Abstract
MicroRNAs 125a and 125b are predicted to be able to bind to the B lymphocyte-induced maturation protein-1 (BLIMP-1) and IFN regulatory protein-4 (IRF-4) transcription factors, which are essential for plasma cell differentiation. A computational survey of the human and mouse genomes revealed that miR-125a and miR-125b are members of a multigene family located in paralogous clusters. The miR-125a cluster on chromosome 19 in humans includes miR-99b and let-7e, whereas the miR-125b cluster on chromosome 21 includes miR-99a and miR-let-7c. Our analysis of the expression profiles for these six miRs during B lineage differentiation indicated that mature miR-125a, miR-125b, miR-99b and let-7e transcripts are preferentially expressed by the actively dividing centroblasts in germinal centers (GC). However, miR-99b and let-7e are not predicted to bind BLIMP-1 or IRF-4 transcripts, and binding to the untranslated region of BLIMP-1 and IRF-4 messenger RNAs could be confirmed only for miR-125b. When the effect of miR-125b over-expression on terminal B cell differentiation was evaluated in an LPS-responsive B cell line, the induction of BLIMP-1 expression and IgM secretion was inhibited in this model system. Furthermore, miR-125b over-expression inhibited the differentiation of primary B cells and compromised the survival of cultured myeloma cells. These findings suggest that miR-125b promotes B lymphocyte diversification in GC by inhibiting premature utilization of essential transcription factors for plasma cell differentiation.
Keywords: BLIMP-1, centroblasts, gene regulation, IRF-4, MicroRNA, plasma cell differentiation
Introduction
Post-transcriptional regulation of gene expression has been shown to involve small RNAs as an important additional level of genetic control (1–4). The microRNA (miRNA) family of small RNA regulators has attracted much attention due to an increasingly appreciated role in development and the pathogenesis of malignancies. miRNAs are single-stranded RNAs of 18–24 nucleotides that bind to complementary seed sites in the 3′ untranslated regions (UTRs) of messenger RNAs (mRNAs) to down-regulate translation and protein production. Since their initial description in Caenorhabditis elegans, almost 4000 miRNAs have been identified in >40 species of animals, plants and viruses. Nearly 700 miRNAs have been identified in the human genome, and more than one-third of all human genes are thought to be regulated by miRNAs (5). miRNAs are often conserved across species and exhibit tissue-specific expression, and their expression or function may be significantly altered in disease states (6, 7).
An important role has been demonstrated for the miR-17–92 cluster during B cell development (8, 9). The pro-apoptotic Bim gene, which has multiple miR-17–92 miRNA-binding sites, is the target of miR-17–92 in B cells (8). Selective B lineage deletion of the Dicer gene, which encodes an enzyme that is essential for miRNA biogenesis, led to an accumulation of pro-B cells and a reduction of pre-B and mature B cells (8). miR-155 has been shown to negatively regulate activation-induced cytidine deaminase, an enzyme that is essential for Ig isotype switching and somatic hypermutation (10, 11). Mice deficient in miR-155 have defective antibody responses to both T-independent and T-dependent antigens; severely reduced IgG1 responses in these mice indicated the defective differentiation of plasma cells that secrete class-switched antibodies (12–14). Conversely, the over-expression of miR-155 resulted in B cell lymphoproliferative disorders in transgenic mice (15).
Despite growing evidence of biological roles for a limited number of miRNAs in immune system development and function in mouse models, the potential for miRNA roles in B cell differentiation in humans has not been analyzed extensively. In a computational search for miRNAs that could modulate essential transcription factors for plasma cell differentiation, B lymphocyte-induced maturation protein-1 (BLIMP-1) (PRDM1) and IFN regulatory protein-4 (IRF-4), we identified highly conserved miR-125a and miR-125b miRNAs in paralogous clusters of related genes in the human and mouse genomes. Our analysis of human tonsillar cells at different stages in B cell differentiation indicated that several members of the miR-125a and miR-125b clusters, miR-125a, miR-125b, let-7e and miR-99b are preferentially expressed in the centroblasts of the germinal centers (GC). These findings led us to examine the potential roles for members of the miR-125 multigene family on terminal B cell differentiation and antibody secretion.
