Abstract
The transcription factor ATF4 regulates the expression of genes involved in amino acid metabolism, redox homeostasis and ER stress responses, and it is overexpressed in human solid tumours, suggesting that it has an important function in tumour progression. Here, we report that inhibition of ATF4 expression blocked proliferation and survival of transformed cells, despite an initial activation of cytoprotective macroautophagy. Knockdown of ATF4 significantly reduced the levels of asparagine synthetase (ASNS) and overexpression of ASNS or supplementation of asparagine in trans, reversed the proliferation block and increased survival in ATF4 knockdown cells. Both amino acid and glucose deprivation, stresses found in solid tumours, activated the upstream eukaryotic initiation factor 2α (eIF2α) kinase GCN2 to upregulate ATF4 target genes involved in amino acid synthesis and transport. GCN2 activation/overexpression and increased phospho-eIF2α were observed in human and mouse tumours compared with normal tissues and abrogation of ATF4 or GCN2 expression significantly inhibited tumour growth in vivo. We conclude that the GCN2-eIF2α-ATF4 pathway is critical for maintaining metabolic homeostasis in tumour cells, making it a novel and attractive target for anti-tumour approaches.
Keywords: amino acid, asparagine, ATF4, GCN2, metabolism
Introduction
Earlier studies in mouse embryonic fibroblasts (MEFs) showed that the basic leucine zipper transcription factor ATF4 is a critical regulator of genes involved in redox balance and maintenance of amino acid metabolism. Consequently, ATF4−/− MEFs require non-essential amino acids (NEAAs) and antioxidants to survive and proliferate (Harding et al, 2003). ATF4 also appears to have multiple functions during development: ATF4 knockout mice exhibit abnormal lens formation, growth retardation, anemia and delayed bone development (Tanaka et al, 1998; Masuoka and Townes, 2002; Yang et al, 2004).
Although ATF4 can be transcriptionally regulated (Siu et al, 2002), it is the translational upregulation of ATF4 that has received the most attention, because of the unusual mode of translational regulation of its mRNA in response to stress through phosphorylation of the eukaryotic initiation factor 2α (eIF2α) (Harding et al, 2000; Lu et al, 2004; Vattem and Wek, 2004). The endoplasmic reticulum kinase PERK (activated by misfolded/unfolded proteins in the ER) or the cytoplasmic kinase GCN2 (activated by amino acid deprivation) phosphorylate eIF2α at Ser51, thereby downregulating global translation. Paradoxically, a group of stress-responsive mRNAs that include ATF4 are translated more efficiently when eIF2α is phosphorylated. In the case of ATF4, this is due to the presence of the two upstream open reading frames located in the 5′UTR of the mRNA. These two elements repress ATF4 translation under unstressed conditions but enable its translation under stressed conditions (eIF2α phosphorylation). This translational regulation model was first characterized in yeast and later found to also exist in mammalian cells (Hinnebusch, 1984; Mueller and Hinnebusch, 1986; Harding et al, 2000; Vattem and Wek, 2004).
GCN2 is a high molecular weight protein kinase activated by uncharged tRNA (Wek et al, 1990, 1995; Ramirez et al, 1992). Activated GCN2 phosphorylates eIF2α to translationally upregulate ATF4, which in turn increases amino acid biosynthetic and transport pathways (Harding et al, 2000, 2003). GCN2 knockout mice are viable and fertile and display no gross phenotypic abnormalities unless fed diets lacking a single amino acid (Zhang et al, 2002; Anthony et al, 2004). Other than maintaining amino acid homeostasis, GCN2 also regulates synaptic plasticity and memory (Costa-Mattioli et al, 2005), feeding behaviour (Hao et al, 2005; Maurin et al, 2005), as well as lipid metabolism (Guo and Cavener, 2007). GCN2 is also activated by UV radiation and mediates NFκB signalling (Deng et al, 2002; Jiang and Wek, 2005).
In the tumour microenvironment, the abnormal development of vasculature results in insufficient blood supply, which is the major reason for the development of acute and chronic hypoxia and has been associated with deprivation of glucose and other nutrients. Earlier, we showed that PERK activation and the resulting eIF2α phosphorylation increase the ability of transformed cells to survive under hypoxia in vitro and in vivo and promote tumour growth (Bi et al, 2005). In the same study, we reported that as a downstream target of PERK and phospho-eIF2α, ATF4 also contributes to hypoxia resistance in MEFs. We and others reported that ATF4 overexpression is elevated in primary tumour tissues and co-localizes with hypoxic regions (Ameri et al, 2004; Bi et al, 2005). However, the precise function of ATF4 in tumour cell survival and proliferation has not been elucidated. In this study, we report that ATF4 is necessary for tumour cells to maintain homeostasis of amino acid metabolism and that activation of GCN2-ATF4-asparagine synthetase (ASNS) pathway promotes tumour cell survival under nutrient (amino acid or glucose) deprivation. GCN2-eIF2α pathway is activated in various human and mouse tumour tissues. Deficiency of ATF4 or GCN2 severely inhibits tumour growth in vivo. Together, these results suggest that GCN2-ATF4-ASNS pathway is a promising target for tumour therapy.
Results
ATF4 expression is required for survival and proliferation of fibrosarcoma and colorectal adenocarcinoma cells in the absence of non-essential amino acid supplementation
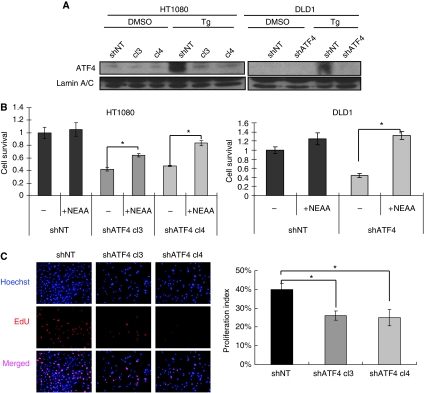
To investigate the function of ATF4 in tumour cell proliferation and survival, plasmids expressing ATF4-specific shRNA (pSM2-shATF4) or non-targeting shRNA (pLKO-shNT) were transfected into HT1080 (human fibrosarcoma) or DLD1 (human colorectal adenocarcinoma) cells. Two established HT1080 shATF4 clones (shATF4.cl3 and shATF4.cl4) and one DLD1 shATF4 clone showed 60–70% reduction of ATF4 mRNA levels compared with corresponding shNT clones (Supplementary Figure S1A). As the basal ATF4 protein levels are low in unstressed cells, we treated cells with the ER stress-inducing agent thapsigargin to upregulate ATF4. Consistent with mRNA levels, both HT1080 and DLD1 shATF4 clones showed no ATF4 induction after treatment with thapsigargin (Figure 1A).
Figure 1.
Knockdown of ATF4 inhibits tumour cell survival and proliferation. (A) ATF4 protein levels from the nuclear fractions of thapsigargin-treated (Tg, 1 μM, 4 h) or DMSO-treated cells. Lamin A/C was used as a loading control. (B) Survival of HT1080 and DLD1 cells cultured in the presence or absence of NEAA (100 μM) measured by MTT assay 48 h after plating (Data represent mean±s.e.m., n=3, *P<0.05). Cell survival was normalized to control (shNT cells without NEAA). (C) Cell proliferation assay. HT1080 cells were plated in DMEM with/without NEAA for 24 h. (Left panel) Fluorescent staining for nuclei (Hoechst, blue) and proliferating cells with EdU (red). (Right panel) Three randomly selected photographs were selected and numbers of EdU positive and total nuclei were counted. Percentage proliferation index was calculated by dividing the number of proliferating nuclei by the total number of nuclei. (Data represent mean±s.e.m., n=3, *P<0.05).
It was reported earlier that ATF4−/− MEFs require the presence of NEAAs and antioxidant such as β-mercaptoethanol (β-ME) to survive (Harding et al, 2003) (Supplementary Figure S2). Similar to SV40-immortalized ATF4−/− MEFs, tumour cells expressing ATF4 shRNA showed significantly reduced survival in the absence of NEAA (Figure 1B). A long-term growth assay suggested that shATF4 clones have defects in cell survival and proliferation rates (Supplementary Figure S1B). In contrast, cell survival of ATF4 knockdown cells was not affected by adding β-ME at concentrations from 25 μM to 0.2 mM (data not shown). Transiently knocking down ATF4 also reduced cell survival, indicating that this effect was not due to clonal effects during selection (Supplementary Figure S1C).
