Abstract
Copy number variants (CNVs) account for a major proportion of human genetic polymorphism and have been predicted to play an important role in genetic susceptibility to common disease. To address this we undertook a large direct genome-wide study of association between CNVs and eight common human diseases. Using a purpose-designed array we typed ~19,000 individuals into distinct copy-number classes at 3,432 polymorphic CNVs, including an estimated ~50% of all common CNVs larger than 500bp. We identified several biological artefacts that lead to false-positive associations, including systematic CNV differences between DNAs derived from blood and cell-lines. Association testing and follow-up replication analyses confirmed three loci where CNVs were associated with disease, IRGM for Crohn's disease, HLA for Crohn's disease, rheumatoid arthritis, and type 1 diabetes, and TSPAN8 for type 2 diabetes, though in each case the locus had previously been identified in SNP-based studies, reflecting our observation that the majority of common CNVs which are well-typed on our array are well tagged by SNPs and so have been indirectly explored through SNP studies. We conclude that common CNVs which can be typed on existing platforms are unlikely to contribute greatly to the genetic basis of common human diseases.
Genome-wide association studies (GWAS) have been extremely successful in associating single nucleotide polymorphisms (SNPs) with susceptibility to common diseases, but published SNP associations account for only a fraction of the genetic component of most common diseases, and there has been considerable speculation about where the “missing heritability” 1 might lie. Chromosomal rearrangements can cause particular rare diseases and syndromes 2, and recent reports have suggested a role for rare copy number variants, either individually or in aggregate, in susceptibility for a range of common diseases, notably neurodevelopmental diseases 3,4,5,6. To date, there have been relatively few reported associations between common diseases and common CNVs, see for example 7,8,9,10,11, which might simply reflect incomplete catalogues of common CNVs or the lack of reliable assays for their large-scale typing. Here we report the results of our direct association study, identify the population properties of the set of CNVs studied, describe novel analytical methods to facilitate robust analyses of CNV data, and document artefacts that can afflict CNV studies.
We designed an array to measure copy number for the majority of a recently-compiled inventory of CNVs from an extensive discovery experiment 12, and several other sources. We then used the array to type 3,000 common controls and 2,000 cases of each of the diseases: bipolar disorder (BD), breast cancer (BC), coronary artery disease (CAD), Crohn's disease (CD), hypertension (HT), rheumatoid arthritis (RA), type 1 diabetes (T1D), and type 2 diabetes (T2D). These eight diseases make a major impact on public ill health 13, cover a range of aetiologies and genetic predispositions, and have been extensively studied via SNP-based GWAS, including our earlier Wellcome Trust Case Control Consortium (WTCCC) study 14.
Pilot experiment, Array Content, Assay, and Samples
Pilot experiment
We undertook a pilot experiment to compare three different platforms for assaying copy-number variation and to assess the merits of different experimental design parameters (full details are given in the SoM). Based on the pilot data, we chose the Agilent Comparative Genomic Hybridisation (CGH) platform, and aimed to target each CNV with 10 distinct probes, although in the analyses below we include any CNV targeted by at least one probe (Supplementary Figure 9). Our analysis of the pilot CGH data indicated that the quality of the copy number signal for genotyping (rather than for discovery) at a CNV is reduced when the reference sample is homozygous deleted, in effect because the reference channel then just measures noise. To minimize this effect we used a fixed pool of DNAs as the reference sample throughout our main experiment.
Array Content
Informed by our pilot experiment, we designed the CNV-typing array in a collaboration with the Genome Structural Variation Consortium (GSV) in which a preliminary set of candidate CNVs was shared at an early stage with the WTCCC. Table 1 summarises the design content of the array, and Figure 1 illustrates the various categories of designed loci unsuitable for association analysis. See online methods for further details.
Table 1. Summary of the discovery source for genomic regions targeted on the WTCCC CNV genotyping array.
GSV CNVs were prioritised according to extent of polymorphism in European discovery samples. See Online Methods for full details of other sources.
| Source of Loci | Number of loci targeted |
Number of loci analysed |
Number of loci polymorphic with good calls |
|
|---|---|---|---|---|
|
| ||||
| CNVs | GSV Discovery Project | 10,835 | 10,217 | 3,096 |
| Affymetrix 500k | 18 | 14 | 12 | |
| Affymetrix 6.0 | 83 | 81 | 47 | |
| Illumina 1M | 82 | 81 | 18 | |
| WTCCC CNV Loci | 231 | 209 | 108 | |
| Novel Sequence | Novel Insert Regions | 292 | 292 | 151 |
|
| ||||
| Total | 11,541 | 10,894 | 3,432 | |
Figure 1. Flow-chart showing which CNVs are included on the array.
The chart shows the reasons for CNVs being removed from consideration (the column of arrows and text to the right of the figure) from those originally targeted on the array and the number of CNVs remaining at each stage of filtering.
Assay
In brief (see SoM for further details) the Agilent assay differentially labels parallel aliquots of the test sample and reference DNA (a pool of genomic UK lymphoblastoid cell-line DNAs from 9 males and 1 female prepared in a single batch for all experiments) and then combines them, hybridises to the array, washes, and scans. Intensity measurements for the two different labels are made at each probe separately for the test and reference DNA. These act as surrogates for the amount of DNA present, with analyses typically relying on the ratio of test to reference intensity measurements at each probe.
Samples
A total of 19,050 case-control samples were sent for assaying: ~2,000 for each of the eight diseases and ~3000 common controls (these were equally split between the 1958 British Birth Cohort (58C) and the UK Blood Services (UKBS) controls). These were augmented by 270 HapMap1 samples (see 12 for additional analyses of the HapMap data) and 610 duplicate samples for QC purposes. About 80% of samples from the WTCCC SNP GWAS were used here. See SoM for further details of sample collections, inclusion criteria etc.
Data Pre-Processing, CNV Calling and Quality Control
Data Pre-Processing
For each sample, raw data from the CNV experiment consist of intensity measurements for the test and reference sample for each probe. There are numerous choices at the data pre-processing stage, including how to normalise data to reduce inter-individual variation, and how to combine the information across the set of probes within a CNV. Several novel analytical tools substantially improved data quality, but no single approach works well for every CNV, so we carried through 16 pre-processing pipelines to maximise the number of CNVs that can be tested for association. See SoM Section 4 for illustrations and a sense of the challenges.
CNV Calling
The objective in CNV calling at each CNV is to assign each assayed sample to a diploid copy-number class, which represents the sum of copy numbers on each allele. This step is analogous to, but typically considerably more challenging than, calling genotypes from SNP-chip data. Available assays for SNPs are more robust and have better signal to noise properties than do available assays for CNVs 15. We used two different statistical methods (“CNVtools” which is available as a Bioconductor package, and “CNVCALL”) in parallel to estimate the number of copy-number classes at each CNV and assign individuals to these classes. See SoM for further details. Figure 2 illustrates three multi-allelic CNVs which have attracted attention in the literature in part due to the difficulties in obtaining reliable data.
Figure 2. Illustrative CNVs.
Histograms of three multiallelic CNVs (one per row) previously reported to be associated with autoimmune diseases: Beta-Defensin (CNVR3771.10), CCL3L1 (CNVR7077.12) and FCGR3B (CNVR383.1), showing 6, 5, and 4 fitted copy number classes respectively. The histogram of normalised intensity ratios is shown for one control and the three autoimmune collections. Histograms are overlaid by the fitted distribution used to model each class (variously the red, blue, light green, cyan, magenta and dark green curves). In all such figures, the area under the fitted curve of a particular colour is the same for all collections at the same CNV.
