Abstract
Y-family DNA polymerases bypass Pt-GG, the cisplatin-DNA double-base lesion, contributing to the cisplatin resistance in tumour cells. To reveal the mechanism, we determined three structures of the Y-family DNA polymerase, Dpo4, in complex with Pt-GG DNA. The crystallographic snapshots show three stages of lesion bypass: the nucleotide insertions opposite the 3′G (first insertion) and 5′G (second insertion) of Pt-GG, and the primer extension beyond the lesion site. We observed a dynamic process, in which the lesion was converted from an open and angular conformation at the first insertion to a depressed and nearly parallel conformation at the subsequent reaction stages to fit into the active site of Dpo4. The DNA translocation-coupled conformational change may account for additional inhibition on the second insertion reaction. The structures illustrate that Pt-GG disturbs the replicating base pair in the active site, which reduces the catalytic efficiency and fidelity. The in vivo relevance of Dpo4-mediated Pt-GG bypass was addressed by a dpo-4 knockout strain of Sulfolobus solfataricus, which exhibits enhanced sensitivity to cisplatin and proteomic alterations consistent with genomic stress.
Keywords: anti-cancer drug, cisplatin, DNA damage, translesion DNA synthesis, Y-family DNA polymerase
Introduction
All living organisms are hindered by DNA damage that generates replication-blocking lesions in DNA. Therefore, there are DNA repair mechanisms to remove lesions and damage-tolerance mechanisms for replication through lesions. Translesion synthesis (TLS) is a DNA damage-tolerance mechanism that uses specialized DNA polymerases (e.g. members of the Y-family) to replicate damaged DNA (Friedberg et al, 2002; Lehmann, 2002). TLS has an important function in cell survival of DNA damage and is highly conserved from bacteria to humans (Goodman, 2002).
cis-Diamminedichloroplatinum (II) (cisplatin) is a widely used chemotherapeutic agent that covalently modifies DNA to block DNA replication in rapidly dividing tumour cells (Davidson et al, 1975; Lippert, 1999). Cisplatin reacts with the N7 atoms of adjacent guanine (G) bases to create a 1,2-intrastrand covalent linkage, cis-Pt-1,2-d(GpG) (Pt-GG) (Supplementary Figure S1), which is the major cisplatin adduct, representing ∼65% of the cisplatin-DNA adducts in cells (Pinto and Lippard, 1985; Sherman et al, 1985; Coll et al, 1990; Takahara et al, 1995; Jamieson and Lippard, 1999). By inhibiting DNA synthesis, cisplatin-DNA adducts slow cell division, possibly activating programmed cell death or apoptosis (Eastman, 1990; Cohen and Lippard, 2001). Thus, cisplatin is an effective anti-cancer drug, especially for cancers of the testis, ovary, bladder, head, neck, and lung. TLS and DNA repair mechanisms have contributed to acquired and intrinsic cisplatin resistance, thereby limiting the drug's application and contributing to cancer recurrence (Siddik, 2003; Zorbas and Keppler, 2005). Cells have been shown to have the ability to replicate past cisplatin-DNA adducts (Gibbons et al, 1991; Mamenta et al, 1994; Vaisman et al, 1998). Some translesional DNA polymerases can bypass cisplatin-DNA adducts (Hoffmann et al, 1995, 1996; Vaisman and Chaney, 2000; Vaisman et al, 2000; Albertella et al, 2005) and are overexpressed in tumour cells (Canitrot et al, 1998; Bergoglio et al, 2001). In addition, TLS is increased in the drug-resistant cells (Gibbons et al, 1991; Mamenta et al, 1994), strongly suggesting that TLS is correlated with resistance to cisplatin (Richon et al, 1987; Gibbons et al, 1991; Roush et al, 1998). Thus, TLS is considered to be one of the main mechanisms of cisplatin resistance in cancer treatment (Vaisman et al, 1998; Suo et al, 1999).
Most of the specialized translesion DNA polymerases are members of the Y-family. These DNA polymerases have a spacious active site that can accommodate bulky and distorted DNA lesions (Yang and Woodgate, 2007). However, a result of the expansion of these enzymes' functional repertoire and substrate specificity is that Y-family DNA polymerases replicate DNA with a higher error frequency (McCulloch and Kunkel, 2008). Hence, although Y-family DNA polymerases promote damage tolerance and cell survival, they also contribute to enhanced cellular mutagenesis. Human Y-family DNA polymerase η bypasses Pt-GG in a relatively efficient and error-free manner (Vaisman et al, 2000; Albertella et al, 2005). Recently, a structural study on yeast DNA polymerase η (yPolη) in complex with Pt-GG DNA provides the first glimpse of Pt-GG lesion bypass during the base insertion stages (Alt et al, 2007). The canonical dCTP is the preferred nucleotide inserted by yPolη opposite both the 3′ and 5′G base of a Pt-GG cross-link (Alt et al, 2007). However, the structures do not show dCTP insertion opposite the 5′G base (Alt et al, 2007). Also notable, the yPolη structures do not have the whole double-base Pt-GG adduct accommodated within the active site. Further study is essential to answer the key question of how the bulky/angular-shaped Pt-GG adduct is translocated through the active site of a DNA polymerase during the lesion bypass.
We report three crystal structures of a model Y-family DNA polymerase, DNA polymerase IV (Dpo4) from Sulfolobus solfataricus, in complex with Pt-GG DNA at 2.9, 1.9, and 2.0 Å resolution, respectively. During the TLS process, DNA polymerases perform two fundamental steps: nucleotide incorporation opposite the damaged DNA base(s) and extension past the lesion. Our three ternary structures encompassed these pertinent steps of TLS: dCTP insertion opposite the 3′ and 5′G bases of the Pt-GG adduct and correct dATP insertion for the extension step immediately downstream of the lesion. To support the structural results, we have investigated the bypass of Pt-GG catalysed by Dpo4 in solution. In addition, the in vivo function of Dpo4 was addressed for the first time through the construction and analysis of a S. solfataricus cell line harbouring a loss of function mutation leading to deficiency of this organism's only lesion bypass polymerase.