Methods
Cells
Human and mouse cell lines were cultured in RPMI-1640 medium containing 100 U ml−1 penicillin, 100 mg ml−1 streptomycin, 2 mM L-glutamine and 10% FCS (Hyclone). Human tonsils were obtained in accordance with policies established by the Emory University Institutional Review Board and with informed consent according to the Declaration of Helsinki. Mononuclear cells in these tissues were isolated by Ficoll-Hypaque gradient centrifugation. Naive B cells in tonsil samples were purified to >90% purity by depletion of CD10+, CD27+, CD38+, CD3+ and CD14+ cells using monoclonal antibodies, antibody-conjugated microbeads or goat anti-mouse IgG-conjugated microbeads (Miltenyi Biotec, Auburn, CA, USA). Stained cells were analyzed on a FACSCyan flow cytometer (BD Biosciences, Mountain View, CA, USA) and plotted using FlowJo software.
Immunofluorescence cell sorting and real-time PCR analysis of mRNA transcripts
Tonsillar B cell sub-populations were purified by immunofluorescent cell sorting with a MoFlow instrument (Cytomation, Fort Collins, CO, USA) as follows: naive cells (CD27−CD38−IgD+CD19+), pre-GC cells (CD38+IgD+CD19+), centroblasts (CD77+CD38+CD19+), centrocytes (CD77−CD38+CD19+), memory B cells (CD27+CD38−CD19+) and plasma cells (CD38++IgD−CD19+). Sorted cells were lysed in TRIzol reagent (Gibco, Grand Island, NY, USA) before preparation of total RNA and first-strand cDNA synthesis using Superscript II system (Invitrogen, Carlsbad, CA, USA). After inactivating the reactions at 50°C for 2 min, real-time PCR was performed by using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) denaturing at 95°C for 10 min, amplification for 40 cycles at 95°C for 15 s and annealing and extension at 60°C for 1 min using an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems). BLIMP-1, IRF-4, c-Myc and β-actin gene-specific primers (kind gift from Goetz Ehrhardt, Emory University) were used for PCR amplification as described previously (16).
Quantitative real-time PCR for miRNA analysis
Sorted B cell subsets were lysed and total miRNA was extracted using an mirVana miRNA Isolation Kit (Ambion, Inc., Austin, TX, USA). miRNA was then measured spectrophotometrically. miRNA analysis was performed as previously described (17). Samples were reverse transcribed and further pre-PCR amplification was performed as described before (18, 19). The pre-PCR mixture was diluted by adding 75 μl of dH2O. The probes for the Taqman reaction (kind gift from Lao, Applied Biosystems) contained 18 nucleotides of RT-RP of each miRNA at the 3′ end, with the fluorescence dye FAM at the 5′ end and a minor groove binder with non-fluorescence quencher, MGB, on the 3′ end. An AB 7900 HT Sequence Detection System in a 384-well plate format was used. Each sample was run in duplicate. Twenty-five miRNA's were tested. RP-2 and RNU6B (Applied Biosystems) were used as a control, and their values were not different between the groups tested. The raw data are represented as a heatmap and were analyzed for fold changes.
Plasmids
A 500-bp sequence containing miRNA 125b was amplified from human peripheral blood mononuclear cell genomic DNA by PCR. The fragment was inserted into the EcoRV restriction site of pBlueScript II SK(+) (Stratagene) and sequenced (Genewiz, Inc., South Plainfield, NJ, USA). The fragment was then sub-cloned by restriction digest with BamHI and XhoI into pCWD, a retroviral expression vector containing a cOP GFP (cGFP) reporter construct upstream of the multiple cloning site (a gift from the laboratory of Rafi Ahmed), or by restriction digest with EcoRI and NotI into pCDH-CMV-MCS-EF1, a lentiviral expression vector containing a cop-GFP marker gene (System Biosciences, Mountain View, CA, USA).
Transfection of cell lines and primary B cells
Bcl1.3B3 murine B cell line was co-transfected at a 2:1 ratio of either control miRNA mimic or miR-125b mimic (Dharmacon, Lafayette, CO, USA) and a GFP plasmid. GFP+ cells were sorted by flow cytometry-based cell sorter and cells were treated with toll-like receptor agonist LPS that induces differentiation of this cell line. Supernatants were harvested 48 h post-LPS stimulation and IgM was quantified by ELISA. Lentiviral vector that encodes for miR-125b precursor mimic and GFP or the control vector was transduced into Bcl1.3B3 cells. Seventy-two hours after transduction, GFP+ cells were sorted and the sorted cells were cultured in the presence or absence of LPS. Supernatants were harvested 48 h post-LPS stimulation and IgM was quantified by ELISA. Retroviral infection of primary B cells was carried out as described previously (20). Splenic B cells were purified by positive selection with B220 magnetic beads (Miltenyi) and cultured as described previously. B cells (106 ml−1) were stimulated with LPS (10 μg ml−1) or CpG (10 μg ml−1; Invivogen) overnight before the retrovirus infection at a multiplicity of infection of 0.2–2 in the presence of 5 μg of Polybrene per milliliter. Cells were infected in the presence of a bicistronic retroviral vector that co-expresses scrambled miR mimic and GFP or with a retroviral vector that co-expresses miR-125b mimic and GFP. A day after infection, cells were cultured in the presence of different doses of LPS or CpG. At 4 days post-infection, cells were analyzed by flow cytometry for expression of GFP, CD19 and CD138 (Syndecan-1).