Quality Control
Following the application of QC metrics to each sample and each CNV (see Online Methods) 17,304 case control samples (of 19,050 initially) were available for association testing. There were 3,432 CNVs with more than one copy-number class which passed QC and were included in subsequent analyses. At these CNVs, concordance of calls between pairs of duplicate samples was 99.7%.
Properties of CNVs
Single class CNVs
Of the 10,894 distinct putative CNVs typed on the array after removal of detectable redundancies, 60% are called with a single copy-number class, and so cannot be tested for association. Following detailed analyses (see Online Methods) we estimate that just under half of these are likely not to be polymorphic. For the remainder, the combination of the experimental assay and analytical methods we have used do not allow separate copy-number classes to be distinguished.
Multi-class CNVs
4,326 CNVs were called with multiple classes. Of these, 3,432 passed quality control filters, which in practice means that the classes were well separated and thus that it was possible to assign individuals to copy-number classes with high confidence. Most of these CNVs (88%) have two or three copy-number classes, consistent with their having only two variants, or alleles, present in the population (we refer to these as bi-allelic CNVs). Note that some loci involving both duplications and deletions could be called with only three classes if both homozygote classes are very rare.
Allele Frequencies
Supplementary Figure 21 shows the distribution of minor allele frequency (MAF) for bi-allelic CNVs passing QC. For example, 44% of autosomal CNVs passing QC had MAF < 5%. This is shifted towards lower MAFs compared to commonly used SNP chips. One consequence is that for given sample sizes association studies will tend to have lower power than for SNP studies. (See Supplementary Figure 22 for power estimates.) Extrapolating from analyses described in 12 gives an estimate that the 3,432 CNVs we directly tested represent 42-50% of common (MAF > 5%) CNVs greater than 0.5kb in length which are polymorphic in a population with European ancestry.
Tagging by SNPs
In the literature discussing the possible role of common CNVs in human disease, there has been controversy over the extent to which CNVs will be in linkage disequilibrium (LD) with SNPs. If LD between CNVs and SNPs were similar to that between SNPs, SNPs typed in GWAS would act as tags not only for untyped SNPs but also for untyped CNVs, and in turn SNP-based GWAS studies would have indirectly explored copy-number variation for association with disease. (See for example 16 and 17 for opposite views.) Our large-scale genotyping of an extensive CNV catalogue allows us to settle this question. In fact, CNVs that are typed well in our experiment are in general well-tagged by SNPs – almost to the same extent that SNPs are well-tagged by SNPs (Supplementary Figure 20). Amongst variable 2- and 3-class CNVs passing QC with MAF > 10%, 79% have r2 > 0.8 with at least one SNP, for those with MAF < 5%, 22% have r2 > 0.8 with at least one SNP. This is consistent with the vast majority having arisen from unique mutational events at some time in the past. It follows that genetic variation, in the form of common CNVs which can be typed on our array, has already been explored indirectly for association with common human disease through the SNP-based GWAS. In passing, we note that the high correlations between our CNV calls and SNP genotypes provides strong indirect evidence that our CNV calls are capturing real variation. It is possible that the CNVs which we cannot type well are systematically different from those we can type, for example in having many more copy-number classes, and hence perhaps that they arise from repeated mutational events in the same region, in which case their LD properties with SNPs could also be systematically different from the CNVs we can type. We have no data that bear on this question, and it seems likely that such CNVs will be difficult to type genome-wide on any currently-available platforms.
Association Testing
We performed association testing at each of the CNVs which passed QC, in two parallel approaches. First, we applied a frequentist likelihood ratio association test that combines calling (using CNVtools) and testing into a single procedure, using an extension of an approach previously described 18. Second, we undertook Bayesian association analyses in which the posterior probabilities from CNVCALL were used to calculate a Bayes Factor to measure strength of association with the disease phenotypes. Important feature of both sets of analyses are that they correctly handle uncertainty in assignment of individuals to copy-number classes, and by allowing for some systematic differences in intensities between cases and controls, that they provide robustness against certain artefacts which could arise from differences in data properties between cases and controls. There were no substantial differences between the broad conclusions from the frequentist and Bayesian approaches.
Our association analyses were based on a model in which a single parameter quantifies the increase in disease risk between successive copy number classes, analogous to that underlying the trend test for SNP data. Various analyses of the robustness of our procedure, adequacy of the model, and lack of population structure were encouraging (see SoM and Online Methods). For example, Supplementary Figure 23 shows quantile-quantile (QQ)-plots for the primary comparison of each case collection against the combined controls, and for the analogous comparisons between the two control groups. These show generally good agreement with the expectation under the null hypothesis.
Careful analysis of our association testing revealed several sophisticated biological artefacts which can lead to false positive associations. These include dispersed duplications, whereby the variation at a CNV is not in the chromosomal location in the reference sequence to which the probes in the CNV uniquely match, and a DNA source effect whereby particular CNVs, and genome-wide intensity data, can look systematically different according to whether the assayed DNA was derived from blood or cell-lines. See Box “Some Artefacts in CNV Association Testing” for illustrations and further details.
Box. Some Artefacts in CNV Association Testing.
Some types of artefacts, such as population structure and calling artefacts, are very similar to those seen in SNP studies. Others, related to differences in data properties between cases and controls, can be potentially more serious for CNVs 26,27. In this box we draw attention to some specific artefacts of biological interest that we observed and which researchers should consider as explanations of putative disease-relevant associations. We note that, for the unwary, some of these artefacts could easily survive “replication” of an association.
Dispersed CNVs
Box Figure 1 shows cluster plots for a particular CNV (CNVR2664.1) which exhibits a strong case-control association signal for breast cancer cases (p = 5×10−143, higher copy number for disease) with a similar signal for rheumatoid arthritis (p = 3×10−27), and a signal in the opposite direction for coronary artery disease (p=4×10−30). The right hand class (green curve) has a higher frequency in BC (and RA), and a lower frequency in CAD. (Area under green curve is the same for each collection.) This turned out to be an artefact caused by differences in sex ratio in the various case and control samples (breast cancer: 100% female; rheumatoid arthritis: 74% female; coronary artery disease: 22% female; controls: 50% female). Comparing breast cancer cases against female controls abolished the signal. The CNV is annotated as being on chromosome 5 and all 10 probes in the CNV map uniquely to chromosome 5 in the human reference sequence. However, we found that SNPs which tagged the variation at this CNV all mapped to the X-chromosome and that the region containing the probes for this CNV is present on the X-chromosome in the Venter genome. We conclude that the CNV is a dispersed duplication, with the variation actually occurring on the X-chromosome, and not on chromosome 5. We found one similar example, of a CNV (CNVR1065.1, featuring in Table 2 as a replicated association) annotated as mapping uniquely to chromosome 2 which shows a strong signal in type 1 diabetes and rheumatoid arthritis. Careful examination shows it to be another dispersed duplication where the polymorphism is located in the HLA, and is well tagged by HLA SNPs known to be associated with both diseases. Supplementary Figure 27 shows the clear evidence from inter-chromosomal linkage disequilibrium that these two loci are dispersed duplications.