Results
Overall structures of the Dpo4–DNA–dNTP ternary complexes
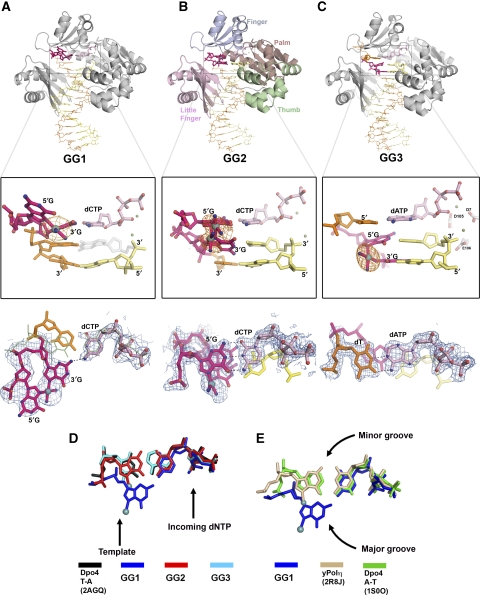
We have co-crystallized Dpo4 with template DNA containing Pt-GG with dNTPs against the 3′G (GG1), the 5′G (GG2) of Pt-GG, and the T base 5′ to the lesion (GG3), respectively (Figure 1). All three of our Dpo4–Pt-GG-DNA–dNTP ternary structures (GG1, GG2, and GG3) share a common crystal form, with one complex per asymmetric unit (Table 1). The overall ternary complexes are essentially identical to the type I complex and other active Dpo4 structures (Ling et al, 2001, 2003, 2004a, 2004b). The type I structure is the first Dpo4–DNA–dNTP ternary complex structure with normal nucleotide incorporation and represents a common active form of Dpo4, as well as most other Y-family DNA polymerases (Supplementary Figure S2). In this common active form, the finger domain is in a closed conformation packing against the catalytic palm domain and contacts the replicating base pair in the active site. Structural superposition of the three complex structures and the type I shows that the Dpo4 remains an identical conformation with root mean square deviations (r.m.s.d.) <0.5 Å on all Cα atoms, pair wisely. The closed finger conformation has been observed in other Dpo4 structures, which were solved in different crystal forms (Ling et al, 2004a; Rechkoblit et al, 2006; Wong et al, 2008). Therefore, the closed finger conformation in our current structures is independent of crystal packing.
Figure 1.
The structures of the GG1 (A), GG2 (B), and GG3 (C) Dpo4–DNA–dNTP ternary complexes. The finger, palm, thumb, and little finger domains distinguished by their respective colours in (B). The Pt-GG lesioned template is represented in magenta, with the platinum atom shown as a cyan ball. The zoomed in boxes of the active site are covered with the Pt-anomalous maps (5 σ) in orange. The catalytic residues are shown in the middle panel of Figure 1C, which present invariantly in all three structures. There are two base conformers of Pt-GG in (B), with each Pt atom at 0.5 occupancy. The top views of the replicating base pairs are covered by a blue 2Fo-Fc maps contoured to 1.0 σ at 2.9, 1.9, and 2.0 Å resolution, respectively. The A-T pair in green sticks superimposed with GG1 (A) is taken from a Dpo4–DNA–dNTP complex structure (PDB: 1S0O), which depicts the regular position of an undamaged purine-pyrimidine base pair in the Dpo4 active site. The grey nucleotide in (A, middle box) is a ghost model for the 3′ primer base that is disordered in GG1. The green spheres are Ca2+ ions. (D) A top view comparison of the GG1 (blue), GG2 (red), and GG3 (cyan) replicating base pairs with an undamaged pyrimindine-purine (T-A) replicating base pair (2AGQ, black). (E) A top view comparison of the Dpo4 GG1 base pair with the yeast pol η GG1 (2R8J, beige) and an undamaged purine-pyrimidine (A-T) (1S0O, green) replicating base pairs.
Table 1. Data collection and refinement statistics.
| GG1 | GG2 | GG3 | |
|---|---|---|---|
| Data collection | |||
| Space group | P21212 | P21212 | P21212 |
| Cell dimensions | |||
| a, b, c (Å) | 98.6, 101.2, 52.4 | 98.2, 103.2, 52.3 | 99.2, 103.7, 52.0 |
| Resolution (Å)a | 50.0–2.90 (3.00–2.90) | 50.0–1.93 (1.96–1.93) | 50.0–2.00 (2.03–2.00) |
| Rsym or Rmerge | 0.061 (0.670) | 0.066 (0.474) | 0.087 (0.722) |
| I/σI | 22.0 (2.2) | 27.4 (2.35) | 24.5 (2.5) |
| Completeness (%) | 98.6 (99.4) | 99.2 (91.6) | 99.8 (95.3) |
| Redundancy | 3.5 (3.4) | 7.6 (5.3) | 8.0 (6.2) |
| Refinement | |||
| Resolution (Å) | 45.0–2.90 | 27.6–1.93 | 29.0–2.00 |
| No. of reflections | 12009 | 40552 | 36729 |
| Rwork/Rfree | 0.285/0.309 | 0.243/0.261 | 0.224/0.235 |
| No. of atoms | |||
| Protein | 2743 | 2728 | 2714 |
| DNA | 586 | 636 | 656 |
| Ligand/ion | 22 | 3 | 4 |
| Water | 49 | 307 | 258 |
| B-factors | |||
| Protein | 60.4 | 32.8 | 34.0 |
| DNA | 76.3 | 34.1 | 38.7 |
| Ligand/ion | 58.8 | 26.1 | 36.8 |
| Water | 55.0 | 42.6 | 41.2 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.008 | 0.015 |
| Bond angles (deg) | 1.73 | 1.71 | 1.64 |
| aValues in parentheses are for highest-resolution shell. | |||
All three GG complexes have the same 18-mer DNA template that contains a site-specific Pt-GG adduct at the 14th and 15th positions from the DNA's 3′ end (Figure 2). Three different 13-mer primers were designed to generate DNA substrates for nucleotide insertions opposite or extension beyond the Pt-GG adduct in the template strand (Figure 2). Dideoxy-terminated primers (ddC or ddG) were used to trap the ternary complexes in the desired reaction stages during TLS of Pt-GG. The single-stranded template residues 5′ to the adduct are completely disordered in both insertion structures GG1 and GG2. The incoming nucleotides in the three GG complexes are superimposed well with the dNTP from the ternary complexes of Dpo4 and undamaged DNA (Figure 1D and E) (Ling et al, 2004a; Vaisman et al, 2005), showing that Dpo4 maintains the incoming dNTP in a similar position and orientation in the active site, regardless of the DNA template position. Thus, the structural differences are localized at the site of the DNA adduct in the template strand. The Pt-GG adduct conformation is varied in the three structures, which deviates from the low-energy Pt-GG conformation as described by the ideal torsion angles α and β (Supplementary Table S1). The torsion angle α defines the degree of Pt out-of-plane bending (C8-N7-C5-Pt, an ideal value of 180° for in-plane conformation), and β represents the torsion angle between the G base planes and the Pt-coordination plane (C8-N7-Pt-cisN, an ideal value of 90°) (Yao et al, 1994). The torsion angles in our three structures deviate up to 10° beyond the range established from the single cis-Pt-GG adduct crystal structures (Sherman et al, 1985) and the protein-free Pt-GG adducted DNA structures (Takahara et al, 1995), and thus are further off from the ideal low-energy state (Yao et al, 1994). The DNA helices bound by Dpo4 are in a straight form with bending angles <18° over the 12-bp helical region, which is in contrast to the curved protein-free Pt-GG adducted DNA (Takahara et al, 1995) (Figure 3). It seems that the helical DNA geometry and the protein binding place additional strains to distort the Pt-GG adduct from its low-energy conformations. The distances between the Pα atoms of the incoming nucleotides and the C3′ atoms at the 3′ end of the primer strands are ∼5 Å, a catalytically competent distance, in the presence of two catalytic divalent metal ions in all three structures (Figure 1).