Search for miRNA target genes and analysis of miRNA clusters
Publicly accessible miRNA sequences were obtained from miRBase (Release 11.0) (21). Two prediction programs TargetScan (22) and RNAhybrid (23) were used to find out the possible targets. To analyze the miRNA clusters, we used the genomic sequences of human (Homo sapiens) (assembly: NCBI Build 36.2, Sep 2006) and mouse (Mus musculus) (assembly: NCBI m36, Dec 2005) from Ensembl genome browser. The homologous sequences were determined using BLASTn algorithm. The sequence alignments were performed using the CLUSTALW program and the alignments were inspected manually to maximize similarity. To find out the possible transcription promoter, we used PROSCAN version 1.7 (available at http://www-bimas.cit.nih.gov/molbio/proscan/index.html) and a neural network-based promoter prediction program NNPP version 2.2 (available at http://www.fruitfly.org/seq_tools/promoter.html).
Luciferase assays
For luciferase reporter experiments, 293T cells were transfected in triplicate with each 3′ UTR luciferase construct using polyethylenimine (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer's instructions. Each experiment was repeated twice. Statistical comparison of luciferase results was performed by 2-tailed t-test.
Statistics analysis
The Student’s t-test was used to evaluate the significance of differences in experimental results.
Results
Cluster analysis of miR-125 genes in the human and mouse genomes
Using the TargetScan (22) and RNAhybrid software programs (23), we found that miR-125a and miR-125b are predicted to bind the 3′ UTR region of the transcripts for the BLIMP-1 and IRF-4 transcription factors (24) (Fig. 1A and B). Both miR-125a and miR-125b possess the seed sequence (CCCUGAG) that is predicted to be able to bind to BLIMP-1 and IRF-4 3′ UTR target sequence (GGGACUC) that is conserved in humans and mice (Fig. 1A and B). Since miRNAs often exist within clusters (25, 26), we conducted an in silico search for neighbours of the miR-125a and miR-125b genes in human and mouse genomic sequences. This analysis indicated that miR-125a and miR-125b are present in clusters, wherein the neighbouring miRNA genes are located within a few kilobases (Fig. 1C). A similarity search using the BLASTn program revealed that the clustering patterns for miR-125a and miR-125b genes are identical in the human and mouse genomes (Fig. 1C). The miR-125a cluster on chromosome 19 of humans includes three miRNA genes: miR-125a, miR-let-7e and miR-99b; the paralogous miR-125b cluster on chromosome 21 also includes three miRNA genes: miR-125b, miR-let-7c and miR-99a. The orthologous miR-125a and miR-125b clusters in the mouse are located on chromosomes 17 and 16, respectively. The genomic organization of both miR-125a and miR-125b clusters thus is highly conserved between humans and mice.
Fig. 1.
Cluster analysis of miR-125 genes in mouse and human genomes. (A) Sequence conservation of miR125a and miR-125b in mice and humans. Alignment of mature miR125a and miR-125b sequences from human and mouse, respectively, is shown. The potential seed sequences are highlighted. (B) The predicted miR125 seed pairing with the 3′ UTRs of BLIMP-1 and IRF-4 transcription factors. The positions in the alignment are based on the human BLIMP-1 and IRF-4 sequences. (C) Genomic organization of miR-125 clusters. The homologous relationships of miR-125b gene clusters are shown for human and mouse with the paralogous miRNA relationship represented by solid and grey shades. The physical distances between miRNAs in the chromosome (Chr) clusters are indicated. The figure is not drawn to scale.
