Abstract
The karyopherin CRM1 mediates nuclear export of proteins and ribonucleoproteins bearing a leucine-rich nuclear export signal (NES). To elucidate the precise mechanism by which NES-cargos are dissociated from CRM1 in the cytoplasm, which is important for transport directionality, we determined a 2.0-Å resolution crystal structure of yeast CRM1:RanBP1:RanGTP complex, an intermediate in the disassembly of the CRM1 nuclear export complex. The structure shows that on association of Ran-binding domain (RanBD) of RanBP1 with CRM1:NES-cargo:RanGTP complex, RanBD and the C-terminal acidic tail of Ran induce a large movement of the intra-HEAT9 loop of CRM1. The loop moves to the CRM1 inner surface immediately behind the NES-binding site and causes conformational rearrangements in HEAT repeats 11 and 12 so that the hydrophobic NES-binding cleft on the CRM1 outer surface closes, squeezing out the NES-cargo. This allosteric mechanism accelerates dissociation of NES by over two orders of magnitude. Structure-based mutagenesis indicated that the HEAT9 loop also functions as an allosteric autoinhibitor to stabilize CRM1 in a conformation that is unable to bind NES-cargo in the absence of RanGTP.
Keywords: CRM1, exportin, NES, nuclear export, RanBP1
Introduction
Macromolecular transport into and out of the nucleus through nuclear pore complexes (NPCs) is a crucial cellular function that regulates many physiological processes including gene expression, cell proliferation and differentiation (Görlich and Kutay, 1999). In most cases, nuclear transport is a signal-mediated active process that relies on concerted interactions between nucleoporins, soluble transport factors and their cargo macromolecules. Transport factors recognize cargo in one compartment, translocate it through NPCs, and then release it in the other compartment before being recycled to participate in further rounds of transport. Assembly/disassembly of cargo:carrier complexes in the appropriate compartment not only imparts directionality to the transport, but also allows accumulation of cargo against a concentration gradient. CRM1 (also known as exportin-1 or Xpo1) is a major nuclear export receptor (exportin) that facilitates export of a broad range of cargo macromolecules containing the leucine-rich nuclear export signal (NES) (Fornerod et al, 1997; Fukuda et al, 1997; Ossareh-Nazari et al, 1997; Stade et al, 1997). The Leu-rich NESs were first identified in HIV Rev (Fischer et al, 1995) and cAMP-dependent protein kinase inhibitor (PKI) (Wen et al, 1995) and have a consensus sequence motif Φ-x2−3-Φ-x2−3-Φ-x-Φ (Φ=L, I, V, F or M; x is any amino acid) (la Cour et al, 2004; Kutay and Güttinger, 2005). The small GTPase Ran regulates loading and unloading of CRM1 with NES-cargos. In the nucleus, CRM1 binds cooperatively to RanGTP and NES-cargo, forming a trimeric complex that translocates to the cytoplasm. In the cytoplasm, the CRM1 nuclear export complex is disassembled by a mechanism involving destabilization of the complex by the Ran-binding proteins RanBP1 or RanBP2 (also known as Nup358; a major component of the cytoplasmic fibril of the NPC) and hydrolysis of Ran-bound GTP promoted by RanGAP.
Recent crystal structures of CRM1 in complex with Snurportin1 (Spn1) (with or without RanGTP) showed that Spn1 binds to CRM1 in a multipartite manner (Monecke et al, 2009; Dong et al, 2009a). CRM1 is a ring-shaped molecule constructed from 21 tandem HEAT repeats. Each HEAT repeat consists of two antiparallel α-helices, designated A and B. The A helices form outer convex surface, whereas the B helices form the inner concave surface. The binary CRM1:Spn1 complex revealed that, in addition to Spn1's import-cargo-binding domain that binds to CRM1 HEAT repeats 12–14, Spn1 has a Leu-rich NES at its N-terminus that adopts a combined α-helical-extended conformation and binds to a hydrophobic cleft on the convex surface of CRM1, between helices 11A and 12A. Although the variable features of Leu-rich NESs (la Cour et al, 2004; Kutay and Güttinger, 2005) suggest that not all Leu-rich NESs bind to the hydrophobic cleft of CRM1 in exactly the same manner as the Spn1's N-terminus, the idea that this cleft is likely to be the general binding site for Leu-rich NESs is supported by mutational studies (Dong et al, 2009a) and also by the observation that the binding site of Leptomycin B, a potent inhibitor of CRM1-mediated nuclear export (Fornerod et al, 1997), is located in this hydrophobic cleft (Dong et al, 2009a). The width of the NES-binding cleft and the constellation of several nonpolar pockets within this cleft, not all of which are used by Spn1, indicate that this cleft can function as a specific yet versatile binding site that accommodates hydrophobic side chains of Leu-rich NESs in adaptable ways. Another recent crystal structure of CRM1 in complex with RanGTP and Spn1 has identified essentially the same interactions between CRM1 and Spn1, and also elucidated structural basis for CRM1:RanGTP interactions in the presence of Spn1 (Monecke et al, 2009).
Structural studies of CRM1 so far revealed only the NES-cargo-bound conformation of CRM1 both in the presence and absence of RanGTP, and so the key question of how cargo is released in the cytoplasm has not been addressed unequivocally. The binding of importin-β and Spn1's import cargo (trimethylguanosine-capped oligonucleotide) to Spn1 competes with the binding of Spn1 to CRM1 (Monecke et al, 2009; Dong et al, 2009b), but this competition mechanism would apply only for Spn1, and cannot be the general mechanism to release NES-cargos in the cytoplasm. Generally, the combined action of RanBP1 (or RanBP2) and RanGAP would be required to release NES-cargos efficiently from CRM1 and RanGTP, although their mechanism of action has remained elusive. It has been shown by RanGAP assays that RanGTP bound to CRM1 and NES-cargo is resistant to RanGAP-stimulated GTP hydrolysis (Askjaer et al, 1999; Maurer et al, 2001) (just like importin-β protects RanGTP from RanGAP; Floer and Blobel, 1996; Görlich et al, 1996), and that RanBP1 releases this inhibition (Askjaer et al, 1999; Maurer et al, 2001). Previous pulldown assays have shown that yeast RanBP1 (Yrb1p) competes with NES-cargo and forms a disassembly intermediate (CRM1:RanBP1:RanGTP complex) (Maurer et al, 2001). The formation of CRM1:RanBP1:RanGTP complex is also important in terms of nuclear export of RanBP1, which is small enough to freely diffuse through NPCs, and so requires an active nuclear export mechanism for its predominant localization to cytoplasm (Künzler et al, 2000; Maurer et al, 2001). In principle, RanBP1 could compete with NES-cargo by either of the following two mechanisms. One is a passive competition mechanism whereby RanBP1 binds to CRM1 and RanGTP in a way that prevents NES binding, only after NES-cargo has spontaneously dissociated from CRM1. The other is an active-displacement mechanism whereby RanBP1 forms a transient quaternary complex with CRM1:NES-cargo:RanGTP and, by inducing conformational changes in CRM1, promotes dissociation of NES-cargo. These two mechanisms can be distinguished by kinetic analysis of NES dissociation. In the passive competition mechanism, RanBP1 does not increase the rate of NES dissociation from CRM1 and RanGTP, whereas in the active-displacement mechanism, RanBP1 increases the off-rate of NES. Thus, to gain insights into the fundamental mechanism underlying export cargo release, we developed a kinetics assay and here we show direct evidence that RanBP1 is able to actively displace NES from CRM1 and RanGTP. Moreover, we determined crystal structure of CRM1:RanBP1:RanGTP complex at 2.0 Å resolution. The structure highlighted an inactive conformation of CRM1 (i.e. a conformation that is unable to bind NES-cargo) and revealed a remarkably efficient mechanism by which the long intra-HEAT9 loop of CRM1 functions as an allosteric autoinhibitor to control NES-cargo loading and unloading.
