Abstract
Background
Chronic obstructive pulmonary disease (COPD) is an important cause of mortality with marked geographic variations in Great Britain. Additional factors beyond cigarette smoking are likely to influence these variations, but direct information on smoking by area is not readily available. We compared methods of jointly modeling the spatial distribution of mortality from COPD and lung cancer, using the latter as a proxy for smoking, to identify areas in which risk factors other than smoking may be important.
Methods
We obtained district-level mortality and population data for men aged 45 years or older in 1981–1999 in Great Britain. Three models were compared: Bayesian ecological regression using observed (model 1) or spatially smoothed (model 2) lung cancer standardized mortality ratio (SMR) as a smoking proxy, and bivariate regression (model 3) treating smoking as a spatial latent variable common to both diseases.
Results
Model selection criteria favored models 2 and 3 over model 1. Between 9% (model 3) and 25% (model 2) of spatial variation in COPD mortality was estimated to be unrelated to smoking. After adjustment for lung cancer as a proxy for smoking, both models showed similar geographic patterns of higher COPD mortality in conurbation and mining areas, historically associated with heavy industry and higher air pollution levels.
Conclusions
Joint modeling of multiple diseases can be used to investigate geographic variations in risk. These models reveal patterns that are adjusted for the effects of shared area-level risk factors for which no direct data are available.
Chronic obstructive pulmonary disease (COPD) is the third-leading cause of death and disability-adjusted life years in adults aged 60 years and older, and the 10th leading cause of mortality worldwide in adults aged 15–59 years.1 Smoking is acknowledged to be a major risk factor for both COPD and lung cancer.2 However, geographic and temporal variations in smoking do not fully account for COPD mortality trends in the UK over the last century,3–5 and urban–rural differences in bronchitis deaths were apparent even before the start of the smoking epidemic.6
The importance of risk factors other than smoking are of increasing interest in COPD research.7 In particular, environmental factors such as air pollution, which has been implicated in COPD mortality trends,5 show strong spatial variations and are amenable to control measures. Smoking is obviously a major confounder. However, there is limited information on geographic variations in smoking, particularly on cumulative tobacco exposure, which is likely to be the most important measure in the development of COPD.8 Perhaps because of this, relatively few studies have examined possible reasons for spatial variations in COPD.
Lung cancer mortality is strongly associated with cumulative smoking,9 and has been used as an indirect indicator of community cumulative smoking exposure to estimate the global burden of mortality associated with tobacco.10 The method was further refined by Ezzati and Lopez.11
Our focus is the detection of geographic variations in COPD mortality in Great Britain, with the aim of identifying areas in which risk factors other than smoking may play an important role. Our hypothesis is that the spatial distribution of areas in which patterns of lung cancer and COPD mortality are discordant reflects the distribution of risk factors other than smoking that influence COPD development and mortality. We use Bayesian statistical methods to model jointly the spatial distribution of mortality from COPD and lung cancer, and compare 3 alternative model formulations. The first 2 are based on an ecologic regression model using COPD mortality as the outcome and lung cancer mortality (either the observed standardized mortality ratio [SMR] or a spatially smoothed estimate) as a covariate representing a proxy for cumulative smoking. In the third model, COPD and lung cancer mortality are treated as a bivariate outcome that depends on both shared and disease-specific spatially correlated latent variables representing unobserved risk factors.
METHODS
Data
Numbers of deaths from COPD and allied conditions (ICD9 490–496) and lung cancer (ICD9 162) between 1981 and 1999 for men 44 years or older were extracted from the national postcoded mortality dataset held by the UK Small Area Health Statistics Unit (SAHSU). Annual population estimates for men in the same age group and time period were also obtained from the database.12 These were then aggregated according to the 459 county-districts in Great Britain. Expected mortality counts for each disease and district were calculated, standardized by 5-year age group using average rates for Great Britain over the 19-year study period.
Ecologic Regression Models
Model 1
Model 1 is a “centered” version of the small-area ecologic regression model proposed by Besag et al13 which includes both spatial and unstructured random effects:
(for areas j adjacent to area i).
y1i and e1i are the observed and expected COPD deaths in each area, respectively, and η1i is an area-level random effect representing the log relative risk (RR) of COPD mortality in area i compared with the national risk. To account for overdispersion, the η1i are assumed to follow independently a normal distribution with variance and mean equal to the linear predictor of the ecologic regression. The logarithm of the observed lung cancer SMR in each area, log(y2i/e2i), is included as a covariate to represent an indirect area-level measure of cumulative smoking, with associated regression coefficient β. We also include a second spatially structured area-level random effect, ψ1i, representing the effects of unmeasured spatially varying risk factors for COPD other than smoking. This is assigned an intrinsic conditional autoregressive prior distribution, which smoothes the latent risk factors toward the average value in neighboring areas, with variance inversely proportional to the number of neighbors, ηi. Identifying the spatial pattern of ψ1i is the main focus of our analysis.
