Abstract
Mycobacterium tuberculosis (Mtb) possesses a proteasome system analogous to the eukaryotic ubiquitin-proteasome pathway. Mtb requires the proteasome to resist killing by the host immune system. The detailed assembly process and the gating mechanism of Mtb proteasome have remained unknown. Using cryo-electron microscopy and X-ray crystallography, we have obtained structures of three Mtb proteasome assembly intermediates, showing conformational changes during assembly, and explaining why the β-subunit propeptide inhibits rather than promotes assembly. Although the eukaryotic proteasome core particles close their protein substrate entrance gates with different amino terminal peptides of the seven α-subunits, it has been unknown how a prokaryotic proteasome might close the gate at the symmetry axis with seven identical peptides. We found in the new Mtb proteasome crystal structure that the gate is tightly sealed by the seven identical peptides taking on three distinct conformations. Our work provides the structural bases for assembly and gating mechanisms of the Mtb proteasome.
Keywords: cryo-electron microscopy, Mycobacterium tuberculosis, 20S proteasome assembly, 20S proteasome gating, X-ray crystallography
Introduction
The 20S proteasome is the core enzyme of the eukaryotic ubiquitin-proteasome pathway that selectively degrades proteins (Voges et al, 1999; Borissenko and Groll, 2007; Goldberg, 2007). The proteasome occurs in all three domains of life, although very rare in bacteria (De Mot et al, 1999). Actinomycete, the only known bacterial lineage with the 20S proteasome, is thought to have acquired the protease complex through a lateral gene transfer event (Gille et al, 2003). In bacteria, a number of other protease complexes, such as HslUV and ClpAP, perform functions similar to that of the proteasomes (De Mot et al, 1999). The Mycobacterium tuberculosis (Mtb) proteasome defends Mtb against nitrosative stress (Darwin et al, 2003), and is essential for survival of the pathogen in its mammalian host (Gandotra et al, 2007). The Mtb proteasome has been established as a viable target for the development of anti-TB agents (Gandotra et al, 2007; Nathan et al, 2008). Towards this direction, a class of oxathiazol-2-one compounds has been identified that selectively inhibits the Mtb proteasome over the human proteasome (Lin et al, 2009).
The prokaryotic 20S proteasome comprises 14 identical copies of α- and β-subunits arranged into a four-ringed α7β7β7α7 cylinder (Voges et al, 1999). The assembly of the prokaryotic 20S proteasome is autonomous, and takes place through a half-proteasome intermediate that contains one α-ring and one β-ring. Apposition of two half proteasomes results in autocleavage of the β-subunit propeptide, yielding a mature proteasome (Voges et al, 1999). In both prokaryotic and eukaryotic 20S proteasomes, the β-propeptide usually promotes the proteasome assembly (Zuhl et al, 1997; Arendt and Hochstrasser, 1999; Kwon et al, 2004). However, the β-propeptide of the Mtb 20S proteasome appears to inhibit rather than promote 20S assembly: growth of the expression host Escherichia coli at 37°C, but not at room temperature, is required for assembly of 20S proteasome and processing of the β-propeptide, and deletion of the β-propeptide lifted the assembly inhibition at the room temperature (Lin et al, 2006). Therefore, the β-propeptide is a thermodynamic barrier for the assembly of the Mtb 20S proteasome. A crucial question is how the Mtb β-propeptide exerts it inhibitory effect on the assembly and maturation of the Mtb proteasomes.
As shown in the crystal structure of the yeast proteasome, the substrate entrance gate in the α-ring of a eukaryotic 20S proteasome is closed, which avoids uncontrolled proteolytic activity (Groll et al, 2000). The gate closure requires dramatic departure from the pseudo seven-fold symmetry of the complex assembly; this requirement is met by the heterogeneous N-terminal sequence of the seven α-subunits enabling these peptides to take on different structures to form a sealed gate, and the gate can be opened by removing the N-terminal peptide of the α3-subunit (Groll et al, 2000). On the contrary, the prokaryotic 20S proteasome, because of the presence of only single type of α-subunit, is considered to have a partial open gate. This notion is supported by several crystal structures of the prokaryotic proteasomes that show disordered α-subunit N-terminal peptides, and by their significant in vitro peptidase activities (Lowe et al, 1995; Groll et al, 2003; Kwon et al, 2004; Zhang et al, 2009). However, the Mtb proteasome, with seven identical α-subunits, was shown to have essentially no in vitro peptidase activity and apparently a closed gate (Lin et al, 2006). Deleting the α-subunit N-terminal octapeptide (α-octapeptide; Met1-Ser2-Phe3-Pro4-Tyr5-Phe6-Ile7-Ser8) opens the gate (Lin et al, 2006). Given the homomeric nature of the Mtb 20S proteasome, another important question is how the prokaryotic 20S proteasome closes the converging space at the seven-fold symmetry axis with only a single type of α-subunit.
We took the combined approach of cryo-electron microscopy (EM) and X-ray crystallography to address these two important questions. By showing and following the β-propeptide position from the Mtb half proteasome, through a novel assembly intermediate we captured, to the finally assembled but immature 20S proteasome, we provide a structural explanation why the β-propeptide is a barrier to the assembly process. By visualizing the N-terminal α-octapeptide in the Mtb 20S proteasome structure, we elucidate how a homomeric proteasome is able to tightly seal its substrate entrance at the seven-fold symmetry axis with seven identical α-octapeptides. Furthermore, we found that in the Mtb 20S open-gate mutant proteasome (20SOG), the first α-helix (H0) is highly flexible, and we further discuss the implication of this observation with regard to the proteasome gate opening mechanism.
Results
The β-propeptides are located outside rather than inside of the Mtb half proteasome as shown by Cryo-EM
Our previous work suggested that at room temperature the Mtb 20S proteasome assembly process is likely paused at the half-proteasome stage (Lin et al, 2006). Therefore, we first set out to analyse the structure of this assembly intermediate. We produced the wild-type (WT) assembly intermediate by culturing the E. coli bacteria expressing the Mtb prcA and prcB genes at 25°C, and isolated the intermediate by affinity column and size exclusion chromatography. The intermediate eluted at about 1 ml later than the mature 20S from the Superose-6 column (Supplementary Figure 1A). The SDS–PAGE gel demonstrates the presence of the β-propeptide (Supplementary Figure 1B). In the side views of the cryo-EM and the negative stain EM images, there are two instead of four rings (Figure 1A; Supplementary Figure 1C). These results show that the assembly process is indeed arrested at the half-proteasome stage.