Reduced cell survival could result from decreased cell proliferation and/or increased cell death. By analysing the levels of fluorescent EdU incorporation in exponentially growing cells, we found that HT1080.shATF4 cells showed a 35% reduction in cell proliferation compared with shNT cells (Figure 1C). Similar findings were seen in DLD1 cells (Supplementary Figure S3A). Adding NEAA to the medium led to full recovery of cell proliferation in both knockdown cell lines (Supplementary Figure S3A). Cell-cycle analysis also showed that shATF4 cells had an increased G1 population compared with shNT cells (Supplementary Figure S3B), indicating that ATF4 knockdown caused G1/S arrest in tumour cells. Addition of NEAA partially reversed the G1 arrest. These data suggest that ATF4 deficiency induces amino acid starvation, which causes G1/S cell-cycle arrest and reduced proliferation.
Knockdown of ATF4 in transformed cells induces apoptosis
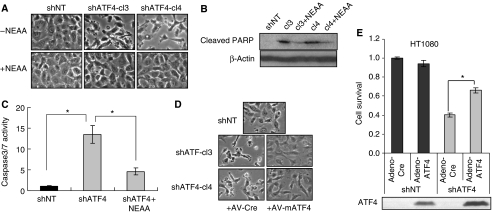
When the ATF4 knockdown cells were cultured in the absence of NEAA, morphological features of apoptosis such as membrane blebbing and cell shrinkage were observed. These apoptotic phenotypes were diminished by the addition of NEAA (Figure 2A). shATF4 cells also had higher levels of cleaved PARP, an apoptosis marker, which was similarly reduced in the presence of NEAA (Figure 2B). To further analyse the levels of apoptosis in shATF4 cells cultured without NEAA, we measured caspase3/7 activities. shATF4 cells exhibited an over 13-fold increase in basal caspase3/7 activity compared with shNT cells; similarly, supplementation with NEAA significantly reduced the caspase activities (Figure 2C).
Figure 2.
Knockdown of ATF4 induces apoptosis in tumour cells. (A) Phase-contrast images of HT1080 shNT and shATF4 cells growing in DMEM with or without NEAA for 24 h. Images shown were taken at × 400 magnification. (B) Immunoblot for cleaved PARP in HT1080 shNT and shATF4 cells. β-Actin was used as a loading control. HT1080 cells were incubated in DMEM with/without NEAA for 24 h. (C) Caspase3/7 activities normalized to total number of cells. HT1080 cells were incubated in DMEM with/without NEAA for 24 h. (D) Phase-contrast images of HT1080 shNT and shATF4 cells infected with the control (Av-Cre) or adenovirus expressing mouse ATF4 (AV-mATF4) indicated at × 400 magnification. After infection, cells were incubated in DMEM without NEAA. Images were taken 24 h after infection. (E) Relative cell survival in infected cells using an MTT assay. Equal numbers of cells were plated in DMEM without NEAA 24 h after infection. MTT assay was performed after 48 h. Cells infected under the same condition were also collected at 24 h after infection for ATF4 immunoblot (bottom panel). (Data represent mean±s.e.m., n=3, *P<0.05).
To exclude the possibility that the defects of shATF4 cells were due to off-target effects of the shRNA or selection-induced mutations, we overexpressed full-length mouse ATF4 that was not targeted by the shRNA against human ATF4 using an adenoviral vector. Overexpression of mATF4 significantly increased cell survival and blocked the apoptotic phenotype of shATF4 cells (Figures 2D and E). These findings further support a pro-survival function of ATF4 in these tumour cells.
Knockdown of ATF4 induces a pro-survival autophagic response
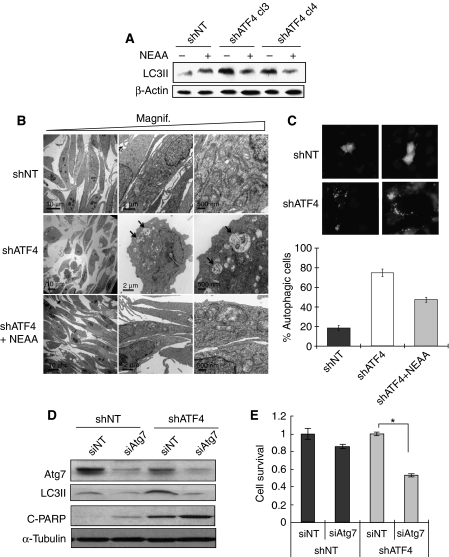
Autophagy is a lysosomal-dependent intracellular degradation process that is activated by certain stresses, primarily by nutrient starvation. As shATF4 cells rely on NEAA to survive, we hypothesized that autophagy might be induced in the HT1080.shATF4 cells as an initial pro-survival response in the absence of NEAA. This hypothesis was supported by the fact that in the absence of NEAA, shATF4 cells had a smaller size compared with shNT cells and cytoplasmic vacuoles (a sign of autophagosome formation) were observed (data not shown). shATF4 cells had elevated levels of the autophagy marker microtubule-associated protein light chain (LC) 3-II compared with shNT cells (Figure 3A), which were reduced to basal levels by adding NEAA, indicating that autophagy was induced in shATF4 cell because of NEAA shortage.
Figure 3.
Knockdown of ATF4 induces protective autophagy in tumour cells. (A) Immunoblot for the autophagy marker cleaved-LC3 from whole cell lysates in HT1080 shNT and shATF4 cells incubated in DMEM with/without NEAA for 24 h. β-Actin was used as a loading control. (B) Electron microscopy imaging. Arrows point to double-membrane-containing autophagosomes. HT1080 cells were incubated in DMEM with/without NEAA for 24 h. (C) Top: HT1080 shNT and shATF4 cells were transfected with pCMV-GFP-LC3 and incubated in DMEM. After 48 h, cells were stained with Hoechst before imaging. Bottom: Quantitation of cells with autophagic (punctate) morphology. Percentage autophagic cells were calculated after normalization to the total number of cells with GFP signal. Data represent mean (±s.e.m., n=3). (D) HT1080 cells were transfected with 100 nM of non-targeting siRNA (siNT) or siRNA targeting Atg7 (siAtg7). Atg7, LC3 and cleaved-PARP levels were analysed by immunoblotting 24 h after transfection. α-Tubulin was used as a loading control. (E) HT1080 cells were transfected with 100 nM non-targeting siRNA (siNT) or siRNA targeting ATG7 (siATG7). After 72 h, cell survival of HT1080 and DLD1 cells was measured by MTT assay. Experiment was carried out in triplicate. (Data represent mean±s.e.m., n=3, *P<0.05).
Under electron microscopy, the HT1080.shNT cells exhibited typical fibroblast morphology with intact ER and mitochondria, whereas the shATF4 cells were smaller, rounded and contained double-membrane autophagosomes, further confirming extensive induction of autophagy in shATF4 cells (Figure 3B, arrows pointing to autophagosomes). Similar to LC3 processing, addition of NEAA reversed the autophagic phenotype. The induction of autophagy was also confirmed by expressing GFP-LC3 in HT1080 cells. In shNT cells, the GFP signal was distributed evenly throughout the cytoplasm. However, in shATF4 cells, the signal was concentrated in green dots or ring-shaped structures, indicating the formation of autophagosomes (Figure 3C). To test whether the autophagy induced in shATF4 cells was a cytoprotective stress response, an siRNA targeting Atg7 (an E1-like ubiquitin conjugating enzyme required for autophagosomes maturation) was used to inhibit autophagy. When Atg7 levels were reduced and autophagy was blocked, the shATF4 cells showed increased apoptosis compared with shNT cells (Figures 3D and E). These results indicate that the autophagy induced by loss of ATF4 in the HT1080 cells promotes survival. The combination of losing ATF4 and the inhibition of autophagy results in a cooperative enhancement of cell death.