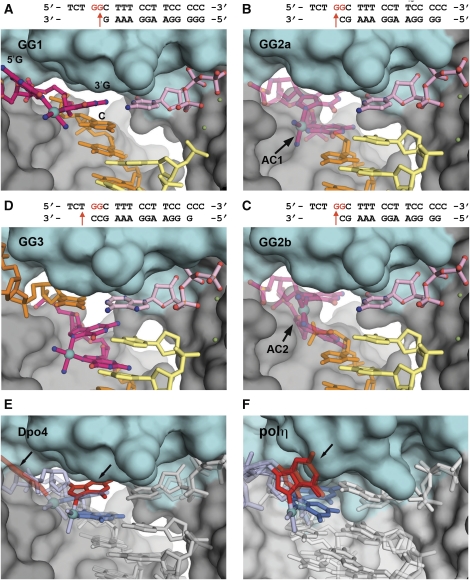
Figure 2.
Pt-GG adducts in the active sites of Dpo4 and ypolη. All panels are close-up views of the enzyme active sites where the finger domain is in cyan. The platinum (Pt) atom is shown as a cyan ball. The DNA is shown as stick-balls, with the template strand in orange and the primer strand in yellow. The sequences of the template/primer DNA in the complexes are shown on the tops of panels (A–D), cross-linked G bases are in red. The arrows indicate the incoming dNTP positions. (A) GG1, where dCTP (pink) is paired opposite the 3′G of Pt-GG (dark pink). (B, C) GG2, two alternate conformations of GG2 (translucent pink, GG2a in (B), GG2b in (C)), where dCTP (pink) is paired opposite the 5′G of Pt-GG. (D) GG3, where dATP is paired with the T base that is 5′ to the Pt-GG lesion. (E) Superposition of the GG1 (semi-transparent, left side) and GG2 (solid, right) structures of Dpo4, where DNA is shown as grey sticks with the 5′G of Pt-GG is in red with arrows, and the 3′G is in blue. The Pt-GG adduct is translocated in different positions and shows different conformations in the two structures. (F) Superposition of the same reaction stage complexes of ypolη with the same colour scheme as (E). In ypolη (F), the Pt-GG adduct remains in a similar position with the 5′G bases (red bases with an arrow) superimposed well in both reaction stages, no translocation of Pt-GG occurs.
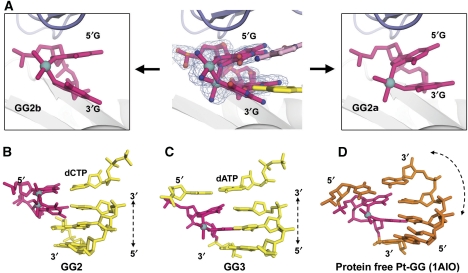
Figure 3.
Pt-GG conformations and helical DNA structures in the presence or absence of Dpo4 enzyme. (A) Alternate conformations of Pt-GG in the GG2 structure. The alternate Pt atom corresponds to discrete bulges of electron density for the 1.9 Å 2Fo-Fc map contoured at 1σ. (B–D) Comparison of the DNA helices in GG2 (B), GG3 (C), and enzyme-free Pt-GG structure (D) (PDB: 1AIO). The Pt-GG in the enzyme-free DNA induces a bend in the DNA helix.
GG1: dCTP insertion opposite the 3′G of Pt-GG (first insertion)
The GG1 structure represents the first insertion stage of Pt-GG lesion bypass. The incoming dCTP is poised for insertion opposite the 3′G base of the Pt-GG adduct at the active site. The position of the platinum atom is identified by an anomalous difference peak, which is shifted into the DNA major groove (Figure 1A). Moreover, the finger domain in the closed conformation wedges between two Pt-coordinated G bases and blocks the 5′G from entering the active site completely (Figure 2A). Thus, the 5′ base of Pt-GG is in close contact with the finger domain and is positioned outside of the active site. As the 3′G and 5′G are covalently linked through cisplatin, the 3′G base is dragged into the major groove by the 5′G, thereby preventing the 3′G from resting in the optimal template position of an undamaged DNA template (Figure 1). Compared with an undamaged DNA template, the 3′G is shifted 4.0 Å towards the major groove (Figure 1A, D, and E). The DNA helix in GG1 keeps a straight form, and the adduct is not completely included into the helical structure yet. In the first insertion structure (GG1), Pt-GG possesses a large roll angle of 135° between the 5′G and 3′G bases due to its close contacts with the finger domain (Figure 2A). In this open angular conformation, the Pt atom is out of plane of the bases with the α angles deviated by 8–37°, and the Pt-coordination plane off the ideal β torsion by 22–60° (Supplementary Table S1). The conformation of Pt-GG in GG1 are dramatically distorted due to the position of the finger domain wedging between the two G bases. In contrast, the Pt-GG adducts in the open ypolη structure are very close to the ideal model with roll angles of ∼90°, α angles of <5° from ideal 180°, and β angles within 30° from ideal 90° (Alt et al, 2007).
The Pt-GG adduct outside of the active site is not stabilized by base stacking and Watson–Crick (WC) base pairing, leading to a disturbed template (poor density, Figure 1A) with high B-factors. The 3′G base of the Pt-GG adduct shifted into the major groove loses base stacking with the upstream −1 template base. Furthermore, the shifted 3′G base does not form a WC base pair with the incoming dCTP that is in the same position as the incoming dNTPs in the structures 2AGQ and 1S0O (Figure 1A, D, and E) which exhibit the regular position of an undamaged base pair in the Dpo4 active site (Ling et al, 2004a; Vaisman et al, 2005). Only one H-bond is maintained in the misaligned base pair, which is between two NH2 groups from the 3′G base (N2) of Pt-GG and the base (N4) of dCTP (Figure 1A). Noticeably, the H-bond is a symmetrical H-bond that is of greater-than-normal strength (Cleland et al, 1998; Kraut et al, 2006; Lone et al, 2007). The symmetric H-bond is formed between donor and acceptor atoms from identical NH2 groups that have equal pKa values (Supplementary Figure S1). The unique H-bond provides a structural basis for the preference of dCTP insertion opposite the lesion in the primer extension assays (see In Vitro Studies). Other bases, such as A, T, and G, do not have such a NH2 group as found in C, to form a symmetrical and stronger H-bond with the major groove-shifted 3′G of Pt-GG in the GG1 structure. Therefore, dCTP is the preferred nucleotide incorporated at the 3′G of Pt-GG, despite the WC base pair being disrupted by the covalent modification of the DNA template base.
The structural perturbation within the DNA template strand is propagated to the 3′ end of the primer strand at the template-primer junction. The template C base upstream (3′) to the Pt-GG lesion loses the base stacking interaction with the 3′G and is over wound by 12° (Figure 2A). Consequently, the 3′G base of the primer as a base pairing partner for the over wound template C base is disordered as the electron density for the G base completely disappears (Figure 1A; Supplementary Figure S3). The disordered primer end likely causes template-primer misalignment, which leads to frameshift mutations. The disordering observed in GG1 is supported by a single deletion by Dpo4 in our solution studies (see In vitro Studies below). The incoming dCTP resides at a catalytically competent position with the distance between its Pα and the 3′ end of the primer being 5.6 Å (Figures 1A and 2A), as the backbone of the primer strand is still structured at its 3′ end (Supplementary Figure S3). However, base stacking interactions were lost between the incoming nucleotide and the disordered primer base, which makes the ternary complex less stable than the structures with undamaged DNA. The lesion template-induced disordering of 3′ terminal primer end would contribute to the impaired catalytic capability of Dpo4 at this stage (see In vitro studies).