Variation in DNA source
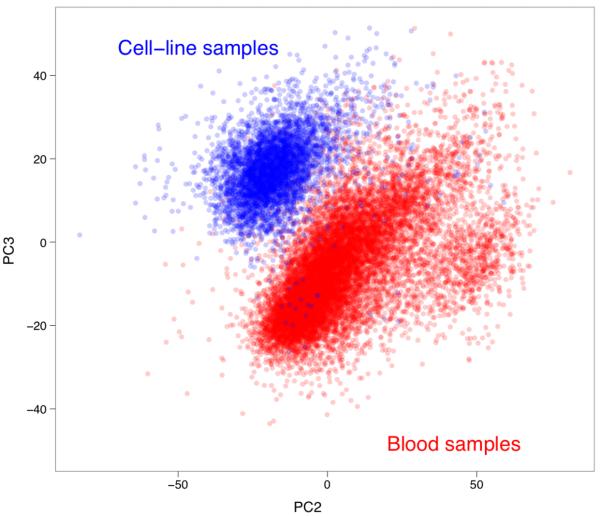
Box Figure 2 shows cluster plots for a different CNV (CNVR866.8) with striking differences in T2D as compared with the UKBS controls (or against just the 58C controls). The plots show histograms of normalised intensity ratios for 6 collections. Examination of the pattern across collections is interesting. The collections in the top row show a single tight peak towards the right of the plot. Those in the bottom row show a single, more dispersed, peak to the left. The collections in the middle row show evidence of both peaks. It turns out that for collections with the tight peak all DNA samples were derived from blood whereas all samples in the two collections with the single dispersed peak had DNA derived from cell lines. The remaining collections contain some DNAs derived from both sources. This CNV (and many others) thus exhibit systematically different behaviour depending on the DNA source. Box Figure 3 shows a plot of the second (PC2) and third (PC3) principal components of the array-wide intensity data (plot created using all samples post QC from all 10 collections using data from all CNVs with each point representing one sample, with the points coloured according to whether that sample was derived from blood (red) or cell-lines (blue)). It is clear that these two components can almost perfectly classify samples according to the source of the DNA.
Lymphoblastoid cell lines are typically grown from transformed B-cells, whereas DNA extracted from blood comes largely from a mixture of white blood cells. One specific feature of B-cells is that each B-cell has been subject to its own pattern of rearrangements around the immunoglobulin genes via the process of V-D-J recombination 28. This suggests a natural candidate for our observed DNA source effect, and indeed the CNV illustrated in Box Figure 2 is located close to one of the immunoglobulin genes, as are the other instances we have found of similar gross DNA source effects. But it is not the whole story. Principal components analysis of genome-wide intensity data with any probe mapping to within 1Mb of an immunoglobulin gene excluded from analysis (Supplementary Figure 29) shows reasonably clear discrimination by DNA source (though less clear than when all probes are included), with many probes, genome-wide, contributing to the discrimination.
Dispersed duplications and DNA source effects represent somewhat interesting biological artefacts. We also observed more prosaic effects. As one example, Supplementary Figure 30 shows that there are systematic effects on probe intensity of the row of the plate in which a sample was run.
Independent replication of putative association signals is a routine and essential aspect of SNP-based association studies. Particularly in view of the differences in data quality between SNP assays and CNV assays, and the wide range of possible artefacts in CNV studies, replication is even more important in the CNV context. Several possible approaches to replication are available. When a CNV is well-tagged by a SNP (or SNPs), replication can be undertaken by assessment of the signal at the tag SNP(s) in an independent sample, either by typing the SNP or by reference to published data. Where no SNP tag is available, direct typing of the CNV in independent samples is necessary, either using a qualitative breakpoint assay or a quantitative DNA dosage assay. In most cases there will be a choice of assays. Interestingly, replication via SNPs was possible for 15 out of 18 of the CNVs for which we undertook replication based on analysis of our penultimate data freeze.
Figure 3 plots p-values for the primary frequentist analysis for each CNV in each collection. Table 2 provides details of the top, replicated, association signals in our experiment after visual inspection of cluster plots to detect artefacts not removed by earlier QC. Cluster plots for each CNV in Table 2 are shown in Supplementary Figures 18 and 19, and Supplementary Files 2 and 3.
Figure 3. Genome-wide association results.
Distribution of −log10(p) along the 23 chromosomes where p is the p-value for the one degree-of-freedom test of association for each disease. The x-axis shows the chromosomes numbered from 1 (on the left) to X (on the right). CNVs included in these plots were filtered on the basis of a clustering quality score (see SoM for details) and manual inspection of the most significant associations. The two apparent associations on chromosome 2 for rheumatoid arthritis and type 1 diabetes result from a dispersed duplication in which the variation is actually located within the HLA locus (see Box).
Table 2. Replicated CNV associations and those at replicated loci.
Only one of the several associated CNVs mapping to the HLA in the reference sequence is shown for each of RA, T1D and CD. Further details of replication assays and methods are given in the supplementary material. AC_000138.1_44 is a novel sequence insertion present in the Venter genome sequence but not in the reference sequence and hence no chromosomal location is presented. Fitted number of classes – the number of diploid copy-number classes. P-value - Combined Controls – the p value from the frequentist association test combining UKBS and 58C as controls. log10(BF) - Combined Controls – the log10 of the Bayes Factor from the Bayesian association analysis combining UKBS and 58C as controls. OR - Combined Controls – The odds ratio estimated for each additional copy of the CNV based on both UKBS and 58C as controls. Extended Reference refers to the analogous quantities calculated in comparing cases of the disease in question with UKBS, 58C, and aetiologically-unrelated cases. Control MAF – The minor allele frequency in controls (UKBS +58C). Case MAF – The minor allele frequency in cases. Minor allele frequency is only estimated for CNVs with 3 or fewer copy number classes.
| Disease | CNV | Chromosome | Start (bp) |
Length (kb) |
Locus | Fitted number ofclasses |
P-value -Combined Controls |
P-value -Extended Reference |
log10(BF) -Combined Controls |
log10(BF) -Extended Reference |
OR -Combined Controls |
OR -Extended Reference |
Control MAF |
Case MAF |
Replication: cases / controls |
Replication P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2D | CNVR5583.1 | 12 | 69,818,942 | 1.0 | TSPAN8 | 3 | 3.9E-05 | 2.5E-06 | 2.8 | 4.3 | 0.85 | 0.85 | 0.40 | 0.36 | 4549 / 5579# | 3.9E-05 |
| CD | CNVR2646.1 | 5 | 150,157,836 | 3.9 | IRGM | 3 | 1.1E-07 | 5.5E-05 | 5.8 | 4.1 | 0.68 | 0.75 | 0.07 | 0.10 | 6894 / 7977# | 7.5E-11 |
| CD | CNVR2647.1 | 5 | 150,183,562 | 20.1 | IRGM | 3 | 1.0E-07 | 4.3E-05 | 6.1 | 3.8 | 0.68 | 0.76 | 0.07 | 0.10 | 6894 / 7977# | 3.9E-10 |
| CD | CNVR2841.20 | 6 | 31,416,574 | 5.1 | HLA | 3 | 1.7E-05 | 1.1E-05 | 3.6 | 3.9 | 0.80 | 0.82 | 0.19 | 0.23 | NA | NA |
| T1D | CNVR2845.46 | 6 | 32,582,950 | 6.7 | HLA | 2 | 8.0E-153 | 2.1E-196 | 125.5 | 154.4 | 0.20 | 0.26 | 0.14 | 0.01 | NA | NA |
| RA | CNVR2845.14 | 6 | 32,609,209 | 4.0 | HLA | 4 | 1.4E-39 | 8.1E-60 | 51.5 | 73.5 | 1.77 | 1.83 | NA | NA | NA | NA |
| RA | CNVR1065.1 | 2⇒6 | 179,004,449 | 0.8 | HLA | 3 | 6.8E-49 | 1.6E-69 | 51.0 | 73.7 | 1.85 | 1.94 | 0.36 | 0.49 | NA | NA |
| T1D | CNVR1065.1 | 2⇒6 | 179,004,449 | 0.8 | HLA | 3 | 1.3E-29 | 1.1E-39 | 28.0 | 38.4 | 1.62 | 1.61 | 0.36 | 0.47 | NA | NA |
| RA |
AC_000138.1 _44 |
NA | NA | 5.6 | HLA | 3 | 8.3E-04 | 1.1E-05 | 1.3 | 2.7 | 0.87 | 0.86 | 0.25 | 0.28 | 3398 / 2743 | 1.1E-03 |
| T1D |
AC_000138.1 _44 |
NA | NA | 5.6 | HLA | 3 | 2.0E-31 | 2.7E-45 | 31.0 | 45.1 | 0.59 | 0.57 | 0.25 | 0.36 | 3883 / 2649 | 7.3E-50 |
| CD | CNVR7113.6 | 17 | 40,930,407 | 33.9 | Chr17inv | 3 | 1.2E-03 | 5.8E-04 | 1.4 | 1.6 | 1.15 | 1.14 | 0.24 | 0.21 | 4978 / 6069# | 8.6E-05 |
| T1D | CNVR7113.6 | 17 | 40,930,407 | 33.9 | Chr17inv | 3 | 1.6E-03 | 7.5E-04 | 1.0 | 1.2 | 1.13 | 1.12 | 0.24 | 0.21 | 7911 / 9395# | 4.6E-06 |
Replication sample includes WTCCC samples
There is one positive control for the diseases we studied, namely the known CNV association at the IRGM locus in Crohn's disease 7. Reassuringly, our study found this association (p= 1 × 10−7, odds ratio (OR) = 0.68; throughout, all ORs are with respect to increasing copy number).