Model 2
Model 1 uses the observed lung cancer SMR as a proxy for cumulative smoking. However, just as with COPD, there is likely to be overdispersion and spatial dependence in the lung cancer data. To account for this, we follow Bernardinelli et al14 and extend the previous ecologic regression model to allow for spatial smoothing and overdispersion of the lung cancer SMRs:
(for areas j adjacent to area i).
(for areas j adjacent to area i).
The COPD model remains unchanged from model 1, with the important exception that the spatially smoothed, rather than observed, lung cancer risks, φi, are now used in the regression equation (with corresponding regression coefficient β*). This is similar to a classic measurement error model in which lung cancer SMR is the observed covariate and φi represents the true covariate, here interpreted as a measure of the underlying cumulative smoking exposure in area i. The model relating observed lung cancer mortality to φi parallels the model for COPD mortality except that no covariates are included in the regression.
Bivariate Regression Model
Model 3
Knorr-Held and Best15 propose an alternative approach to the joint spatial modeling of small-area risks of 2 related diseases. Rather than treating 1 disease as a proxy for unmeasured risk factors affecting the other disease, their model treats the 2 diseases symmetrically and assumes that the area-specific relative risks of each depend on a shared latent component plus additional latent components specific to one or other disease. These latent components act as surrogates for unmeasured risk factors that affect both or only one of the diseases, respectively. Our model 3 is an adaptation of this model using conditional autoregression priors for the latent components:
COPD specific component: (for areas j adjacent to area i).
Lung Cancer specific component: (for areas j adjacent to area i) Shared component: (for areas j adjacent to area i).
Note that model 3 excluding the lung cancer specific component ψ2i is equivalent to model 2 with β* > 0, and β* = δ2.16 In this case, the shared component, , is just a scaled version of the true unknown area-level smoking proxy, and δ2 is equivalent to the regression coefficient in the measurement error version of the ecologic regression model. The advantage of model 3 over model 2 that it treats both diseases symmetrically, and the addition a lung cancer–specific spatial residual allows for the possibility that not all of the spatial variation in lung cancer mortality is shared with COPD. Thus, we can interpret the shared component, , as a latent variable representing cumulative smoking (plus any other shared spatial risk factors) in each area, and the 2 disease-specific components represent unmeasured risk factors associated with only one of the diseases. As before, our main interest is in the spatial pattern of the COPD-specific component, ψ1i.
Prior Distributions
Diffuse or weakly informative prior distributions were chosen for all variance parameters and regression coefficients, and sensitivity of our inference to alternative choices of these priors was assessed (Appendix).
Bayesian Analysis
Models were estimated using Markov chain Monte Carlo methods in WinBUGS.17 Convergence was checked by visually inspecting trace plots of sampled parameter values against iterations for 2 chains per model. Quoted results were based on posterior sample sizes sufficient to give Monte Carlo standard errors less than 5% of the posterior standard deviation for parameters of interest. The eAppendix gives Win-BUGS code for models 2 and 3 (http://links.lww.com/A938).
Model fit was evaluated by comparing the posterior mean of the standardized deviance to a χ2 distribution on 918 (total number of observations).18 Models were compared using the deviance information criterion (DIC)18 which can be interpreted similarly to AIC (Akaike information criteria)– that is, the model with the smallest AIC is preferred– but is appropriate for use with Bayesian hierarchical models.
RESULTS
Descriptive Analysis
A total of 361,194 COPD deaths and 483,637 lung cancer deaths were identified. Both COPD and lung cancer showed a similar spread of SMRs across districts, with slightly more variability for COPD, mainly due to some extreme SMRs based on small numbers (Table 1). Lung cancer and COPD SMRs were highly correlated across districts (r = 0.76), reflecting their similar geographic patterns, with higher SMRs in conurbation areas of central and south-western Scotland, northeastern, northwestern and north-central England, west Midlands, south Wales, London, and the Thames estuary (Fig. 1).
TABLE 1.
Distribution of Counts (No.) for COPD and Lung Cancer Deaths and Population for Men Aged 45+ Years, and SMRs, by District for Great Britain 1981–1999
| Total | Min | 5th Percentile | Median | 95th Percentile | Max | |
|---|---|---|---|---|---|---|
| Counts | ||||||
| COPD deaths | 361,194 | 5 | 192 | 577 | 2,137 | 6,773 |
| Lung cancer deaths | 483,637 | 12 | 279 | 748 | 2,737 | 10,357 |
| Population | 183,049,913 | 8,140 | 117,472 | 332,344 | 927,514 | 3,090,359 |
| SMRs | ||||||
| COPD | 32.6 | 61.0 | 90.9 | 143.3 | 179.6 | |
| Lung cancer | 58.0 | 68.9 | 91.6 | 135.5 | 180.7 |
FIGURE 1.
Maps of district-level COPD and lung cancer SMRs for Great Britain 1981–1999. (A) SMR for COPD; and (B) SMR for lung cancer.