Expression of the miR-125 gene family during human B cell differentiation
To examine the expression patterns for miRNAs in the miR-125a and miR-125b clusters during different stages of B lineage differentiation, we isolated subsets of CD19+ B cells from human tonsils on the basis of their expression of IgD, CD27, CD77 and CD38. For all the members of the miR-125a cluster, miR-125a, miR-let-7e and miR-99b, we found that mature transcripts were elevated preferentially in centroblasts of the GC (Fig. 2A–F), whereas when transcripts for members of the miR-125b cluster were examined, preferential centroblast expression was observed exclusively for the miR-125b transcripts (Fig. 2B). Notably, this analysis also suggested that the miR-125b transcript levels are ∼1000-fold higher than the miR-125a transcript levels in the centroblasts. The expression of miR-let-7c and miR-99a miRs was not detected in any of the B cell subsets, although these miRs were found to be expressed in T cells and/or macrophages (Fig. 2E and F). The coordinate expression of the miR-125a, miR-let-7e and miR-99b genes by the centroblasts may reflect their shared transcriptional orientation and close proximity within the transcription unit of a non-protein-coding RNA gene (NCRNA 00085) (Supplementary Figure 1 is available at International Immunology Online), given that miRNA genes located in the transcription unit with the same transcription orientation are often co-transcribed with the host gene (27, 28). In contrast, a potential independent promoter and a c-Myc-binding site were identified 12 Kb upstream of the miR-125b gene in humans. Transcription factors, including c-Myc and TP53, have been shown to regulate the expression of miRNAs through binding to their promoters, thereby contributing to a feedback loop wherein transcription factors regulate miRNA gene expression and the miRNAs regulate transcription factors post-transcriptionally by mRNA binding (29, 30). The differential expression patterns observed for the 125a and 125b miRs indicate that these closely related miRs could inhibit the differentiation of GC centroblasts, with a dominant role for miR-125b being favored by its much higher levels of expression.
Fig. 2.
Expression profiling of miR-125 genes in human B cells. Real-time PCR analysis indicates that miR-125a (A), miR-125b (B), miR-let-7e (C) and miR-99b (D) are preferentially expressed by centroblasts in the GC. miR-let-7c (E) and miR-99a (F) are expressed by T cells and macrophages, respectively. Three independent tonsils were examined and a representative plot is shown. Values represent arbitrary numbers derived from Ct values normalized to RP-2 house keeping control. Statistical analysis of miRNA expression was done by the Student's t-test and P value <0.05 are indicated by asterisks. Bars indicate SEM.
Analysis of miR-125b versus miR-125a binding to BLIMP-1 and IRF-4 transcripts
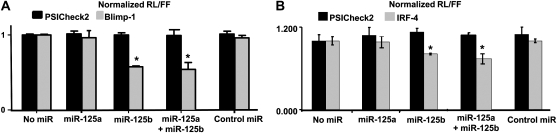
Because both miR-125a and miR-125b possess the seed sequence that is predicted to be able to bind to BLIMP-1 and IRF-4 3′ UTR target sequences, we used a luciferase reporter assay to test whether miR-125a and miR-125b could bind to the 3′ UTR of BLIMP-1 and IRF-4. This analysis confirmed the binding of miR-125b to the 3′ UTR of BLIMP-1 and IRF-4 mRNAs (31) but did not affirm the predicted binding of miR-125a (Fig. 3A and B). Moreover, when expressed together, miR-125a did not enhance or compete with miR-125b binding to the 3′ UTR of BLIMP-1 and IRF-4 mRNAs (Fig. 3A and B). Ectopic miR-125b expression achieved by transfection of an miR-125b duplex into HEK-293T cells suppressed the activity of a Renilla luciferase construct containing the miR-125b miRNA response element (MREs) of mouse BLIMP-1 by ∼50% (P < 0.01) and the activity of a R. luciferase construct containing the miR-125b MREs of mouse IRF-4 by ∼20% (P < 0.05). In contrast, miR-125a had a negligible effect alone or when combined with miR-125b (Fig. 3A and B). Thus, while miR-125a and miR-125b are unique among the members of their respective clusters in their predicted ability to bind to BLIMP-1 or IRF-4 transcripts, the actual binding to the 3′ UTR was verifiable only for miR-125b.
Fig. 3.
miR-125b but not miR-125a binds to 3′ UTR of BLIMP-1 and IRF-4 mRNAs. Dual luciferase activity of reporter plasmids with the wild-type 3′ UTR of BLIMP-1 (panel A) or IRF-4 (panel B) fused to the luciferase gene upon miR-125a or miR-125b precursor co-transfection of the same reporter vector with the non-targeting control. Results are expressed as relative luciferase (RL) units normalized to firefly luciferase (FF) units in triplicate analyses from two experiments. Statistical comparisons between the co-transfected miRNA and the non-targeting control for the same reporter vector were performed and P values <0.05 are indicated by *. Bars correspond to the SEM. Statistical comparisons were performed by 2-tailed t-test.
