoxM1 target (Tan et al, 2007), was not affected by FoxM1 depletion (Fig 1B,C), suggesting that the function of FoxM1 during the DNA-damage-induced arrest is specifically required for expression of mitotic inducers such as cyclin A/B and Plk1. This finding contrasts with previous observations that suggest repression of FoxM1 function by the DNA-damage checkpoint. However, we could observe a reduction in FoxM1 protein levels after activation of the DNA-damage checkpoint in G2 (Fig 1D), consistent with the idea that active p53 can repress FoxM1 expression (Barsotti & Prives, 2009; Pandit et al, 2009). Nonetheless, transactivation of a luciferase reporter containing a Forkhead-response element by ectopically expressed FoxM1 was not repressed by DNA damage and FoxM1 activity remained high during recovery (Fig 1D). A similar trend was observed in cells lacking exogenous FoxM1, and this was due largely to the activity of endogenous FoxM1 (supplementary Fig S2 online). Thus, FoxM1 is active during a DNA-damage-induced arrest and is required for continued expression of cyclin A/B and Plk1.
Figure 1.
FoxM1 is transcriptionally active during a DNA-damage-dependent arrest and checkpoint recovery. (A) Scheme of the recovery-induced experimental setting. (B) U2OS cells were transfected with siRNA control (GAPDH) and two independent siRNAs targeting FoxM1. After thymidine treatment for 24 h, cells were released for 6 h and doxorubicin (dox) was applied for 1 h. Caffeine (caff) was added 18 h after doxorubicin washout. The protein level of FoxM1 and FoxM1 targets was determined by western blotting at the indicated time points. (C) Reverse transcription PCR of FoxM1 target genes 18 h after doxorubicin treatment. Cells were transfected with either GAPDH or siFOXM1 #1 and treated as described in (B). The graphs in panels (B) and (C) represent the relative protein or mRNA level, respectively, of the FoxM1 targets in siFoxM1 #1-treated cells as compared with that in control cells 18 h after doxorubicin treatment. (D) U2OS cells were transfected with an empty vector (mock) or a plasmid encoding FoxM1 wild type, and co-transfected with the pBP-1 luciferase reporter plasmid. Relative luciferase expression was measured at the indicated time points during the DNA-damage response. Expression of endogenous FoxM1 was analysed by western blotting. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; siRNA, small inhibitory RNA.
FoxM1 is required for checkpoint recovery
Next we wondered if FoxM1 was required for cell-cycle re-entry following a DNA-damage-induced G2 arrest. We depleted FoxM1 from U2OS cells and monitored the percentage of cells that entered mitosis after caffeine addition. Importantly, FoxM1 depletion caused a dramatic reduction in the percentage of cells that could re-enter the cell cycle after addition of caffeine (Fig 2A,B). A similar defect in caffeine-induced recovery was observed in FoxM1-deficient mouse embryonic fibroblasts (MEFs) as compared with wild-type MEFs, and in HCT116 cells (supplementary Fig S3 online). Furthermore, spontaneous recovery in the absence of caffeine was also strongly inhibited in FoxM1-depleted cells arrested in G2 by γ-irradiation (Fig 2C).
Figure 2.
FoxM1 is essential for checkpoint recovery. (A) U2OS cells transfected with an siRNA against GAPDH and with two siRNAs against FoxM1 were synchronized and treated as in Fig 1 and the amount of mitotic cells was determined by pH3 positivity by FACS. The graph shows the percentage of mitotic cells at the indicated time points. The level of FoxM1 depletion was assessed by western blot analysis. (B) Cells transfected and treated as in (A) were followed by time-lapse microscopy after caffeine addition and scored for entry into mitosis. (C) Cells were transfected and synchronized as described in (B) and subjected to 5 or 10 Gy γ-irradiation. Immediately after irradiation, paclitaxel was added for 24 h before harvesting and the mitotic index was determined by pH3 staining. (D) U2TR cells stably expressing inducible RNAi-resistant FoxM1 wild type were co-transfected with GFP-spectrin, pBabe-Puro and empty pSuper (pS) or pS-FoxM1. The cells were synchronized and treated as described in (A) and doxycycline was added immediately (early) or 18 h (late) after doxorubicin washout. Cells were harvested 8 h after caffeine addition and the percentage of mitotic cells was determined by scoring GFP-positive cells for pH3 reactivity by FACS. The data on the graph are the average of three independent experiments and the error bars represent the standard deviation. The protein levels of FoxM1 and its targets were assessed by western blot analysis of cells selected with puromycin at the indicated time points. (E) Cells were transfected and synchronized as described in (D) and fixed at the indicated time points after caffeine addition. The mitotic index of GFP-positive cells was determined by FACS analysis. Cyclin B protein levels were determined by immunofluorescence and automated image analysis was performed as described under Methods. At least 200 GFP-positive cells were counted per well. Measurements were performed in triplicate and error bars represent the standard deviation. FACS, fluorescence-activated cell sorting; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFP, green fluorescent protein; RNAi, RNA interference; siRNA, small inhibitory RNA.