Bayesian Analysis
Results for all models were robust to different priors on the variance parameters, with posterior mean estimates of interest for a given model being within 3 times Monte Carlo sampling error under the different prior assumptions. For brevity, we only report results using prior set 1.
Variance Components
A key difference between the models is the way in which the total between-district variation in RR of death from COPD and lung cancer is partitioned between spatially varying, shared, and disease-specific latent risk factors. To compare this, we calculated posterior distributions of the empiric variances of the various latent variables in each model.
For each disease, the between-district variance of the overall log RRs was similar across all 3 models (Table 2, row 1), and comparable to the between-district variance in the log SMRs (0.070 for COPD and 0.042 for lung cancer). Maps (not shown) of the posterior mean of the overall RRs for each disease were virtually identical across models and were similar to the maps of SMRs (Fig. 1). This indicates that the overall risk estimates for each district are robust to the choice of model and that little overall smoothing is performed by the Bayesian models. This is expected given the large mortality counts in most areas, which provide strong information about the overall area-specific risks.
TABLE 2.
Posterior Medians and 95% Credible Intervals for Variance Components and Relative Risk of COPD Mortality Associated With Cumulative Smoking Proxy Plus Measures of Model Fit
| Parameter | Model 1 | Model 2 | Model 3 | |||
|---|---|---|---|---|---|---|
| (1) Total between-district variance in (log) risk | ||||||
| COPD | Var(ηli) | 0.065 (0.063–0.068) | Var(ηli) | 0.065 (0.063–0.067) | Var(ηli) | 0.065 (0.063–0.067) |
| Lung cancer | 0.042 | Var(η2i) | 0.041 (0.039–0.043) | Var(η2i) | 0.041 (0.039–0.043) | |
| (2) Overdispersion variance | ||||||
| COPD | 0.0039 (0.001–0.007) | 0.0006 (0.0001–0.0023) | 0.0006 (0.0001–0.0026) | |||
| Lung cancer | — | — | 0.0031 (0.0013–0.0049) | 0.0031 (0.0013–0.0049) | ||
| (3) Between-district variance of shared (log) risk |
0.039 (0.034–0.045) | 0.054 (0.046–0.063) | 0.061 (0.047–0.069) | |||
| (4) Between-district variance of disease-specific residual (log) risk | ||||||
| COPD | Var(ψi) | 0.019 (0.016–0.023) | Var(ψli) | 0.018 (0.015–0.021) | Var(ψli) | 0.0060 (0.0002–0.022) |
| Lung cancer | — | 0 | — | 0 | Var(ψ2i) | 0.0065 (0.0002–0.013) |
| (5) Fraction of total spatial variation shared with other disease | ||||||
| COPD | 66.9% (61.1%–72.5%) | 75.0% (70.0%-79.5%) | 91.0% (71.4%–99.7%) | |||
| Lung cancer | — | 100% | — | 100% | 83.9% (70.8%–99.5%) | |
| (6) RR of COPD mortality associated with unit increase in smoking proxya |
exp(β) | 2.62 (2.44–2.80) | exp(β*) | 3.33 (2.94–3.74) | exp(δ2) | 3.74 (3.05–4.41) |
| (7) Deviance | 904.3 | 914.0 | 913.8 | |||
| (8) DIC | 1775.0 | 1694.2 | 1694.4 | |||
A difference of 1 unit in the value of the smoking proxy between 2 areas can be thought of as equivalent to a difference of 1 in the log relative risk of lung cancer in those 2 areas (see text for further explanation).
Our main interest, however, is in how the overall geographic variation in COPD and lung cancer mortality is partitioned into shared and disease-specific components, and here the models showed some differences. For all models, the between-area variances of the spatially structured shared and specific latent components (Table 2, rows 3–4) were much larger than the unstructured overdispersion variance (Table 2, row 2), suggesting strong spatial dependence in mortality from both diseases, although there was more overdispersion in COPD risk in model 1 than models 2 and 3 (Table 2, row 2).
About 67% and 75% of the spatial variation in COPD was captured by the shared term in models 1 and 2, respectively (Table 2, row 5). The difference in these percentages is mainly due to the smaller between-area variance of the shared term under model 1 (Table 2, row 3), suggesting that not accounting for measurement error in the proxy smoking covariate masks some of the shared pattern of variation in COPD and lung cancer mortality.
Unlike models 1 and 2, in model 3, discordant spatial patterns can be captured by the lung cancer residual and the COPD residual. This resulted in a smaller between-area variance for the latter (Table 2, row 4) and a slightly higher between-area variance for the shared term (Table 2, row 3) compared with model 2, with about 91% of the spatial variation in COPD now captured by latent spatial risk factors shared with lung cancer (although the credible interval for this percentage is wider than for model 2, with which it overlaps; Table 2, row 5). Slightly less (84%) of the spatial variation in lung cancer was also captured by these shared risk factors under model 3.