BLIMP-1 and IRF-4 inhibition by miR-125b over-expression in B and myeloma cell lines
Since BLIMP-1 and IRF-4 transcripts are potential targets for miR-125b, we re-examined their expression levels during B lineage differentiation. Consistent with previous reports (32–34), we found in this analysis of the different stages of B lineage cells isolated from human tonsils that BLIMP-1 and IRF-4 transcripts are selectively expressed by plasma cells (Fig. 4A and B). To evaluate the effects of miR-125b on BLIMP-1 expression, we introduced precursors of miR-125b into the murine Bcl1.3B3 B lymphoma and the human U266 multiple myeloma cell lines. Stable lentiviral vector-mediated expression of an miR-125b mimic resulted in reduced expression of the BLIMP-1 protein in both cell lines, findings consistent with our observations in transient transfection experiments (Fig. 5A–C). Introduction of the synthetic miR-125b mimic into the U266 human myeloma cell line led to reduced expression of Syndecan-1 (CD138), a cell surface molecule expressed by myeloma and primary plasma cells (Fig. 6A). In addition, an exaggerated death rate was noted for the myeloma cells in these experiments (Fig. 6B); evidence of apoptotic cell death was observed for ∼80% of the miR-125b mimic transfectants versus ∼25% for control mimic transfectants. These findings suggest that miR-125b can modulate B cell differentiation and myeloma cell survival via its ability to down-regulate BLIMP-1 and IRF-4 translation.
Fig. 4.
Expression pattern of BLIMP-1 and IRF-4 transcription factors in plasma cells derived from human tonsils. (A) BLIMP-1, a transcription factor predicted to be bound by miR-125b is expressed only by plasma cells as determined by real-time PCR analysis. (B) IRF-4, a transcription factor predicted to be bound by miR-125b is expressed only by plasma cells as determined by real-time PCR analysis. Three independent tonsils were examined and a representative plot is shown. Values represent arbitrary numbers derived from Ct values normalized to RP-2 house keeping control.
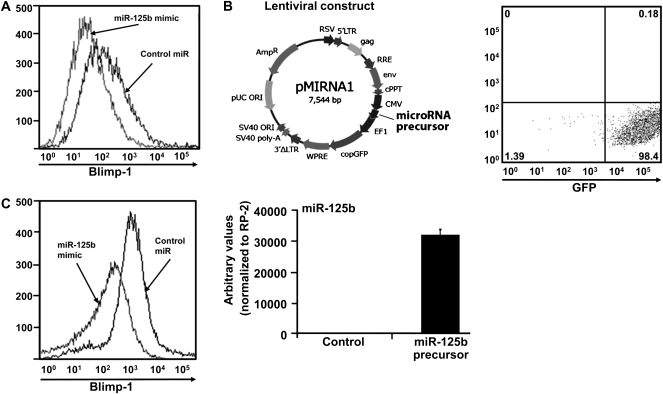
Fig. 5.
BLIMP-1 protein levels in B cell lines over-expressing miR-125b. (A) Flow cytometric analysis of intracellular BLIMP-1 expression in Bcl1.3B3 mouse cells infected with control virus or miR-125b-expressing virus treated with 20 μg ml−1 of LPS. (B) miR-125b precursor mimic cloned into a lentiviral vector that co-expresses GFP or the control vector were transduced into U266 myeloma cell line (top left panel). GFP+ cells were sorted 72 h after infection and cultured in vitro (top right panel). Detection of lentivirus-mediated miR-125b expression in U266 cells by real-time PCR (bottom left panel). (C) Flow cytometric analysis of intracellular BLIMP-1 expression in U266 myeloma cells infected with control virus or miR-125b-expressing virus.
Fig. 6.
Effects of miR-125b over-expression in a myeloma cell line. (A) CD138 expression in U266 myeloma cells transfected with control mimic or miR-125b mimic (left panel) and CD138 expression in U266 myeloma cells infected with control virus or miR-125b-expressing lentivirus (right panel); (B) annexin V staining of myeloma cells transfected with control mimic or miR-125b mimic. Cells were stained for propidium iodide (PI) and annexin V. Annexin V+ PI− cells are plotted as a histogram.