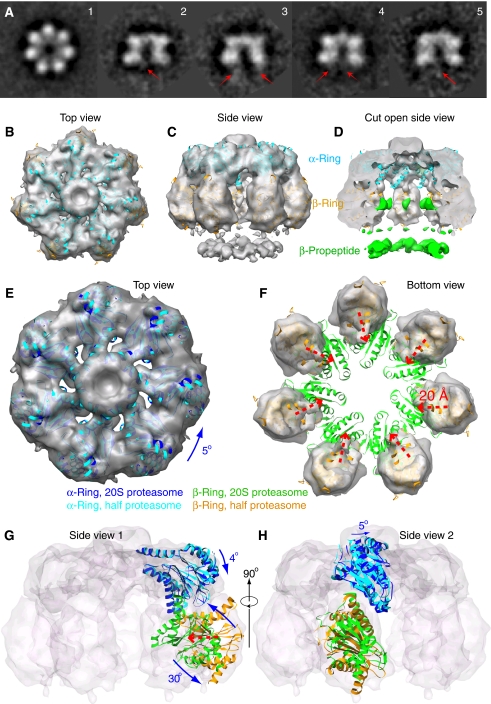
Figure 1.
The 3D cryo-EM structure of the Mtb half proteasome. (A) A gallery of 2D class averages of the cryo-EM images of the Mtb half proteasome particles. The red arrows in the side views (panels 2–5) indicate averaged residual density of the flexible β-propeptides located at the bottom under the β-ring. (B–D) Surface rendered cryo-EM 3D map of the Mtb half proteasome docked individually with the α-subunit (in cyan) and β-subunit (in brown) crystal structures in the top, side, and cut-open views. The EM density map is shown partially transparent. The discontinuous densities, coloured green in the cut-open view, are attributed to the flexible β-propeptides, and disappear at display threshold higher than 1.4 σ (D). (E, F) Computationally segmented α-ring (E) and β-ring electron density (F), superimposed with individually docked α-subunits (E, cyan) and β-subunits (F, yellow). Also superimposed are the α-ring (in blue) and β-ring structure (in green) of the mature Mtb 20S proteasome to illustrate the required structural re-arrangement. (G, H) Two orthogonal side views with electron density map shown as transparent surface rendering in grey. One α-subunit and one β-subunit structure in half proteasome and full proteasome positions are shown in carton representation. (E–H) The translation and rotation of the individual α- and β-subunits in the Mtb half proteasome required for reaching the full Mtb 20S proteasome structure. See text for details.
Importantly, in most of the averaged side views such as shown in Figure 1A and Supplementary Figure 1C, there are weak and apparently flexible densities underneath the β-ring, as marked by the red arrows. These weak densities are likely from the β-propeptides. Each β-propeptide is 56-residues long, which makes the total mass of the seven β-propeptides to ∼43 kDa. We determined the 3D cryo-EM map of the WT half proteasome at ∼10 Å resolution (Supplementary Figure 1D–F; Figure 1B–H). The 3D map was calculated by imposing the seven-fold symmetry. The most striking feature is the presence of a ‘floating' ring of densities below the β-ring (Figure 1C and D). This additional ring of densities is consistent with the weak densities observed in the 2D averages (Figure 1A), and is attributed to the β-propeptides. We note that because of its exterior location, the β-propeptide is likely flexible, so the observed density in this seven-fold averaged 3D map represents only partial density of the β-propeptides.
In the Rhodococcus proteasome, the β-propeptide is proposed to help β-subunit folding, and it binds the α-subunit at the α/β heterodimer stage before the formation of the half proteasome. As a consequence, the β-propeptide should be hidden inside the half proteasome (Zuhl et al, 1997; Witt et al, 2006). Consistent with this proposal, no density was observed outside the β-ring in the 3D EM reconstruction of the Archaeoglobus fulgidus half proteasome whose β-propeptide also promotes assembly (Mullapudi et al, 2004). The outside location of the Mtb half proteasome implies that the β-propeptide would hinder the association of two half proteasomes because two β-rings face each other in the 20S proteasome, and that the β-propeptide has to translocate to the interior of the chamber during assembly such that it could be eventually cleaved through autoproteolysis. Therefore, the exterior location of the β-propeptide at the half-proteasome stage likely accounts for it being the assembly barrier of the Mtb 20S proteasome.
The crystal structures of the α- and β-subunits were docked as 14 separate rigid bodies into the cryo-EM map (PDB ID: 2FHG) (Figure 1B–H). The fit between the EM density and the structure is excellent with a correlation coefficient of ∼0.85. There is a small density region inside the β-ring, which is unoccupied. We interpreted this region of density as part of the β-propeptide, and thus coloured the density the same as the exterior β-propeptide ring in green (Figure 1D). To facilitate comparison of the half-proteasome structure with the mature Mtb 20S, we further docked half of the mature Mtb 20S crystal structure (one α-ring and one β-ring) as a single rigid body into the half-proteasome EM map (Figure 1E–H). As the radius of the β-ring in half-proteasome map is markedly larger than that of the 20S proteasome, only the α-ring portion of the crystal structure was used for correlation-based computational alignment with the EM density. The resulting position is regarded as the α- and β-ring structure of the Mtb 20S proteasome. From the docking, it is clear that there is significant re-arrangement for both α- and β-subunits in the Mtb half proteasome as compared with the Mtb 20S proteasome. For α-subunit, the re-arrangement can be approximated by a ∼5° rotation around the central seven-fold symmetry axis (the blue curved arrows in Figure 1E, top view), and a second rotation of ∼4° around an axis perpendicular to the main seven-fold symmetry axis (Figure 1G, side view 1). For β-subunit, the re-arrangement is more extensive, requiring an inward shift of ∼20 Å (the dashed red arrows in Figure 1F, bottom view), and a rotation of nearly 30° around an axis that connects two neighbouring β-subunits (Figure 1G). Large re-arrangements, including a 28° rotation, of the β-subunit in the A. fulgidus half proteasome as compared with the A. fulgidus 20S structure were also reported (Mullapudi et al, 2004). However, in the A. fulgidus half proteasome, the 28° rotation is within the plane of the β-ring; in the Mtb half proteasome, the 30° rotation is perpendicular to the β-ring (Figure 1G). We suggest that the difference in β-subunit re-arrangement between Mtb versus A. fulgidus half proteasome is due to the different β-propeptide position: in the Mtb half proteasome, the perpendicular rotation of the β-subunit likely enables the translocation of the β-propeptides to the interior chamber during the apposition of two half proteasomes.