Relative Risk of COPD Mortality Associated With the Smoking Proxy
In model 1, the lung cancer log SMR is used as a proxy for cumulative smoking in each area. A difference of 1 between 2 areas corresponds to an exp (1) = 2.7-fold difference in lung cancer SMR. The estimated relative risk of COPD mortality for 2 such areas was found to be of similar magnitude (2.62; 95% credible interval [CI] = 2.44–2.80). Allowing for measurement error in this proxy (model 2) led to the usual deattenuation of the regression coefficient (RR = 3.33; 95% CI = 2.94–3.74). An even stronger association between COPD mortality and the shared latent risk factor was found in model 3 (3.74; 3.05–4.41). This trend in the relative risk estimates reflects the increase in percentage of spatial variation in COPD mortality explained by the shared latent risk factors (smoking proxy) across models 1–3.
Model Fit
All 3 models provided an adequate fit based on the deviance (P > 0.05; Table 2, row 7). Models 2 and 3 were more parsimonious, however, with virtually identical DIC values that were substantially smaller than for model 1 (Table 2, row 8).
Geographic Pattern of COPD Risk
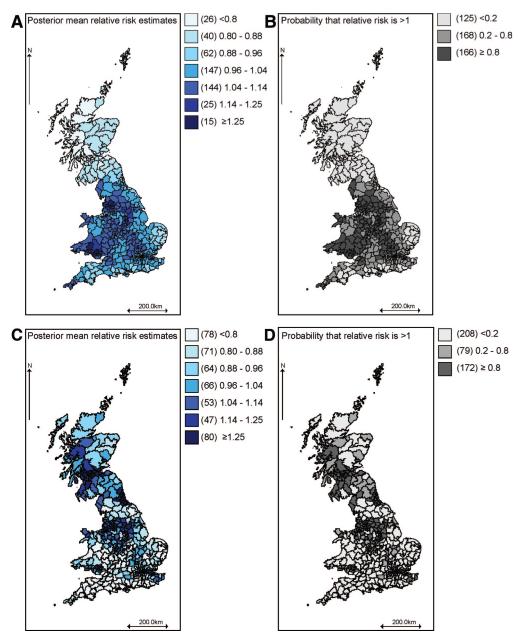
Geographic variations in COPD mortality not explained by the latent smoking covariate are captured by the spatially smoothed residual relative risk, exp(ψ1i). The posterior means of this term estimated using model 2 are shown in Figure 2A. To assess statistical significance of these estimates, the posterior probability that the residual relative risk in each area exceeds 1 is also mapped (Fig. 2B). Areas highlighted in dark (light) gray have at least 80% probability of an excess (reduced) risk of COPD mortality compared with the national average attributed to the presence (absence) of risk factors other than the shared component, the smoking proxy) in that area. The 80% cut-off has been found to give reasonable sensitivity and specificity in simulation studies.19
FIGURE 2.
Maps of posterior mean relative risk estimates for the COPD-specific and latent covariates in model 2 and associated probabilities of excess risk. (A) Residual relative risk of COPD mortality not explained by the latent smoking proxy; (B) Probability that residual relative risk of COPD exceeds 1; (C) Relative risk of COPD mortality associated with latent smoking proxy; and (D) Probability that smoking-related relative risk of COPD mortality exceeds 1 .
Figure 2C shows a map of the posterior mean of exp(βφi) from model 2, interpreted as the relative risk of COPD mortality (relative to the national age-standardized average) associated with the smoking proxy (lung cancer risk) in area i. Figure 2D shows the posterior probability that these smoking proxy–related relative risks in each area exceed 1.
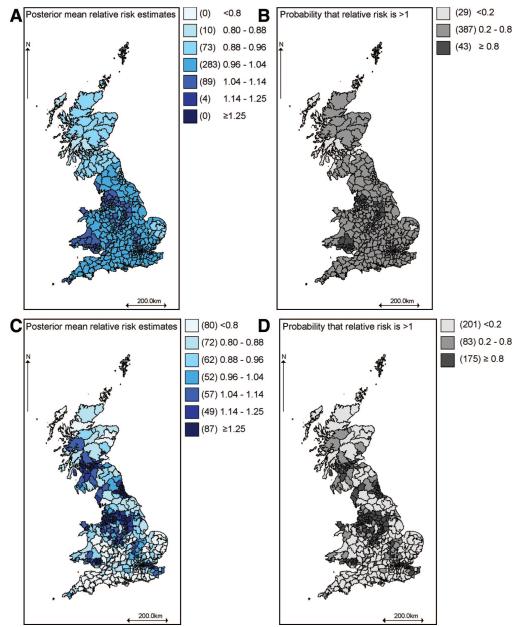
Maps of the corresponding quantities estimated using model 3 are shown in Figure 3. The geographic patterns in models 2 and 3 are similar, but (comparing Fig. 2B with 3B) model 2 produced a larger number of areas for which there was greater than 80% probability of an excess or reduced risk of COPD due to factors not shared with lung cancer.
FIGURE 3.