Addition of Asn in trans or expression of ASNS, rescues the survival of shATF4 cells
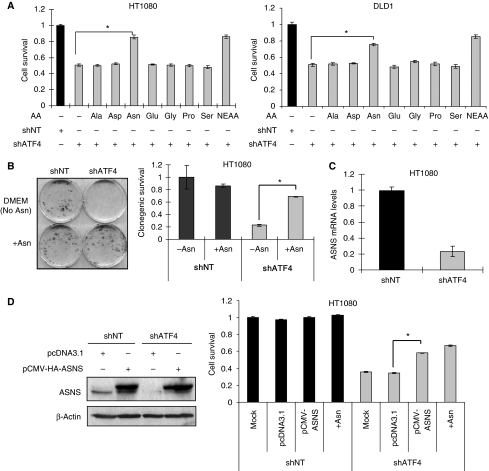
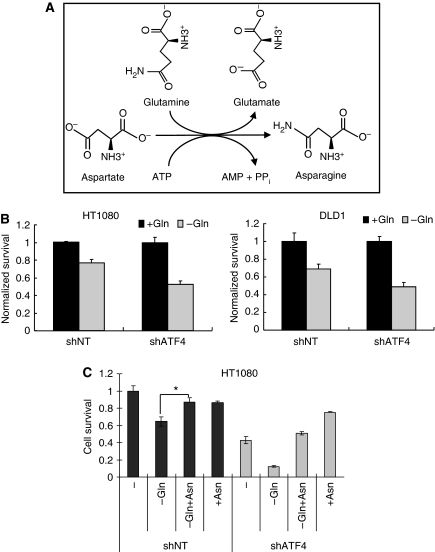
The mixture of NEAAs used in the experiments described above was comprised of seven amino acids: Ala, Asp, Asn, Glu, Gly, Pro and Ser. Each amino acid was added to a final concentration of 100 μM in DMEM. Regular DMEM contains 400 μM Gly and Ser but not the other five amino acids (Supplementary Table S1). To determine which amino acid(s) was/were responsible for mediating the pro-survival effects of NEAA, individual amino acids were added into DMEM at a 100 μM final concentration in cultured shATF4 cells. The results indicated that Asn, but not any other individual amino acid, could rescue the survival of shATF4 cells (Figure 4A). An even more substantial pro-survival effect of Asn was observed in a long-term clonogenic survival assay (Figure 4B).
Figure 4.
Supplementation with Asn or overexpression of ASNS rescues survival of shATF4 cells. (A) Survival of HT1080 (left) or DLD1 (right) cells grown in DMEM with indicated amino acids supplemented at a concentration of 100 μM for 48 h. (B) Clonogenic survival of HT1080 shNT/shATF4 cells. Left: Picture of a representative experiment. Right: Clonogenic survival of HT1080 shNT/shATF4 cells. (Data represent mean±s.e.m., n=4, *P<0.05.) (C) ASNS mRNA levels in HT1080 shNT/shATF4 cells measured using real-time RT–PCR. (Data represent mean±s.e.m., n=3, *P<0.05.) (D) ASNS expression and cell survival in transfected HT1080 cells. (Left panel) Top bands represent HA-tagged ASNS, lower bands represent endogenous ASNS. (Right panel) Survival of HT1080 shNT and shATF4 cells after indicated treatments. After 24 h, transfected cells were harvested for immunoblot or plated for MTT assay (48 h). (Data represent mean±s.e.m., n=3, *P<0.05).
It has been reported that the ASNS gene is directly regulated by ATF4 through binding to its promoter (Siu et al, 2002; Gjymishka et al, 2009). Indeed, we also found that ASNS expression was reduced by >70% in shATF4 cells compared with shNT cells (Figure 4C). Moreover, adding Asn to HT1080.shATF4 cells rescued NEAA deprivation-induced G1 arrest (Supplementary Figure S3B), a finding that is consistent with reports that ASNS deficiency can induce a G1 arrest (Greco et al, 1987; Gong and Basilico, 1990). Furthermore, overexpression of ASNS in shATF4 cells partially rescued cell survival (Figure 4D) and adding Asn repressed both apoptosis and autophagy in shATF4 cells (Figure 6A). This result suggests that ASNS is an important enzyme for maintaining intracellular asparagine levels, which are crucial for tumour cell survival and cell-cycle progression.
The amino acid glutamine (Gln) serves not only as a substrate for nucleotide and protein synthesis, a precursor for the synthesis of Asn, but also as an important energy source for tumour cells (Reitzer et al, 1979; DeBerardinis et al, 2007). To produce Asn, ASNS transfers the amino group from Gln to Asp (Figure 5A). As ASNS requires Gln to synthesize Asn, we wanted to test whether shATF4 cells were more sensitive to Gln deprivation than shNT cells. To test this, shNT and shATF4 cells were cultured in DMEM with/without 4 mM glutamine. MTT assays showed that shATF4 cells showed about 50% reduction in survival in the absence of Gln, whereas the shNT cells exhibited only a 25% reduction after 48 h incubation (Figure 5B). Interestingly, adding Asn (100 μM final concentration) to Gln-deprived cells could partially rescue cell survival (Figure 5C), suggesting that producing Asn may also be an important function of Gln, at least in this tumour cell line. In summary, ATF4 deficiency severely inhibits tumour cell survival in vitro, which is primarily due to Asn deprivation.
Figure 5.
Knockdown of ATF4 increases sensitivity to glutamine deprivation. (A) The biosynthetic reaction catalysed by ASNS. (B) Survival of HT1080 (left panel) or DLD1 (right panel) cells grown in DMEM with/without 4 mM L-glutamine for 48 h. (C) Survival of HT1080 cells in DMEM with/without Gln or Asn for 48 h. Gln: 4 mM, Asn: 100 μM. (Data represent mean±s.e.m., n=3, *P<0.05).
Activation of the GCN2-eIF2α pathway under amino acid deprivation promotes cell survival, upregulates p21 (cip1/waf1) and activates autophagy
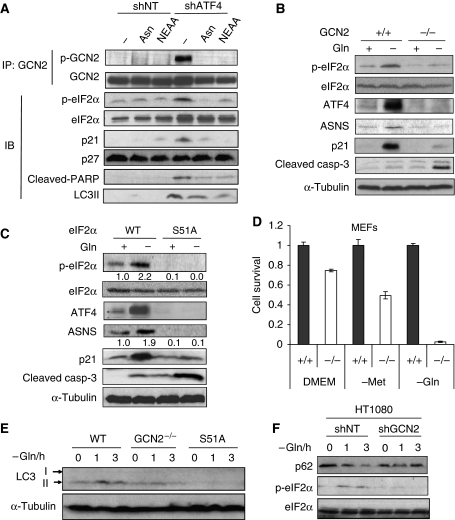
We hypothesized that if shATF4 cells are deficient in the biosynthesis of NEAAs, this should lead to the activation of the upstream kinase GCN2, completing an autoregulatory feedback loop. Indeed, we found that GCN2 was phosphorylated in HT1080.shATF4 cells and adding Asn or NEAA repressed this phosphorylation (Figure 6A), suggesting that knocking down ATF4 reduces ASNS expression, causing an Asn deficiency, which activated GCN2. eIF2α, the substrate of GCN2, was also phosphorylated in shATF4 cells in response to NEAA and similar to GCN2, its phosphorylation was repressed by addition of Asn or NEAA in trans. The CDK inhibitors p21 and p27 have a critical function in G1/S cell-cycle arrest in response to stress, and it had been reported that they can be induced by amino acid deprivation (Leung-Pineda et al, 2004). shATF4 cells constitutively expressed high levels of p21, which were substantially reduced by adding NEAA or Asn; however, p27 levels were unaffected (Figure 6A). This is consistent to an earlier report that ATF4-null primary mouse bone marrow stromal cells have increased p21 but not p27 expression (Zhang et al, 2008). The induction of p21 is likely responsible for the G1/S cell-cycle arrest in shATF4 cells.
Figure 6.
Activation of GCN2-eIF2α pathway under amino acid deprivation promotes cell survival, upregulates ATF4 and p21, and activates autophagy. (A) HT1080 shNT and shATF4 cells were incubated in the media indicated for 24 h. Whole cell lysates were harvested for immunoblot (IB) or immunoprecipitation (IP) with the indicated antibodies. (B) GCN2+/+ and GCN2−/− MEFs were incubated with/without 4 mM Gln for 24 h and immunoblotting was performed. (C) eIF2α wt or eIF2α S51A mutant MEFs were incubated with/without 4 mM Gln for 24 h and immunoblotting was performed with indicated antibodies. Numbers below the blots of p-eIF2a and ASNS indicate fold change in levels normalized to those of α-tubulin. Analysis was performed using the Scion Image version of the NIH Image shareware image analysis program. (D) GCN2+/+ and GCN2−/− MEFs were incubated with or without Met or Gln for 48 h. Cell survival was analysed using MTT assay. (Data represent mean±s.e.m., n=3, *P<0.05.) (E) Wild-type, GCN2−/− and eIF2α S51A mutant MEFs were cultured without 4 mM Gln for 1 h or 3 h, cell lysates were subjected to immunoblotting. (F) HT1080 cells stably transfected with shNT or shGCN2 plasmid were cultured without Gln for 1 or 3 h, cell lysates were subjected to immunoblotting with indicated antibodies.