Characterization of miR-125b effects on B cell differentiation
The functional role for miR-125b in B cell differentiation was evaluated further by delivery of synthetic miRNA mimics and by lentiviral vector-mediated over-expression in cells of a B cell line that LPS can induce to undergo plasma cell differentiation and antibody secretion. In these experiments, the introduction of miR-125b mimic inhibited the LPS induction of terminal differentiation as reflected by diminished IgM secretion in miR-125b mimic-expressing cells compared with control mimic-expressing cells (Fig. 7A and B). The reduction in IgM secretion was ∼70% for transient transfections and ∼50% for lentivirus-mediated infections. Since miR-125b could be shown to regulate antibody production in these cell lines, we examined the effect of miR-125b by retroviral vector-mediated over-expression in primary B cells from the spleen, which were then stimulated with either LPS or CpG. In keeping with our observations in the B cell line, retroviral-mediated over-expression of an miR-125b mimic in murine primary splenic B cells impaired terminal differentiation as reflected by a reduction in the numbers of LPS-induced CD19loCD138hi plasma cells (Fig. 7C). A similar inhibitory effect was observed for CpG-induced plasma cell differentiation (Fig. 7D). The inhibition of plasma cell differentiation by miR-125b over-expression was not attributable to non-specific cellular toxicity, since no differences were seen in the number of viable cells after miR-125b versus control miR transduction (Supplementary Figure 2 is available at International Immunology Online). These findings indicate that miR-125b can inhibit B cell differentiation into plasma cells without demonstrable effect on B cell viability.
Fig. 7.
miR-125b effects on the induction of plasma cell differentiation and antibody secretion. (A) Bcl1.3B3 murine B cell line was co-transfected at a 2:1 ratio of either control miRNA mimic or miR-125b mimic and a GFP plasmid. GFP+ cells were sorted by flow cytometry-based cell sorter and cells were treated with toll-like receptor agonist LPS (10 μg ml−1) that induces differentiation of this cell line. Supernatants were harvested 48 h post-LPS stimulation and IgM was quantified by ELISA. Statistical analysis of IgM quantification was done by the Student's t-test and P < 0.05 was considered significant comparing control miR mimic and miR-125b mimic in the presence of LPS. This experiment was repeated twice and a representative experiment is shown. (B) Lentiviral-mediated stable expression of miR-125b precursor mimic in Bcl1.3B3 mouse cells and cultured in the presence or absence of LPS. Supernatants were harvested 48 h post-LPS stimulation and IgM was quantified by ELISA. Statistical analysis of IgM quantification was done by the Student's t-test and P < 0.05 was considered significant comparing control virus-expressing cells and miR-125b mimic-expressing cells in the presence of LPS. (C) Retroviral-mediated expression of miR-125b precursor mimic in splenic B cells cultured in the presence or absence of different doses of LPS. Cells were harvested on day 6 and analyzed for CD19 and CD138 expression by FACS analysis of GFP+ cells. This experiment was repeated four times and the plot derived from all these experiments is shown. Significant differences between control miR and 125b miR in four experiments showed P values <0.05 and correspond to the SEM. (D) Retroviral-mediated expression of miR-125b precursor mimic in splenic B cells cultured in the presence or absence of 1 μg ml−1 of CpG. Cells were harvested on day 6 and analyzed for CD19 and CD138 expression by FACS analysis of GFP+ cells. This experiment was repeated thrice and the plot derived from these experiments is shown. Differences between control miR and 125b miR in three experiments showed P values <0.05 and bars correspond to the SEM.
Discussion
The coordination of antigen-induced B cell proliferation, somatic V region hypermutation and Ig isotype switching in GC is essential for optimal humoral responses in higher vertebrates. Progression through the distinctive phases in GC differentiation requires a finely tuned program of gene regulation during which extrinsic signals derived from T cell interaction and cytokines modulate the up-regulation or down-regulation of specific transcription factors, including BCL6 and BLIMP-1. Aberrations in the complex gene expression patterns that coordinate the GC proliferation and differentiation programs may lead to B cell malignancies and autoimmune diseases (35–37). The results of our studies suggest that miR-125b serves an important checkpoint role to guard against premature GC B cell differentiation in humans by binding and sequestering BLIMP-1 and IRF-4.