Maps of posterior mean relative risk estimates for the COPD-specific and latent covariates in model 3 and associated probabilities of excess risk. (A) Residual relative risk of COPD mortality not explained by the latent smoking proxy; (B) Probability that residual relative risk of COPD exceeds 1; (C) Relative risk of COPD mortality associated with latent smoking proxy; and (D) Probability that smoking-related relative risk of COPD mortality exceeds 1.
DISCUSSION
These analyses present an extension of standard disease mapping techniques that allows a more detailed investigation of spatial patterns of disease and their potential causes. Marked spatial variations were seen in COPD mortality risks in the UK after adjustment for lung cancer risks, which were used as a proxy for smoking because of limited spatial information on cumulative smoking rates. A standard ecologic regression model used lung cancer SMRs directly as a proxy for smoking, but additional benefit—in terms of both improved model fit and interpretation—was demonstrated by modeling this covariate using 2 alternative latent variable formulations. Both the latter models gave similar overall fit and geographic patterns, and so the final choice of model must also be guided by other considerations.
Model Selection
The fraction of spatial variance unrelated to the smoking proxy was higher with model 2 than with model 3 (25% vs. 9%), along with greater confidence that the relative risks for COPD deaths related to factors other than smoking were above or below 1 in many areas. The latter indicates that model 2 estimated the area-specific risks more precisely than model 3. However, model 2 makes the strong assumption that all of the spatial variation in the second disease (proxy covariate) is shared with the disease under primary investigation. This is probably reasonable here, because the literature would support an interpretation that risk factors for lung cancer could be considered a subset of those for COPD.
Model 3 fits separate spatial residuals for both diseases and so estimates 3 spatial latent variables per area. With only 2 observations (diseases) per area, a strong signal is needed about all 3 components to fully identify the model. However, for our data, we found that most of the risk was partitioned into the shared component, suggesting a relatively weak residual signal. This may explain more uncertain risk estimates from this model, suggesting model 2 is more appropriate here. Application of these models to other disease pairs may lead to stronger residuals in both diseases, in which case model 3 may be preferable–for example, in mapping COPD and cardiovascular disease, because the relative risks associated with smoking are higher for COPD than cardiovascular disease.20 Model 3 also extends more naturally to situations in which more than 2 diseases are hypothesized to share common spatially varying risk factors or confounders.16
Methodologic Issues
The choice of comparison disease in this situation is crucial and needs to consider both spatial and temporal factors and, when interpreting results, what risk factors may be shared. Many, if not most, respiratory physicians would regard lung cancer rates as a suitable proxy for community cumulative tobacco exposure because over 90% of lung cancer in developed countries is estimated to be attributable to smoking.10 However, if there is spatial clustering of other risk factors for lung cancer unrelated to COPD, such as occupational exposure to asbestos concentrated in traditional ship building areas in the UK,21 then lung cancer mortality may be a poor proxy for cumulative smoking in those areas. This would particularly affect models 1 and 2, which do not allow for a lung cancer–specific spatial residual. Further, if there are important shared causes of lung cancer and COPD other than smoking, these will also be at least partially captured by the shared component, in which case the COPD specific residual may underestimate the risk associated with nonsmoking causes.
Another implicit assumption is that the exposure–response function affecting both diseases is similar enough in terms of lag periods (or latency) and nature of exposure (cumulative, intensity or duration) for the model to generate meaningful results. This is reasonable for COPD and lung cancer—both diseases show an increase in risk with increased levels of smoking; lung cancer rates have been seen to lag smoking rates by about 20 years22 or the product of average tar content and adult tobacco consumption per capita by 25–30 years.23 Lag effects of smoking on COPD mortality are rarely explicitly stated but are suggested at 25 years.24 As long as there is reasonable similarity between the exposure–response function affecting both diseases and disease counts are accumulated over a long time frame (20 years in the present analysis) relative to any potential differences in lags, a meaningful partitioning of risk into shared and nonshared amounts should be possible. However, distortions to the model may occur if the exposure–response function varies—for example, if the increased risks related to smoking waned over long periods of time in COPD whereas that for lung cancer increased. Unfortunately, such detailed exposure–response information is rarely available, especially for diseases taking many decades to develop. In situations in which differential lags were an issue, one could simply lag the data appropriately (assuming the lag period was well established) and then apply the same methods as used here. Alternatively, if lags were not known, a space-time version of our models could be used25 with inclusion of an explicit lag parameter.
ICD9 codes for “COPD and allied conditions” included those for asthma (asthma codes contributed <5% of deaths). Asthma codes were included as it can be difficult to distinguish between asthma and COPD in older age groups and studies of death certificates suggest misclassification of COPD deaths to asthma and vice versa.26,27 Misclassification of deaths recorded to COPD and allied conditions or lung cancer will introduce bias if it varies substantially spatially. Variation over time is only important to this method if this results in spatial variations that would not be averaged out over the time period under study. The extent to which misclassification varies spatially in Great Britain is difficult to assess. However, factors such as the central control of all medical education and specialist training and sole handling of death certificate coding by the Office for National Statistics will tend to minimize systematic variations. Random variations in certification practice should be mitigated as each spatial unit (district) will typically contain several hospitals and several hundred general practitioners.