We next generated a tetracycline-inducible cell line expressing an RNA interference (RNAi)-insensitive variant of FoxM1. Induced expression of the RNAi-resistant FoxM1 was able to revert the recovery defect of cells depleted of endogenous FoxM1, showing that the effect on recovery is truly dependent on FoxM1 (Fig 2D). Furthermore, FoxM1 expression restored the levels of the mitotic inducers cyclin B and Plk1 during the G2 arrest (Fig 2D). Importantly, whereas the rescue was near complete when FoxM1 was induced directly after inflicting the DNA damage, FoxM1 was not so efficient to revert the recovery defect when expressed at the time of caffeine addition (Fig 2D). Cells in which FoxM1 expression was induced late showed significant delay to enter mitosis (∼4 h) if compared with FoxM1 being active already during the arrest, and this delay correlated with the expression of its transcriptional targets (Fig 2E; supplementary Fig S4 online). Therefore, these results indicate that FoxM1 activity during a DNA-damage-induced arrest is required to efficiently re-enter the cell cycle after checkpoint inactivation.
Cdk keeps FoxM1 active during a DNA-damage response
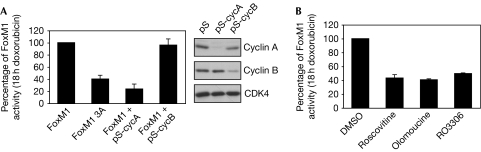
As cyclin–Cdk activity is inhibited as a consequence of DNA-damage checkpoint activation, we wondered how FoxM1 activity is maintained during the DNA-damage-induced G2 arrest. Surprisingly, we found that RNAi-mediated depletion of cyclin A, but not cyclin B, strongly inhibited FoxM1 transcriptional activity after doxorubicin treatment (Fig 3A; supplementary Fig S5A online), similar to our previous results in an unperturbed G2 (Laoukili et al, 2008a). In addition, a triple alanine mutant of known Cdk-phosphosites in FoxM1 (FoxM1 T600A/T611A/S6383A; Luscher-Firzlaff et al, 2006; Laoukili et al, 2008a) failed to transactivate the FoxM1 reporter in DNA-damaged cultures in U2OS cells and in HCT116 cells (Fig 3A; supplementary Fig S5B online). This strongly suggests that Cdk-dependent phosphorylation of FoxM1 is maintained during the DNA-damage response. To confirm this, we analysed in vivo the phosphorylation of exogenously expressed FoxM1 by mass spectrometry. Most phosphorylated sites that we identified in this manner corresponded to Cdk-phosphorylation consensus sites (supplementary Fig S6 online). Moreover, most of these sites, such as T600, continued to be phosphorylated in cells arrested in G2 by doxorubicin treatment. Indeed, we could confirm the requirement of Cdk activity by treating doxorubicin-arrested cells with the Cdk inhibitors roscovitine, olomoucine and RO3306 (Fig 3B). All inhibitors caused a reduction in the FoxM1-mediated transactivation of the Forkhead luciferase reporter in DNA-damaged cells, showing that Cdk1- and/or Cdk2-associated kinase activities, although suppressed after checkpoint activation, are necessary to keep FoxM1 active throughout the checkpoint response.
Figure 3.