Results and discussion
RanBDs of RanBP1 or RanBP2 accelerate NES release from CRM1 and RanGTP
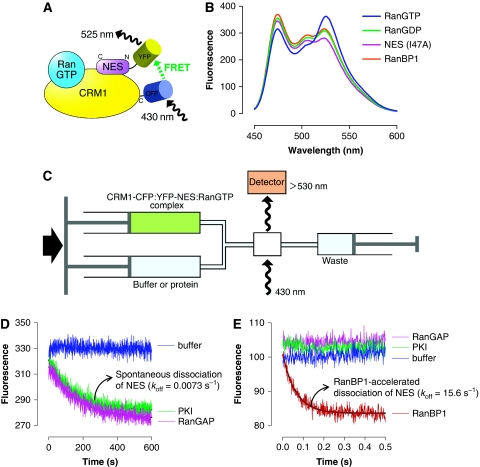
To study the kinetics of NES dissociation from CRM1 and RanGTP, we developed an assay based on fluorescence resonance energy transfer (FRET) (Figure 1A). We used the Leu-rich NES of PKI (human PKIα residues 38–47; LALKLAGLDI), which is a representative Leu-rich NES (Wen et al, 1995), for this assay. Although the structure of CRM1:PKI:RanGTP complex has not been determined, it is most likely that the PKI NES binds to the NES-binding cleft on the CRM1 outer surface of HEAT repeats 11 and 12, because mutations of yeast CRM1 residues I532/L536/F572/F583 in the hydrophobic NES-binding cleft to alanines dramatically weakened the binding of PKI to CRM1 in the presence of RanGTP (Supplementary Figure S1). A CFP-YFP FRET signal was observed when CRM1-CFP was added to YFP-NES in the presence of RanGTP (Figure 1B). This signal decreased when RanGDP was used instead of RanGTP, and no FRET signal was observed when a critical hydrophobic residue (Ile47) in the NES was mutated to alanine (Figure 1B). Thus, this FRET system specifically detects the binding of an NES to CRM1 in the presence of RanGTP. The rate of spontaneous NES dissociation from yeast CRM1 and RanGTP (koff of 0.0073/s; half-life of 94.5 s) was estimated from the decrease in the CRM1-CFP:YFP-NES:RanGTP complex's FRET signal in a 40-fold molar excess of the NES-cargo (PKI) to prevent rebinding (Figure 1D) and was not increased at higher PKI concentrations. Although addition of yeast RanGAP (Rna1p) to the CRM1-CFP:YFP-NES:RanGTP complex did not increase the NES off-rate (Figure 1D), yeast RanBP1 (Yrb1p) dramatically increased the NES off-rate by three orders of magnitude (koff∼15.6/s; half-life of 44.2 ms; Figure 1E). These kinetic data support the idea that RanGAP does not activate RanGTPase while RanGTP is in a ternary complex with CRM1 and NES-cargo (Askjaer et al, 1999; Maurer et al, 2001), and also provide the first direct evidence that RanBP1 displaces NES-cargo from CRM1 and RanGTP by an active mechanism, rather than by passive competition. Similarly, human RanBP1 increased the NES off-rate from human CRM1 and RanGTP by two orders of magnitude (Table 1). Although the C-terminal extension of human RanBP1 has been proposed to function as a Leu-rich NES (Richards et al, 1996), the Ran-binding domain (RanBD) of human RanBP1 was sufficient for this active displacement (Table 1). Consistent with this observation, any of the four RanBDs of human RanBP2 accelerated release of NESs from human CRM1:RanGTP by two orders of magnitude (Table 1). Taken together, these kinetic measurements indicate that the active displacement of NESs by RanBD of RanBP1/RanBP2 is an evolutionarily conserved mechanism, which is so effective that the majority of the nuclear export cargo would be rapidly dissociated from CRM1 by this mechanism on translocation across the nuclear pore (Supplementary Figure S2).
Figure 1.
NES dissociation kinetics measured by a FRET-based assay. (A) Design of FRET constructs to detect the specific binding of Leu-rich NES to CRM1 and RanGTP. CFP was fused to the C-terminus of CRM1, whereas YFP was fused to the N-terminus of an NES (PKI residues 38–47; LALKLAGLDI; a representative Leu-rich NES). (B) Steady-state fluorescence spectra. Blue: 0.2 μM yeast CRM1-CFP, 1.0 μM YFP-NES, 3.0 μM yeast RanGTP. Green: 0.2 μM yeast CRM1-CFP, 1.0 μM YFP-NES, 3.0 μM yeast RanGDP. Magenta: 0.2 μM yeast CRM1-CFP, 1.0 μM YFP-NES (I47A), 3.0 μM yeast RanGTP. Orange: 0.2 μM yeast CRM1-CFP, 1.0 μM YFP-NES, 3.0 μM yeast RanGTP, 4.0 μM yeast RanBP1. (C) Setup of stopped-flow experiments to measure the time-dependent decrease in the FRET signal after rapid mixing. (D, E) 0.4 μM yeast CRM1-CFP, 2.0 μM YFP-NES (PKI residues 38–47; LALKLAGLDI) and 6.0 μM yeast RanGTP were preincubated, rapidly mixed with equal volume of either buffer alone (blue) or 80 μM PKI (green) or 8 μM yeast RanGAP (magenta) or 8 μM yeast RanBP1 (red), and then the time-dependent change in the CFP-YFP FRET signal was recorded. The green trace represents the spontaneous dissociation of NES from CRM1 and RanGTP (koff ∼0.0073/s). The red trace represents the RanBP1-accelerated release of NES from CRM1 and RanGTP (fitted exponential curve is superimposed on the stopped-flow trace; koff ∼15.6/s). Yeast RanBP1 dramatically increased the NES off-rate by ∼2000-fold. These data provided direct evidence that RanBP1 uses an active-displacement mechanism to dissociate NES-cargo from CRM1 and RanGTP.