This type of study is ecologic in nature and therefore prone to the ecologic fallacy, where risk estimates seen at the group level may not reflect risk estimates at the individual level.28 However, the purpose of this analysis is not to obtain estimates of the risk to individuals of dying of COPD due to smoking, but to identify geographic differences in COPD mortality after attempting to adjust for cumulative smoking.
Interpretation
We consider that the proportion of geographic variation in COPD mortality shared with lung cancer mortality is mainly smoking-related, but will also include smaller contributions from other shared risk factors such as air pollution, diet, or occupational exposures.2 In situations in which a shared risk factor is a stronger predictor of one disease than the other, we would expect the shared risk component estimated from our models to reflect only partially the spatial pattern of that underlying risk factor. The differential or excess variation in the disease having the stronger association with the shared risk factor will be captured by the specific component for that disease. Thus, if putative risk factors such as air pollution or diet are stronger predictors of COPD mortality than of lung cancer mortality, we would expect the COPD-specific risks estimated from our models to reflect at least partially spatial variation associated with these factors.
Our results can thus be interpreted as providing descriptive estimates of geographic variations in COPD mortality after adjusting for a proxy that primarily reflects cumulative smoking. Higher adjusted COPD mortality risks were seen in the conurbation areas of England (Greater London, Manchester, Liverpool, Leeds, Sheffield, and Birmingham) and in mining areas in southern Wales (Figs. 2, 3). These areas are generally known to have high levels of risk factors other than smoking that have been linked to COPD in previous studies, including (historical) air pollution levels, occupational risk factors, and mining and heavy industry.5 Higher COPD-specific mortality risks were not seen in central Scotland (including Glasgow and Edinburgh), which might also be expected to have high rates of these exposures. One suggested explanation is that this could be related to competing risks due to higher cardiovascular mortality in Scotland,29 reducing the numbers of persons surviving to die of COPD.30
In addition to providing a descriptive summary of geographic variations in COPD mortality, the area-specific adjusted COPD risks estimated in this study could also be used to investigate further the relative importance of specific etiologic factors by examining the relationship between these estimates and ecologic data on, for example, air pollution, deprivation, fruit and vegetable consumption, and other modifiable risk factors of public health interest. By using appropriate aggregated individual-level regression models that account for the within-area distribution of such risk factors,31 such an approach may provide insights about the relative importance of individual-level risks of COPD associated with factors not shared with lung cancer. This forms the basis of research in progress.32 It is likely that any associations thus identified would be underestimates because as explained previously, the COPD-specific risk estimates may only partially capture the spatial variations associated with factors such as air pollution and diet, if these are also risk factors for lung cancer. Working out the size of the relative underestimation could be attempted through an extensive simulation study, which might be appropriate in certain circumstances— for example, if such studies were used to support expensive policy interventions.
Other Possible Applications of the Model
The model can be applied to any diseases with shared risk factors, for example, cancers with related dietary risk factors33 or childhood diabetes and leukemia in relation to viral infections in childhood.34 An infectious disease example might be to use rates of a reportable sexually transmitted infection as a proxy for sexual behavior in studying the etiology of another sexually transmitted infection. Alternatively, the model could be applied to the same disease in 2 different time periods, to detect emerging disease clusters. The shared component would capture stable patterns of variation in disease risk over time, whereas the specific components would capture patterns present in one or other time period, which might indicate a time-localized disease cluster or changes in risk factors such as a new hazard. In health policy research the model could be used to identify areas with high values of the shared component for different causes of avoidable mortality, with a view to targeting suitable interventions in those areas.
CONCLUSIONS
Joint modeling of 2 diseases can be used to investigate geographic patterns of disease related to shared or specific risk factors when direct information on those risk factors is not readily available. Such information provides a richer perspective on spatial variations in disease risk than a standard disease mapping analysis. The precise interpretation of the shared and specific patterns must be guided by what is already known about the 2 diseases and their risk factors, but can be used, for example, to help inform etiologic debate about specific causes of disease, generate new hypotheses, or to aid policy formulation and evaluation or resource allocation.
ACKNOWLEDGMENTS
We thank Paul Aylin for discussion and interpretation of data quality and Kees de Hoogh for provision of UK district polygons and the Small Area Health Statistics Unit for the data.
Supported by grants 075883 and 066901 from the Wellcome Trust (to A.H.).
APPENDIX: PRIOR DISTRIBUTIONS
Uniform distributions were assumed for α1 and α2. Normal distributions with large variance (100,000)—which are locally uniform across the plausible range of values—were chosen for β and β*. Following Knorr-Held and Best,15 the log of the scaling parameter δ in model 3 was assigned a normal prior with mean 0 and variance 0.17, which is a diffuse prior with high probability that δ2 (the relative risk associated with the shared latent covariate) is between one-fifth and 5.