GG2: dCTP incorporation against the 5′G of Pt-GG (second insertion)
An incoming dCTP is inserted opposite the 5′G of Pt-GG in the GG2 ternary complex that captures the second insertion stage. For the first time, this structure shows the two G bases of Pt-GG entering a polymerase active site simultaneously (Figure 1B). The adduct is in two alternate conformations (GG2a and GG2b) that are tilted about 10° with respect to each other (Figures 2B, C, and 3A). In the anomalous difference map, two discrete peaks show the alternate positions of the Pt atom (Figure 1B). The presence of the alternate conformers agrees with the bulged electron density map around the Pt-GG adduct (Figure 3A). The DNA helix keeps a straight form as observed in other Dpo4–DNA complex structures (Figure 3B). As it translocated into the active site (Figure 2E), the adduct adopts depressed roll (ρ) angles as low as 22° between the cross-linked bases, substantially less than ∼90° roll angles observed in a single Pt-GG (Sherman et al, 1985). In other words, the roll angle is converted from 135° in GG1 to 22° in GG2 to fit Pt-GG into the active site (Figure 2E). Accordingly, the α and β torsion angles of Pt-GG deviate up to 60° from the low-energy conformation (Supplementary Table S2). A reduction in the roll angles and use of alternate conformations make the cross-linked Pt-GG more compatible with the base stacking system of a straight helix (Figure 3B). This set-up reduces the perturbation caused by an angular adduct on the DNA helical structure and helps the lesion fit into the active site.
The 5′G of Pt-GG forms a WC base pair with the incoming dCTP in the active site (Figure 1B). The replicating base pair is in the regular position and superimposed well with the undamaged DNA replicating base pairs (Figure 1D). The finger domain is in contact with the replicating base pair (Figure 2B and C), which makes the 5′G base of Pt-GG less tilted from the base stacking system than the 3′G base (Figure 3A and B). The α-phosphate of dCTP was 4.5 Å from the C3′ atom of ddC at the 3′ end of the primer strand, indicating that the incoming nucleotide is poised for catalysis. The WC base paired replicating base pair explains the preference of dCTP over other dNTPs in the second insertion step (see In Vitro Studies). However, the alternate conformations and the geometric strain on the angular Pt-GG bases reflect a high energetic state for the GG2 complex, which contributes to the reduced nucleotide incorporation in the second insertion (Brown et al, 2008). Overall, the energetically unfavourable adduct conformation and the Pt-GG conformational conversion associated entropy cost may account for the lowest efficiency of nucleotide incorporation in the second insertion during the Pt-GG bypass (Brown et al, 2008).
GG3: nucleotide incorporation after the bypass of the Pt-GG adduct (extension step)
In the GG3 structure, the incoming dATP is base paired with the template T base 5′ to the lesion, illustrating primer extension after the Pt-GG lesion (Figure 1C). The Pt-GG adduct is in a depressed conformation with roll angle of 26° to fit into the straight DNA helix in GG3 (Figures 1C and 2D). As the Pt-GG adduct has moved out of the active site in GG3, the adduct is less strained than the adduct in GG2. The α/β torsion angles of Pt-GG in GG3 are a few degrees less distorted from the low-energy conformation than the lesion in GG2 (Supplementary Table S1). A single anomalous peak shows one position of the platinum atom with a well-defined lesion structure (Figure 1C). The replicating base pair exhibits normal WC base pairing in the active site and is superimposed well with the standard replicating base pair in Dpo4 (Figure 1D). The replicating base pair has the template T and dATP in plane, which is confined by the closed finger domain (Figure 2D). However, the base pair (5′G*:C) beneath the replicating base pair is significantly disrupted due to the tilted orientation of the 5′G base from the angular Pt-GG (Figure 3C). The G base buckles 15° and propels 18° from the base pair partner C base (Figures 1C and 3C). The disturbed base pair is not in the active site as a replicating base pair, so the structural distortion appears to be less disruptive to DNA replication as in GG1 and GG2. The crosslinked G base 18° tilting from the template T base in the active site, however, does disturb the replicating base pair as a poor stacking partner (Figure 3C), which corresponds to the nucleotide incorporation reduction observed in the extension stage (Brown et al, 2008). Overall, the GG3 complex structure was similar to that of Dpo4 with undamaged DNA (Ling et al, 2001, 2003, 2004a, 2004b; Vaisman et al, 2005), which maintains an ideal WC base paring for the replicating base pair in the active site (Figure 1C). The DNA structural disturbance caused by Pt-GG in GG3 is mainly on the base pair which has moved out of the active site. Therefore, the Pt-GG lesion would impair catalysis less in the extension than that in the insertion stages (Brown et al, 2008).
In vitro studies of Pt-GG bypass by Dpo4
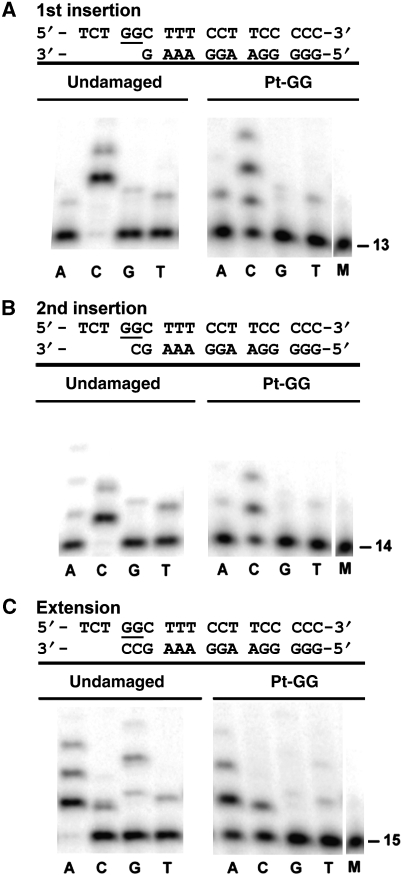
To examine Dpo4-mediated bypass of Pt-GG under conditions similar to crystal formation, we performed a running start assay with a short heteroduplex 10/18-mer at 23°C (Supplementary Figure S4). The template was modified at positions 14 and 15 with cisplatin, identical to the one used for crystallographic studies. There was significant accumulation of intermediate products (13- and 14-mers) corresponding to nucleotide incorporation opposite the two G bases of the Pt-GG adduct (13-mer is pre-first insertion, 14-mer is pre-second insertion), identical to the observation for the assays with longer substrates at 37°C (Brown et al, 2008). Thus, these results support our structural observations that the Pt-GG disturbs the replicating base pair in the active site in GG1 and GG2, which causes Dpo4 to pause during nucleotide incorporation opposite the Pt-GG adduct. Interestingly, the full product is 1 nt shorter in the lesion DNA than that of the undamaged DNA in the control panel (Supplementary Figure S4). The shorter product is likely caused by −1 frameshift mutation due to template-primer misalignment. The misalignment has been observed in the Dpo4 structures in which template-primer junctions are disturbed in the active site (Ling et al, 2001, 2004a; Bauer et al, 2007). The single deletion observed in our running start assay supports the template-primer junction instability caused by the Pt-GG adduct, such as the disordered 3′ end primer end in GG1. The frameshift mutations has been also observed in Pt-GG replication by human DNA polymerase η in vitro (Bassett et al, 2002), which may be a common mechanism in TLS of Pt-GG.