As GCN2 is the molecular sensor of amino acid deprivation that induces translational upregulation of ATF4, we tested whether GCN2 activation promotes tumour cell survival when a single amino acid is removed from the culture media. SV40 immortalized, Ras-transformed GCN2+/+ and GCN2−/− MEFs were cultured in DMEM with or without Gln. Under Gln deprivation, GCN2+/+ cells showed enhanced eIF2α phosphorylation and upregulation of ATF4, ASNS and p21, whereas the GCN2−/− cells failed to activate this pathway and had increased levels of cleaved caspase3 (Figure 6B). These results showed that the induction of p21 under amino acid starvation depends on GCN2 activation.
As eIF2α is currently the sole known substrate of GCN2, we wanted to further investigate whether the induction of ATF4 and p21 was dependent on eIF2α phosphorylation. To test this, eIF2α wild-type or eIF2α S51A mutant MEFs (a Ser-Ala mutation blocks eIF2α phosphorylation) were incubated in DMEM with or without Gln. Similar to the GCN2−/− cells, eIF2α S51A mutant cells were unable to induce ATF4, ASNS or p21 in the absence of Gln, but had increased levels of apoptosis (Figure 6C).
In full DMEM (i.e. +Gln, +Met), GCN2−/− cells showed 25% reduction in cell survival compared with wild-type cells after 48 h incubation, whereas Met or Gln deprivation further reduced the cell survival of GCN2−/− cells to around 50% or 4%, respectively (Figure 6D). In summary, the activation of GCN2-eIF2α-ATF4 pathway is necessary for tumour cell survival under amino acids starvation.
It was reported earlier that GCN2 activation and eIF2α phosphorylation induce autophagy in yeast (Talloczy et al, 2002). We also observed a correlation between GCN2 activation with LC3 cleavage in HT1080.shATF4 cells (Figure 6A), suggesting that GCN2 could be the molecular switch that senses amino acid shortage and induces autophagy in mammalian cells. To test this, wild-type, GCN2−/− and eIF2α S51A mutant MEFs were incubated in Gln-free media. GCN2−/− cells had significant lower LC3 processing compared with wild-type cells in response to Gln starvation. eIF2α S51A mutant cells could not induce LC3 processing at all (Figure 6E). To confirm the function of GCN2 in autophagy induction in human tumour cells, HT1080 cells stably transfected with shNT or shGCN2 plasmid, were incubated in Gln-free media. Autophagy was analysed by blotting for an autophagy marker p62/SQSTM1, a long-lived protein that is rapidly degraded during autophagy progression (Klionsky et al, 2008). p62 was degraded in shNT cells on Gln deprivation but stabilized in shGCN2 cells (Figure 6F). In conclusion, a functional GCN2-eIF2α pathway is required for amino acid starvation-activated autophagy in transformed cells.
Activation of phospho-eIF2α-ATF4 pathway under glucose deprivation depends on GCN2
Induction of ATF4-ASNS pathway by glucose deprivation has been observed in tumour cell lines (Siu et al, 2002; Cui et al, 2007). The upstream event of ATF4 translational upregulation, eIF2α phosphorylation, can also be induced by glucose starvation (Gomez et al, 2004), and it was suggested that this may be PERK dependent (Gomez et al, 2008). In yeast, which does not have PERK, eIF2α phosphorylation is dependent on GCN2 under glucose starvation (Yang et al, 2000). Given that the carbon backbone of amino acids can enter glycolysis or citric acid cycle to produce ATP, and that GCN2 is activated by uncharged tRNAs, we hypothesized that tumour cells may use amino acids as alternative energy source under glucose deprivation. The reduced amino acid pool should then lead to GCN2 activation, eIF2α phosphorylation and ATF4 induction to increase amino acid synthesis/uptake.
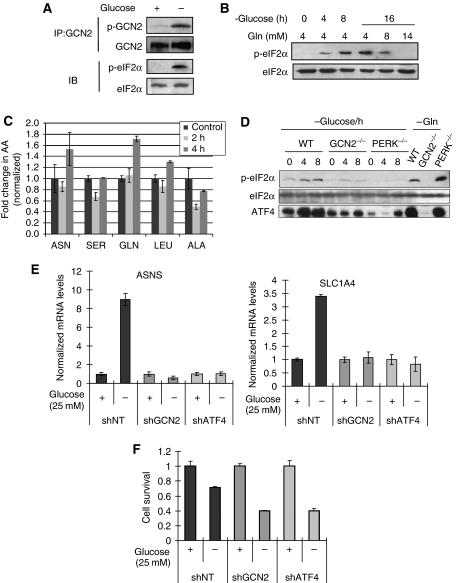
To test this, HT1080 cells were incubated in DMEM with or without 25 mM glucose for 16 h. GCN2 phosphorylation was detected after glucose deprivation, and was associated with eIF2α phosphorylation (Figure 7A). Moreover, adding excess glutamine to the media potently suppressed eIF2α phosphorylation under glucose deprivation (Figure 7B), strongly suggesting that eIF2α phosphorylation induced by glucose deprivation was due to reduced concentrations in one or more amino acids. To test this more directly, we measured intracellular amino acid levels using liquid chromatography-coupled mass spectrometry (LC-MS). Analysis of the levels of five amino acids (Asn, Ser, Gln, Leu and Ala) after normalization to total protein revealed that 2 h after glucose deprivation, the levels of Ser and Ala were significantly reduced by 37 and 50%, respectively (Figure 7C). The levels of the other amino acids were not significantly affected at this time point. After 4 h of glucose deprivation (when eIF2α phosphorylation is increased), the levels of Ser and Ala returned to near control levels, whereas the levels of Asn and Gln were increased compared with control cells in glucose-replete DMEM, reflecting activation of a recovery pathway (e.g. induction of autophagy, increased amino acid uptake from the media or both). This notion was further supported by the finding that under glucose deprivation, HT1080 cells consumed Gln at a much faster rate compared with that under glucose-replete media (Supplementary Figure S4E).
Figure 7.
Glucose deprivation activates the GCN2-eIF2α-ATF4 pathway. (A) HT1080 cells were incubated with/without 25 mM glucose for 16 h. Whole cell lysates (WL) were harvested for immunoblot (IB) or immunoprecipitation (IP). (B) HT1080 cells were incubated in glucose-free DMEM for up to 16 h with indicated concentrations of glutamine added and lysates were subjected to immunoblotting. (C) Cells were deprived of Glucose for 2 and 4 h and the levels of individual amino acids were analysed by LC-MS. (D) Wild-type, GCN2−/− and PERK−/− MEFs were incubated in glucose-free DMEM for up to 8 h, cells were harvested for immunoblot. Glutamine deprivation treatment was for 4 h. (E) HT1080 cells were incubated with/without 25 mM glucose for 8 h, total RNA was extracted for real-time PCR for the ATF4 targets ASNS (left) or SLC1A4 (right). (F) HT1080 cells were cultured with/without 25 mM glucose, 2 × 104 cells/well. shATF4 cells were supplemented with 1 × NEAA. After 24 h, cell survival was measured using MTT assay and survival was normalized to that of the high glucose group.
To test whether the eIF2α phosphorylation in response to glucose deprivation depends on GCN2 or PERK, wild-type, GCN2−/− and PERK−/− MEFs were glucose deprived. Unlike wild-type cells, both GCN2−/− and PERK−/− MEFs had substantially reduced eIF2α phosphorylation and ATF4 induction on glucose deprivation (Figure 7D), suggesting that both GCN2 and PERK contribute to eIF2α phosphorylation and ATF4 upregulation on glucose deprivation. It is possible that both kinases are activated under glucose starvation by different stress signals. It was reported that both PERK and GCN2 contribute to ER stress-induced cell-cycle arrest, suggesting that there could be crosstalk between two kinases (Hamanaka et al, 2005).