A novel Mtb 20S proteasome assembly intermediate formed by apposition of two half proteasomes
As it protrudes outside the β-ring in the Mtb half proteasome, the β-propeptide will hinder the apposition of two half proteasomes. We wondered how two half proteasomes are able to eventually assemble into the 20S proteasome; this assembly step would likely be different from that of proteasomes from other species where their respective β-propeptide is inside the half-proteasome chamber and the β-rings of two half proteasomes are ready to interface with each other. We sought to address this question by attempting to capture and to visualize the downstream in vitro assembly intermediates. We found that incubation at 37°C for 3 h totally converted the half proteasome into the fully assembled mature Mtb proteasome, whereas extended incubation (16 h) at 31°C did not (data not shown). Shorter incubation at lower temperature resulted in a mixture of particles that can be roughly divided into three species, based on EM observation: the first two species correspond to the half proteasome and the fully assembly 20S proteasome, and the third species is composed of two half proteasomes, but this species is clearly different from the fully matured 20S (Figure 2A). The major distinction of the third species from the mature 20S is the left–right asymmetry; the mature 20S proteasome is symmetric. The asymmetric 20S particles constitute a novel assemble intermediate not seen earlier. We hereby term this intermediate as the Mtb 20S intermediate I (20S IM-I).
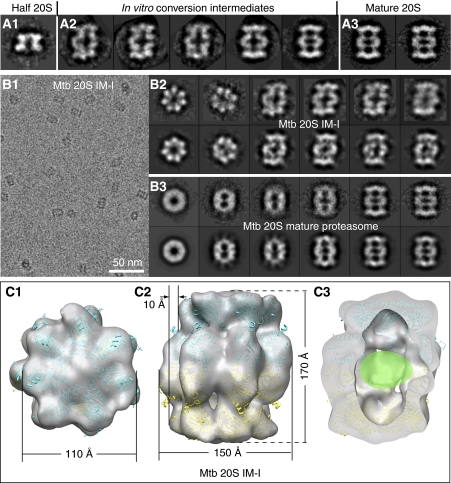
Figure 2.
A novel proteasome assembly intermediate as shown by cryo-EM and image classification. (A) A comparison of the side views of the freshly purified Mtb WT half proteasome (left panel, A1); in vitro conversion intermediates (middle panel, A2); and the fully assembly mature Mtb 20S proteasome (right panel, A3). (B) Cryo-EM characterization of the inhibitor-treated and in vitro assembled 20S proteasome intermediate (20S IM-I). The left panel (B1) shows a raw micrograph of the 20S IM-I embedded in vitreous ice, and the right panels show the reference-free 2D averages of the 20S IM-I (B2, top) and the purified 20S mature particles (B3, top). The corresponding reprojections from the 3D reconstructions are shown in the bottom panels for comparison (b2 bottom, the Mtb 20S IM-I; b3 bottom, the Mtb 20S mature proteasome). (C) Surface-rendered, 20% transparent top, side, and cut-open views of the cryo-EM map of the Mtb 20S IM-I at ∼25 Å resolution. The atomic model of the half 20S, derived from docking the α- and β-subunits into 3D cryo-EM map of the Mtb half proteasome, fits well in the density of the Mtb 20S IM-I. The β-propeptide densities, not resolved in the low-resolution map, is likely inside the central chamber, as indicated by a green oval in (C3).
The incubation mixture is unsuitable for 3D EM structural determination of the Mtb 20S IM-I, because of the presence of three types of particles. Interestingly, we found that a homogeneous preparation of the Mtb 20S IM-I can be obtained by incubating the purified WT half proteasome at 20°C for 6 h in the presence of 1 mM each of three proteasome inhibitors MLN273, GL1, Z-VLRBA (Adams, 2003; Lin et al, 2009) (Supplementary Figure 2; Figure 2B and C), although we do not know why these inhibitors are able to arrest the in vitro conversion at this particular stage. We note that 20S IM-I might represent only one of a series of possible assembly intermediates. The structural difference of Mtb 20S IM-I from the Mtb 20S is very obvious in the averaged 2D cryo-EM images (Figure 2B): though the seven-fold symmetry is always obscured in the top view of the Mtb 20S because of the rotationally staggered α- and β-rings, the symmetry is very clear in the top view of the Mtb 20S IM-I, indicating that the α- and β-rings are approximately aligned rotationally along the length of the cylinder. This architectural difference in subunit arrangement is also visible in the side views of the cryo-images (Figure 2B), but is much more obvious in the side views of the negative stain EM images (Supplementary Figure 2B). This is so because negative staining EM shows the surface feature of a 3D object whereas cryo-EM shows the true projection.