We identified three loci – HLA for Crohn's disease, rheumatoid arthritis, and type 1 diabetes; IRGM for Crohn's disease; and TSPAN8 for type 2 diabetes – at which CNVs appeared associated with disease, all of which we convincingly replicated through previously typed SNPs that tag the CNV, and a fourth locus (CNV7113.6), at which there is suggestive evidence for association and replication in both Crohn's disease and type 1 diabetes.
We observed CNVs in the HLA region associated variously with Crohn's disease (CNVR2841.20, p= 1.2 × 10−5, OR = 0.80), rheumatoid arthritis (CNVR2845.14, p= 1.4 × 10−39, OR = 1.77), and type 1 diabetes (CNVR2845.46, p= 8 × 10−153, OR = 0.2). Copy number variation has previously been documented on various HLA haplotypes 19 and due to the extensive linkage disequilibrium in the region it is perhaps not unexpected to have found CNV associations in our direct study. Linkage disequilibrium across the HLA region has hampered attempts to fine-map causal variation across this locus, and we have no evidence that suggests that the HLA CNVs associated with autoimmune diseases in this study represent signals independent of the known associated haplotypes.
We identified two distinct CNVs 22kb apart upstream of the IRGM gene, both of which are associated with Crohn's disease. The longer CNV (CNVR2647.1, p= 1.0 × 10−7, OR = 0.68) has previously been identified 7 as a possible causal variant on an associated haplotype first identified through SNP GWAS 14, and acted as our positive control but the association of the smaller CNV (CNVR2646.1, p= 1.1 × 10−7, OR = 0.68, located <2kb downstream from a different gene, MST150) is a novel observation. While direct experimental evidence links the associated haplotypes with variation in expression of the IRGM gene, it does not bear on the question of which of the two CNVs or the associated SNPs might be driving this variation 7. Our conditional regression analyses on the two CNVs and SNPs on this haplotype do not point significantly to any one of these as being more strongly associated.
SNP variation in the TSPAN8 locus was recently shown to be reproducibly associated with type 2 diabetes 20, but the potential role of a CNV is a novel observation. This CNV (CNVR5583.1, p= 3.9 × 10−5, OR = 0.85) potentially encompasses part or all of an exon of TSPAN8 and so is a plausible causal variant. The most significantly associated SNP identified in the recent meta-analysis is only weakly correlated with the CNV as originally tested (r2 =0.17), and so the CNV may simply be weakly correlated with the true causal variant. Closer examination of probe-level data at this CNV suggests a series of different events (including an inverted duplication and a deletion) resulting in more complex haplotypes than those tested for association by our automated approach. With this more refined definition of haplotypes the signal is somewhat stronger. See SoM for details.
CNVR7113.6 lies within a cluster of segmentally duplicated sequences that demarcate one end of a common 900kb inversion polymorphism on chromosome 17 that has previously been shown to be associated with number of children and higher meiotic recombination in females 21. The CNV shows weak evidence for association with Crohn's disease (p= 1.8 × 10−3, OR = 1.15) and type 1 diabetes (p= 1.1 × 10−3, OR = 1.13), but is in extremely high LD (r2=1) with SNPs known to tag the inversion, and so is in tight LD with a long haplotype spanning many possible causal variants. This CNV encompasses at least one spliced transcript, but no high confidence gene annotations. Fine-mapping the causal variant within such a long, tightly-linked, haplotype is likely to prove challenging.
In addition to the loci in Table 2, we undertook replication on thirteen other loci, detailed in Supplementary Table 13, for which there was some evidence of association (p<1×10−4 or log10(Bayes Factor [BF])> 2.1) in our analysis of the penultimate data freeze. Replication results were negative for all these loci. Several other loci for which there is weak evidence (p < 1×10−4 or log10(BF) > 2.6) for association in our final data analysis are listed in Supplementary Table 14.
To further investigate the potential role of CNVs as pathogenically relevant variants underlying published SNP-associations we took 94 association intervals in T1D, CD, and T2D (excluding the HLA), and for the index SNP in each association interval assessed its correlation with our calls at 3,432 CNVs. We identified two index SNPs as being correlated with an r2 of greater than 0.5 with a called CNV. The SNPs were: rs11747270 with both CNVR2647.1 and CNVR2646.1 (IRGM), and rs2301436 and CNVR3164.1 (CCR6), both for Crohn's disease. Both of these association intervals were also identified in an independent analysis using CNV calls on HapMap samples by Conrad et al. 12.
As a further test of our approach, we examined three multi-allelic CNVs which have attracted attention in the literature, both for the challenges of obtaining reliable data, and for putative associations with a range of autoimmune diseases: CCL3L1 (our CNVR7077.12); Beta-Defensins (CNVR3771.10), and FCGR3A/B (CNVR383.1) 10,22,23,24. Encouragingly, all three CNVs pass QC and give good data. Figure 2 shows cluster plots for these CNVs in our experiment. The best calls for the three CNVs required the use of two analysis pipelines (sets of choices about normalisation and probe summaries) different from our standard pipeline. None of the CNVs shows significant association with the three autoimmune diseases in our study after allowance for multiple testing. In particular, we do not see formally significant evidence to replicate the reported association for CCL3L1 and rheumatoid arthritis 24 (nominal p = 0.058).
We also assessed whether CNVs which delete all or part of exons might be enriched amongst disease susceptibility loci, even if our study were not well-powered enough to see statistically significant evidence of association for individual CNVs. To do so, we compared the 53 exonic deletion CNVs 12 which passed QC with collections of CNVs of the same size, matched for MAF and numbers of classes. We used a (two-sided) Wilcoxon signed-rank test 25 to ask whether the strength of signal for association (measured by Bayes Factors) was systematically different for the exon-deletion CNVs as compared to the matched CNVs. We found no evidence that deletion of an exon systematically changed evidence for association (see SoM). In a related analysis, we compared CNVs passing QC which were well tagged by SNPs (r2 > 0.8) to those passing QC which were not, again matching for MAF and number of classes (excluding low MAF CNVs and those failing Hardy-Weinberg equilibrium tests to avoid calling artefacts). There was no evidence that CNVs passing QC which are not well tagged by SNPs are enriched for stronger signals of association compared to those which were well tagged (see SoM).