To study the function of GCN2 for tumour cells under nutrient deprivation, we generated GCN2 knockdown cells (HT1080.shGCN2). The reduced GCN2 expression was confirmed by both real-time PCR and immunoblot (Supplementary Figures S4A and B). Unlike shNT cells, ASNS expression cannot be induced in shGCN2 cells by Gln deprivation (Supplementary Figure S4C). The mRNA levels of three downstream target genes of ATF4, ASNS, SLC1A4 and SLC7A5 (two amino acid transporters) were significantly increased by glucose deprivation in shNT cells, but not in shGCN2 or shATF4 cells (Figure 7E; Supplementary Figure S4D). As several ATF4 target genes are involved in amino acid transportation and synthesis, it is likely that GCN2 senses the reduction of amino acid levels under glucose deprivation and phosphorylates eIF2α to reduce global translation, but at the same time upregulates ATF4 to supply amino acids. We hypothesized that if this was true, the activation of the GCN2-ATF4 pathway should protect cells under glucose deprivation. Indeed, knocking down GCN2 or ATF4 sensitized tumour cells to low glucose (Figure 7F), indicating that the integrity of the GCN2-ATF4 pathway is required for cell survival under glucose deprivation.
GCN2-ATF4 pathway contributes to tumour growth in vivo
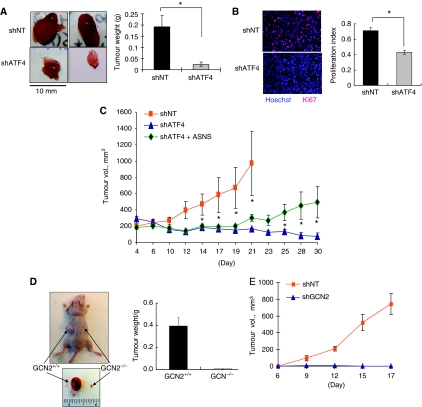
The fact that ATF4 deficiency causes significant reduction in cell survival in vitro suggests that ATF4 might have a function in tumour growth. To test this, equal numbers of HT1080 shNT or shATF4 cells were injected in the flanks of nude mice and tumour growth was monitored over a 3–4-week period. shNT cells grew rapidly and formed large tumours. However, the shATF4 cells formed fewer tumours that were significantly smaller compared with those from shNT cells (Figure 8A). Immunofluorescence analysis of cell proliferation in vivo using the Ki67 antigen as a marker, showed that, consistent with the in vitro data, cells in shATF4 tumours had a significantly lower proliferation rate (Figure 8B). Also consistent with the in vitro data, overexpression of ASNS in shATF4 cells led to partial, but significant rescue of tumour growth (Figure 8C). Similarly, the absence of GCN2 in Ras-transformed MEFs or knockdown of GCN2 in HT1080 cells, blocked tumour growth (Figures 8D and E). These findings suggest that xenograft tumour growth requires a functioning GCN2-ATF4 pathway.
Figure 8.
Inhibition of GCN2-ATF4 pathway blocks tumour growth in vivo. (A) Left panel: Xenografted tumours from HT1080 shNT/shATF4 cells in nude mice injected in each side with 2 × 106 cells. Tumours grew for 3 weeks. Right panel: Average tumour weight at conclusion of experiment (N=10, *P<0.05, one-tailed Student's t-test). (B) Left panel: Immunofluorescent staining for Ki67 (red) in tumour sections. Nuclei were counterstained with Hoechst 33342 (blue). Tumours were photographed at × 200 magnification. Right panel: The quantification of Ki67 signal. (C) Growth of HT1080 shNT, shATF4 and shATF4+ASNS tumour xenografts. Nu/Nu mice were injected in each side with 2 × 106 HT1080 cells (N=8). Four days after injection, tumour volume was measured every 2–3 days. Error bars represent s.e. values. *P<0.05, Student's two-tailed t-test. (D) Growth of K-RasV12-transformed GCN2+/+ and GCN2−/− MEFs in vivo. Nu/Nu mice were injected in each side with 2.5 × 106 MEFs. Tumours grew for 9 days. At the end of the experiment, tumours were excised, photographed (left) and weighed (right). (N=4, *P<0.05, one-tailed Student's t-test). (E) Growth of HT1080 shNT and shGCN2 tumour xenografts. Nu/Nu mice were injected in each side with 2 × 106 shNT (N=7) or shGCN2 cells (N=10). Six days after injection, tumour volume was measured every 2–3 days.
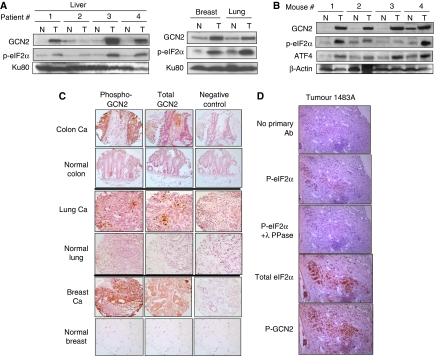
To investigate whether the GCN2-eIF2α pathway is activated in primary tumours, clinical samples of human liver, breast and lung tumours with corresponding normal tissue controls were homogenized and lysates subjected to immunoblotting. Three out of four liver tumours and the samples form breast and lung tumours exhibited substantial GCN2 overexpression and increased phospho-eIF2α levels compared with normal tissues (Figure 9A). Similar results were obtained from spontaneous mouse tumours. Analysis of the components of this pathway from tumours and corresponding normal breast tissue from mammary tumour-prone MMTV-Neu mice was performed. All mouse breast tumours exhibited significant GCN2 overexpression as well as increased levels of eIF2α phosphorylation and ATF4 upregulation compared with normal tissue (Figure 9B).
Figure 9.
The GCN2/p-eIF2α pathway is activated in human and mouse spontaneous tumours. (A) Immunoblots for GCN2 and p-eIF2α in human tumour tissue (T) and corresponding normal tissue (N) samples. Ku80 was used as a loading control. (B) Immunoblots for GCN2, p-eIF2α and ATF4 in normal (N) and tumour (T) samples from a mammary tumour-prone MMTV-Neu mouse strain. Immunoblotting for β-actin was used as a loading control. (C) Expression of p-GCN2 and total GCN2 in human normal and tumour tissues from the NIH TARP consortium. Staining using only the secondary antibody was used as a negative control. Images were taken under × 40 magnification. (D) Expression of P-GCN2, P-eIF2α, total GCN2 and total eIF2α in serial sections of human colorectal metastatic cancer to liver. Specificity of p-eIF2α staining is demonstrated by decreased signal in sample treated with λ phosphatase (λ-PPase) before antibody incubation.
Despite extensive efforts, we could not detect phospho-GCN2 in tissue homogenates by immunoblot, even after GCN2 immunoprecipitation, probably because of loss of the phospho-group during tissue homogenization or immunoprecipitation. Therefore, we performed immunohistochemical staining for total and phospho-GCN2 levels using paraffin-embedded human tumour tissue arrays. Colon and breast carcinoma tissues exhibited strong staining of total and phospho-GCN2. Less but substantial staining of total and phospho-GCN2 were detected in lung carcinoma; in contrast, normal tissues showed very little or no staining for total or phospho-GCN2 (Figure 9C). Interestingly, phospho-GCN2 staining was not homogeneous across the tumour section, suggesting that local microenvironmental factors might be affecting its phosphorylation. Further analysis of phospho-GCN2, phospho-eIF2α and corresponding total proteins in serial sections of human samples of colorectal cancer metastases to the liver, revealed substantial overlap in the signal for GCN2 and eIF2α phosphorylation (Figure 9D; Supplementary Figure S6). Interestingly, we also found substantially higher levels of total eIF2α in the tumour cells compared with the normal surrounding stroma. The signal for phospho-eIF2α was weaker and present in a subset of cells that showed strong signal for total eIF2α. In summary, the upregulation of GCN2-eIF2α-ATF4 signalling module in tumour compared with normal tissues implies that there must exist a tumour-specific requirement for GCN2 activation in tumours for adaptation to a nutrient-deprived microenvironment.
Discussion
The GCN2-eIF2α-ATF4 signalling module has been described as a vital regulator of protein synthesis and amino acid metabolism in response to amino acid deprivation in eukaryotes, from yeast to mammals. Yet, the consequences of ablating components of this pathway on survival and proliferation of transformed cells under physiological and stress conditions in vitro and in vivo have not been adequately described. Rapidly proliferating transformed cells have been shown to increase their nutrient uptake in excess of their bioenergetic needs and to divert metabolic programs towards pathways that support macromolecular biosynthesis to support their rapid growth (DeBerardinis et al, 2008). Our study supports a model in which inhibition of ATF4 or GCN2 leads to suboptimal growth and survival of tumour cells and xenografts because of an imbalance between amino acid/energy requirements and biosynthetic pathway function and identifies Asn as a key component of this regulatory mechanism.