We determined the 3D cryo-EM map of the Mtb 20S IM-I at 25 Å resolution (Figure 2C). The two-fold symmetry was imposed because obviously there were two half proteasomes in the structure, and because the two-fold symmetry was present in certain averaged side views (Figure 2B; Supplementary Figure 2). The resolution was low, probably because of the meta-stable, thus, has partially flexible nature of the intermediate. The Mtb 20S IM-I structure is about 10 Å taller than the mature 20S. The two half proteasomes bind to each other with a ∼10 Å off-axis shift (Figure 2C). Surprisingly, the pseudo-atomic model of the Mtb half proteasome, as shown in Figure 1, fits well with the cryo-EM map of the Mtb 20S IM-I. This means that the Mtb 20S IM-I corresponds to a rigid body and off-axis apposition of the two Mtb half proteasomes. In other words, at the resolution we are concerned (25 Å), there were no detectable α- or β-subunit re-arrangements within the half proteasome as the two half proteasomes initially associate (Figure 2C). At this stage of assembly, the β-propeptide is expected to be inside the central chamber of the Mtb 20S IM-I (Figure 2C). However, there is no isolated density for the β-propeptides inside the EM map, probably because of their disorder and the low resolution of EM map. As the α- and β-subunits have not re-arranged in the Mtb 20S IM-I, it is thus likely that translocation of the β-propeptide precedes and is independent from the final α- and β-subunits re-arrangement described above (Figure 1E–H). We suggest that translational staggering of the two half proteasomes in the 20S IM-I might be a mechanism to avoid the steric clash because of the protruding β-propeptides in each half proteasome.
The β-propeptides become ordered and ascend to the anterior chamber on α- and β-subunits re-arrangement into their final positions
After assembling into the Mtb 20S IM-I, the α- and β-subunits are expected to re-arrange into their respective final positions. We asked how the β-propeptides respond to the subunit re-arrangement. We introduced a point mutation at the active site Thr-1 in the β-subunit with Ala (T1A); mutation at the active site prevents β-propeptide removal by the Thr-1-mediated autoproteolysis (Groll et al, 2003). The T1A mutant was purified as the half proteasome, based on the gel filtration peak position and our negative stain EM characterization (Supplementary Figure 3). However, this mutant half proteasome assembled into the full but immature proteasome, hereby termed the 20S IM-II, during crystallization at high concentration (10 mg/ml) and at either 4 or 21°C. The crystal structure of the 20S IM-II determined at 2.5 Å resolution was very similar to the mature 20S full proteasome (Figure 3A and B). The β-propeptide is intact in the 20S IM-II, but now has ascended into the antechamber, a space between the α- and the β-rings, becomes partially ordered, and forms hydrophobic interaction with the α-subunit (Figure 3; Supplementary Figure 4A). In the crystal structure, 30 out of the 58 β-propeptide residues were traced (Figure 3C). Interestingly, the β-propeptide takes on a conformation similar to that of the β-propeptide in the Rhodococcus K33A mutant 20S proteasome (Supplementary Figure 4B) (Kwon et al, 2004). This is quite remarkable, considering the opposite effect these β-propeptides exert on their respective proteasome assembly process and the different positions they started with during assembly. We suggest that the ordered β-propeptide structure might be required to drive the re-arrangement of the α- and β-subunits, and for the final cleavage of the β-propeptides in the prokaryotic proteasomes. We also determined the 2.2 Å crystal structure of the WT mature Mtb 20S proteasome (see description below), which is at a significantly higher resolution than the previously reported 3.0 Å structure (Hu et al, 2006). Superposition of the 20S IM-II with the new WT 20S structure at the β-subunit region shows a 1.5 Å inward shift of the α-subunit on final maturation.
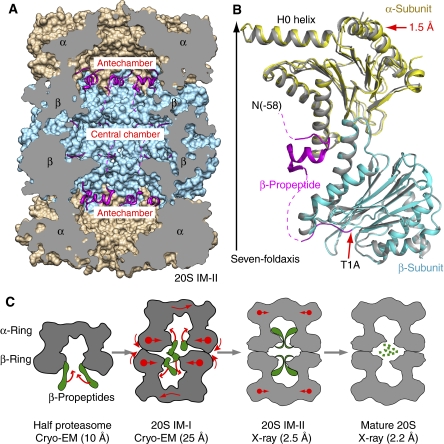
Figure 3.
The β-propeptide is ordered and ascended to the antechambers in the 20S T1A mutant proteasome (20S IM-II). (A) The crystal structure of the Mtb 20S IM-II shown in a cut-open side view. The β-propepetides are shown as ribbons in magenta, and the α- and β-rings are rendered in surface view and shown in brown and cyan, respectively. (B) Superposition of one α/β heterodimer of 20S IM-II (in yellow and cyan, respectively) with that of the mature Mtb 20S proteasome (in grey). The β-propeptide is shown in magenta. The dashed curves indicate unresolved residues. (C) Proposed structural changes accompanying the assembly of the Mtb 20S proteasome, starting from the half proteasome to the metastable 20S assembly intermediate I (20S IM-I), towards the assembled but immature proteasome intermediate II as represented by the T1A mutant structure (20S IM-II), and finally to the mature 20S proteasome. The β-propeptides are illustrated in green.
On the basis of the cryo-EM structures of the Mtb half proteasome (10 Å resolution) and the 20S IM-I (25 Å), and the crystal structures of the 20S IM-II (2.5 Å) and the mature Mtb 20S proteasome (2.2 Å), we propose the following assembly process of the Mtb 20S proteasome starting at the stage of the half proteasome (Figure 3C). The two half proteasomes initially bind with their β-rings facing each other, but with their seven-fold symmetry axes shifted laterally by ∼10 Å we assume that the lateral shift may either facilitate apposition of the two β-rings by avoiding steric clash, or as a mechanism for triggering the retraction of the protruding β-propeptides. In the next step that converts the 20S IM-I to the 20S IM-II, three events must occur: (1) ascending of the β-propeptide into the antechamber and formation of the ordered structure on interaction with the α-subunit; (2) re-arrangement of the α- and β-subunits; and (3) alignment of the two half proteasomes. These events are likely coupled, that is, the ordering of β-propeptide drives the re-arrangement of α- and β-subunits into their final positions, and the subunit arrangement results in the alignment of the two half proteasomes. At this assembly stage, the complex has evolved into the nearly final structure and the enzymatically active configuration has been established. Therefore, the last step is mainly autodegradation of the β-propeptides into the small fragments. Accompanying the removal of β-propeptides from the antechamber, the α-rings shrink radially by 1.5 Å, finally forming the matured Mtb 20S proteasome.