Regulation of FoxM1 activity during a DNA-damage-induced G2 arrest. (A) U2OS cells were transfected with FoxM1 wild type, FoxM1 3A (FoxM1 T600A/T611A/S638A), together with short-hairpin RNA-targeting vectors against cyclin A (pS-cycA) or cyclin B (pS-cycB). The cells were synchronized in G2 and transactivation of BP-1 reporter by FoxM1 was determined 18 h after doxorubicin washout. The efficiency of cyclin depletion was analysed by western blotting of puromycin-selected cells. (B) Cells transfected with FoxM1 and the pBP-1 luciferase reporter were treated with doxorubicin and incubated with the indicated Cdk inhibitors for 18 h before harvesting. The graph shows the relative luciferase activity. The data shown in the graphs (A,B) are the average of three independent experiments and the error bars represent the standard deviation. Cdk, cyclin-dependent kinase; pS, pSuper.
Cdk activity confers checkpoint recovery competence
Our data suggest that Cdk activity needs to be maintained at a minimal level during the DNA-damage response to allow eventual cell-cycle re-entry. To test this, we treated cells with Cdk inhibitors for various time periods and investigated whether cells could re-enter the cell cycle after removal of the inhibitor. Interestingly, if G2 cells were treated with Cdk inhibitors for periods up to 8 h, the arrest was fully reversible, whereas treatment for 16 h led to irreversible G2 arrest and cells were no longer able to re-enter the cell cycle after washing-out the inhibitor (Fig 4A; supplementary Fig S7A online). When Cdk inhibitors were added during a DNA-damage-induced arrest, checkpoint recovery was abolished completely, even though the inhibitor was removed at the time of caffeine addition (Fig 4B; supplementary Fig S7B online). Time-lapse microscopy analysis confirmed that, whereas about 50% of control cells entered mitosis 8 h after caffeine addition, more than 95% of cells pretreated with either roscovitine or RO3306 remained arrested (Fig 4B). In HCT116 cells, Cdk inhibition in unperturbed cells was almost fully reversible even after 16 h of treatment, suggesting differences in Cdk activity between different cell lines (supplementary Fig S7C online). However, similar to U2OS cells, treatment with the Cdk inhibitor for 16 h during the doxorubicin-induced G2 arrest completely blocked recovery after addition of caffeine and removal of the inhibitor (supplementary Fig S7C,D online). This is consistent with the idea that cells need to maintain a minimal level of Cdk activity to allow recovery after checkpoint termination. Interestingly, the protein levels of cyclin A/B and Plk1 were reduced in cells treated with Cdk inhibitors during the checkpoint-dependent arrest (Fig 4B). This suggests that the loss of recovery competence induced by Cdk inhibitors could be due to the loss of expression of mitotic inducers such as cyclin A/B and Plk1. This is similar to our recent observations with the phosphatase Wip1, which we showed maintains recovery competence by counteracting p53-dependent repression of cyclin B and Plk1 (Lindqvist et al, 2009), indicating that sustained expression of these genes has a crucial role in recovery competence.
Figure 4.
Cdk inhibition induces loss of recovery competence after DNA damage. (A) U2OS cells were synchronized by thymidine treatment (24 h). Roscovitine was added 6 h after release from the thymidine block, for the indicated periods of time. The mitotic index was determined 12 h after roscovitine washout in the presence of nocodazole. (B) U2OS cells were synchronized as described in (A) and treated with doxorubicin for 1 h. After doxorubicin washout, roscovitine and RO3306 were added for 18 h. Subsequently, the Cdk inhibitors were washed out extensively and caffeine was added for 8 h. Mitotic entry after caffeine addition was analysed by time-lapse microscopy. Lysates from cells harvested 18 h after doxorubicin and Cdk inhibitor treatment were subjected to immunoblotting with the indicated antibodies. (C) U2TR cells stably expressing the inducible RNAi-resistant FoxM1ΔN/ΔKEN mutant were treated as described in (B). Doxycycline was added during release from the thymidine block. Cells were collected 18 h after doxorubicin treatment for western blot analysis and the amount of mitotic cells was determined 8 h after caffeine addition. The graph shows the average of three independent experiments and the error bars represent the standard deviation. Cdk, cyclin-dependent kinase; RNAi, RNA interference.