To confirm that WC base paring in the active site is disturbed by the Pt-GG lesion in solution, we performed the nucleotide incorporation assays using individual dNTP nucleotides (Figure 4). Less product formation was observed when Dpo4 was replicating cisplatin-DNA compared with undamaged DNA (Figure 4), supporting the structural disturbance of the replicating base pairs in the GG1, GG2, and GG3 structures. During Pt-GG lesion bypass, Dpo4 preferentially inserted a canonical dCTP opposite the Pt-GG lesion in the following order: dCTP≫dATP>dTTP>dGTP (Figure 4), a preference that is identical to the earlier results conducted at 37°C with a longer DNA substrate (Brown et al, 2008). These results are consistent with the structures in which the replicative base pair is either in a WC base pair (GG2) or in a misaligned WC base pair with a strong and symmetric H-bond (GG1). Interestingly, the dCTP misincorporation appears more pronounced than other dNTP misincorporations in Pt-GG adducted DNA replication (Figure 4, right panels). The dCTP mismatches are shown as a strong band in the extension (Figure 4C) and extra bands after two insertions (Figure 4A and B). The pronounced C base mismatches are likely associated with the unstable replicating base pairs in the lesion DNA structures, which increases the probability of template-primer misalignment. Template-primer misalignment is sequence dependent (Ling et al, 2004a; Garcia-Diaz and Kunkel, 2006; Bauer et al, 2007), thus, a two G base lesion, such as Pt-GG, in the template may promote pronounced C misincorporation specifically.
Figure 4.
Specificity of Dpo4-mediated nucleotide incorporation was tested for the (A) GG1 (first insertion), (B) GG2 (second insertion), and (C) GG3 (extension) reaction stages with a dNTP (A, C, G, or T). M represents the marker for the primer strand. In each panel, nucleotide incorporation assays were terminated after 10 min for undamaged DNA (left) and after 60 min for Pt-GG DNA (right).
Earlier kinetic studies have suggested that two distinct populations appear during nucleotide incorporation against the 3′G and 5′G of the Pt-GG lesion by Dpo4 (Brown et al, 2008). We confirmed that two distinct kinetic populations also existed with Dpo4 by performing similar kinetic assays with short primer-template DNA at room temperature (data not shown). Biphasic kinetics is consistent with the multiple conformations and mobility of the DNA substrate observed in the GG1 and GG2 structures.
In vivo studies of Pt-GG bypass
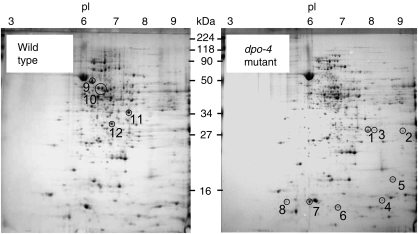
TLS by the human Y-family enzymes is a putative mechanism of intrinsic cisplatin resistance (Rabik and Dolan, 2007). However, the overlapping functions among the 16 human DNA polymerases complicate the contribution by Y-family DNA polymerases (Shachar et al, 2009). S. solfataricus is an ideal model organism, as it encodes only one Y-family DNA polymerase, Dpo4, which has TLS function in vitro. To better understand how a Y-family enzyme participates in the mechanism of drug resistance, we constructed a dpo-4 knockout mutant of S. solfataricus by targeted gene disruption as described earlier (Worthington et al, 2003; Schelert et al, 2004) and characterized the consequence of Dpo4 deficiency. The dpo-4 chromosomal mutation was confirmed using PCR to show that the mutant strain carried a disrupted dpo-4 allele due to lacS gene insertion; and by western blot analysis to show the absence of detectable levels of Dpo4 in cell extracts (Supplementary Figures S5 and S6). Physiological studies that compared the dpo-4 mutant with its otherwise isogenic wild-type parent were conducted and showed that Dpo4 was not essential for life as the growth rate, cell yield, efficiency of plating, and cell morphology were all normal in the dpo-4 mutant strain (data not shown). These data also indicate that Dpo4 did not contribute substantially to replicative synthesis of the chromosome. However, chemical challenge using cisplatin reveals that Dpo4 deficiency elicited significant biological effects. To directly evaluate the in vivo function of Dpo4 towards cisplatin, a dose-response study was conducted using both the wild-type and dpo-4 disruption mutant cell lines. The 50% lethal concentration (LC50) values of cisplatin were 23 μg/ml for the wild-type and 16 μg/ml for the dpo-4 mutant (Supplementary Figure S7). The reduction in LC50 dose of cisplatin in the dpo-4 strain shows that Dpo4 has a significant function in the tolerance of cisplatin. Proteomic analysis revealed that Dpo4 deficiency perturbed the abundance of many proteins (Figure 5; Supplementary Table S2). For example, proteins with increased abundance in the dpo-4 mutant include peptidyl-prolyl isomerase (SSO0758), fibrillarin (SSO0940), and single-stranded DNA binding protein (SSO2364). Together, increased abundance of these proteins suggests that Dpo4 deficiency elicits some form of genomic stress and is consistent with heightened sensitivity to cisplatin drug challenge.
Figure 5.
Analysis of wild-type and dpo-4 mutant proteomes. Proteins from cell extracts prepared from exponentially growing cell lines, were fractionated by 2D SDS–PAGE and their identities were determined by tandem MS/MS sequencing of peptides derived by trypsin hydrolysis, with subsequent matching to the S. solfataricus genome sequence. Values for isoelectric point (pI) and mass (MW) are indicated across the top and sides of the figure, respectively. Proteins with increased abundance in the dpo-4 mutant are circled in the dpo-4 panel; proteins with decreased abundance are circled in the wild-type panel. Proteins are numbered in correspondence with Supplementary Table S2.
Discussion
Structural snapshots capture unique Pt-GG conformational transitions in an active Y-family polymerase
Our structures provide the first complete picture of how a Y-family DNA polymerase traverses the two G bases of a Pt-GG cross-link and catalyses primer extension beyond the damaged site. The adduct in the template strand is shown in three distinct positions relative to the incoming nucleotide at the active site, clearly illustrating the translocation of the lesion template during bypass. Particularly, all three DNA-bound Dpo4 structures are in a common closed conformation in which the finger, thumb, and little finger domains grip DNA tightly (Ling et al, 2001; Nair et al, 2006; Lone et al, 2007; Kirouac and Ling, 2009). Interestingly, Pt-GG undergoes significant conformational changes, as it travels through the active site from GG1 to GG3. With the conformational change, the 5′G base of Pt-GG is able to enter into the active site in GG2 and GG3, which have the entire double-base adduct accommodated in a straight DNA helix. The unique conformational change of Pt-GG is not observed in the earlier ypolη structures (Figure 2F) (Alt et al, 2007). The second insertion ypolη structure with dATP does not translocate the 5′G into its active site and keeps the Pt-GG adduct in similar conformations and positions in both insertion stages (Figure 2F). In addition, our structures with an extension stage further highlight the importance of the depressed Pt-GG conformation to the DNA helix in the productive ternary complexes after the first insertion.