How is the gate of Mtb 20S proteasome tightly closed by the seven identical α-subunits N-terminal octapeptides?
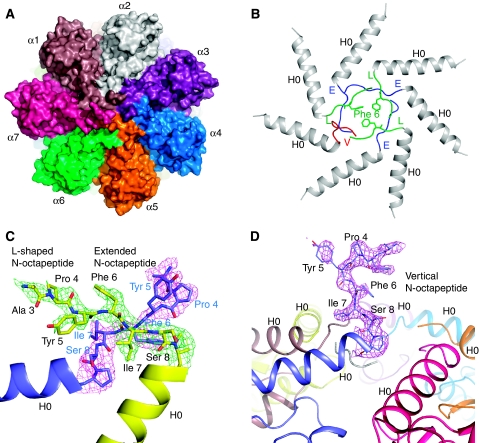
Surprisingly, the seven α-subunits N-terminal octapeptides (α-octapeptides) had well-defined electron densities in the 2.5 Å crystal structure of the T1A mutant structure (20S IM-II) (Figure 4). These ordered α-octapeptides tightly sealed the substrate gate (Figure 4A). Five α-octapeptides each has six ordered residues, missing only the very first two residues. The α-octapeptide generally adopts one of the two conformations: the ‘L' conformation, as it forms the ‘L'-like shape with the following H0 helix, or the extended (‘E') conformation because in this conformation the α-octapeptide extends almost linearly out from the H0 helix (Figure 4B). The ‘L'-conformation and the ‘E'-conformation are, in general, adjacent to each other: the ‘L' α-octapeptide crosses above its ‘E'-conformation neighbour with its respective bulky hydrophobic residues Tyr5-Phe6-Ile7 stacking against each other (Figure 4C). This alternating pattern of ‘L' and ‘E' repeats three times in the first six α-octapeptides until the last one. Here, two ‘L'- or two ‘E'-conformed α-octapeptides cannot be arranged next to each other without steric clash. In the crystal structure, the seventh α-octapeptide assumes a third conformation that protrudes almost vertically out of the proteasome top surface, being stabilized by crystal packing (Figure 4D). This upward-projecting conformation of the seventh α-octapeptide in Mtb proteasome is reminiscent of the N-terminal tails of α6 and α7 at the gate of the yeast 20S proteasome (Groll et al, 2000). In this gate-closed Mtb 20S IM-II structure, only 2–3 extreme N-terminal residues of the α-octapeptides are disordered. In comparison, there are 3–8 disordered residues (∼5 residues on average) at the closed gate of the yeast 20S (Groll et al, 2000). The final constriction at the Mtb 20S proteasome gate is formed by three hydrophobic side chains of the Phe-6 of the three ‘L' configured α-octapeptides (Figure 4B).
Figure 4.
The Mtb proteasome has a tightly closed gate in the crystal structure of the T1A mutant 20S proteasome. (A) A space-filling surface view of the T1A mutant 20S proteasome shows that the substrate entrance channel in the centre of the top view is totally sealed. (B) Ribbon representation of the end structure of the T1A mutant 20S. The α-octapeptides take the ‘E' (coloured in blue) and ‘L' conformation (coloured in green) alternatively, except for the last one that protrudes upwards (labelled as ‘V' conformation, coloured in red), being almost perpendicular to the end surface of the Mtb 20S cylinder. (C) The 2Fo−Fc electron density of the α-octapeptides in two adjacent α-subunits rendered at 1.0 σ. The ‘L' configuration is in green, and the ‘E' configuration in purple. This picture is viewed from the top of the proteasome cylinder. Note residue Phe-3 is modelled as Ala-3 because of the absence of the side chain density. (D) The α-octapeptide that assumes the up-standing (‘V') conformation. Superimposed in purple is its 2Fo−Fc density contoured at 1.0 σ. The residue Ser-2 is modelled as Ala-2 because of the absence of the side chain density. The picture is viewed from the side of the proteasome cylinder.
Is the closed gate observed in the Mtb 20S IM-II structure described above similar to the WT Mtb 20S? Although our EM reconstructions showed significant density blocking the gate (Hu et al, 2006; Lin et al, 2006), our previous 3.1 Å structure of the WT Mtb 20S did not resolve the α-octapeptides, leaving a 23 Å pore at the substrate entrance (Hu et al, 2006). In the improved 2.2 Å structure of the WT Mtb 20S proteasome, we indeed observed electron densities of the α-octapeptides (Supplementary Figure 5). The WT 20S has weaker and often discontinuous densities for the α-octapeptides such that a complete modelling is not possible. Nevertheless, these electron densities suggest that the α-octapeptides are similarly arranged in WT Mtb 20S as compared with those in the T1A mutant 20S structure. The 1.5 Å larger radius at the α-ring of the Mtb 20S IM-II than the mature 20S might have allowed for better ordering of the α-octapeptide in the 20S IM-II.
This is the first time a prokaryotic proteasome gate has been shown. The interesting question of how to seal a converging space at the seven-fold symmetry axis with seven identical peptides in a prokaryotic 20S proteasome appears to have been solved in the Mtb proteasome by having the same α-octapeptide to assume different conformations. In the Mtb 20S proteasome, the same α-octapeptide takes on three markedly different conformations. This gate closure strategy is different from that used in the eukaryotic 20S proteasome in which each of the seven N-terminal α-peptides has different primary sequence and length, thus takes on different structure to fill up the converging space at the yeast proteasome gate region (Groll et al, 2000).