The function of ATF4 in adaptation of transformed cells to nutritional stress
We and others have shown that ATF4 is overexpressed in several human tumour tissues and is upregulated in response to hypoxic/anoxic stress (Ameri et al, 2004; Bi et al, 2005). Moreover, dysregulation of ATF4 expression has been implicated in the induction of chemoradioresistance: ATF4 mRNA levels correlate with tumour cell resistance to DNA-interacting drugs such as cisplatin (Levenson et al, 2000) and recent work has suggested that the circadian regulator protein Clock binds to the E-box in the ATF4 promoter and transcriptionally upregulates ATF4 in response to cisplatin, which induces enzymes involved in glutathione metabolism and contributes to chemoresistance (Igarashi et al, 2007). Collectively, these findings suggest that ATF4 has an important function in cellular resistance to chemotherapeutic agents and genotoxic stress, perhaps through the upregulation of target genes that promote production of reducing compounds.
Our studies show that inhibition of ATF4 in the absence of any stress, sensitizes tumour cells to NEAA deprivation, a result that is consistent with earlier studies performed in MEFs (Harding et al, 2003). In contrast to ATF4−/− MEFs that required β-ME, tumour cells expressing ATF4 shRNA did not need this antioxidant to survive. This difference could be due to at least two possible mechanisms: (1) the low level of ATF4 remaining in tumour cells still satisfies the cellular needs for antioxidant activity; (2) tumour cells either overexpress antioxidant enzymes or are more tolerant to oxidative stresses. However, ATF4 knockdown tumour cells did show reduced survival in the absence of NEAA, indicating that these cells were not able to synthesize certain amino acids that are crucial for survival and proliferation, thus cell-cycle arrest, apoptosis and autophagy were induced.
Loss ATF4 and induction of autophagy
Autophagy, a mechanism in which cells digest their own proteins and organelles, is believed to enable cancer cells to survive under nutrient starvation and has multiple functions in tumour progression. In this study, we found evidence that loss of ATF4 leads to a lower apoptotic threshold; yet paradoxically also appears to stimulate autophagy. However, this result can be explained under the prism of energy homeostatic mechanisms: initially, loss of ATF4 (and thereby a substantial portion of amino acid biosynthetic and transport capacity) leads to nutrient deprivation and initiation of the autophagic response. This notion is supported by morphological and molecular evidence showing formation of double-membrane-engulfed cytoplasmic vacuoles and elevated levels of the autophagic marker cleaved LC3 in HT1080.shATF4 cells in the absence of NEAA. These effects are readily reversed when knockdown cells are supplemented with NEAA, suggesting that amino acid deprivation is the key activator of autophagy. Despite the activation of this pro-survival mechanism, however, the inability of HT1080.shATF4 cells to synthesize amino acids (and more specifically Asn) ultimately leads to the activation of apoptotic pathways leading to cell death and loss of clonogenic survival.
The pro-survival function of the initial autophagic response is further supported by the fact that inhibition of Atg7, an enzyme responsible for fusion of peroxisomal and vacuolar membranes, prevents the induction of autophagy and results in increased apoptosis in shATF4 cells. It has been reported that after Bortezomib treatment, ATF4 promotes autophagy by upregulating LC3B levels and this confers protection against Bortezomib-induced apoptosis (Milani et al, 2009). In this work, we have shown that ATF4 deficiency leads to higher levels of processing of LC3. Therefore, although ATF4 upregulation because of proteasomal inhibition can positively affect autophagy, lack of ATF4 can also indirectly promote autophagy through a distinct mechanism involving Asn depletion and GCN2 activation.
Asparagine is a key effector of ATF4-dependent amino acid homeostasis
An analysis of the effect of individual amino acids on shATF4 tumour cells showed that supplementation of Asn, and no other individual amino acid, was sufficient to rescue cell survival, a finding that was later validated by our studies of ASNS. Among the known downstream targets of ATF4 are proteins involved in amino acid transport and metabolism, including ASNS. ASNS catalyses the ATP and glutamine-dependent conversion of L-aspartate to L-asparagine, a NEAA that is necessary for protein synthesis and cell growth (Richards and Shuster, 1998). HT1080 cells, like several other human tumour cell lines, grow in DMEM, which is rich in Gln (4 mM). Gln deprivation, significantly reduces survival of HT1080 and DLD1 shATF4 cells, which could be partially rescued if Asn was added, suggesting that in tumour cells, the high requirement of Gln (a vital source of energy and nitrogen) (Sauer et al, 1982; DeBerardinis et al, 2007), may be (at least partially) due to the biosynthesis of Asn. According to our unpublished microarray data, the expression of glutaminase, an enzyme that catalyses Gln deamidation (the first reaction in glutamine catabolism), is upregulated by ATF4, suggest that ATF4 may also promote tumour cells to take use of Gln.
Interestingly, L-asparaginase, an enzyme that catalyses the biodegradation of Asn, is a common chemotherapeutic drug for childhood acute lymphoblastic leukaemia (ALL) and forms of acute myeloblastic leukaemia (Cooney and Handschumacher, 1970; Ertel et al, 1979; Richards and Kilberg, 2006). To compensate for the lack of exogenous Asn available to L-asparaginase treated cells, leukaemic and stromal cells upregulate ASNS synthesis and activity, which may contribute to the development of L-asparaginase resistance in the tumour cells (Hutson et al, 1997; Aslanian et al, 2001). Moreover, a causal relationship between L-asparaginase activity and ASNS expression has been observed in ovarian cancer cell lines (Lorenzi et al, 2008).
However, further investigation is still needed into the precise function of Asn in tumour cell survival and tumour growth: it is unclear whether Asn is just a substrate for protein synthesis, or whether it has additional, yet unidentified functions in tumour cell metabolism and proliferation. The ability of ASNS expression to rescue (at least partially) survival and growth of cells with reduced ATF4 levels, coupled with the clinical efficacy of L-asparaginase in ALL, highlight the importance of this pathway in the maintenance of amino acid homeostasis. Our study also supports the need for further screening of cancer cells lines and strains, which might be susceptible to L-asparaginase treatment and to balance the administration of L-asparaginase with the potential development of drug resistance.
GCN2 forms an autoregulatory loop to compensate for ATF4 deficiency
GCN2, a protein kinase activated by uncharged tRNAs that occur under amino acid deprivation, triggers the repression of protein synthesis as well as the upregulation of amino acid biosynthesis/transportation through increased translation of ATF4 mRNA. Both GCN2 and its substrate, eIF2α, showed increased phosphorylation in shATF4 cells, which was reduced when cells were treated with Asn or NEAA. Under glutamine deprivation, GCN2+/+ cells exhibited increased eIF2α phosphorylation, and ATF4, ASNS and p21 induction; in contrast, GCN2−/− cells were not able to activate this pathway and underwent apoptosis. Moreover, the induction of ATF4 and p21 under amino acid deprivation depends on eIF2α phosphorylation, suggesting that p21 may be translationally induced by phosphorylated eIF2α. Under genotoxic stress, the induction of p21 contributes to cell survival by blocking cell-cycle progression and allowing sufficient time for the repair of damaged DNA (McDonald et al, 1996). Although it remains to be formally shown, the induction of p21 under amino acid starvation may similarly have a cytoprotective function, by inhibiting proliferation and thus promoting conservation of energy otherwise expended for protein synthesis.
Our finding that eIF2α phosphorylation and ATF4 induction under low glucose is GCN2 dependent, also shows that GCN2 is a molecular switch that senses nutrient deprivation in the tumour microenvironment to downregulate protein synthesis (eIF2α phosphorylation), slow down cell-cycle progression (p21 induction) and increase amino acid uptake (ATF4 induction). These responses could help tumour cells conserve energy and maintain homeostasis of amino acid metabolism.
Another pro-survival function of GCN2-eIF2α pathway under amino acid deprivation may be the induction of autophagy. Consistent to earlier study performed in yeast (Talloczy et al, 2002), phosphorylation of eIF2α is required for autophagy induction in MEFs, though further investigation is needed to determine the mechanism. The LC3II levels in GCN2−/− cells are lower compared with wild-type cells, but higher than those in eIF2α mutant cells. This may be due to the crosstalk from other eIF2α kinases (PERK, PKR, etc.).