Table 1. The off-rates of an NES (PKI residues 38–47; LALKLAGLDI) from CRM1 and RanGTP.
| Constructs | koff (s−1) | ||
|---|---|---|---|
| Spontaneous dissociation of NES | |||
| CRM1 Yeast CRM1 | Ran Yeast Ran | 0.0073±0.0006 | |
| Yeast CRM1 (residues 377–413 deleted) | Yeast Ran | 0.013±0.002 | |
| Yeast CRM1 (V441D) | Yeast Ran | 0.010±0.002 | |
| Yeast CRM1 (V441A/L442A) | Yeast Ran | 0.014±0.001 | |
| Yeast CRM1 (V443D) | Yeast Ran | 0.009±0.001 | |
| Yeast CRM1 (P754D) | Yeast Ran | 0.011±0.001 | |
| Human CRM1 | Canine Ran | 0.0055±0.0007 | |
| Dissociation of NES in the presence of RanGAP | |||
| Yeast CRM1, yeast Ran, yeast RanGAP (Rna1p) | 0.00558±0.0002 | ||
| RanBD-accelerated dissociation of NES | |||
| CRM1 | Ran | RanBD | |
| Yeast CRM1 | Yeast Ran | Yeast RanBP1 | 15.6±0.3 |
| Yeast CRM1 | Yeast Ran | Yeast RanBP1 (residues 71–201) | 14.6±0.4 |
| Yeast CRM1 (residues 377–413 deleted) | Yeast Ran | Yeast RanBP1 (residues 11–201) | 29.5±0.6 |
| Yeast CRM1 (V441D) | Yeast Ran | Yeast RanBP1 | 0.288±0.005 |
| Yeast CRM1 (V441A/L442A) | Yeast Ran | Yeast RanBP1 | 0.502±0.007 |
| Yeast CRM1 (V443D) | Yeast Ran | Yeast RanBP1 | 0.30±0.01 |
| Yeast CRM1 (P754D) | Yeast Ran | Yeast RanBP1 | 13.4±0.2 |
| Human CRM1 | Canine Ran | Human RanBP1 | 2.55±0.03 |
| Human CRM1 | Canine Ran | RanBD of human RanBP1 (residues 12–159) | 1.04±0.02 |
| Human CRM1 | Canine Ran | RanBD1 of human RanBP2 (residues 1155–1321) | 1.28±0.03 |
| Human CRM1 | Canine Ran | RanBD2 of human RanBP2 (residues 2002–2146) | 2.57±0.01 |
| Human CRM1 | Canine Ran | RanBD3 of human RanBP2 (residues 2302–2441) | 1.20±0.01 |
| Human CRM1 | Canine Ran | RanBD4 of human RanBP2 (residues 2895–3056) | 1.89±0.09 |
| Data represent mean±s.d. based on three experiments. Constructs are full length, wild type, unless indicated otherwise. | |||
Structure of yeast CRM1:RanBP1:RanGTP complex
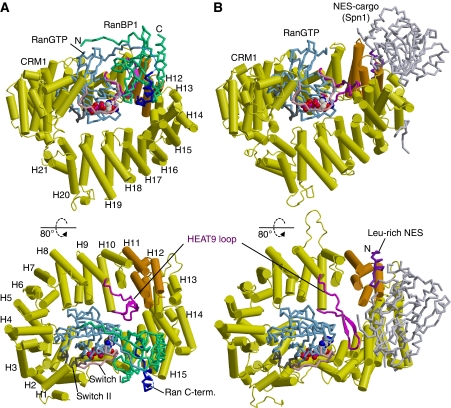
To understand how RanBP1 works to displace NES-cargo, we determined the crystal structure of yeast CRM1:RanBP1:RanGTP complex (Xpo1p:Yrb1p:Gsp1pGTP complex) at 2.0 Å resolution (Figure 2A). We used yeast proteins for crystallization because yeast CRM1:RanBP1:RanGTP complex is stable enough to be purified by gel filtration (Maurer et al, 2001) and so was a good target for structural study to take a snapshot just after displacement of NES-cargo by RanBP1. We obtained diffraction quality crystals by using the Q71L mutant of full-length Ran to stabilize the GTP-bound conformation (Klebe et al, 1995), and also by deleting CRM1 residues 377–413 (a long, poorly conserved loop that is susceptible to proteolysis; Petosa et al, 2004) and RanBP1 residues 1–10. Biochemical experiments confirmed the functionality of these deletion mutants (see Supplementary Figure S3; Table 1). The structure was solved by molecular replacement and refined to a crystallographic R-factor of 17.5% (Rfree=22.1%) (Table 2). Although RanBP1 residues 11–70 did not have defined electron density, the RanBP1 residues 71–201, clearly visible in the electron density map, were sufficient to increase the koff of PKI NES by three orders of magnitude (Table 1). Therefore, the crystal structure includes all of the RanBP1 residues necessary to displace NES. In the ternary CRM1:RanBP1:RanGTP complex, the ring-shaped CRM1 wraps around the RanBP1:RanGTP complex (Figure 2A), which had almost the same structure as the binary RanBD1:RanGMPPNP complex (Vetter et al, 1999) with Cα root mean squared deviation of 1.3 Å. Comparison of the CRM1:RanBP1:RanGTP complex (Figure 2A) and CRM1:Spn1:RanGTP complex (Figure 2B) revealed functionally important differences in CRM1 conformation. In particular, the hydrophobic NES-binding cleft between helices 11A and 12A on the CRM1 outer surface (Monecke et al, 2009; Dong et al, 2009a) is closed in the CRM1:RanBP1:RanGTP complex as detailed below, consistent with the function of RanBP1 in NES dissociation. In the CRM1:RanBP1:RanGTP complex, RanBP1 is located far away (∼40 Å) from the NES-binding site, indicating that RanBP1 uses an allosteric mechanism to displace NES-cargo.
Figure 2.
Overview of crystal structures. (A) Yeast CRM1:RanBP1:RanGTP complex (this study; PDB accession code: 3M1I). (B) CRM1:Spn1:RanGTP complex (Monecke et al, 2009; PDB accession code: 3GJX). The HEAT repeats of CRM1 are labelled H1–H21. CRM1 is coloured in yellow, except that HEAT9 loop (that shows large movement between the two structures) and HEAT repeats 11–12 (that constitute the NES-binding site) are highlighted in magenta and orange, respectively. Ran is coloured in cyan (switch I, switch II and the C-terminal tail are highlighted in pink, grey and blue, respectively; GTP is shown as space-filling spheres). RanBP1 and Spn1 are coloured in green and light grey, respectively. Leu-rich NES at the N-terminus of Spn1 (residues 1–16), highlighted in purple, binds to the outer surface of CRM1, away from Ran, whereas RanBP1 makes intimate contacts with RanGTP and binds to a site that is distinct from the NES-binding site. The structures suggest that RanBP1, together with the C-terminal tail of Ran, induces a large movement of HEAT9 loop that in turn causes conformational rearrangements in HEAT repeats 11–12 so that the hydrophobic NES-binding cleft on the CRM1 outer surface closes, squeezing out the NES-cargo (see Figures 3 and 4 and Supplementary Movies S1–S3 for details).