Discussion
We have undertaken a genome-wide association study of common copy-number variation in eight diseases, by developing a novel array targeting most of a recently discovered set of CNVs. Our findings inform understanding of the genetic contributions to common disease, offer methodological insights into CNV analysis, and provide a resource for human genetics research.
One major conclusion is that considerable care is needed in analysing copy-number data from array CGH experiments. Choices of normalisation, probe summary, and probe weighting can make major differences to data quality and utility in association testing. Strikingly, the optimal choices vary greatly across the CNVs we studied.
A second major conclusion is that CNV association analyses are susceptible to a range of artefacts which can lead to false positive associations. Some are a consequence of the less-robust nature of the data compared to SNP-chips. But others, such as systematic differences depending on DNA source (eg. blood v. cell lines), and dispersed duplications, are more subtle. Several artefacts could survive replication studies. Simultaneously studying eight diseases helped greatly in identifying these artefacts and stringent QC was invaluable in eliminating false positive associations. At least for currently available CNV-typing platforms, we recommend considerable care in interpreting putative CNV associations combined with independent replication, on a different experimental platform.
Despite the important technical challenges and potential artefacts discussed above, we have demonstrated that high-confidence CNV calls can be assigned in large, real-world case-control samples for a substantial proportion of the common copy number variation estimated to be present in the human genome. We have identified directly several CNV loci that are associated with common disease. Such loci could contribute to disease pathogenesis. However, the loci identified are well tagged by SNPs and, hence, the associations can be, and were, detected indirectly via SNP association studies.
There is a striking difference between the number of confirmed, replicated associations from our CNV study (3 loci) and that from the comparably-powered WTCCC1 SNP-GWAS of seven diseases and its immediate follow-up (~24 loci). (In assessing the importance of CNVs in disease, it is the absolute number of associations, rather than the proportion among loci tested, which is important.) Following 12 we estimated that our study directly tests approximately half of all autosomal CNVs >500bp long, with MAF >5%. For such CNVs, our power averages over 80% for effects with odds ratios >1.4, and ~50% for odds ratio =1.25 (Supplementary Figure 22). We conclude that at least for the eight diseases studied, and probably more generally, there are unlikely to be many associated CNVs with effects of this magnitude.
Might there be many more common disease-associated CNVs each of small effect, in the way we now know to be the case with SNP associations for many diseases? The total number of CNVs over 500bp with MAF > 5% is limited (estimated to be under 4,000 12), so unless many of these simultaneously affect many different diseases (something for which we saw no evidence outside HLA) there would seem to be insufficient such CNVs for hundreds to be associated with each of many common diseases. In addition, most common CNVs (MAF > 5%) are well tagged by SNPs, and thus amenable to indirect study by SNP GWAS. Examining the large meta-analyses of SNP GWAS for Crohn's disease, type 1 and type 2 diabetes, there were 95 published associated loci of which only 3, including HLA, had the property that CNVs correlated with the associated SNPs; two of these were detected in our direct study.
We conclude that common copy number variants typable on current platforms are unlikely to play a major role in the genetic basis of common diseases, either through particular CNVs having moderate or large effects (odds ratios > 1.3 say) or through many such CNVs having small effects. In particular, such common CNVs seem unlikely to account for a substantial proportion of the “missing heritability” for these diseases. Amongst the CNVs we could type well, those not well-tagged by SNPs have the same overall association properties as those which are well-tagged. We saw no enrichment of association signals amongst CNVs involving exonic deletions.
We have argued elsewhere 14 that the concept of “genome-wide significance” is misguided, and that under frequentist and Bayesian approaches it is not the number of tests performed, but rather the prior probability of association at each locus, which should determine appropriate p-value thresholds. Here, to reduce the possibility of missing genuine associations, we deliberately set relaxed thresholds for taking CNVs into replication studies. Having completed these analyses the hypothesis that, a priori, an arbitrary common CNV is much more likely than an arbitrary common SNP to affect disease susceptibility is not supported by our data.
Limitations
Our findings should be interpreted within the context of several limitations. First, despite our successes in robustly testing some of the previously noted challenging CNVs in the genome, for some CNVs we could not reliably assign copy number classes from our assay. We estimate that somewhat under half of these were not polymorphic in our data, being either false positives in the discovery experiment, or very rare in the UK population. For the remainder, we were also unable to perform reliable association analyses based directly on intensity measurements (that is, without first assigning individuals to copy number classes; data not shown). Such CNVs might plausibly be systematically different from those we do type successfully, in which case it is not possible to extrapolate from our results to their potential role in human disease. Second, we note that we have not studied CNVs of sequences not present in the reference assembly, high-copy number repeats such as LINE elements, or most polymorphic tandem repeat arrays and our findings may not generalize to such variation. Finally, our experiment was powered to detect associations with common copy number variation and our observations and conclusions do not necessarily generalize to the study of rare copy number variants. Different approaches will be necessary to investigate the contribution of such variation to common disease.
Methods Summary
A detailed description of materials and methods is given in Online Methods with further details in SoM.
Pilot Study
A total of 384 samples spanning a range of DNA quality were assayed for 156 previously-identified CNVs on each of three different platforms: Agilent CGH, NimbleGen CGH and Illumina iSelect. The pilot experiment contained many more probes per CNV than we anticipated using in the main study, and replicates of these probes, to allow an assessment of data quality as a function of the number of probes per CNV and of the merits of replicating probes predicted in advance to perform well, compared to using distinct probes.
Sample Selection
Case samples came from previously established UK collections. Control samples came from two sources: half from the 1958 Birth Cohort and half from a UK Blood Service sample. Approximately 80% of samples had been included within the WTCCC SNP GWAS study. The 610 duplicate samples were drawn from all collections.
Array Design
The main study used an Agilent CGH array comprising 105,072 long oligonucleotide probes. Probes were selected to target CNVs identified mainly through the GSV discovery experiment 12 with some coming from other sources. Ten non-polymorphic regions of the X-chromosome were assayed for control purposes.
Array Processing
Arrays were run at Oxford Gene Technology (OGT). The samples were processed in batches of 47 samples drawn from two different collections, with each batch containing one control sample for QC purposes. These batches were randomised to protect against systematic biases in data characteristics between collections.
Data Analysis
Primary data and low-level summary statistics were produced at OGT. All substantive data analyses were undertaken within the consortium. Plates failing QC metrics were rerun as were 1,709 of the least well-performing samples. Details of the common CNVs assayed in this study, including any tag SNP, are given in supplementary data (http://www.wtccc.org.uk/wtcccplus_cnv/supplemental.shtml).
Methods
Pilot Experiment
Full details are given in the SoM, but in brief a total of 384 samples from four different collections spanning the range of DNA quality encountered in our previous WTCCC SNP-based association study 14 were assayed for 156 previously-identified CNVs on each of three different platforms: Agilent Comparative Genomic Hybridization (CGH), and NimbleGen CGH (run in service laboratories) and Illumina iSelect (run at the Sanger Institute). The pilot experiment contained many more probes per CNV (40-90 depending on platform) than we anticipated using in the main study, and replicates of these probes, to allow an assessment of data quality as a function of the number of probes per CNV and of the merits of replicating probes predicted in advance to perform well, compared to using distinct probes.