Unique mechanism for double-base lesion bypass
The Pt-GG bypass differs from the only other double-base bypass in complex with Dpo4 for the cis-syn cyclobutane pyrimidine dimer lesion (CPD, TT dimer). For CPD lesion bypass, the entire TT dimer enters Dpo4's active site during the two-stage bypass, without conformational changes occurring in the dimer (Ling et al, 2003). The rigid and compact nature of the CPD allows Dpo4 to accommodate the entire double-base lesion in the active site for both insertion stages. In Pt-GG bypass, Dpo4 has to alter the conformation of the adduct to accommodate the Pt-GG adduct in its active site. The structural differences reflect the varying catalytic efficiencies of Dpo4 bypassing different double-base lesions at the two insertion stages. Dpo4 bypasses the 5′T (second insertion) more efficiently than the 3′T (first insertion) because the double-base lesion does not fit the ‘closed off' the active site well at the first insertion stage (Ling et al, 2003). In contrast, the insertion efficiency at the second insertion in Pt-GG bypass is much lower (180-fold) than the first insertion (Brown et al, 2008). The structural basis for lower dCTP incorporation against the 5′G at the second insertion would be strong geometric strains on the adduct when the angular adduct moves into the active site and the translocation-coupled conformational conversion, which is likely to be entropically costly. Thus, TLS for different double-base lesions catalysed by the same enzyme occurs through different bypass mechanisms, as revealed by the structural analyses.
Of the human Y-family enzymes, polη bypasses cisplatin-DNA adducts error freely while polκ performs error-prone TLS across Pt-GG (Bassett et al, 2003; Shachar et al, 2009). Human polκ keeps a similar closed conformation on the finger domain as Dpo4 and other mammalian Y-family polymerases (Supplementary Figure S2). A polη structure is only available from the yeast homologue (Alt et al, 2007), in which the finger domain is 16° open from the common closed conformation. The open conformation of ypolη seems unable to translocate the Pt-GG adduct within the active site (Figure 1D). Moreover, based on steady-state kinetic data, the extension step for hpolη is more difficult than Dpo4 in the incorporation opposite the lesion (Bassett et al, 2003), which is different from Dpo4 that has lowest replication efficiency at the second insertion. This discrepancy may be caused by structural differences observed in the two polymerases. The open conformation of ypolη does not confine the entire adduct into the active site (Alt et al, 2007), which may affect the extension step. A recent study suggested that multiple polymerases participate in the bypass of Pt-GG in mammalian cells, in which Y-family polymerases, such as polη, polκ, insert nucleotides opposite the lesion, and a B-family polymerase ζ extends the primer after insertion to the lesion (Shachar et al, 2009). The slow extension step by hpolη may be remedied by hpolζ in mammalian cells.
The mobile adducts destabilize the replicating base pair
Earlier biphasic kinetic data support that the Dpo4–DNA complex exists as at least two distinct populations (Brown et al, 2008). Pt-GG in different structural environments adopts different conformations with varied roll angles and α/β torsion angles (Sherman et al, 1985; Takahara et al, 1995; Ohndorf et al, 1999). The two guanine bases in the cis-Pt(NH3)2[d(pGpG)] (a single Pt-GG) crystal structure has a perpendicular conformation with a roll angle of ∼90° and α/β angles differ from the ideal values in a narrow range of 5–30° (Sherman et al, 1985). When a Pt-GG adduct is incorporated in the middle of a duplex DNA, the roll angle is compressed to 30°, with the torsion angles α/β deviated from ideal values up to 50° in protein-free adducted DNA helix (Takahara et al, 1995). The distorted and angular Pt-GG causes the 12-base pair duplex bending to a 43° curve (Figure 3D; Takahara et al, 1995). In GG2 and GG3, the DNA helices are straight with bending angle <18° over a 12-bp helical region, which further compresses the Pt-GG adduct with roll angle as low as 22° (Figure 3B and C). The Pt-GG adduct is distorted with the α/β torsion angles up to 60° to fit into the active site and helical DNA, 10° more distortion than the ones observed in the Pt-GG adducts in protein-free DNA (Takahara et al, 1995). The α/β torsion angles in the Dpo4 complexes are energetically unfavourable based on the molecular simulation (Yao et al, 1994). Particularly, the additional depression on the roll angle is reinforced by the close contact of the finger domain on the replicating base pair in GG2 (Figure 2B and C). The depression, combined with alternate conformations, brings the adduct to nearly parallel conformations in GG2 (Figure 3B). The geometric strains from the distortion would bring the adduct to a high-energy state that takes alternate conformations in GG2. Overall, Pt-GG is highly mobile in the two insertion structures due to its solvent exposure (GG1) and geometric strains (GG2). A similar mobile Pt-GG adduct also exists in the cisplatinated nucleosome structure where the intrastrand platinum adducts are packed into helical DNA (Wu et al, 2008). At 3.4 Å resolution, elongated Pt-anomalous density indicates the inherent mobility of the platinum atom covalently bonded to the parallel purine bases, which contrasts with the spherical anomalous selenium peaks in the same structure (Wu et al, 2008).
In summary, destabilization of the adduct perturbs the DNA structure, so that catalytic efficiency and fidelity are reduced in Dpo4-mediated bypass of Pt-GG (Brown et al, 2008). The misaligned G*:dCTP in GG1 and alternate G*:dCTP base pair in GG2 make replicating base pairs unstable, which would increase the probability of mutations and decrease the populations of the productive complexes of Dpo4–DNA. The mobility is consistent with the following observations in solution: (1) the pronounced C base misincorporation in the primer extension assays (Figure 4); (2) 72- and 860-fold reduction in incorporation efficiency (kp/Kd) at the first and second insertion steps, respectively, and six-fold reduction at the extension step, relative to control undamaged DNA (Brown et al, 2008); and (3) two orders of magnitude of fidelity reduction in Pt-GG adducted DNA replication compared with undamaged DNA replication by Dpo4 (Brown et al, 2008). Interestingly, the symmetric H-bonding in the misaligned G*:dCTP at the first insertion (GG1) and WC H-bonding in the second insertion (GG2) still provide specificity for selective dCTP incorporation against the Pt-GG lesion though enhanced misincorporation occurs in the disturbed base paring environment.