Table 2. Crystallographic statistics.
| Crystal | Yeast CRM1: RanBP1:RanGTP |
|---|---|
| Data collection | |
| Space group | P43212 |
| Unit cell dimensions (Å, degree) | a=b=106.21, c=303.60, α=β=γ=90 |
| Resolution range (Å)a | 45.33–2.00 (2.11–2.00) |
| Wilson B-factor | 35.7 |
| Total observationsa | 1 675 198 (183 853) |
| Unique reflectionsa | 117 954 (16 944) |
| Completeness (%)a | 99.9 (100.0) |
| Rmerge (%)a | 8.4 (83.4) |
| Mean I/σa | 20.4 (2.9) |
| Multiplicitya | 14.2 (10.9) |
| Refinement | |
| Number of used reflections | 111 903 |
| Rcryst/Rfree (%) | 17.5/22.1 |
| Total number of non-H atoms | 11 649 |
| Number of water molecules | 784 |
| r.m.s.d from ideal bond length (Å) | 0.026 |
| r.m.s.d. from ideal bond angles (degree) | 2.039 |
| aParentheses refer to final resolution shell. | |
HEAT9 loop functions as an allosteric autoinhibitor of NES binding
The structure of CRM1:RanBP1:RanGTP complex suggested that a long hairpin loop connecting the A- and B-helices of HEAT repeat 9 (hereafter referred to as HEAT9 loop) is important in the long-range communication between RanBP1 and the NES-binding site. In CRM1:RanBP1:RanGTP complex, RanBP1 interacts with the switch I loop and the acidic C-terminal tail of Ran, as observed in the RanBD1:RanGMPPNP complex (Vetter et al, 1999). The C-terminal DEDDADL amino-acid motif of Ran forms salt bridges with Arg154RanBP1, Arg177RanBP1, Lys39Ran, Lys154Ran and Arg757CRM1 (Figure 3A). CRM1 may stabilize these electrostatic interactions by shielding these charged residues from bulk solvent. In the CRM1:Spn1:RanGTP complex, the cluster of acidic residues at the tip of HEAT9 loop interacts with the switch I loop of Ran and the inner surface of HEAT repeats 14 and 15 (Monecke et al, 2009) (Figure 3B). Therefore, the binding of RanBP1 would displace HEAT9 loop because the acidic C-terminal residues of Ran would clash with HEAT9 loop and also substitute for the acidic residues of HEAT9 loop in interacting with the Ran switch I loop and the CRM1 inner surface. Unexpectedly, the structure of the CRM1:RanBP1:RanGTP complex showed that the displaced HEAT9 loop in turn binds to the inner surface of HEAT repeats 11 and 12, that is immediately adjacent to the NES-binding site, and the way HEAT9 loop interacted with HEAT repeats 11 and 12 provided structural insight into NES release mechanism (Figure 4). The highly conserved hydrophobic residues (Val441, Leu442, Val443 and Ile451 in yeast CRM1) of the HEAT9 loop are involved in this interaction and, although the hydrophobic side chains of the corresponding residues are mostly exposed to solvent in CRM1:Spn1:RanGTP complex (see Supplementary Figure S4 for sequence alignment of yeast and mouse CRM1), these residues pack intimately against the nonpolar patch on the CRM1 inner surface in CRM1:RanBP1:RanGTP complex. The intimate packing of hydrophobic side chains at the interface is probably optimized by rotations of the side chains of Met594 (on 12B helix) and Met556 (on 11B helix) towards HEAT9 loop, and movement of Phe583 (on 12A helix) towards HEAT11, concomitant with rotation of 12B helix and movement of 12A helix closer to 11A helix (indicated by arrows in Supplementary Figure S5A). As a consequence, HEAT repeats 11 and 12 pack intimately against each other, and there is no room to accommodate hydrophobic side chains of Leu-rich NES between the outer helices 11A and 12A in CRM1:RanBP1:RanGTP complex (Figures 4B and C; see also Supplementary Movies S1–S3).
Figure 3.
RanBP1 induces movement of HEAT9 loop by recruiting the C-terminal tail of Ran. Close-up view into the region where the C-terminal tail of Ran clashes with HEAT9 loop is shown. (A) Yeast CRM1:RanBP1:RanGTP complex. (B) CRM1:Spn1:RanGTP complex (CRM1 is mouse, whereas Spn1 and Ran are human) (Monecke et al, 2009). Colouring is the same as in Figure 2. Dashed lines in light blue indicate hydrogen bonds or salt bridges. GTP is shown as space-filling spheres. Residue numbering differs between species. Comparison of the two structures suggests that the C-terminus of Ran (blue in (A)) would clash with HEAT9 loop (magenta) and displace it on RanBP1 binding.
Figure 4.
Structural rearrangements of the NES-binding cleft associated with RanBP1 binding and NES release. This figure illustrates how RanBP1-induced movement of HEAT9 loop forces CRM1 into a conformation that is incompatible with NES binding. Only HEAT repeats 9, 11 and 12 of CRM1 are shown for clarity. Left, CRM1:Spn1:RanGTP complex (Monecke et al, 2009); right, yeast CRM1:RanBP1:RanGTP complex. (A) Top view. HEAT9, magenta; HEAT11 and HEAT12, yellow; Leu-rich NES of Spn1, purple. Hydrophobic interaction between highly conserved hydrophobic residues of HEAT9 loop (Val441, Leu442, Val443, Ile451 of yeast CRM1, which correspond to Val430, Leu431, Val432, Val440 of mouse CRM1, respectively) and the inner surface of HEAT repeats 11 and 12 appears to induce movements of Met594, Met556, Phe583 of yeast CRM1 (Met583, Met545, Phe572 of mouse CRM1, respectively), leading to closure of the hydrophobic NES-binding cleft between outer helices 11A and 12A. (B) Same view as (A), but HEAT repeats 11 and 12 are shown in surface representation (the residues that directly interact with Leu-rich NES are white, whereas the other residues are yellow). (C) Side view. Colouring is the same as in (B). Only the side chains of Leu-rich NES are shown for clarity. Supplementary Movies S1–S3 show animation of the structural changes; Supplementary Figure S5 shows superposition of the two structures.
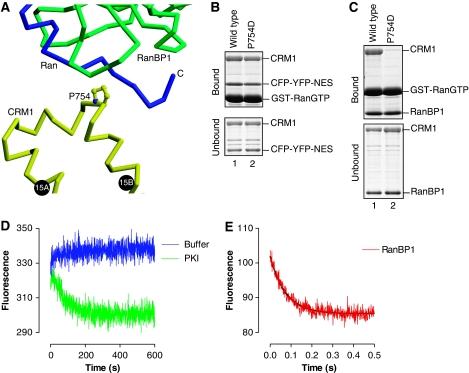
We used Ran and CRM1 mutants to evaluate functional significance of the conformational changes of HEAT9 loop. When the C-terminal residues of Ran (212–216) were deleted, RanBP1 did not compete effectively with the PKI NES in a pulldown assay, and the RanΔC mutant formed a stable quaternary complex with CRM1, an NES-cargo and RanBP1 (Figure 5A). This supports the idea that the displacement of HEAT9 loop by the C-terminal residues of Ran is important for RanBP1-induced NES release. Furthermore, although CRM1 mutants V441D or V441A/L442A or V443D normally bound to PKI (an NES-cargo) in the presence of RanGTP (Figure 5B), these mutations strikingly reduced the rate of RanBP1-accelerated NES dissociation by ∼30–50-fold (Figures 5C and D; Table 1), confirming that the hydrophobic interactions between HEAT9 loop and the inner surface of HEAT repeats 11 and 12 are functionally important in the acceleration of cargo release by RanBP1.