Our in silico analysis of the mouse and human genomes indicated that the miR-125a and miR-125b genes are members of multigene families located on separate chromosomes. However, we found interesting differences in the transcriptional control and expression patterns for members of the two miR-125 gene clusters during differentiation of B lineage cells in humans. All the miR-125a cluster members (miR-99b, miR-let-7e and miR-125a) were expressed preferentially by centroblasts, suggesting that they could be controlled by a single promoter element. This possibility is also supported by the location of the miR-125a cluster within the transcription unit of a non-protein-coding RNA gene (NCRNA 00085) with the same transcription orientation for all three miR-125a family members. While miR-125b, too, was expressed preferentially by the centroblasts (31, 38), its neighbouring miR-99a and miR-let-7c genes were not co-transcribed. In keeping with the demonstration that miR-125b is regulated, albeit weakly, via an upstream c-Myc-binding site in an EBV-immortalized B cell line (30), we identified a c-Myc consensus-binding site 12 Kb upstream of human miR-125b gene. The selective expression of miR-125b in centroblasts of GCs therefore could reflect the function of BCL6 as a direct transcriptional repressor of c-Myc, in view of the mutually exclusive expression patterns for BCL6 and c-Myc (32). The reciprocal expression of BCL6 and BLIMP-1 during the transition of GC B cells to the ensuing stage of plasma cell differentiation is well known (39, 40). BCL6 is essential for GC responses through promoting cell cycle progression, somatic hypermutation and B cell maturation, whereas BLIMP-1 and IRF-4 are essential for plasma cell differentiation (20, 35, 41). Our data thus suggest that miR-125b facilitates BCL6 maintenance of the GC phenotype by inhibiting BLIMP-1 and IRF-4 expression. Although currently available methods do not detect mature miRNA–mRNA complexes (19, 42), the effect of centroblast miR-125b transcription may extend into the GC centrocyte stage due to persistence of the mature miR-125b transcript complexes with BLIMP-1 and IRF-4 mRNAs, thereby maintaining the check on premature plasma cell differentiation.
Our studies confirm and extend the observations in two recent studies indicating that miR-125a and miR-125b transcripts are expressed preferentially during the GC centroblast stage (31, 38). We found the level of miR-125b expression by centroblasts to be much higher (∼1000-fold) than that for miR-125a. Moreover, while miR-125b binding to BLIMP-1 and IRF-4 was readily demonstrable, in the case of miR-125a, we failed to find physical or functional evidence for binding to the 3′ UTR of BLIMP-1 and IRF-4 mRNAs. In this regard, a canonical 7–8mer sequence alone may be insufficient for target recognition in that the surrounding nucleotide composition may influence target recognition by miRNAs; Watson–Crick base pairing at nucleotides 12–17 adjacent to seed sequence is one such determinant of miRNA targeting (43). It is therefore possible that miR-125a is unable to target BLIMP-1 and IRF-4 mRNAs because of limitations imposed by determinants beyond seed sequence pairing. When the effects of miR-125b over-expression on BLIMP-1 and IRF-4 expression were examined, we observed a reduction of BLIMP-1 expression and concomitant inhibition of plasma cell differentiation of primary B cells and antibody secretion by a B cell line. These new findings in support of a functional role of miR-125b in B cells extend the earlier observations made by others of miR-125b binding to BLIMP-1 and IRF-4 mRNAs and the reduction of BLIMP-1 and IRF-4 protein expression following miR-125b over-expression in a B cell line (31). BLIMP-1 mRNA binding, albeit to a lesser degree than miR-125b, has been shown for two other miRs, miR-9 and miR-30, that also are preferentially expressed by centroblasts (38, 44). These miRs could potentially augment the inhibitory effect proposed for miR-125b during the GC phase of B cell responses. The requirement of both BLIMP-1 and IRF-4 for differentiation of B cells into plasma cells (45), the reciprocal expression patterns in centroblasts and plasma cells for mature miR-125b versus BLIMP-1 and IRF-4 and our demonstration that miR-125b inhibits B cell differentiation strongly support the possibility that miR-125b contributes to a fail-safe mechanism to block premature GC B cell differentiation. Nevertheless, the cellular details of this inhibition remain speculative because of the technical limitation in detecting BLIMP-1 and IRF-4 mRNAs that are bound to miR-125b within individual centroblasts or centrocytes and the unavailability of an in vitro system capable of recapitulating the differentiation of GC B cells into memory B cells and plasma cells.
Over-expression of BLIMP-1 and IRF-4 have been observed in multiple myeloma and autoimmune diseases (46, 47). The observations that miR-125b over-expression inhibits expression of both BLIMP-1 and IRF-4 and enhances cell death of myeloma cells could reflect an inhibitory influence on key cell survival pathways. In this context, IRF-4 appears to be essential for the survival of myeloma cell lines (46). Therefore, in addition to its role in inhibiting premature B cell differentiation by GC B cells, impaired miR-125b expression could also contribute to the pathogenesis of multiple myelomas.