The Agilent CGH platform performed best in our pilot and we settled on an array which comprised 105,072 long oligonucleotide probes. Based on the pilot data we aimed to target each CNV with 10 distinct probes. Actual numbers of probes per CNV on the array varied from this for several reasons (see SoM and Supplementary Figure 9), and we included in our analyses any CNV with at least one probe on the array.
Array Content, Assay, and Samples for the Main Experiment
Array Content
The GSV discovery experiment 12 involved 20 HapMap Utah residents with European ancestry (CEU) and 20 HapMap Yoruban (YRI) individuals, and 1 Polymorphism Discovery Resource individual, assayed via 20 NimbleGen arrays containing a total of 42,000,000 probes tiled across the assayable portion of the human reference genome. We prioritised CNVs for our experiment based on their frequency in the discovery sample, with those identified in CEU individuals given precedence. A total of 10,835 out of 11,700 CNVs were included from the list provided by the GSV, with those not included on the array being present in discovery in only 1 YRI individual and not overlapping genes or highly conserved elements. This list was augmented by any common CNVs not present among the GSV list found from analyses of Affymetrix SNP 6.0 data in HapMap 2 samples (83 CNVs), Illumina 1M data in HapMap 3 samples (82 CNVs), analyses of Affymetrix 500K samples (18 CNVs) 7,29,30, and from our own analyses of WTCCC1 SNP data (231 CNVs). In addition, we sought to identify copy number variants not present in the human reference sequence through analyses of published 31,32 novel sequence insertions (292 CNVs in total). Thus in total, our array targeted 11,541 putative CNVs. Ten non-polymorphic regions of the X-chromosome were also assayed for control purposes.
Most loci targeted on the CNV-typing array derive from microarray-based CNV discovery, which is inherently imprecise. In contrast to SNP discovery by sequencing, arrays do not provide nucleotide level resolution, nor do they locate additional copies of a sequence in the genome. As a result, when CNVs called in different individuals overlap, but are not identical, these could be called as one or two different CNVs, and where discovered CNVs involve probes which map to multiple places in the reference genome, they might be called as CNVs in each of these locations. Interpretation of counts of CNVs from discovery experiments is thus not straightforward. Data on CNVs across thousands of individuals provides added power to refine CNV definitions and derive a non-redundant set of CNVs. In addition, our CNV-typing array draws together CNVs from different sources and additional redundancy between these, while minimised during array design, can be identified and removed. Analyses of the final array design revealed 434 of the 11,541 CNVs to be redundant because they were targeted by exactly the same probes as other CNVs on the array, and analysis of our array data revealed a further 213 of 562 CNVs to be redundant from instances where overlapping CNVs passing QC were called as distinct in discovery yet had effectively identical copy-number calls. See SoM section 3.1 for further details on array content.
Assay
Arrays were run at Oxford Gene Technology (OGT), with each plate containing one control sample for QC purposes. Primary data and low-level summary statistics were produced at OGT. All substantive data analyses were undertaken within the consortium. Plates which failed pre-specified QC metrics were rerun on the array, and in addition we repeated 1709 of the least well-performing samples, chosen according to our own QC analyses. See SoM for further details.
Samples
The WTCCC CNV study analysed cases from 8 common diseases (Breast Cancer (BC), Bipolar Disorder (BD), Coronary Artery Disease (CAD), Crohn's Disease (CD), Hypertension (HT), Rheumatoid Arthritis (RA), Type I Diabetes (T1D), and Type 2 Diabetes (T2D)) and two control cohorts (1958 Birth Cohort (58C) and the UK Blood Service collection (UKBS)). The number of subjects from each cohort that were analysed and the numbers that passed each phase of the quality control (QC) procedures within this study are shown in Supplementary Table 7. For BD, CAD, CD, HT, RA, T1D, T2D, and the two control cohorts, a large proportion of the subjects studied in this experiment were the same as those in the WTCCC1 SNP genome-wide association study (GWAS) (Supplementary Table 2). Where sufficient DNA was not available for the original WTCCC1 individuals, additional new samples from the same cohorts were used, selected using the same approaches used for the WTCCC1 samples. Any samples that failed any of the relevant QC metrics in WTCCC1 were excluded from consideration for this experiment. The BC cohort was not included in the WTCCC1 SNP GWAS study.
Data Pre-Processing, CNV Calling and Quality Control
Data Pre-Processing
For each of the targeted loci, the subset of probes that target the locus of interest (at least 1bp overlap) whilst also targeting the least number of additional CNVs was selected for assaying (see SoM section 4.2 for more details). A total of 16 different analysis “pipelines” were used to create one-dimensional intensity summaries for each CNV. First, a range of different methods were used to create single intensity measurements for each probe from the red channel (test DNA) and green channel (reference DNA) intensity data. This included different methods for normalization of the signals (see SoM section 4.3 for details). Secondly, some pipelines incorporated a new method called probe variance scaling (PVS) which weights probes based on information derived from duplicate samples (see SoM section 4.5 for details). Thirdly, some pipelines used the first principal component of the normalized probe intensities to summarise the probe-level data to CNV-level data, whereas other pipelines used the mean of the probe intensities. Finally, some pipelines additionally used a linear discriminant function (LDF) to further refine the summaries based on information from an initial round of genotype calling (see SoM section 4.4 for details).
CNV Calling
Algorithmic details of the two calling methods used (“CNVtools” and “CNVCALL”) are provided in SoM section 6. Each method was applied separately to the intensity summaries created from each of the 16 pre-processing pipelines for each CNV locus.
Quality Control
Samples were excluded on the basis of sample handling errors, evidence of non-European ancestry, evidence of sample mixing, manufacturer's recommendations on data quality, outlying values of various pre-calling and post-calling quality metrics, and identity or close relatedness to other samples (see SoM section 5.1 for further details). To choose which pipeline to use for a given CNV we used the pipeline which gave the highest number of classes, and the highest average posterior probability in cases where more than 1 pipeline gave the same maximum number of classes. CNVs were excluded that had identical probe sets to other CNVs, that were called with 1 class in all pre-processing pipelines, that had low average posterior calls in all pre-processing pipelines, or that had a high calls correlation with an overlapping CNV (see SoM section 5.2 for further details).
Properties of CNVs
Single class CNVs
Supplementary Table 15 shows the proportion of the single-class CNVs from the GSV discovery project broken down according to the number of individuals and population(s) in which they were discovered. Of the GSV CNVs discovered in CEU, 52% are single class in our data, whereas a higher proportion (74%) of GSV CNVs discovered exclusively in YRI are single class, as would be expected. CNVs at which distinct copy number classes cannot be distinguished might result because: (i) although polymorphic, the signal to noise ratio at that CNV does not allow reliable identification of distinct copy-number classes; (ii) the copy-number variant has an extremely low population frequency; or (iii) the CNV was a false positive in a discovery experiment and is in fact monomorphic. In a genuinely polymorphic CNV, the intensity measurements within a pair of duplicates should be more similar than between a random pair of distinct individuals. Intensity comparisons between duplicates and random pairs of individuals thus allow estimates of the proportion of single-class CNVs which are not copy-number variable in our data (see SoM). These estimates range from ~23% for single-class CNVs discovered in two or more CEU individuals, to ~50% of single-class CNVs discovered exclusively in YRI (see SoM for details). We estimate 18% of GSV CNVs discovered in CEU do not exhibit polymorphism in our UK sample. This figure is similar to the GSV estimate for false positives in discovery of 15% 12. Overall, considering CNVs on the array from all sources, we estimate that 26% do not exhibit polymorphism, so that just under half of the CNVs which appear in our data to have a single class are likely not to be polymorphic. Not all of these will be false positives in discovery, some represent CNVs which are either unique to the individual in which they were discovered or are extremely rare in the UK population.