Crystal structure of the Mtb open-gate mutant proteasome shows significant flexibility of the α-subunit H0 helix
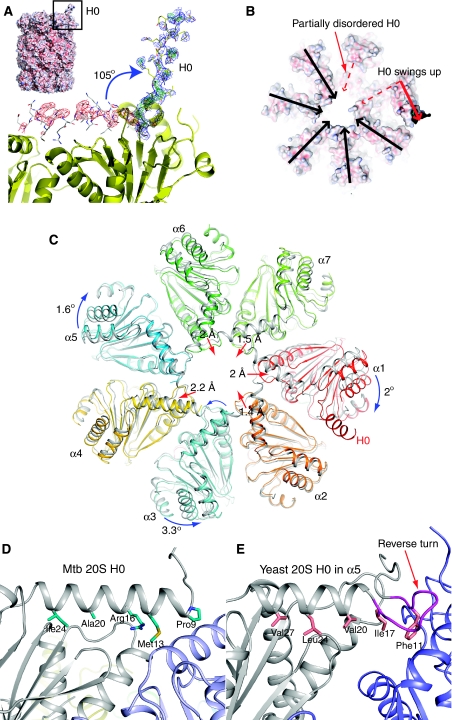
Deletion of the α-octapeptide produces an Mtb 20S proteasome with significant in vitro peptidase activity. This activity suggests that the substrate gate is at least partially open, and we thus called the construct Mtb 20SOG (Lin et al, 2006). Negative stain EM of the Mtb 20SOG indeed showed two opened ends of the cylindrical complex, whereas the ends are closed in the WT Mtb 20S (Lin et al, 2006). We asked whether there were other important structural differences that were not shown because of the low resolution of the EM maps. We solved a new crystal structure of the Mtb 20SOG treated with the 1 mM GL1. The inhibitor GL1 was shown earlier to covalently modify the active site residue Thr-1 of Mtb 20S and introduced structural changes around the substrate-binding pocket in the β-subunit, and such mechanism of inhibition was not observed in the human proteasome (Lin et al, 2009). Surprisingly, we found in the new structure that the amphipathic H0 helix of an α-subunit, usually lay horizontally on top of the proteasome cylinder, swung up by 105° to an almost vertical position (Figure 5A and B). The dislocation of the H0 helix caused partial disorder of the H0 helix in an adjacent α-subunit (Figure 5B). Furthermore, each of the seven α-subunits underwent certain translational and/or rotational movements as compared to those of the WT 20S (Figure 5C). These extensive changes in α-ring suggest that there is considerable flexibility built in the Mtb 20S proteasome structure.
Figure 5.
The H0 helix in one α-subunit is displaced in the crystal structure of the Mtb 20SOG. (A) The electron density map of the displaced H0 helix, which is shown in the yellow stick-and-ball mode. The normal H0 position is shown in the grey stick-and-ball mode. The electron density map was calculated using a model with H0 in its normal position. The green and red meshes are Fo−Fc different density map contoured at ±3.0 σ, respectively, and the blue mesh is 2Fo−Fc electron density map contoured at 1.0 σ. Inserted at the top left corner is a surface representation of the Mtb 20SOG. (B) Top surface view of the Mtb 20SOG. The upward swinging of H0 helix in one α-subunit (dashed and solid red arrows) results in partial disorder of the H0 in its neighbouring α-subunit (dashed red arrow). (C) Multiple subunit movements in the α-ring of the 20SOG structure (in colour) as shown by comparing with the WT 20S α-ring (in grey). The α-subunit harbouring the displaced H0 helix (labelled α1) shifted outwards by ∼2 Å (the red arrow head), and rotated clockwise 2° (the blue curved arrow). In response to the outward movement of the α1-subunit the neighbouring α2-, α7-, and α6-subunits shifted towards the centre (the red arrows). Finally, responding to the changes at the right side of the α-ring (α1, α2, α6, and α7), the α5 and α3 at the left side of the ring each slightly rotated in an opposite direction, and the α4-subunit moved outwards by 2 Å. (D) The interactions of the H0 helix with the underlying structure at its normal position in the Mtb 20S proteasome as compared with the H0 helix in the α5-subunit of the yeast 20S proteasome (E). The reverse turn in the yeast α-subunit, which is absent in the Mtb α-subunit, is shown in purple in (E).
We asked whether the observed H0 displacement is merely a crystallization artefact. Careful examination of how H0 interacts with its underlying structure suggests otherwise (Figure 5D and E). The H0 interfacial amino acid residues in Mtb 20S, (Ile24, Ala20, Arg16, Met 13, and Pro9) are much less hydrophobic, as compared with corresponding residues of H0 in the yeast proteasome (Val27, Leu24, Val20, Ile17, Phe11), which maintains strong hydrophobic interactions. Therefore, the H0 helix in the Mtb 20S interacts only weakly with its underneath structure, and appears to be primed for movement. This notion corroborates with our previous observation that the Mtb 20SOG are more readily to form complex with the proteasome ATPase Mpa than the WT proteasome, and the association between Mpa and 20SOG is highly flexible, presumably because of the flexible H0 helix onto which the Mpa binds (Wang et al, 2009).
Discussion
Proteasome maturation is coupled with its assembly process (Groll et al, 2003; Witt et al, 2006). Through cryo-EM and X-ray crystallography, we have shown the underlying mechanism for the inhibitory effect of the Mtb β-propeptide. Being outside the half proteasome, the Mtb β-propeptide does not help β-subunit folding, and presents a kinetic and dynamic barrier for subsequent apposition of two half proteasomes. Apparently, the Mtb β-subunit is able to fold on itself, in the absence of the β-propeptide or the α-subunit, as shown by solution of the crystal structure of the β-subunit alone (PDB ID: 2JAY). In the Rhodococcus 20S, the β-propeptide helps folding of the β-subunit, and promotes the 20S assembly (Zuhl et al, 1997). However, the Rhodococcus β-propeptide when supplied in trans allowed far more rapid dimerization of half proteasomes compared with normal β-subunits with the β-propeptide in cis (Zuhl et al, 1997). This observation is consistent with the idea that the β-propeptide has to get repositioned or processed for stable dimerization to occur. Such notion is reminiscent of the negative effect of β-propeptide on dimerization of Mtb half 20S. Therefore, despite the existence of individual characteristics of the proteasomes, certain crucial steps in the assembly process might be shared by the two actinomycete proteasomes. Further support to the notion is provided by the highly similar structures of the Rhodococcus preholoproteasome and its counterpart in Mtb (the Mtb 20S IM-II), particularly at the ordered propeptide region (Supplementary Figure 4B). The observed novel assembly intermediate, the Mtb 20S IM-I, is likely formed to facilitate the translocation of β-propeptide to the interior chamber. Such a distinct 20S assembly intermediate was not reported before. It is currently unclear whether other intermediates exist during assembly of Mtb 20S.