In conclusion, our studies indicate the conservation of miR-125a and miR-125b multigene family members in mice and humans. While several members of the miR-125 gene family were shown to be expressed preferentially by the GC centroblasts during B lineage differentiation in humans, our findings point to a particularly important role for miR-125b in promoting B cell responses in GC by targeting BLIMP-1 and IRF-4 transcription factors to inhibit premature plasma cell differentiation.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
National Institutes of Health Grant (AI39816 to M.D.C.) from the National Institute of Allergy and Infectious Diseases and the Georgia Research Alliance; Biotechnology and Biological Sciences Research Council; fellowship from the Medical Research Council, UK, to M.T.; fellowship from the Cancer Research UK to D.H.
Supplementary Material
Acknowledgments
We thank Ms Sommer Durham and Mr Robert Karaffa for help with FACS sorting (supported by Emory University School of Medicine). Author contributions: M.G. and M.D.C. designed research; M.G. performed research; C.L.H., D.H., M.T., S.D., C-M.L. and S.J. contributed new reagents/analytic tools; M.G.H., C.L.H., S.D. and M.D.C. analyzed data; and M.G., C.L.H., S.D. and M.D.C. wrote the paper.
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Maatouk D, Harfe B. MicroRNAs in development. Scientific World Journal. 2006;6:1828. doi: 10.1100/tsw.2006.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl Acad. Sci. USA. 2005;102:16961. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Connell RM, Rao DS, Chaudhuri AA, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008;205:585. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl Acad. Sci. USA. 2008;105:3903. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koralov SB, Muljo SA, Galler GR, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorsett Y, McBride KM, Jankovic M, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng G, Hakimpour P, Landgraf P, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 13.Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrhardt GR, Hijikata A, Kitamura H, Ohara O, Wang JY, Cooper MD. Discriminating gene expression profiles of memory B cell subpopulations. J. Exp. Med. 2008;205:1807. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lao K, Xu NL, Sun YA, Livak KJ, Straus NA. Real time PCR profiling of 330 human micro-RNAs. Biotechnol. J. 2007;2:33. doi: 10.1002/biot.200600119. [DOI] [PubMed] [Google Scholar]
- 18.Mestdagh P, Feys T, Bernard N, et al. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36:e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calame KL, Lin KI, Tunyaplin C. Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 2003;21:205. doi: 10.1146/annurev.immunol.21.120601.141138. [DOI] [PubMed] [Google Scholar]
- 25.Jiang D, Yin C, Yu A, et al. Duplication and expression analysis of multicopy miRNA gene family members in Arabidopsis and rice. Cell Res. 2006;16:507. doi: 10.1038/sj.cr.7310062. [DOI] [PubMed] [Google Scholar]
- 26.Das S. Evolutionary origin and genomic organization of micro-RNA genes in immunoglobulin lambda variable region gene family. Mol. Biol. Evol. 2009;26:1179. doi: 10.1093/molbev/msp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 28.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 29.He X, He L, Hannon GJ. The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 30.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malumbres R, Sarosiek KA, Cubedo E, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein U, Tu Y, Stolovitzky GA, et al. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl Acad. Sci. USA. 2003;100:2639. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ci W, Polo JM, Cerchietti L, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Laar JM, Melchers M, Teng YK, et al. Sustained secretion of immunoglobulin by long-lived human tonsil plasma cells. Am. J. Pathol. 2007;171:917. doi: 10.2353/ajpath.2007.070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008;8:22. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 36.Kuppers R, Klein U, Hansmann M-L, Rajewsky K. Cellular origin of human B-cell lymphomas. N. Engl. J. Med. 1999;341:1520. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 37.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 2005;5:251. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 38.Basso K, Sumazin P, Morozov P, et al. Identification of the human mature B cell miRNome. Immunity. 2009;30:744. doi: 10.1016/j.immuni.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo TC, Shaffer AL, Haddad J, Jr., Choi YS, Staudt LM, Calame K. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J. Exp. Med. 2007;204:819. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 2004;173:1158. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 41.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Jima DD, Jacobs C, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angelin-Duclos C, Cattoretti G, Lin K-I, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with blimp-1 expression in vivo. J. Immunol. 2000;165:5462. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- 46.Shaffer AL, Emre NC, Lamy L, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin FR, Kuo HK, Ying HY, Yang FH, Lin KI. Induction of apoptosis in plasma cells by B lymphocyte-induced maturation protein-1 knockdown. Cancer Res. 2007;67:11914. doi: 10.1158/0008-5472.CAN-07-1868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.