Three sets of priors for the variance parameters were chosen as follows:
Set 1
Conjugate inverse gamma priors with shape parameter 0.5 and scale parameter 0.0005 for each of , , , , λ2 and λ*2. This prior was proposed by Kelsall and Wakefield35 as being a suitable diffuse prior for the variance components in a Bayesian disease mapping model, and implies that, a priori, each variance has a prior mode at 0.0005/(0.5 + 1) = 0.00033 and infinite expectation and variance.
Set 2
Following a recent suggestion by Gelman36 we use standard half-normal priors (standard normal distributions that are truncated on the left at zero) for the square root of each variance parameter. This prior has a mode at zero, expectation 0.79, and variance 0.36.
Set 3
Same as set 1 for the overdispersion parameters and , and the variances of the disease-specific spatial residuals and , but inverse gamma priors with shape parameter 0.5 and scale parameter 0.0005 × K on the variances of the latent covariate (λ2 in model 2 and λ*2 in model 3). This gives a prior mode for λ2 or λ*2 that is K times larger than the prior modes of and and reflects our prior beliefs that the majority of the spatial variation in COPD and lung cancer mortality will reflect shared risk factors (specifically smoking). We consider values of K = 2, 4, and 10.
Footnotes
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
REFERENCES
- 1.World Health Organization . The World Health Report 2003. Shaping the Future. World Health Organization; Geneva, Switzerland: 2003. [Google Scholar]
- 2.Doll R, Darby S, Whitley E. Trends in mortality from smoking-related diseases. In: Charlton J, Murphy M, editors. The Health of Adult Britain 1841–1994. Vol 1. The Stationery Office; London: 1997. pp. 128–155. [Google Scholar]
- 3.Reid DD, Fletcher CM. International studies in chronic respiratory disease. Br Med Bull. 1971;27:59–64. doi: 10.1093/oxfordjournals.bmb.a070816. [DOI] [PubMed] [Google Scholar]
- 4.Law MR, Morris JK. Why is mortality higher in poorer areas and in more northern areas of England and Wales? J Epidemiol Community Health. 1998;52:344–352. doi: 10.1136/jech.52.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marks G, Burney P. Diseases of the respiratory system. In: Charlton J, Murphy M, editors. The Health of Adult Britain 1841–1994. The Stationery Office; London, UK: 1997. pp. 93–113. [Google Scholar]
- 6.Collis EL. The general and occupational prevalence of bronchitis and its relation to other respiratory diseases. J Ind Hyg. 1923;5:264–276. [Google Scholar]
- 7.Marsh S, Aldington S, Shirtcliffe P, Weatherall M, Beasley R. Smoking and COPD: What really are the risks? Eur Respir. 2006;28:883–884. doi: 10.1183/09031936.06.00074806. [DOI] [PubMed] [Google Scholar]
- 8.Pena VS, Miravitlles M, Gabriel R, et al. Geographic variations in prevalence and underdiagnosis of COPD: results of the IBERPOC Multicentre Epidemiological Study. Chest. 2000;118:981–989. doi: 10.1378/chest.118.4.981. [DOI] [PubMed] [Google Scholar]
- 9.Peto R, Lopez AD, Boreham J, Thun M, Heath C., Jr . Mortality From Smoking in Developed Countries 1950–2000: Indirect Estimates From National Vital Statistics. Oxford University Press; Oxford: 1994. [Google Scholar]
- 10.Peto R. Influence of dose and duration of smoking on lung cancer rates. In: Zaridze DG, Peto R, editors. Tobacco: A Major International Health Hazard. IARC; Lyon, France: 1986. [Google Scholar]
- 11.Ezzati M, Lopez AD. Estimates of global mortality attributable to smoking in 2000. Lancet. 2003;362:847–852. doi: 10.1016/S0140-6736(03)14338-3. [DOI] [PubMed] [Google Scholar]
- 12.Arnold RA. Small area health statistics unit procedures for estimating small area populations. In: Arnold R, Elliott P, Wakefield J, et al., editors. Population Counts in Small Areas: Implications for Studies of Environment and Health. HMSO; London: 1999. pp. 10–24. [Google Scholar]
- 13.Besag J, York J, Mollié A. Bayesian image restoration with two applications in spatial statistics. Ann Inst Stat Math. 1991;43:1–59. [Google Scholar]
- 14.Bernardinelli L, Pascutto C, Best NG, Gilks WR. Disease mapping with errors in covariates. Stat Med. 1997;16:741–752. doi: 10.1002/(sici)1097-0258(19970415)16:7<741::aid-sim501>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 15.Knorr-Held L, Best NG. A shared component model for detecting joint and selective clustering of two diseases. J R Stat Soc [A] 2001;164:73–86. [Google Scholar]