Conclusions
This work provides a new molecular model for TLS over DNA lesions. Our structures reveal that Pt-GG bypass is a unique, dynamic process in which the Pt-GG adduct undergoes conformational changes, as it is translocated through the ‘close-off' active site of the Y-family polymerase. The angular double-base lesion adopts a depressed conformation to fit into the active site and the helical structure of DNA during the second insertion and extension stages. The incompatibility of angular adducts with helical DNA structure and the rigid active site, along with the stress caused by the depressed conformation, make the Pt-GG adduct in energetically unfavourable conformations. The disturbed and deformed DNA template leads to instability of the replicating base pair, resulting in low replication efficiency and reduced fidelity during Pt-GG bypass. Particularly, in the second insertion, the entropically costly conformational conversion may additionally impede nucleotide incorporation against the 5′G of Pt-GG. The in vivo observations support that the Y-family polymerase Dpo4 has an important function in tolerance of cisplatin, implicating possible contributions of Y-family polymerases to cisplatin resistance. The knowledge of how platinum cross-linked lesions are replicated by a Y-family DNA polymerase may help the development of cross-linking platinum agents for cancer therapy.
Materials and methods
Protein purification, DNA preparation, and crystallization
Dpo4 was expressed and purified as described earlier (Ling et al, 2001). DNA oligonucleotide preparation was similar to that in the earlier reported work (Brown et al, 2008). A hanging drop vapor diffusion method was used to grow crystals of the Dpo4–DNA–dNTP ternary complex by using our earlier conditions with some modifications (Bauer et al, 2007). The detailed procedures are reported in the Supplementary data.
Data collection and structure determination
Diffraction data for the GG1, GG2, and GG3 crystals were collected at beamline 24-ID-C (Argonne National Laboratory, IL) and processed using DENZO and SCALEPACK (Otwinowski and Minor, 1997). All crystal structures were solved with molecular replacement program PHASER (McCoy et al, 2005), with the type I structure as search model (PDB: 1JX4). Iterative cycles of simulated annealing, positional refinement, and B-factor refinement were performed using CNS (Brunger et al, 1998), along with manual rebuilding of the model using the graphics program COOT (Emsley and Cowtan, 2004). The GG1, GG2, and GG3 structures were refined to 2.9, 1.9, and 2.0 Å resolution, respectively.
In vitro assays
All the in vitro assays were carried out as described earlier (Brown et al, 2008; Wong et al, 2008), with reaction temperature at 23°C. The detailed procedures are reported in the Supplementary data.
Construction of the S. solfataricus dpo-4 mutant
The dpo-4 knockout mutant of S. solfataricus was constructed as described earlier (Worthington et al, 2003; Schelert et al, 2004). The detailed procedures are reported in the Supplementary data.
Molecular biology and proteomic methods
DNA cloning, DNA sequencing, PCR, and plasmid transformation of Escherichia coli were performed as described (Rockabrand et al, 1998; Rolfsmeier et al, 1998; Haseltine et al, 1999). DNA concentrations were measured using a DyNA Quant 200 fluorometer (Hoefer). Protein concentrations were measured using the BCA Protein Assay Reagent Kit (Pierce). Chemiluminescent western blot analysis using anti-Dpo4 polyclonal sera was performed using the ECL system (Amersham Biosciences) as described (Rockabrand et al, 1998). Proteins were fractionated by two-dimensional SDS–PAGE as described (Hajduch et al, 2005). Protein samples were prepared using 50 ml of mid-exponential phase cultures collected by centrifugation at 3000 g for 15 min at 26°C. Pelleted cells were extracted as described (Hajduch et al, 2005) and protein was precipitated for 1 h by exposure to five volumes of ice-cold 0.1 M ammonium acetate in 100% methanol at 22°C. Proteins were sequenced using tandem mass spectrometry (MS/MS) and peptides were identified by local BLAST against the S. solfataricus proteome as described (Worthington et al, 2003). Confirmed hits required a threshold peptide matching value (N) of at least five.
In vivo cisplatin assay
Cell lines were cultured at 80°C with aeration in a minimal salts medium (Allen, 1959), at pH 3.0 with tryptone 0.2% (wt/vol) as sole carbon and energy source. Growth was monitored at a wavelength of 540 nm using a Cary 50 spectrophotometer (Varian). Cisplatin was prepared as fresh aqueous solutions (2 mg/ml) before use and was added at early logrithmic phase.
Coordinates
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.rcsb.org, with accession codes 3M9M, 3M9N, and 3M9O for the structures GG1, GG2, and GG3, respectively.
Supplementary Material
Acknowledgments
This work was supported by the Terry Fox Foundation (NCIC-015411) to HL. JAB was supported by an International PEO Scholar Award. ZS was supported by the National Science Foundation Career Award (MCB-0447899). PB is supported by National Institutes of Health (NIGMS 1R01GM090209-01). TN is supported by the Ministry of Education, Culture, Sports, Science, and Technology, Japan (MEXT, 18201010). We thank the staff at 24-ID of APS in Argonne National Laboratory for beamline support. TN thanks Masami Yamada and Mariarita De Felice for DNA constructs. PB thanks Karl Dana and Edith Soo for mutant analysis. We thank Dr Lynn Weir for critical reading of the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albertella MR, Green CM, Lehmann AR, O'Connor MJ (2005) A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res 65: 9799–9806 [DOI] [PubMed] [Google Scholar]
- Allen MB (1959) Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch Mikrobiol 32: 270–277 [DOI] [PubMed] [Google Scholar]
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T (2007) Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science 318: 967–970 [DOI] [PubMed] [Google Scholar]
- Bassett E, Vaisman A, Havener JM, Masutani C, Hanaoka F, Chaney SG (2003) Efficiency of extension of mismatched primer termini across from cisplatin and oxaliplatin adducts by human DNA polymerases beta and eta in vitro. Biochemistry 42: 14197–14206 [DOI] [PubMed] [Google Scholar]
- Bassett E, Vaisman A, Tropea KA, McCall CM, Masutani C, Hanaoka F, Chaney SG (2002) Frameshifts and deletions during in vitro translesion synthesis past Pt-DNA adducts by DNA polymerases beta and eta. DNA Repair (Amst) 1: 1003–1016 [DOI] [PubMed] [Google Scholar]
- Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H (2007) A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc Natl Acad Sci USA 104: 14905–14910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergoglio V, Canitrot Y, Hogarth L, Minto L, Howell SB, Cazaux C, Hoffmann JS (2001) Enhanced expression and activity of DNA polymerase beta in human ovarian tumor cells: impact on sensitivity towards antitumor agents. Oncogene 20: 6181–6187 [DOI] [PubMed] [Google Scholar]
- Brown JA, Newmister SA, Fiala KA, Suo Z (2008) Mechanism of double-base lesion bypass catalyzed by a Y-family DNA polymerase. Nucleic Acids Res 36: 3867–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Canitrot Y, Cazaux C, Frechet M, Bouayadi K, Lesca C, Salles B, Hoffmann JS (1998) Overexpression of DNA polymerase beta in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc Natl Acad Sci USA 95: 12586–12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland WW, Frey PA, Gerlt JA (1998) The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem 273: 25529–25532 [DOI] [PubMed] [Google Scholar]
- Cohen SM, Lippard SJ (2001) Cisplatin: from DNA damage to cancer chemotherapy. Prog Nucleic Acid Res Mol Biol 67: 93–130 [DOI] [PubMed] [Google Scholar]