Figure 5.
Structure-based mutants verified the allosteric mechanism of NES release. (A) A pulldown assay showed that the C-terminal tail of Ran is required for RanBP1-mediated dissociation of NES from CRM1 and RanGTP. Immobilized GST-PKI (60 μg) was incubated with CRM1 (68 μg) and RanGTP (48 μg), unbound CRM1 and Ran washed away and then the beads were incubated without (lanes 1 and 3) or with (lanes 2 and 4) RanBP1 (130 μg). Bound and unbound fractions were analysed by SDS–PAGE. Lanes 1 and 2, full-length Ran; lanes 3 and 4, a C-terminal deletion mutant of Ran (residues 1–211). As the band of RanBP1 overlapped with that of GST-PKI, RanBP1 in the bound fraction was detected by western blotting; the other proteins were detected by Coomassie staining. (B–D) Mutational analysis demonstrated that the conserved hydrophobic residues in HEAT9 loop are important for loading and unloading of NES-cargo. (B) Pulldown assays to analyse the effects of mutating hydrophobic residues of HEAT9 loop on the binding of yeast CRM1 to PKI (NES-cargo) in the absence (lanes 1–4) or presence (lanes 5–8) of RanGTP. Immobilized GST-PKI (64 μg) was incubated with CRM1 (76 μg) without (lanes 1–4) or with (lanes 5–8) RanGTP (78 μg). Bound and unbound proteins were analysed by SDS–PAGE and Coomassie staining. SDS–PAGE of input proteins shows that the same amount of CRM1 (either wild type or mutant) was used in each experiment. The mutation(s) designed to disrupt the hydrophobic interactions between HEAT9 loop and the inner surface of HEAT repeats 11 and 12 strengthened the binding of CRM1 to PKI (NES-cargo) in the absence of RanGTP. (C, D) The mutations of the hydrophobic residues of HEAT9 loop also substantially reduced the rate of RanBP1-accelerated NES release. Preincubated yeast CRM1-CFP:YFP-NES (PKI residues 38–47):RanGTP mixture (0.4 μM CRM1 (wild type (red) or V441D (green) or V441A/L442A (orange) or V443D (magenta))-CFP, 2.0 μM YFP-NES, 6.0 μM RanGTP) was rapidly mixed with buffer alone (blue), or with 8 μM RanBP1.
In addition to making intimate contacts with RanGTP, RanBP1 also binds to CRM1 directly at HEAT15, burying a surface area of 237 Å2 (Figure 6A). This binding is accompanied by a change in the superhelical path followed by CRM1 HEAT repeats 12–19 compared with CRM1:Spn1:RanGTP complex, with the largest movement (∼11 Å) being centred at HEAT15, on top of which RanBP1 binds (Supplementary Figure S6). Because of this conformational change, the direct contact between the switch I loop of Ran and CRM1 HEAT repeats 17 and 18, as well as the direct contact between the switch II loop of Ran and CRM1 HEAT19 observed in CRM1:Spn1:RanGTP complex (Monecke et al, 2009), are completely lost in CRM1:RanBP1:RanGTP complex, whereas RanBP1 induces only minor changes in the interactions between RanGTP and CRM1 N-terminal region (HEAT repeats 1–8). The RanBP1-induced loss of the contacts between CRM1 and the switch loops of Ran might facilitate dissociation of RanGTP from CRM1, enabling the access of RanGAP to Ran and GTP hydrolysis. CRM1 appears to use the conformational flexibility of HEAT repeats (Conti et al, 2006) to accommodate RanBP1. We used the CRM1 mutant P754D to explore the importance of the direct contact between RanBP1 and CRM1. The P754D mutation was engineered to disrupt the shape complementarity at the RanBP1:CRM1 interface, and a pulldown assay showed that the P754D mutation impaired the stability of CRM1:RanBP1:RanGTP complex as expected (Figure 6C), although the P754D mutation did not affect the rate of RanBP1-accelerated release of NES from CRM1 and RanGTP (Figures 6D and E; Table 1). Therefore, stable binding of RanBP1 to CRM1 may not be required for NES displacement. The observation that RanBP1 formed a stable quaternary complex with CRM1, an NES-cargo (PKI) and RanΔC (1–211) (Figure 5A), also indicated that it is the movement of HEAT9 loop, and not the change in superhelical shape of CRM1 required to accommodate RanBP1, that is primarily responsible for the active displacement of NES-cargo by RanBP1.
Figure 6.
Functional significance of the interaction interface between CRM1 and RanBP1 in CRM1:RanBP1:RanGTP complex. (A) A close-up view of the interface between RanBP1 and CRM1. Yellow, CRM1 (only HEAT15 is shown for clarity); green, RanBP1; blue, the C-terminal region of Ran. Pro754CRM1 makes van der Waals contact with RanBP1. (B) A pulldown assay of the assembly of CRM1:NES:RanGTP complex. Immobilized 93 μg GST-RanGTP was incubated with 39 μg CRM1 (wild type or P754D mutant) and 20 μg CFP-YFP-NES (a Leu-rich NES of MVM NS2 protein; Askjaer et al, 1999). (C) A pulldown assay of the assembly of CRM1:RanBP1:RanGTP complex. Immobilized 93 μg GST-RanGTP was incubated with 39 μg CRM1 (wild type or P754D mutant) and 20 μg RanBP1. (D, E) NES dissociation kinetics. Preincubated CRM1(P754D)-CFP:YFP-NES (PKI residues 38–47):RanGTP mixture (0.4 μM CRM1(P754D)-CFP, 2.0 μM YFP-NES, 6.0 μM RanGTP) was rapidly mixed with buffer alone (blue) or with 80 μM PKI (green) or with 8 μM RanBP1 (red).
Except for a salt bridge between Asp447CRM1 and Lys130RanBP1 (Figure 3A), all of the extensive interactions that determine the conformation of HEAT9 loop in CRM1:RanBP1:RanGTP complex are made between CRM1 residues. It therefore seems likely that the HEAT9 loop has an inherent property to inhibit NES binding, not by directly masking the NES-binding site, but by stabilizing the closed conformation of the NES-binding cleft through interactions with the CRM1 inner surface, and that RanBP1 exploits this autoinhibitory function of HEAT9 loop to actively displace NES-cargo. The structure of CRM1:Spn1:RanGTP complex (Monecke et al, 2009) is unusual in that the well-conserved hydrophobic residues of the HEAT9 loop are mostly exposed to solvent. In the absence of RanGTP, lack of interaction between Ran and CRM1 probably causes the HEAT9 loop to assume the energetically favourable conformation observed in the CRM1:RanBP1:RanGTP complex where stabilization energy is provided by the hydrophobic interaction with the inner surface of HEAT repeats 11 and 12.