Multi-class CNVs
Conrad et al 12 have estimated that 83% of the bi-allelic CNVs they genotyped represent deletions, with the remainder being duplications. Supplementary Table 7 compares the number of copy-number classes estimated by the two calling algorithms used in the analyses for each of the CNVs passing QC. Most differences in numbers of called classes between the algorithms arise from CNVs where one class is very rare, and is handled differently by the algorithms (for example called as a separate class in one algorithm but classed as outlier samples or merged with a larger class by the other).
These 3,432 CNVs include 80% of the CNVs genotyped on the Affymetrix 6.0 array that are common (MAF > 5%) in a population with European ancestry 33, conversely only 15% of the common CNVs we called could be called using the Affymetrix 6.0 array.
Allele Frequencies
We calculated minor allele frequencies (MAFs) for 2- and 3-class CNVs by assuming these CNVs were biallelic and using the expected posterior genotype counts (see SoM section 7.3 for further details).
Tagging by SNPs
In order to determine how well-tagged the CNVs analysed in our experiment were by SNPs, we carried out correlation analyses using control samples that were common to the current studies and other WTCCC studies. We analysed three different collections of SNPs. We used imputed HapMap2 SNP calls in the WTCCC1 study which used the Affymetrix 500k array, and actual calls from the WTCCC2 study using both the Affymetrix 6.0 array and a custom Illumina 1.2M array. In all cases we used samples from the UKBS collection (see SoM section 7.1 for further details).
Geographical Variation
Geographical information, at the level of 13 pre-defined regions of the UK, was available for 82% of the samples in our study and we undertook analyses for differences in copy-number class frequencies between regions. The results, shown in Supplementary Figure 24 confirm that there is no major genome-wide population structure, but that, unsurprisingly, there is differentiation at CNVs within HLA. It does not seem easy to determine whether other regions with low p-values in this test represent genuine departures from the null hypothesis of no differentiation, rather than chance effects, though we note that the third most regionally differentiated CNV outside the HLA (CNVR7722.1, p = 3 × 10−5, 12-df) is a deletion located within the gene LILRA3, which may act as soluble receptor for class I MHC antigens, and so would be consistent with the observed HLA stratification. This deletion is also the subject of a reported disease association 34 in multiple sclerosis, a finding which may require some caution given the level of geographical stratification at this CNV in our data. See SoM section 9.1 for further details.
Association Testing
Diagnostic plots such as quantile-quantile (QQ) and cluster plots were created using R. Cluster plots were visually inspected for all CNVs with putative associations.
Principal component analysis (PCA) was applied to the summarised intensity levels for all CNVs, and for all samples which passed QC. Plots of the first ten principal components were coloured by various sample parameters and these revealed some of the artefacts described in the box.
Where possible, replication was carried out by using data from other studies for SNPs that tag the CNVs of interest. Where there was no SNP tag available, breakpoint or direct quantitative CNV assays were designed (see SoM section 9 for further details).
We used a two-sided Wilcoxon signed-rank test to test for differences between distributions of Bayes factors between different subsets of CNVs (those which delete all or part of an exon vs. those which do not, and CNVs that are well-tagged by SNPs vs. those which are not well-tagged). Further details are given in SoM section 9.5)
Testing for population stratification
All our samples are from within the UK, and we have excluded any for which the genetic data suggests evidence of non-European ancestry. All collections in this study, apart from breast cancer, were involved in the WTCCC SNP GWAS, and across these collections, 80% of samples coincided between the two studies. Analysis of the WTCCC SNP data 14 established that population structure was not a major factor confounding association testing. Similar analyses using SNP data available for the breast cancer samples yielded similar results (data not shown). These SNP results reinforce the evidence from the QQ-plots in Supplementary Figure 23 and our geographical analyses of the CNV data.
Expanded reference group analysis
In addition to our primary case-control analyses, following 14 we also undertook expanded reference group analyses, in which copy-number class frequencies in cases for a particular disease are compared with those for controls and the other diseases with no aetiological or known genetic connection (see Supplementary Table 10 for details).
Other Analyses
We used information on variability between duplicate samples to determine whether CNVs called with 1 class show signals of polymorphism (details are given in SoM section 9.2).
We used estimates of the number of common autosomal CNVs segregating in a population of European ancestry from Conrad et al. to estimate the coverage of common autosomal CNVs in our study (see SoM section 9.3 for further details).
We designed a series of PCR primers to further analyse the complex signals associated with CNVR5583.1 found in the TSPAN8 region. Further details are given in SoM section 9.4.
Additional References
- 29.International HapMap Project. < http://hapmap.ncbi.nlm.nih.gov/>.
- 30.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nature Genetics. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 34.Koch S, et al. Association of multiple sclerosis with ILT6 deficiency. Genes Immun. 2005;6:445–447. doi: 10.1038/sj.gene.6364187. [DOI] [PubMed] [Google Scholar]
Supplementary Material
Box Figure 1.
Box Figure 2.
Box Figure 3.
Acknowledgements
The principal funder of this project was the Wellcome Trust. Many individuals, groups, consortia, organizations and funding bodies have made important contributions to sample collections and coordination of the scientific analyses. Details are provided in SoM section 11. We are indebted to all those who participated within the sample collections.
Footnotes
Summary information for the CNVs studied, including genomic locations, numbers of classes and SNP tags on different platforms is available at http://www.wtccc.org.uk/wtcccplus_cnv/supplemental.shtml. Full data are available, under a data access mechanism, from the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/page.php).
The authors declare no competing financial interests.
MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Heath Park, Cardiff, CF14 4XN, UK.y
The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA UK.
Department of Statistics, University of Oxford, 1 South Parks Road, Oxford, OX1 3TG, UK.
The Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory, Department of Medical Genetics, Cambridge Institute for Medical Research, University of Cambridge, Wellcome Trust/MRC Building, Cambridge CB2 0XY, UK.
Departments of Health Sciences and Genetics, University of Leicester, 217 Adrian Building, University Road, Leicester, LE1 7RH, UK.
Genetics of Complex Traits, Peninsula College of Medicine and Dentistry University of Exeter, EX1 2LU, UK.
Department of Haematology, University of Cambridge, Long Road, Cambridge, CB2 0PT, UK.
National Health Service Blood and Transplant, Cambridge Centre, Long Road, Cambridge CB2 0PT, UK.
Multidisciplinary Cardiovascular Research Centre (MCRC), Leeds Institute of Genetics, Health and Therapeutics (LIGHT), University of Leeds, Leeds, LS2 9JT, UK.
arc Epidemiology Unit, Stopford Building, University of Manchester, Oxford Road, Manchester, M13 9PT, UK.
Oxford Centre for Diabetes, Endocrinology and Medicine, University of Oxford, Churchill Hospital, Oxford OX3 7LJ, UK.
Department of Medical and Molecular Genetics, King's College London School of Medicine, 8th Floor Guy's Tower, Guy's Hospital, London, SE1 9RT, UK.
Centre for Endocrinology, Diabetes and Metabolism, Institute of Biomedical Research, University of Birmingham, Birmingham, B15 2TT, UK.
Department of Cardiovascular Sciences, University of Leicester, Glenfield Hospital, Groby Road, Leicester LE3 9QP, UK.
IBD Genetics Research Group, Addenbrooke's Hospital, Cambridge, CB2 0QQ, UK.