Next, we investigated whether enforced expression of a constitutive active mutant of FoxM1 could restore the expression of FoxM1 target genes in Cdk-inhibited cells. For this, we used an amino-terminal-deleted form of FoxM1 (FoxM1ΔN/ΔKEN), which is active independently of cyclin A–Cdk activity and resistant to Cdh1-dependent degradation (Laoukili et al, 2008a,b,Laoukili et al, 2008a,b). Indeed, we found that expression of FoxM1ΔN/ΔKEN in DNA-damaged cells increased the expression of cyclin A/B and Plk1 in the presence of the Cdk inhibitor RO3306 (Fig 4C). Moreover, expression of FoxM1ΔN/ΔKEN restored, at least partly, the ability of the cells to enter mitosis after full Cdk inhibition (Fig 4C). These data suggest that FoxM1 is one of the Cdk targets that can contribute to maintaining recovery competence. However, it might not be the only target, as expression of FoxM1ΔN/ΔKEN could not completely restore the expression of cyclin A/B and Plk1. For example, it is not unlikely that Cdk activity is required to maintain the activity of additional transcription factors during the DNA-damage response. Similar to FoxM1, activity of NF-Y and B-myb is controlled by cyclin–Cdk complexes and both are involved in the transcriptional regulation of genes required for G2–M progression (Ziebold et al, 1997; Chae et al, 2004). Interestingly, B-myb was recently shown to be required for checkpoint recovery (Mannefeld et al, 2009), suggesting that it might cooperate with FoxM1 to regulate recovery.
Together, our data show that cells require residual Cdk activity during the DNA-damage response to maintain FoxM1-dependent transcription. This mechanism ensures that cells can rapidly re-enter the cell cycle once the checkpoint is silenced. These findings are compatible with recent studies showing that Cdk1 or Cdk2 complexes are not mere targets of the DNA-damage checkpoint, but are actively engaged during the ongoing damage response. In budding yeast, Cdk1 (Cdc28) is required for resection of double-strand break ends (Ira et al, 2004), and in mammalian cells treatment with roscovitine has been shown to compromise Chk1 phosphorylation after ionising radiation (Jazayeri et al, 2006). More recently, it has been reported that depletion of Cdk1 impairs efficient phosphorylation of ATM/ATR substrates and recruitment of BRCA1 to the DNA-damage foci (Johnson et al, 2009). Thus, the DNA-damage response must fine-tune Cdk activity in such a way that levels are low enough to maintain a checkpoint-dependent arrest, but high enough to initiate a proper checkpoint response and to maintain the transcription of G2 genes to allow efficient recovery after repair of the damage.
Methods
Cell culture, transfections and drugs. U2OS cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 6% fetal calf serum and antibiotics. FoxM1-inducible cell lines were generated by transfection of U2TR cells (that is, U2OS cells expressing the tetracycline repressor) with pcDNA4/TO plasmids (Invitrogen, Carlsbad, CA, USA), and stable clones were selected in DMEM with 6% Tet system-approved fetal bovine serum (Clontech, Saint-Germain-en-Laye, France), antibiotics and zeocin (400 μg/ml). FoxM1 expression was induced by the addition of doxycycline (1 μg/ml). Thymidine, puromycine, paclitaxel, nocodazole, doxorubicin and caffeine were purchased from Sigma and used at 2.5 mM, 2 μg/ml, 1 μM, 250 ng/ml, 0.5 μM and 5 mM, respectively. Roscovitine (Sigma), olomoucine (Biomol) and RO3306 (Calbiochem, Darmstadt, Germany) were used at 25, 200 and 10 μM, respectively. Cells were transfected with plasmid DNA by using the standard calcium phosphate transfection protocol. siRNA oligonucleotides were transfected with HiPerFect (Qiagen) following the manufacturer's protocol.
Plasmids and oligonucleotides. pBP-1 has been described previously by Furuyama et al (2000). FoxM1 wild type, FoxM1 3A (T600A/T611A/S638A), pSuper (pS)-cyclin A and pS-cyclin B have also been described (Laoukili et al, 2008a). FoxM1 wild type and FoxM1ΔN/ΔKEN were converted into RNAi-insensitive mutants by site-direct mutagenesis. siRNAs targeting glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and FoxM1 were purchased from Dharmacon (Lafayette, CO, USA): siFoxM1 #1 targets the sequence CCTTTCCCTGCACGACATG (Wonsey & Follettie, 2005) and siFoxM1 #2 is an On-target SMART pool. The short-hairpin RNA plasmid targeting FoxM1 (pS-FoxM1) was generated by annealing oligonucleotides corresponding to the target sequence of siFoxM1 #1 into pS (Brummelkamp et al, 2002).
Antibodies. FoxM1 (C-20), cyclin A2 (H-432), cyclin B1 (GNS1) and actin (I-19) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-α-tubulin and anti-BRCA2 were from Sigma and Calbiochem, respectively. Plk1 and pS10-histone H3 antibodies were from Upstate (Billerica, MA, USA).