Consistent with the idea of autoinhibition by HEAT9 loop, the CRM1 mutation(s) in HEAT9 loop (V441D or V441A/L442A or V443D) substantially increased the binding of CRM1 to PKI (an NES-cargo) in the absence of RanGTP (Figure 5B). In this context, it is noteworthy that there is no clear electron density for HEAT9 loop in the crystal structure of CRM1:Spn1 heterodimer (Dong et al, 2009a). It is likely that HEAT9 loop must be dissociated from HEAT repeats 11 and 12 to open the hydrophobic NES-binding cleft and enable stable binding of Leu-rich NES. The autoinhibitory function of HEAT9 loop can also explain, at least in part, why RanGTP increases the affinity of NES to CRM1. The interaction between the switch I loop of RanGTP and HEAT9 loop observed in CRM1:Spn1:RanGTP complex (Monecke et al, 2009) would take HEAT9 loop away from the inner surface of HEAT repeats 11 and 12, and thereby relieve the inhibition of NES binding. However, HEAT9 loop may not be solely responsible for inhibiting NES binding in the absence of RanGTP. The C-terminus of CRM1 possibly functions as another autoinhibitory segment, because deletion of the C-terminal residues of CRM1 following the helix 21A increases the affinity of CRM1 to an NES-cargo in the absence of RanGTP (Dong et al, 2009b). Both HEAT9 loop and the C-terminus of CRM1 might act in concert to ensure that the binding of an NES-cargo to CRM1 depends on RanGTP. It will be important to solve the structure of free CRM1 at high resolution to fully understand why RanGTP stabilizes the binding of NES to CRM1.
Primarily nuclear RanBDs of RanBP3 (Yrb2p) or Nup50 (Nup2p) do not displace NES from CRM1 and RanGTP
It has been suggested that another RanBD-containing protein RanBP3 (Yrb2p) functions as a cofactor for CRM1-mediated nuclear export (Taura et al, 1998; Englmeier et al, 2001; Lindsay et al, 2001). In contrast to RanBP1 (Yrb1p) and RanBP2 that are localized in the cytoplasm, RanBP3 (Yrb2p) is localized primarily in the nucleus, and has been proposed to promote the formation of CRM1:NES-cargo:RanGTP complex in the nucleus and may accompany the export complex through NPCs (Lindsay et al, 2001). The RanBD of RanBP3 binds to RanGTP much more weakly than RanBP1 (Mueller et al, 1998), indicating that the RanBDs of RanBP1 and RanBP3 function quite differently. We found by the FRET-based kinetics assay that the RanBD of human RanBP3 or yeast Yrb2p does not induce dissociation of NES (Figures 7A and B), in agreement with the observation that RanBP3 forms a stable complex with CRM1, NES-cargo and RanGTP (Englmeier et al, 2001; Lindsay et al, 2001). The FRET assay also showed that the RanBD of human Nup50 (or yeast Nup2p), a RanBD-containing nucleoporin mainly localized at the nuclear side of the NPCs (Guan et al, 2000; Solsbacher et al, 2000), does not displace NES from CRM1 and RanGTP (Figures 7C and D). We therefore propose that the RanBDs can be classified into two types in terms of their effects on NES dissociation from CRM1 and RanGTP: the cytoplasmic RanBP1-type RanBDs (found in RanBP1 (Yrb1p) and RanBP2) function as disassembly factors and accelerate NES release, whereas the primarily nuclear RanBP3-type RanBDs (found in RanBP3 (Yrb2p) and Nup50 (Nup2p)) do not inhibit the binding of NES.
Figure 7.
Primarily nuclear RanBDs of RanBP3 (Yrb2p) or Nup50 (Nup2p) do not displace NES-cargo from CRM1 and RanGTP. NES dissociation kinetics was measured by a FRET-based assay. (A) Preincubated human CRM1-CFP:YFP-NES (PKI residues 38–47):RanGTP mixture (0.4 μM CRM1-CFP, 2.0 μM YFP-NES, 6.0 μM RanGTP) was rapidly mixed with 80 μM PKI (green) or with 8 μM RanBD of human RanBP3b (residues 312–448) (orange), and then the time-dependent change in CFP-YFP FRET signal was recorded. (B) Preincubated yeast CRM1-CFP:YFP-NES (PKI residues 38–47):RanGTP mixture (0.4 μM CRM1-CFP, 2.0 μM YFP-NES, 6.0 μM RanGTP) was rapidly mixed with 80 μM PKI (green) or with 8 μM RanBD of yeast Yrb2p (residues 195–327) (orange). (C) Preincubated human CRM1-CFP:YFP-NES (PKI residues 38–47):RanGTP mixture (0.4 μM CRM1-CFP, 2.0 μM YFP-NES, 6.0 μM RanGTP) was rapidly mixed with 80 μM PKI (green) or with 8 μM RanBD of human Nup50 (residues 320–468) (orange). (D) Preincubated yeast CRM1-CFP:YFP-NES (PKI residues 38–47):RanGTP mixture (0.4 μM CRM1-CFP, 2.0 μM YFP-NES, 6.0 μM RanGTP) was rapidly mixed with 80 μM PKI (green) or with 8 μM RanBD of yeast Nup2p (residues 566–720) (orange).
The NMR structure of the RanBD of human RanBP3 (Zhang HP et al, PDB accession code: 2CRF) provides possible explanation why the RanBD of RanBP3 does not displace NES. The NMR structure shows that the loop of RanBP1 (residues 148–153) that contacts the C-terminal tail of Ran in the CRM1:RanBP1:RanGTP complex is much shorter in RanBP3 and located away from Ran (Supplementary Figure S7). Moreover, the other residues of RanBP1 that interact with the C-terminal tail of Ran are not strictly conserved in RanBP3 (Yrb2p) and Nup50 (Nup2p) (Supplementary Figure S8), and the conspicuous basic patch on RanBP1 (the binding site for the acidic C-terminal residues of Ran) is much less basic on RanBP3 (Supplementary Figure S9). Therefore, the interaction between the RanBP3-type RanBDs and the C-terminus of Ran is probably not strong enough to displace the HEAT9 loop. This would account for the lack of the NES-displacement activity of the primarily nuclear RanBP3-type RanBDs.
Variations of structural mechanisms by which RanBP1 acts on nuclear export complexes
The crystal structures of nuclear export complexes solved so far suggest that the RanBDs of RanBP1/RanBP2 are used generally in the disassembly of almost all of the nuclear export complexes, although the precise way RanBDs are used differs in each export pathway. Earlier study showed that the C-terminus of Ran is required for the RanBP1-mediated disassembly of CAS(Cse1p):importin-α(Kap60p):RanGTP complex, probably because the C-terminus of Ran clashes with the HEAT repeats of CAS (Cse1p) on RanBP1(Yrb1p) binding (Matsuura and Stewart, 2004). The structure of exportin-t:tRNA:RanGTP complex (Cook et al, 2009) suggests that RanBP1 would clash with exportin-t and thereby destabilize the tRNA export complex in the cytoplasm (Supplementary Figure S10), consistent with the observation that RanBP1 (Yrb1p) relieves the inhibition of RanGAP-mediated RanGTP hydrolysis by exportin-t (Los1p) and tRNA (Hellmuth et al, 1998; Kutay et al, 1998). However, another export receptor for small RNA, namely exportin-5, appears to be a rare exception and its complex with export cargo and RanGTP may not be destabilized by RanBP1, because superposing RanBP1:RanGTP complex and the exportin-5:pre-microRNA:RanGTP complex (Okada et al, 2009) suggests that RanBP1 and the C-terminus of Ran would not clash with exportin-5 nor with its cargo (pre-microRNA) (Supplementary Figure S11). Rapid release of pre-microRNA from exportin-5 on translocation across the NPCs may not be favourable, because naked small RNAs could be easily degraded (unlike stably folded tRNA). Exportin-5 might resist the RanBP1 (or RanBP2)-mediated cargo release to protect pre-microRNA from degradation until the ternary export complex encounters the processing enzyme Dicer for the final maturation of microRNA in the cytoplasm (Bernstein et al, 2001; Köhler and Hurt, 2007).