University of Aberdeen, Institute of Medical Sciences, Foresterhill, Aberdeen AB25 2ZD, UK.
SGDP, The Institute of Psychiatry, King's College London, De Crespigny Park, Denmark Hill, London SE5 8AF, UK.
Clinical Pharmacology Unit, University of Cambridge, Addenbrookes Hospital, Hills Road, Cambridge CB2 2QQ, UK.
Section of Cancer Genetics, Institute of Cancer Research, 15 Cotswold Road, Sutton SM2 5NG, UK.
Department of Psychiatry, University of Birmingham, National Centre for Mental Health, 25 Vincent Drive, Birmingham, B15 2FG, UK.
BHF Glasgow Cardiovascular Research Centre, University of Glasgow, 126 University Place, Glasgow, G12 8TA, UK.
Gastrointestinal Unit, Division of Medical Sciences, School of Molecular and Clinical Medicine, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU, UK.
Academic Unit of Genetic Medicine, University of Southampton, Southampton, UK.
Endoscopy Regional Training Unit, Torbay Hospital, Torbay TQ2 7AA, UK.
Academic Unit of Musculoskeletal Disease, University of Leeds, Chapel Allerton Hospital, Leeds, West Yorkshire LS7 4SA, UK.
MRC Centre for Causal Analyses in Translational Epidemiology, Department of Social Medicine, University of Bristol, Bristol, BS8 2BN, UK.
Department of Medical Genetics, Manchester Academic Health Science Centre (MAHSC), University of Manchester, Manchester M13 0JH, UK.
School of Neurology, Neurobiology and Psychiatry, Royal Victoria Infirmary, Queen Victoria Road, Newcastle upon Tyne, NE1 4LP, UK.
The Centre for Applied Genomics and Program in Genetics and Genomic Biology, The Hospital for Sick Children, MaRS Centre- East Tower, 101 College St, Room 14-701, Toronto, Ontario M5G 1L7, Canada.
Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, Uppsala 75185, Sweden.
Institute for Digestive Diseases, University College London Hospitals Trust, London NW1 2BU, UK.
University Hospital Birmingham NHS Foundation Trust, Birmingham, B15 2TT, UK.
Genetics of Complex Traits, Peninsula College of Medicine and Dentistry, University of Exeter, Magdalen Road, Exeter, EX1 2LU, UK.
Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, Boston MA02115, USA.
University of Oxford, Institute of Musculoskeletal Sciences, Botnar Research Centre, Oxford, OX3 7LD, UK.
Centre for Diabetes and Metabolic Medicine, Barts and The London, Royal London Hospital, Whitechapel, London, E1 1BB, UK.
Bone Research Group, Department of Medicine and Therapeutics, University of Aberdeen, Aberdeen, AB25 2ZD, UK.
Clinical Pharmacology and Barts and The London Genome Centre, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK.
Institute of Cellular Medicine, Musculoskeletal Research Group, 4th Floor, Catherine Cookson Building, The Medical School, Framlington Place, Newcastle upon Tyne, NE2 4HH, UK.
Gastroenterology Unit, Radcliffe Infirmary, University of Oxford, Oxford, OX2 6HE, UK.
Centre National de Genotypage, 2, Rue Gaston Cremieux, Evry, Paris 91057, France.
Department of Gastroenterology & Hepatology, University of Newcastle upon Tyne, Royal Victoria Infirmary, Newcastle upon Tyne NE1 4LP, UK.
ALSPAC Laboratory, Department of Social Medicine, University of Bristol, BS8 2BN, UK.
Division of Nutritional Sciences, King's College London School of Biomedical and Health Sciences, London SE1 9NH, UK.
NIHR-Leeds Musculoskeletal Biomedical Research Unit, University of Leeds, Chapel Allerton Hospital, Leeds, West Yorkshire LS7 4SA, UK.
Department of General Internal Medicine, Ninewells Hospital and Medical School, Ninewells Avenue, Dundee DD1 9SY, UK.
INSERM UMR915, L'Institut du Thorax, Nantes, 44035, France.
Clinical and Academic Rheumatology, Kings College Hosptal National Health Service Foundation Trust, Denmark Hill, London SE5 9RS, UK.
University of Toronto, St. Michael's Hospital, 30 Bond Street, Toronto, Ontario M5B 1W8, Canada.
University of Bath, Claverdon, Norwood House, Room 5.11a Bath Somerset BA2 7AY, UK.
Diabetes Research Group, School of Clinical Medical Sciences, Newcastle University, Framlington Place, Newcastle upon Tyne NE2 4HH, UK.
Medicine and Therapeutics, Aberdeen Royal Infirmary, Foresterhill, Aberdeen, Grampian AB9 2ZB, UK.
School of Medicine and Biomedical Sciences, University of Sheffield, Sheffield, S10 2JF, UK.
Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, Nuffield Orthopaedic Centre, University of Oxford, Windmill Road, Headington, Oxford, OX3 7LD, UK.
UBC Institute of Mental Health, 430-5950 University Boulevard Vancouver, British Columbia, V6T 1Z3, Canada.
Department of Clinical Neurosciences, University of Cambridge, Addenbrooke's Hospital, Hills Road, Cambridge, CB2 2QQ, UK.
Department of Molecular Genetics, University of Toronto, Toronto, Ontario M5S 1A8, Canada.
Division of Community Health Sciences, St George's, University of London, London SW17 0RE, UK.
Diamantina Institute of Cancer, Immunology and Metabolic Medicine, Princess Alexandra Hospital, University of Queensland, Ipswich Road, Woolloongabba, Brisbane, Queensland, 4102, Australia.
Cardiovascular Medicine, University of Oxford, Wellcome Trust Centre for Human Genetics, Roosevelt Drive, Oxford OX3 7BN, UK.
Genetics of Diabetes, Peninsula College of Medicine and Dentistry, University of Exeter, Barrack Road, Exeter, EX2 5DW, UK.
Clinical and Molecular Genetics Unit, Institute of Child Health, University College London, 30 Guilford Street, London WC1N 1EH, UK.
The Wellcome Trust, Gibbs Building, 215 Euston Road, London NW1 2BE, UK.
Oxford NIHR Biomedical Research Centre, Churchill Hospital, Oxford, OX3 7LJ, UK.
Leicester NIHR Biomedical Research Unit in Cardiovascular Disease, Glenfield Hospital, Leicester, LE3 9QP, UK.
References
- 1.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev. 2007;17:182–192. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008 doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Cid R, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollox EJ, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diskin SJ, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray CJ, Lopez AD. Evidence-based health policy--lessons from the Global Burden of Disease Study. Science. 1996;274:740–743. doi: 10.1126/science.274.5288.740. [DOI] [PubMed] [Google Scholar]
- 14.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 16.Locke DP, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 18.Barnes C, et al. A robust statistical method for case-control association testing with copy number variation. Nat Genet. 2008;40:1245–1252. doi: 10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton R, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 22.Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mamtani M, et al. CCL3L1 gene-containing segmental duplications and polymorphisms in CCR5 affect risk of systemic lupus erythaematosus. Ann Rheum Dis. 2008;67:1076–1083. doi: 10.1136/ard.2007.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney C, et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67:409–413. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 25.Wilcoxon F. Individual Comparisons by Ranking Methods. Biometrics Bulletin. 1945;1:80–83. [Google Scholar]
- 26.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 27.Field SF, et al. Experimental aspects of copy number variant assays at CCL3L1. Nat Med. 2009;15:1115–1117. doi: 10.1038/nm1009-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.