- 16.Held L, Natario I, Fenton SE, Rue H, Becker N. Towards joint disease mapping. Stat Methods Med Res. 2006;14:61–82. doi: 10.1191/0962280205sm389oa. [DOI] [PubMed] [Google Scholar]
- 17.Spiegelhalter DJ, Thomas A, Best NG, Lunn D. WinBUGS Bayesian inference using Gibbs sampling manual. Version 1.4. Imperial College London and MRC Biostatistics Unit; Cambridge: 2002. Available at: http:/www.mrc-bsu.cam.ac.uk/bugs. [Google Scholar]
- 18.Spiegelhalter DJs, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with discussion) J R Stat Soc [Ser B] 2002;64:583–639. [Google Scholar]
- 19.Richardson S, Thomson A, Best N, Elliott P. Interpreting posterior relative risk estimates in disease-mapping studies. Environ Health Perspect. 2004;112:1016–1025. doi: 10.1289/ehp.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.US Department of Health and Human Services . In: Reducing the Health Consequences of Smoking: 25 Years of Progress. A Report of the Surgeon General. CDC, editor. US Department of Health and Human Services; Atlanta, GA: 1989. [Google Scholar]
- 21.McElvenny DM, Darnton AJ, Price MJ, Hodgson JT. Mesothelioma mortality in Great Britain from 1968 to 2001. Occup Med (Lond) 2005;55:79–87. doi: 10.1093/occmed/kqi034. [DOI] [PubMed] [Google Scholar]
- 22.Alberg AJ, Samet JM. Epidemiology of lung cancer. Chest. 2003;123:21S–49S. doi: 10.1378/chest.123.1_suppl.21s. [DOI] [PubMed] [Google Scholar]
- 23.Shibuya K, Inoue M, Lopez AD. Statistical modeling and projections of lung cancer mortality in 4 industrialized countries. Int J Cancer. 2005;117:476–485. doi: 10.1002/ijc.21078. [DOI] [PubMed] [Google Scholar]
- 24.Izumi T. Chronic obstructive pulmonary disease in Japan. Curr Opin Pulm Med. 2002;8:102–105. doi: 10.1097/00063198-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Richardson S, Abellan JJ, Best N. Bayesian spatio-temporal analysis of joint patterns of male and female lung cancer in Yorkshire (UK) Stat Methods Med Res. 2006;15:385–407. doi: 10.1191/0962280206sm458oa. [DOI] [PubMed] [Google Scholar]
- 26.Jones K, Berrill WT, Bromly CL, Hendrick DJ. A confidential enquiry into certified asthma deaths in the North of England, 1994–96: influence of co-morbidity and diagnostic inaccuracy. Respir Med. 1999;93:923–927. doi: 10.1016/s0954-6111(99)90061-6. [DOI] [PubMed] [Google Scholar]
- 27.Smyth ET, Wright SC, Evans AE, Sinnamon DG, MacMahon J. Death from airways obstruction: accuracy of certification in Northern Ireland. Thorax. 1996;51:293–297. doi: 10.1136/thx.51.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothman KJ, Greenland S. Modern Epidemiology. Lippincott-Raven Publishers; Philadelphia: 1998. [Google Scholar]
- 29.Morris RW, Whincup PH, Lampe FC, Walker M, Wannamethee SG, Shaper AG. Geographic variation in incidence of coronary heart disease in Britain: the contribution of established risk factors. Heart. 2001;86:277–283. doi: 10.1136/heart.86.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feenstra TL, van Genugten ML, Hoogenveen RT, Wouters EF, Rutten-van Mölken MP. The impact of aging and smoking on the future burden of chronic obstructive pulmonary disease. A model analysis in the Netherlands. Am J Respir Crit Care Med. 2001;164:590–596. doi: 10.1164/ajrccm.164.4.2003167. [DOI] [PubMed] [Google Scholar]
- 31.Richardson S, Stucker I, Hemon D. Comparison of relative risks obtained in ecological and individual studies: some methodological considerations. Int J Epidemiol. 1987;16:111–120. doi: 10.1093/ije/16.1.111. [DOI] [PubMed] [Google Scholar]
- 32.Hansell AL, Aylin P, Best N. Are annual average ambient air pollution levels associated with spatial variations in COPD mortality in the UK?. Proceedings of the American Thoracic Society 3 Abstracts Issue (April); 2006. A46. [Google Scholar]
- 33.Tzala E. Multivariate Analysis of Spatial and Temporal Variation in Cancer Mortality in Greece. Imperial College; London: 2004. [PhD thesis] [Google Scholar]
- 34.Feltbower RG, Manda SO, Gilthorpe MS, et al. Detecting small-area similarities in the epidemiology of childhood acute lymphoblastic leukemia and diabetes mellitus, Type 1:a Bayesian approach. Am J Epidemiol. 2005;161:1168–1180. doi: 10.1093/aje/kwi146. [DOI] [PubMed] [Google Scholar]
- 35.Kelsall J, Wakefield J. Discussion of “Bayesian Models for Spatially Correlated Disease and Exposure Data” by Best et al. In: Bernardo JM, Berger JO, David AP, et al., editors. Bayesian Statistics 6. Oxford University Press; Oxford: 1991. p. 151. [Google Scholar]
- 36.Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Analysis. 2006;1:515–533. [Google Scholar]