Our structural studies span the assembly process from the Mtb half proteasome to the final mature 20S proteasome; the assembly step preceding the half proteasome in Mtb has not been characterized. Therefore, it is not known whether the α-subunits first form the α-ring onto which the β-subunits assemble, as in the archaeal 20S proteasomes, from Thermoplasma acidophilum and A. fulgidas (Zwickl et al, 1994; Groll et al, 2003); or the α- and β-subunits first form heterodimer and then seven α/β dimers assemble into the half proteasome, as in the Rhodococcus 20S proteasome (Sharon et al, 2007). As the α- and β-genes had to be expressed with a single promoter in E. coli to produce a mature Mtb proteasome (Lin et al, 2006), we suggest that in Mtb, the α- and β-subunits first form heterodimer, similar to that of the Rhodococcus 20S proteasome.
This study shows a tightly sealed substrate gate in the Mtb 20S proteasome, supporting our previous conclusion that the Mtb 20S is gated (Lin et al, 2006). All previously characterized prokaryotic 20S proteasomes are thought to either lack a gating mechanism or loosely gated because of a partial closure of the gate (Voges et al, 1999; Groll et al, 2003). Indeed those 20S proteasomes have marked in vitro proteolytic activity towards oligopeptides (Seemuller et al, 1995; Tamura et al, 1995; Mc Cormack et al, 1997), and the N-terminal α-peptides surrounding the gate are disordered in the crystal structures (Lowe et al, 1995; Groll et al, 2003; Kwon et al, 2004). The closed gate of the Mtb 20S proteasome necessitates a gate opening mechanism. In archaea and eukaryotes, the 11S proteasomal activators or the proteasomal ATPases are responsible for gate opening, and the gating mechanism involves a three-residue YDR motif and a reverse turn at the α-subunit N-terminal region (Forster et al, 2005; Rabl et al, 2008). These two structural elements are universally conserved in the eukaryotic and archaeal proteasomes, but are absent in the Mtb proteasome (Lin et al, 2006). On the other hand, the Mtb proteasome ATPase Mpa contains the conserved carboxyl terminal HbYX motif, particularly the penultimate Tyr-608, that was shown to be required for the in vivo proteasome-mediated protein degradation (Darwin et al, 2005). Therefore, it remains to be analysed whether the Mtb 20S proteasome uses a distinct mechanism for gate opening.
Our new crystal structure of the inhibitor GL1-treated Mtb 20SOG showed surprisingly that the H0 helix around the substrate gate is highly flexible. Furthermore, we found previously that the Mtb proteasomal ATPase Mpa binds more readily to the Mtb 20SOG than the WT 20S, and Mpa capping on the Mtb 20SOG is also highly flexible (Wang et al, 2009). Mostly recently, the Mtb 20SOG in the presence of Mpa was shown to have significantly higher in vitro activity towards degradation of the pupylated PanB than the WT Mtb 20S (Striebel et al, 2010). Taken together, these observations suggest that the H0 flexibility might contribute to the gating mechanism of the Mtb 20S proteasome. In one scenario, the proteasomal ATPase may completely displace one of the seven H0 helices, leaving the gate wide open. The remaining six H0 helices would be able to better interact with the hexameric Mpa with smaller and localized adjustment, thus circumventing the symmetry mismatch problem. Alternatively, if the proteasomal ATPase only binds partially to the CP, which appears to be the case for Mpa, a relatively small positional and rotational adjustment of those H0 helices that actually bind to the ATPase might be sufficient to accommodate the six-folded surface of the proteasomal ATPase.
We further observed in the structure of the Mtb 20SOG that the α-subunits can move as rigid bodies. Such flexibility of the entire α-subunit in the α-ring is reminiscent of the 26S human proteasome in which the gate opening by the ATPases involves the radial and outward movement of the entire α-subunits (da Fonseca and Morris, 2008). The large-scale α-ring opening in the inhibitor-induced transition state of the enzyme complex, visible at single molecular level by atomic force microscopy, was recently reported in the yeast 20S proteasome (Osmulski et al, 2009). These observations reinforce the emerging concept that the 20S proteasomes are actually highly dynamic. Such notion is also consistent with the recent finding that the 20S proteasomes have the potential to keep entire substrates in store for continual degradation, which presumably requires considerable structural plasticity of the proteasome to house the folded substrate protein (Sharon et al, 2006).
Materials and methods
Expression and purification of WT and T1A mutant Mtb 20S proteasome
See Supplementary data.
EM and 3D reconstruction
The WT Mtb half proteasome, the Mtb 20S assembly intermediates, and the Mtb WT full-length 20S were analysed by both negative stain and cryo-EM with underfocus values ranged from 1.0–1.5 μm for the negatively stained particles and 1.5–3.5 μm for the cryo-particles. The software packages EMAN (Ludtke et al, 1999) and SPIDER (Frank et al, 1996) were used for image processing. Surface rendering and docking of atomic structures into the EM maps were carried out in UCSF Chimera (Pettersen et al, 2004). See Supplementary data for more details.
Crystallization, data collection, and structure solution
The protein samples at a concentration of 10 mg/ml were set up in crystallization plates with the sitting drop method at 4°C. For the Mtb 20S IM-II (the starting sample was the T1A mutant half proteasome), the well solution contained 60 mM sodium citrate (pH 5.8), 100 mM glycine, and 14% PEG 6000. For the WT 20S full proteasome, the well solution contained 60 mM sodium citrate (pH 5.7), 12.5% PEG 6000. For the Mtb 20SOG, the well solution contained 1 mM GL1 inhibitor. For cryo-crystallography, the crystal solution was replaced in several concentration steps with the cryo-protecting solution containing the original mother liquor and 35% dimethylformamide. X-ray diffraction data were collected at National Synchrotron Light Source in Brookhaven National Laboratory, and processed and scaled with the HKL2000 package (Otwinowski and Minor, 1997). The Mtb proteasome structure (PDB ID: 2FHG) was used for molecular replacement. Conventional crystallographic rigid body, simulated annealing, positional, and temperature factor refinement was carried out with CNS (Brunger, 2007). Fourteen non-crystallographic symmetry restraints were applied during the refinements. After each refinement, atomic model was manually rebuilt or adjusted in Coot (Emsley and Cowtan, 2004). After the Rfree of the model was reduced to below 26%, solvent molecules were built into the model. The data processing, refinement, and model statistics are given in Supplementary Table 1.