Conclusion
In summary, we have presented direct evidence that RanBDs of RanBP1 (Yrb1p) or RanBP2 (but not RanBP3 (Yrb2p) or Nup50 (Nup2p)) increase the rate of dissociation of NES-cargo from CRM1 and RanGTP. The 2.0-Å crystal structure of CRM1:RanBP1:RanGTP complex, in conjunction with biochemical assays using structure-based mutants, provided mechanistic details of the first step in the disassembly of CRM1 nuclear export complex in the cytoplasm, and highlighted crucial function of HEAT9 loop as an allosteric autoinhibitor of NES binding.
Materials and methods
Expression and purification of recombinant proteins for biochemical assays
Standard techniques were used for the manipulation of recombinant DNA (Sambrook and Russel, 2001). All proteins were expressed in Escherichia coli strain BL21-CodonPlus(DE3)RIL (Stratagene). His/S-CRM1 (yeast Xpo1p), His/S-RanBP1 (yeast Yrb1p), His/S-Ran (canine) and His-CFP-YFP-MVM NS2 NES was expressed as described (Matsuura and Stewart, 2004). His/S-CRM1(yeast Xpo1p)-CFP, His/S-RanBP1 (human) and His/S-RanBD of RanBP2 (human) were expressed from pET30a-TEV (Matsuura and Stewart, 2004). His/S-PKIα (human) and His/S-RanGAP (yeast Rna1p) were expressed from pET30a (Novagen). His-Ran (yeast Gsp1p) was expressed from pET15b as described (Matsuura et al, 2003). His-YFP-PKI NES (residues 38–47) was expressed from pET15b. His-tagged proteins were purified over Ni-NTA (Novagen) and gel filtration over Superdex75 or Superdex200 (GE Healthcare). GST-PKIα (human) and GST-Ran (yeast Gsp1p) were expressed from pGEX-TEV (Matsuura and Stewart, 2004) and pGEX-4T-1, respectively, and purified over Glutathione-sepharose (GE Healthcare). GST-CRM1 (human)-CFP was expressed from pGEX-TEV (Matsuura and Stewart, 2004) and initially purified over Glutathione-sepharose (GE Healthcare). After removal of GST-tag by His-TEV protease, human CRM1-CFP was finally purified over Superdex200 (GE Healthcare). Mutants were created using the Quickchange system (Stratagene). All DNA constructs were verified by DNA sequencing.
NES dissociation kinetics
NES dissociation kinetics were measured at 20°C in buffer A (10 mM Tris–HCl (pH 7.5), 200 mM NaCl, 5 mM Mg(OAc)2, 0.1 mM AEBSF, 2 mM β-mercaptoethanol) using either a Hi-Tech Scientific SF-61DX2 stopped-flow spectrophotometer or a JASCO FP-6500 spectrofluorometer. CFP was excited at 430 nm, with YFP emission monitored using a 530-nm high-pass filter (in the case of stopped-flow measurements) or at 530 nm (in the case of measurements using the spectrofluorometer). In all, 0.4 μM CRM1-CFP, 2.0 μM YFP-NES (PKI residues 38–47; LALKLAGLDI) and 6.0 μM RanGTP were preincubated at 20°C for 15 min, and then rapidly mixed with equal volume of either buffer A (to check for photobleaching) or protein (80 μM PKI or 8 μM RanBD or 8 μM RanGAP). Protein concentrations are those before mixing in the measurement cell. As photobleaching was negligible, data were fitted to single exponential by nonlinear regression using GraphPad Prism to determine NES off-rate. The NES off-rates are summarized in Table 1. Steady-state fluorescence spectra were recorded using a JASCO FP-6500 spectrofluorometer at 20°C in buffer A. Samples were incubated for 15 min before measurements.
GST pulldown assay
Pulldown assays were performed in binding buffer (10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM Mg(OAc)2, 0.05% Tween20, 2 mM β-mercaptoethanol) as described (Matsuura and Stewart, 2004). GST fusions were immobilized on 10 μl of packed Glutathione-sepharose beads and each binding reaction was performed by incubating the beads with reaction mixtures in a total volume of 50 μl for 1 h at 4°C. The amounts of proteins used are indicated in the figure legends. Beads were then spun down and supernatant saved (unbound fraction). Beads were washed twice with 1 ml of binding buffer and bound proteins eluted with SDS-sample buffer (bound fraction). Bound and unbound fractions were analysed by SDS–PAGE and Coomassie staining. In case where the band of RanBP1 overlapped with GST-PKI, RanBP1 was detected by western blotting (Figure 5A, bound fraction) using S-protein HRP conjugate (Novagen) that binds to S-tag fused to the N-terminus of RanBP1.
Preparation of yeast CRM1:RanBP1:RanGTP complex for crystallization
Untagged Ran (Saccharomyces cerevisiae, full-length Gsp1p, Q71L mutant) and GST-RanBP1 (S. cerevisiae, Yrb1p residues 11–201, TEV site inserted between GST and Yrb1p) were coexpressed from a bicistronic vector based on pET30a (Novagen), whereas His/S-CRM1 (S. cerevisiae, Xpo1p, full length except that residues 377–413 were deleted) was expressed separately from pET30a-TEV (Matsuura and Stewart, 2004), in E. coli strain BL21-CodonPlus(DE3)RIL (Stratagene). The Q71L mutation of yeast Ran, which corresponds to Q69L of metazoan Ran, inhibits GTP hydrolysis and stabilizes the GTP-bound form of Ran (Klebe et al, 1995). The residues 377–413 of yeast CRM1 correspond to a long loop connecting HEAT repeats 8 and 9. This loop is susceptible to proteolysis (Petosa et al, 2004) and the amino-acid sequence of this loop is poorly conserved across species. This (presumably disordered) loop is particularly long in S. cerevisiae. Truncation of this loop was vital to obtain diffraction quality crystals. We confirmed that the deletion mutants used for crystallization are functional in vitro (Supplementary Figure S3). After harvesting, the two sets of cells were mixed, resuspended in buffer B (30 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM Mg(OAc)2, 10 mM imidazole, 0.1 mM GTP, 7 mM β-mercaptoethanol, 1 mM AEBSF) and lysed by sonication on ice. All subsequent purification steps were performed at 4°C. Clarified lysates were loaded onto Ni-NTA resin (Novagen), washed with buffer B containing 25 mM imidazole and eluted with buffer B containing 30 mM NaCl and 250 mM imidazole. Tween20 was added to the eluate to a final concentration of 0.05%. After incubating the Ni-NTA eluate with Glutathione-sepharose resin (GE Healthcare), the resin was washed with buffer C (10 mM Tris–HCl (pH 7.5), 100 mM NaCl, 5 mM Mg(OAc)2, 0.05% Tween20, 7 mM β-mercaptoethanol). The GST- and His/S-tags were cleaved off RanBP1 and CRM1 by incubating the resin overnight with His-TEV protease (0.18 mg/ml) in buffer C containing 0.1 mM GTP and 0.2 mM AEBSF. The CRM1:RanBP1:RanGTP complex, released from the resin, was passed through Ni-NTA and finally purified by gel filtration over Superdex200 (GE Healthcare) in buffer D (10 mM Tris–HCl (pH 7.5), 100 mM NaCl, 5 mM Mg(OAc)2, 2 mM β-mercaptoethanol). GTP was added to the gel filtration fractions to a final concentration of 0.1 mM, and the complex was concentrated to 12.5 mg/ml using a Millipore concentrator (Mol. wt. cutoff 10 000).