A function for GCN2 and ATF4 in tumour progression
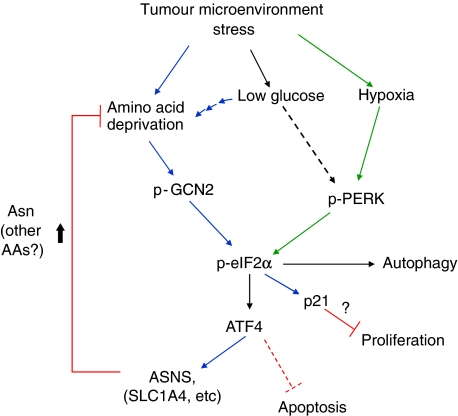
Several of these in vitro findings were recapitulated in vivo by comparing tumour growth in ATF4 or GCN2 knockdown cells with their WT counterparts. shATF4 cells formed fewer tumours, which were significantly smaller and grew slower compared with the rapid formation of large shNT tumours. The shATF4 tumours exhibited reduced proliferation rates based on the analysis of Ki67 staining. Similarly, Ras-transformed GCN2 knockout MEFs were unable to form large tumours in vivo. We showed earlier that deficiency in PERK or eIF2α phosphorylation results in decreased resistance to hypoxic stress both in vitro and in vivo (Koumenis et al, 2002; Bi et al, 2005). Although it is possible that GCN2 also contributes to resistance to hypoxia in vivo, GCN2−/− MEFs were as sensitive to hypoxia as WT MEFs in vitro; moreover, GCN2−/− MEFs still phosphorylate eIF2α and induce ATF4 under hypoxia (Supplementary Figure S5). On the other hand, PERK−/− MEFs still phosphorylate eIF2α and induce ATF4 under Gln deprivation (Figure 7D). Therefore, we propose that in the tumour microenvironment, GCN2 and PERK cooperate to provide resistance to distinct forms of stress (Figure 10). Our findings further suggest that yet another stress commonly found in tumours, low glucose, requires both PERK and GCN2 to activate eIF2α phosphorylation; therefore, hypoxia, glucose and amino acid deprivation each appear to activate this pathway though distinct and sometimes overlapping pathways (Figure 10). The precise function and contribution of each component of the pathway to resistance to stress and tumour growth will require further analysis of activation of each pathway with specific markers and correlation to each stress in vivo. As a first step towards this analysis, we have shown that overexpression of ASNS in the tumour resulted in partial rescue of tumour growth (Figure 8C). These results show that expression of ASNS is, at least partially, responsible for the growth defect in the ATF4 knockdown cells.
Figure 10.
A model for the function of GCN2, ATF4, ASNS and Asn in conferring tumour cell protection from microenvironment stress. Blue arrows indicate signalling primarily in response nutrient deprivation stress and green arrows indicate signalling primarily from hypoxic stress. Common pathways are indicated by black arrows. ATF4 likely has additional functions in response to hypoxia that are not depicted here.
The lack of complete rescue of the phenotype is likely due to suboptimal presence of Gln, the precursor for Asn synthesis and ASNS substrate throughout the tumour growth or contribution of ATF4 to other processes, such as angiogenesis. Indeed, an angiogenesis defect was also attributed to suboptimal growth of PERK−/− tumours previously shown by our group and more recently suggested by another study in mouse insulinomas (Gupta et al, 2009).
The slower tumour growth in ATF4 and GCN2-deficient cells reflects the tumour cells' dependency on the integrity of this GCN2-eIF2α-ATF4 pathway under nutrient deprivation; this reliance for tumour cell growth makes the GCN2-eIF2α-ATF4 pathway a biological target for efficient cancer therapies. GCN2−/− mice do not exhibit gross morphological or functional abnormalities unless they are fed a diet lacking certain essential amino acids, such as Leu (Anthony et al, 2004). As ATF4−/− mice exhibit abnormalities such as micropthalmia and anemia (mostly attributed to the antioxidant function of ATF4), GCN2 might offer a better therapeutic target than ATF4. We would predict that normal tissues with sufficient nutrient supply would not be as affected by a specific GCN2 inhibitor compared with tumour tissues, which would be under nutrient deprivation (because of suboptimal blood flow and increased metabolic demands) and thereby be more dependent on functional GCN2 for survival and proliferation.
Materials and methods
Cell culture and generation of stable cell clones
HT1080 and DLD1 cells were cultured in DMEM (4.5 g/l glucose, 4 mM Gln) supplemented with penicillin, streptomycin, 10% fetal calf serum. To establish stable ATF4 knockdown cell lines, HT1080 and DLD1 cells were transfected with pLKO-shNT or pSM2-shATF4 plasmids (OpenBiosystems) using Lipofectamine2000 (Invitrogen) and selected with puromycin (2 μg/ml and reduced to 0.5 μg/ml for maintenance). All cells were supplemented with NEAA and 55 μM β-ME after transfection. Stable GCN2 knockdown cell line was produced in the same manner. All the MEFs were cultured in the same conditions as HT1080.shATF4 cells. Individual amino acids (Sigma) were dissolved in water to make 10 mM (100 × ) stock solutions.
Real-time PCR
RNA was isolated from cells after TRI-Reagent protocol (Invitrogen). Reverse transcription was performed using AMV Reverse Transcriptase (Promega). Real-time PCR was performed on Applied Biosystems 7300 Real-Time PCR System using Power SYBR® Green PCR Master Mix.
Cell survival, proliferation and apoptosis assays
MTT assay was performed using Cell Proliferation Kit I (Roche Diagnostics). Cell proliferation was assayed with Click-iT EdU Flow Cytometry Kit (Invitrogen). Caspase3/7 activities were measured using Caspase-Glo 3/7 Kit (Promega). For clonogenic survival assays, cells were plated at a density of 500 cells per plate, incubated for 12 days and fixed with 10% methanol/10% acetic acid and stained with 0.4% crystal violet. As HT1080 cells do not form well-defined colonies, 300 μl 33% acetic acid was added to each dish to solubilize the stain, which was transferred to a 96-well plate and absorbance was read at 540 nm. The average normalized surviving fraction from four independent experiments and the s.e. are reported.
Plasmids and viral vector
pCMV-mATF4 vector and Adeno-mATF4 virus was a gift from Dr Guozhi Xiao (Department of Medicine, University of Pittsburgh). pCMV-HA-ASNS vector was purchased from OriGene. ATF4 and GCN2 shRNA vectors are from Open Biosystems.
Glutamine (Gln) assay
Gln concentration in the media was measured using a glutamine assay kit (Sigma). Gln consumption was calculated from the difference between Gln concentration before and after 16 h incubation. The result was normalized to the number of surviving cells.
Intracellular amino acid concentration
Cells were glucose starved (DMEM, no glucose, 10% FBS, L-glutamine) for 2 or 4 h. Control cells received same media with 4.5 g/l glucose. Cells were washed with ice-cold PBS, 4% perchloric acid (with 20 μM internal standard) was added to each dish, and the cells were collected and resuspended in the perchloric acid solution. The solution was frozen at −80 °C for 5 min, thawed, and the precipitated protein was collected in a microcentrifuge. The protein pellet was dissolved in 1 M NaOH and protein assay was performed (Lowry, Bio-Rad). The supernatant was neutralized to pH 7–8 with KOH. Samples were incubated on ice for 30 min, potassium perchlorate was precipitated and the resulting supernatant was submitted for amino acid analysis performed by LC separation of the o-pthalaldehyde derivatives and fluorescent detection at the Mass Spec Core Facility, The Children's Hospital of Philadelphia. Concentrations were determined as μg/ml and normalized to total protein.
Animals
Athymic NCR-Nu/Nu male mice of ages 6–8 weeks (NCI at Frederick) were used. Animals were housed and cared for at the University of Pennsylvania, Stemmler Animal facility. All animal experiments were performed in accordance with NIH guidelines and with the approval of the University of Pennsylvania Animal Use Committees (IACUC). Nu/Nu mice were subcutaneously injected with HT1080 cells (2 × 106 cells/tumour) or MEFs (2.5 × 106 cells/tumour). When tumours became cumbersome or necrotic (∼3 weeks for HT1080 or 9 days for MEFs), mice were killed; tumours were excised, photographed, weighed, frozen and embedded in OCT freezing medium.
Tumour samples
Snap frozen, human tumour and normal tissues (liver, breast and lung) were obtained from the Tumour Tissue and Biospecimen Bank facility at the University of Pennsylvania, School of Medicine. Collection and processing of human specimens was performed in accordance to the regulations of the Abramson Cancer Center and the Department of Pathology and Laboratory Medicine. For immunohistochemistry see Supplementary data.