Reporter assays. Luciferase activity was determined by using the dual luciferase kit (Promega) according to the manufacturer's instructions. Relative luciferase was expressed as the ratio of firefly luciferase activity to control Renilla luciferase activity.
Reverse transcription–PCR. Total RNA was isolated by using the Qiagen RNeasy kit, according to the manufacturer's instructions. cDNA was synthesized from 1 μg of total RNA by using Superscript II reverse transcriptase (Invitrogen) and oligo-(dT) primers. The resultant cDNA was used as a template for PCR amplification using specific primers.
Recovery assays and FACS analysis. Checkpoint recovery assays and fluorescence-activated cell sorting (FACS) analysis were performed as described by van Vugt et al (2004).
Automated image analysis. Cells were grown in 96-well plates (Viewplate-96; Perkin Elmer, Waltham, MA, USA). At the indicated time points, cells were fixed with 4% formaldehyde and methanol, and were stained with 4′,6-diamidino-2-phenylindole and the indicated antibodies. Image acquisition was performed by using a Cellomics ArrayScan VTI (Thermo Scientific, Waltham, MA, USA) with a 20 × 0.40 NA objective. Image analysis was performed by using a Cellomics ArrayScan HCS Reader (Thermo Scientific).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank A. Lindqvist for critical reading of the paper and the members of the Medema laboratory for helpful discussions. This work was supported by the Dutch Cancer Society (UU-2007-3826) and the Netherlands Genomic Initiative of NWO.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barsotti AM, Prives C (2009) Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 28: 4295–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2007) DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 19: 238–245 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Chae HD, Yun J, Bang YJ, Shin DY (2004) Cdk2-dependent phosphorylation of the NF-Y transcription factor is essential for the expression of the cell cycle-regulatory genes and cell cycle G1/S and G2/M transitions. Oncogene 23: 4084–4088 [DOI] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N (2000) Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G et al. (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Johnson N, Cai D, Kennedy RD, Pathania S, Arora M, Li YC, D'Andrea AD, Parvin JD, Shapiro GI (2009) Cdk1 participates in BRCA1-dependent S phase checkpoint control in response to DNA damage. Mol Cell 35: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH (2005) FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 7: 126–136 [DOI] [PubMed] [Google Scholar]
- Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, Heck AJ, Medema RH (2008a) Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol Cell Biol 28: 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH (2008b) FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle 7: 2720–2726 [DOI] [PubMed] [Google Scholar]
- Lindqvist A, de Bruijn M, Macurek L, Bras A, Mensinga A, Bruinsma W, Voets O, Kranenburg O, Medema RH (2009) Wip1 confers G2 checkpoint recovery competence by counteracting p53-dependent transcriptional repression. EMBO J 28: 3196–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher-Firzlaff JM, Lilischkis R, Luscher B (2006) Regulation of the transcription factor FOXM1c by cyclin E/CDK2. FEBS Lett 580: 1716–1722 [DOI] [PubMed] [Google Scholar]
- Mannefeld M, Klassen E, Gaubatz S (2009) B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res 69: 4073–4080 [DOI] [PubMed] [Google Scholar]
- Pandit B, Halasi M, Gartel AL (2009) p53 negatively regulates expression of FoxM1. Cell Cycle 8: 3425–3427 [DOI] [PubMed] [Google Scholar]
- Smits VA, Medema RH (2001) Checking out the G2/M transition. Biochim Biophys Acta 1519: 1–12 [DOI] [PubMed] [Google Scholar]
- Tan Y, Raychaudhuri P, Costa RH (2007) Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol 27: 1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Stark GR (2001) Regulation of the G2/M transition by p53. Oncogene 20: 1803–1815 [DOI] [PubMed] [Google Scholar]
- van Vugt MA, Bras A, Medema RH (2004) Polo-like kinase 1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell 15: 799–811 [DOI] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH (2005) Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol 25: 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonsey DR, Follettie MT (2005) Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res 65: 5181–5189 [DOI] [PubMed] [Google Scholar]
- Ziebold U, Bartsch O, Marais R, Ferrari S, Klempnauer KH (1997) Phosphorylation and activation of B-Myb by cyclin A–Cdk2. Curr Biol 7: 253–260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.