X-ray crystallography
Crystals of yeast CRM1:RanBP1:RanGTP complex were grown at 20°C from 6.2 mg/ml protein by hanging drop vapour diffusion against 0.1 M Bis-Tris (pH 6.6), 0.2 M NH4NO3 and 18% PEG3350. Rod-shaped crystals appeared in 2–3 weeks and grew to a maximum dimension of 0.3 × 0.3 × 0.5 mm in 1 month. Crystals were serially transferred to 0.1 M Bis–Tris (pH 6.6), 0.2 M NH4NO3, 23% PEG3350 and 12% glycerol in three steps and flash frozen by plunging into liquid nitrogen. Initially, a 2.5-Å resolution data set was collected at 100 K at Photon Factory beamline BL5A and processed using MOSFLM and CCP4 programs (Collaborative Computational Project Number 4, 1994). The crystal had P43212 symmetry (a=b=106.21 Å, c=303.60 Å) with one complex in the asymmetric unit. The structure was solved by molecular replacement using MOLREP (Vagin and Teplyakov, 1997). The initial models were RanBD1:Ran complex (Vetter et al, 1999), human CRM1 C-terminal fragment (residues 707–1027) (Petosa et al, 2004) and a model of yeast CRM1 residues 479–655 built by homology modelling using the program MODELLER (Sali and Blundell, 1993) based on a sequence alignment between CRM1 and Kapβ2 (Petosa et al, 2004) and a crystal structure of Kapβ2 (Lee et al, 2006). The density map calculated using the phases from molecular replacement showed interpretable α-helical densities for CRM1 N-terminal region. Iterative cycles of model building using COOT (Emsley and Cowtan, 2004) and refinement using REFMAC5 (Murshudov et al, 1997) reduced Rfree to 32.2% (Rcryst 24.9%). At this stage, a 2.0-Å resolution data set was collected at 100 K at SPring-8 beamline BL41XU, and the structure was further refined against this data set. A TLSMD analysis (Painter and Merritt, 2006) was used to define TLS groups for the final cycles of refinement. The final model had an Rfree of 22.1% (Rcryst 17.5%) and contained CRM1 residues 1–376 and 414–1058, RanBP1 residues 71–201, Ran residues 10–188 and 197–217, MgGTP and 784 water molecules. In a Ramachandran plot, 93.9% of residues lie in the favoured regions, 5.9% in the allowed regions, 0.1% in the generously allowed regions and 0.1% in the disallowed regions. Structural figures and movies were produced using Molscript (Kraulis, 1991), Raster3D (Merritt and Bacon, 1997), PyMOL (DeLano, 2008), CCP4MG (Potterton et al, 2004) and UCSF Chimera (Pettersen et al, 2004).
Accession codes
Atomic coordinates and structure factors for the yeast CRM1:RanBP1:RanGTP complex have been deposited at the Protein Data Bank under accession code 3M1I.
Supplementary Material
Acknowledgments
We thank Hidemi Hirano, Tatsuo Hikage and Hirofumi Onishi for excellent technical assistance; the staff of Photon Factory and SPring-8 for help during data collection; Yuichiro Maeda for discussion; Murray Stewart for critical reading. This work was supported in part by the Sumitomo Foundation and Grants-in-Aid for Young Scientists from the MEXT, Japan. MK was supported by JSPS Research Fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M (1999) RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol 19: 6276–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Conti E, Müller CW, Stewart M (2006) Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol 16: 237–244 [DOI] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E (2009) Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2008) The PyMOL molecular graphics system http://www.pymol.org
- Dong X, Biswas A, Chook YM (2009b) Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol 16: 558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM (2009a) Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 458: 1136–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Cryst D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Englmeier L, Fornerod M, Bischoff FR, Petosa C, Mattaj IW, Kutay U (2001) RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep 2: 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475–483 [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G (1996) The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem 271: 5313–5316 [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308–311 [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 15: 5584–5594 [PMC free article] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L (2000) Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol 20: 5619–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt E, Simos G (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A (1995) Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34: 639–647 [DOI] [PubMed] [Google Scholar]
- Köhler A, Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst 24: 946–950 [Google Scholar]
- Künzler M, Gerstberger T, Stutz F, Bischoff FR, Hurt E (2000) Yeast Ran-binding protein1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a CRM1 (Xpo1)-dependent pathway. Mol Cell Biol 20: 4295–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Güttinger S (2005) Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15: 121–124 [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536 [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Süel KE, Louis TH, Zhang Z, Chook YM (2006) Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126: 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG (2001) Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol 153: 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M (2003) Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J 22: 5358–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M (2004) Structural basis for the assembly of a nuclear export complex. Nature 432: 872–877 [DOI] [PubMed] [Google Scholar]
- Maurer P, Redd M, Solsbacher J, Bischoff FR, Greiner M, Podtelejnikov AV, Mann M, Stade K, Weis K, Schlenstedt G (2001) The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p). Mol Biol Cell 12: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol 277: 505–524 [DOI] [PubMed] [Google Scholar]
- Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R (2009) Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 324: 1087–1091 [DOI] [PubMed] [Google Scholar]
- Mueller L, Cordes VC, Bischoff FR, Ponstingl H (1998) Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett 427: 330–336 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T (2009) A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326: 1275–1279 [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C (1997) Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278: 141–144 [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA (2006) TLSMD web server for the generation of multi-group TLS models. J Appl Cryst 39: 109–111 [Google Scholar]
- Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuerwald U, Soler-López M, Baudin F, Mattaj IW, Müller CW (2004) Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell 16: 761–775 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M (2004) Developments in the CCP4 molecular-graphics project. Acta Cryst D 60: 2288–2294 [DOI] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Carey KL, Macara IG (1996) A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol 134: 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbour: Cold Spring Harbour Press [Google Scholar]
- Solsbacher J, Maurer P, Vogel F, Schlenstedt G (2000) Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol Cell Biol 20: 8468–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Taura T, Krebber H, Silver PA (1998) A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA 95: 7427–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A (1999) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398: 39–46 [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.