Cell-cycle analysis
HT1080 cells were cultured in regular DMEM, DMEM with 100 μM Asn or DMEM with 100 μM NEAA for 24 h. Cell-cycle analysis was performed as described earlier (Javvadi et al, 2008).
Statistics
All statistics were performed using unpaired two-tailed Student's t-test unless otherwise specified. A P-value of 0.05 was chosen as the threshold for statistical significance.
Supplementary Material
Acknowledgments
We thank Dr Guozhi Xiao (University of Pittsburgh) for the pCMV-mATF4 vector and Adeno-mATF4 virus; Dr Marc Yudhoff (Children's Hospital of Philadelphia, PA) for consultations for analysing intracellular amino acid levels; Dr Craig B Thompson, Dr M Celeste Simon and Dr Serge Y Fuchs (University of Pennsylvania) for critically reviewing the manuscript and helpful comments. This work was supported by NIH grant CA94214 to CK and by NIH grants DK47119 and ES08681 to DR.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ameri K, Lewis CE, Raida M, Sowter H, Hai T, Harris AL (2004) Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood 103: 1876–1882 [DOI] [PubMed] [Google Scholar]
- Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, McNurlan MA, Wek RC (2004) Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem 279: 36553–36561 [DOI] [PubMed] [Google Scholar]
- Aslanian AM, Fletcher BS, Kilberg MS (2001) Asparagine synthetase expression alone is sufficient to induce l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem J 357: 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24: 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney DA, Handschumacher RE (1970) L-asparaginase and L-asparagine metabolism. Annu Rev Pharmacol 10: 421–440 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, Bruno M, Bidinosti M, Ben Mamou C, Marcinkiewicz E, Yoshida M, Imataka H, Cuello AC, Seidah N, Sossin W, Lacaille JC, Ron D, Nader K, Sonenberg N (2005) Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 436: 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Darmanin S, Natsuisaka M, Kondo T, Asaka M, Shindoh M, Higashino F, Hamuro J, Okada F, Kobayashi M, Nakagawa K, Koide H, Kobayashi M (2007) Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. Cancer Res 67: 3345–3355 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA 104: 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N (2002) Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol 12: 1279–1286 [DOI] [PubMed] [Google Scholar]
- Ertel IJ, Nesbit ME, Hammond D, Weiner J, Sather H (1979) Effective dose of L-asparaginase for induction of remission in previously treated children with acute lymphocytic leukemia: a report from Childrens Cancer Study Group. Cancer Res 39: 3893–3896 [PubMed] [Google Scholar]
- Gjymishka A, Su N, Kilberg MS (2009) Transcriptional induction of the human asparagine synthetase gene during the unfolded protein response does not require the ATF6 and IRE1/XBP1 arms of the pathway. Biochem J 417: 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez E, Powell ML, Bevington A, Herbert TP (2008) A decrease in cellular energy status stimulates PERK-dependent eIF2alpha phosphorylation and regulates protein synthesis in pancreatic beta-cells. Biochem J 410: 485–493 [DOI] [PubMed] [Google Scholar]
- Gomez E, Powell ML, Greenman IC, Herbert TP (2004) Glucose-stimulated protein synthesis in pancreatic beta-cells parallels an increase in the availability of the translational ternary complex (eIF2-GTP.Met-tRNAi) and the dephosphorylation of eIF2 alpha. J Biol Chem 279: 53937–53946 [DOI] [PubMed] [Google Scholar]
- Gong SS, Basilico C (1990) A mammalian temperature-sensitive mutation affecting G1 progression results from a single amino acid substitution in asparagine synthetase. Nucleic Acids Res 18: 3509–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A, Ittmann M, Basilico C (1987) Molecular cloning of a gene that is necessary for G1 progression in mammalian cells. Proc Natl Acad Sci USA 84: 1565–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cavener DR (2007) The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab 5: 103–114 [DOI] [PubMed] [Google Scholar]
- Gupta S, McGrath B, Cavener DR (2009) PERK regulates the proliferation and development of insulin-secreting beta-cell tumors in the endocrine pancreas of mice. PLoS One 4: e8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA (2005) PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell 16: 5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, Koehnle TJ, Gietzen DW (2005) Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science 307: 1776–1778 [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (1984) Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA 81: 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson RG, Kitoh T, Moraga Amador DA, Cosic S, Schuster SM, Kilberg MS (1997) Amino acid control of asparagine synthetase: relation to asparaginase resistance in human leukemia cells. Am J Physiol 272: C1691–C1699 [DOI] [PubMed] [Google Scholar]
- Igarashi T, Izumi H, Uchiumi T, Nishio K, Arao T, Tanabe M, Uramoto H, Sugio K, Yasumoto K, Sasaguri Y, Wang KY, Otsuji Y, Kohno K (2007) Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene 26: 4749–4760 [DOI] [PubMed] [Google Scholar]
- Javvadi P, Segan AT, Tuttle SW, Koumenis C (2008) The chemopreventive agent curcumin is a potent radiosensitizer of human cervical tumor cells via increased reactive oxygen species production and overactivation of the mitogen-activated protein kinase pathway. Mol Pharmacol 73: 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Wek RC (2005) GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J 385: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT et al. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol 22: 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V, Pan Y, Chen H, Kilberg MS (2004) Induction of p21 and p27 expression by amino acid deprivation of HepG2 human hepatoma cells involves mRNA stabilization. Biochem J 379: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson VV, Davidovich IA, Roninson IB (2000) Pleiotropic resistance to DNA-interactive drugs is associated with increased expression of genes involved in DNA replication, repair, and stress response. Cancer Res 60: 5027–5030 [PubMed] [Google Scholar]
- Lorenzi PL, Llamas J, Gunsior M, Ozbun L, Reinhold WC, Varma S, Ji H, Kim H, Hutchinson AA, Kohn EC, Goldsmith PK, Birrer MJ, Weinstein JN (2008) Asparagine synthetase is a predictive biomarker of L-asparaginase activity in ovarian cancer cell lines. Mol Cancer Ther 7: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuoka HC, Townes TM (2002) Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99: 736–745 [DOI] [PubMed] [Google Scholar]
- Maurin AC, Jousse C, Averous J, Parry L, Bruhat A, Cherasse Y, Zeng H, Zhang Y, Harding HP, Ron D, Fafournoux P (2005) The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab 1: 273–277 [DOI] [PubMed] [Google Scholar]
- McDonald ER III, Wu GS, Waldman T, El-Deiry WS (1996) Repair defect in p21 WAF1/CIP1−/− human cancer cells. Cancer Res 56: 2250–2255 [PubMed] [Google Scholar]
- Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, Harris AL (2009) The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res 69: 4415–4423 [DOI] [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG (1986) Multiple upstream AUG codons mediate translational control of GCN4. Cell 45: 201–207 [DOI] [PubMed] [Google Scholar]
- Ramirez M, Wek RC, Vazquez de Aldana CR, Jackson BM, Freeman B, Hinnebusch AG (1992) Mutations activating the yeast eIF-2 alpha kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol Cell Biol 12: 5801–5815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer LJ, Wice BM, Kennell D (1979) Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem 254: 2669–2676 [PubMed] [Google Scholar]
- Richards NG, Kilberg MS (2006) Asparagine synthetase chemotherapy. Annu Rev Biochem 75: 629–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards NG, Schuster SM (1998) Mechanistic issues in asparagine synthetase catalysis. Adv Enzymol Relat Areas Mol Biol 72: 145–198 [DOI] [PubMed] [Google Scholar]
- Sauer LA, Stayman JW III, Dauchy RT (1982) Amino acid, glucose, and lactic acid utilization in vivo by rat tumors. Cancer Res 42: 4090–4097 [PubMed] [Google Scholar]
- Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS (2002) ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem 277: 24120–24127 [DOI] [PubMed] [Google Scholar]
- Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B (2002) Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA 99: 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tsujimura T, Takeda K, Sugihara A, Maekawa A, Terada N, Yoshida N, Akira S (1998) Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells 3: 801–810 [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA 101: 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Ramirez M, Jackson BM, Hinnebusch AG (1990) Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol 10: 2820–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 15: 4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wek SA, Wek RC (2000) Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol 20: 2706–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G (2004) ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 117: 387–398 [DOI] [PubMed] [Google Scholar]
- Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, Jefferson LS, Cavener DR (2002) The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol 22: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yu S, Galson DL, Luo M, Fan J, Zhang J, Guan Y, Xiao G (2008) Activating transcription factor 4 is critical for proliferation and survival in primary bone marrow stromal cells and calvarial osteoblasts. J Cell Biochem 105: 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.