- Coll M, Sherman SE, Gibson D, Lippard SJ, Wang AH (1990) Molecular structure of the complex formed between the anticancer drug cisplatin and d(pGpG): C222(1) crystal form. J Biomol Struct Dyn 8: 315–330 [DOI] [PubMed] [Google Scholar]
- Davidson JP, Faber PJ, Fischer RG Jr, Mansy S, Peresie HJ, Rosenberg B, VanCamp L (1975) ‘Platinum-pyrimidine blues' and related complexes: a new class of potent antitumor agents. Cancer Chemother Rep 59: 287–300 [PubMed] [Google Scholar]
- Eastman A (1990) Activation of programmed cell death by anticancer agents: cisplatin as a model system. Cancer Cells 2: 275–280 [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Wagner R, Radman M (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296: 1627–1630 [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Kunkel TA (2006) Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem Sci 31: 206–214 [DOI] [PubMed] [Google Scholar]
- Gibbons GR, Kaufmann WK, Chaney SG (1991) Role of DNA replication in carrier-ligand-specific resistance to platinum compounds in L1210 cells. Carcinogenesis 12: 2253–2257 [DOI] [PubMed] [Google Scholar]
- Goodman MF (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71: 17–50 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine C, Montalvo-Rodriguez R, Carl A, Bini E, Blum P (1999) Extragenic pleiotropic mutations that repress glycosyl hydrolase expression in the hyperthermophilic archaeon Sulfolobus solfataricus. Genetics 152: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JS, Pillaire MJ, Garcia-Estefania D, Lapalu S, Villani G (1996) In vitro bypass replication of the cisplatin-d(GpG) lesion by calf thymus DNA polymerase beta and human immunodeficiency virus type I reverse transcriptase is highly mutagenic. J Biol Chem 271: 15386–15392 [DOI] [PubMed] [Google Scholar]
- Hoffmann JS, Pillaire MJ, Maga G, Podust V, Hubscher U, Villani G (1995) DNA polymerase beta bypasses in vitro a single d(GpG)-cisplatin adduct placed on codon 13 of the HRAS gene. Proc Natl Acad Sci USA 92: 5356–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson ER, Lippard SJ (1999) Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev 99: 2467–2498 [DOI] [PubMed] [Google Scholar]
- Kirouac KN, Ling H (2009) Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase iota. EMBO J 28: 1644–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut DA, Sigala PA, Pybus B, Liu CW, Ringe D, Petsko GA, Herschlag D (2006) Testing electrostatic complementarity in enzyme catalysis: hydrogen bonding in the ketosteroid isomerase oxyanion hole. PLoS Biol 4: e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR (2002) Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat Res 509: 23–34 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W (2003) Replication of a cis-syn thymine dimer at atomic resolution. Nature 424: 1083–1087 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107: 91–102 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W (2004a) Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell 13: 751–762 [DOI] [PubMed] [Google Scholar]
- Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W (2004b) Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc Natl Acad Sci USA 101: 2265–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert B (1999) Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug. Zürich: Helvetica Chimica [Google Scholar]
- Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK (2007) Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell 25: 601–614 [DOI] [PubMed] [Google Scholar]
- Mamenta EL, Poma EE, Kaufmann WK, Delmastro DA, Grady HL, Chaney SG (1994) Enhanced replicative bypass of platinum-DNA adducts in cisplatin-resistant human ovarian carcinoma cell lines. Cancer Res 54: 3500–3505 [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA (2008) The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res 18: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2006) An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure 14: 749–755 [DOI] [PubMed] [Google Scholar]
- Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ (1999) Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 399: 708–712 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pinto AL, Lippard SJ (1985) Binding of the antitumor drug cis-diamminedichloroplatinum(II) (cisplatin) to DNA. Biochim Biophys Acta 780: 167–180 [DOI] [PubMed] [Google Scholar]
- Rabik CA, Dolan ME (2007) Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 33: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ (2006) Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol 4: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon VM, Schulte N, Eastman A (1987) Multiple mechanisms of resistance to cis-diamminedichloroplatinum(II) in murine leukemia L1210 cells. Cancer Res 47: 2056–2061 [PubMed] [Google Scholar]
- Rockabrand D, Livers K, Austin T, Kaiser R, Jensen D, Burgess R, Blum P (1998) Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J Bacteriol 180: 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfsmeier M, Haseltine C, Bini E, Clark A, Blum P (1998) Molecular characterization of the alpha-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 180: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush AA, Suarez M, Friedberg EC, Radman M, Siede W (1998) Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol Gen Genet 257: 686–692 [DOI] [PubMed] [Google Scholar]
- Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P (2004) Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol 186: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z (2009) Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman SE, Gibson D, Wang AH, Lippard SJ (1985) X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: cis-[Pt(NH3)2(d(pGpG))]. Science 230: 412–417 [DOI] [PubMed] [Google Scholar]
- Siddik ZH (2003) Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22: 7265–7279 [DOI] [PubMed] [Google Scholar]
- Suo Z, Lippard SJ, Johnson KA (1999) Single d(GpG)/cis-diammineplatinum(II) adduct-induced inhibition of DNA polymerization. Biochemistry 38: 715–726 [DOI] [PubMed] [Google Scholar]
- Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ (1995) Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature 377: 649–652 [DOI] [PubMed] [Google Scholar]
- Vaisman A, Chaney SG (2000) The efficiency and fidelity of translesion synthesis past cisplatin and oxaliplatin GpG adducts by human DNA polymerase beta. J Biol Chem 275: 13017–13025 [DOI] [PubMed] [Google Scholar]
- Vaisman A, Ling H, Woodgate R, Yang W (2005) Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J 24: 2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Masutani C, Hanaoka F, Chaney SG (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry 39: 4575–4580 [DOI] [PubMed] [Google Scholar]
- Vaisman A, Varchenko M, Umar A, Kunkel TA, Risinger JI, Barrett JC, Hamilton TC, Chaney SG (1998) The role of hMLH1, hMSH3, and hMSH6 defects in cisplatin and oxaliplatin resistance: correlation with replicative bypass of platinum-DNA adducts. Cancer Res 58: 3579–3585 [PubMed] [Google Scholar]
- Wong JH, Fiala KA, Suo Z, Ling H (2008) Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J Mol Biol 379: 317–330 [DOI] [PubMed] [Google Scholar]
- Worthington P, Hoang V, Perez-Pomares F, Blum P (2003) Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 185: 482–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Droge P, Davey CA (2008) Site selectivity of platinum anticancer therapeutics. Nat Chem Biol 4: 110–112 [DOI] [PubMed] [Google Scholar]
- Yang W, Woodgate R (2007) What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci USA 104: 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Plastaras J, Marzilli LG (1994) A molecular mechanics AMBER-type force field for modeling platinum complexes of guanine derivatives. Inorg Chem 33: 6061–6077 [Google Scholar]
- Zorbas H, Keppler BK (2005) Cisplatin damage: are DNA repair proteins saviors or traitors to the cell? Chembiochem 6: 1157–1166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.