Supplementary Material
Acknowledgments
The crystallographic data were deposited in PDB with accession numbers 3MKA, 3MFE, and 3MI0 for Mtb T1A mutant 20S, OG 20S with displaced H0, and the 2.2 Å resolution WT 20S proteasome, respectively. We thank staff of Beam lines X25 and X29 at the National Synchrotron Light Source, Brookhaven National Laboratory for technical assistance in data collection. We thank Carl Nathan for support and encouragement. This work is supported by NIH grant AI070285 and Brookhaven National Laboratory LDRD grant 10-016 to HL.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams J (2003) The proteasome: structure, function, and role in the cell. Cancer Treat Rev 29: 3–9 [DOI] [PubMed] [Google Scholar]
- Arendt CS, Hochstrasser M (1999) Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J 18: 3575–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borissenko L, Groll M (2007) 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem Rev 107: 687–717 [DOI] [PubMed] [Google Scholar]
- Brunger AT (2007) Version 1.2 of the Crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Morris EP (2008) Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J Biol Chem 283: 23305–23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF (2003) The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302: 1963–1966 [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF (2005) Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol 55: 561–571 [DOI] [PubMed] [Google Scholar]
- De Mot R, Nagy I, Walz J, Baumeister W (1999) Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol 7: 88–92 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Forster A, Masters EI, Whitby FG, Robinson H, Hill CP (2005) The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell 18: 589–599 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S (2007) In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med 13: 1515–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C, Goede A, Schloetelburg C, Preissner R, Kloetzel PM, Gobel UB, Frommel C (2003) A comprehensive view on proteasomal sequences: implications for the evolution of the proteasome. J Mol Biol 326: 1437–1448 [DOI] [PubMed] [Google Scholar]
- Goldberg AL (2007) Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans 35: 12–17 [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D (2000) A gated channel into the proteasome core particle. Nat Struct Biol 7: 1062–1067 [DOI] [PubMed] [Google Scholar]
- Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R (2003) Investigations on the maturation and regulation of archaebacterial proteasomes. J Mol Biol 327: 75–83 [DOI] [PubMed] [Google Scholar]
- Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H (2006) Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol 59: 1417–1428 [DOI] [PubMed] [Google Scholar]
- Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK (2004) Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J Mol Biol 335: 233–245 [DOI] [PubMed] [Google Scholar]
- Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, Weich N, Nathan C (2006) Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol 59: 1405–1416 [DOI] [PubMed] [Google Scholar]
- Lin G, Li D, de Carvalho LP, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H, Nathan C (2009) Inhibitors selective for mycobacterial versus human proteasomes. Nature 461: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268: 533–539 [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Mc Cormack T, Baumeister W, Grenier L, Moomaw C, Plamondon L, Pramanik B, Slaughter C, Soucy F, Stein R, Zuhl F, Dick L (1997) Active site-directed inhibitors of Rhodococcus 20 S proteasome. Kinetics and mechanism. J Biol Chem 272: 26103–26109 [DOI] [PubMed] [Google Scholar]
- Mullapudi S, Pullan L, Bishop OT, Khalil H, Stoops JK, Beckmann R, Kloetzel PM, Kruger E, Penczek PA (2004) Rearrangement of the 16S precursor subunits is essential for the formation of the active 20S proteasome. Biophys J 87: 4098–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Gold B, Lin G, Stegman M, de Carvalho LP, Vandal O, Venugopal A, Bryk R (2008) A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis (Edinb) 88: S25–S33 [DOI] [PubMed] [Google Scholar]
- Osmulski PA, Hochstrasser M, Gaczynska M (2009) A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure 17: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Carter Jr CW, Sweet RM (eds), Vol. 276, pp 307–326. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell 30: 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W (1995) Proteasome from Thermoplasma acidophilum: a threonine protease. Science 268: 579–582 [DOI] [PubMed] [Google Scholar]
- Sharon M, Witt S, Felderer K, Rockel B, Baumeister W, Robinson CV (2006) 20S proteasomes have the potential to keep substrates in store for continual degradation. J Biol Chem 281: 9569–9575 [DOI] [PubMed] [Google Scholar]
- Sharon M, Witt S, Glasmacher E, Baumeister W, Robinson CV (2007) Mass spectrometry reveals the missing links in the assembly pathway of the bacterial 20 S proteasome. J Biol Chem 282: 18448–18457 [DOI] [PubMed] [Google Scholar]
- Striebel F, Hunkeler M, Summer H, Weber-Ban E (2010) The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup's N-terminus. EMBO J 29: 1262–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, Tanaka K, De Mot R, Baumeister W (1995) The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol 5: 766–774 [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Wang T, Li H, Li D, Lin G, Nathan C, Darwin KH, Li H (2009) Structural insights on the mycobacterium tuberculosis proteasomal ATPase Mpa. Structure 17: 1377–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt S, Kwon YD, Sharon M, Felderer K, Beuttler M, Robinson CV, Baumeister W, Jap BK (2006) Proteasome assembly triggers a switch required for active-site maturation. Structure 14: 1179–1188 [DOI] [PubMed] [Google Scholar]
- Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y (2009) Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell 34: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhl F, Seemuller E, Golbik R, Baumeister W (1997) Dissecting the assembly pathway of the 20S proteasome. FEBS Lett 418: 189–194 [DOI] [PubMed] [Google Scholar]
- Zwickl P, Kleinz J, Baumeister W (1994) Critical elements in proteasome assembly. Nat Struct Biol 1: 765–770 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.