Abstract
Complex Object Parametric Analyzer and Sorter (COPAS) devices are large-object, fluorescence-capable flow cytometers used for high-throughput analysis of live model organisms, including Drosophila melanogaster, Caenorhabditis elegans, and zebrafish. The COPAS is especially useful in C. elegans high-throughput genome-wide RNA interference (RNAi) screens that utilize fluorescent reporters. However, analysis of data from such screens is relatively labor-intensive and time-consuming. Currently, there are no computational tools available to facilitate high-throughput analysis of COPAS data. We used MATLAB to develop algorithms (COPAquant, COPAmulti, and COPAcompare) to analyze different types of COPAS data. COPAquant reads single-sample files, filters and extracts values and value ratios for each file, and then returns a summary of the data. COPAmulti reads 96-well autosampling files generated with the ReFLX adapter, performs sample filtering, graphs features across both wells and plates, performs some common statistical measures for hit identification, and outputs results in graphical formats. COPAcompare performs a correlation analysis between replicate 96-well plates. For many parameters, thresholds may be defined through a simple graphical user interface (GUI), allowing our algorithms to meet a variety of screening applications. In a screen for regulators of stress-inducible GFP expression, COPAquant dramatically accelerated data analysis and allowed us to rapidly move from raw data to hit identification. Because the COPAS file structure is standardized and our MATLAB code is freely available, our algorithms should be extremely useful for analysis of COPAS data from multiple platforms and organisms. The MATLAB code is freely available at our web site (www.med.upenn.edu/lamitinalab/downloads.shtml).
Keywords: C. elegans, computational method, high-throughput screening, flow cytometry, Wormsorter
Automation has been a great boon to the field of high-throughput screening. The Complex Object Parametric Analyzer and Sorter (COPAS) platform (Union Biometrica, Holliston, MA, USA) is a tool that allows for rapid quantification of the fluorescence, size, and optical density of small biological specimens, such as Caenorhabditis elegans, Drosophila, and zebrafish. The COPAS utilizes micro-fluidic approaches to draw intact live organisms through a fluorescence-compatible flow cell at extremely high rates (~50 animals per second) and quantifies the size [measured as object time-of-flight (TOF)], object optical density (EXT), and fluorescence emissions from up to three separate fluorescent channels for each animal. Because of its complete optical transparency, rapid growth rates, and amenability to forward and reverse genetic approaches, C. elegans is an excellent model system for COPAS-based high-throughput phenotypic and genetic studies (1–7). In many cases, these studies are enabled by the expression of fluorescent reporter transgenes (5,7,8), which often exhibit significant animal-to-animal variability. Because of this inherent variability in reporter expression, quantification of fluorescence by the COPAS within a population of animals is a more accurate phenotypic assessment than subjective visual inspection of individual animals (8). While the COPAS excels at the rapid collection of population-based data, the number of individual samples analyzed during a large-scale screen can easily reach into the thousands. Efficient analysis of such large COPAS data sets requires the use of automated computational tools, which have so far not been developed.
Currently, the COPAS can collect data in two modes, a single-sample mode and an autosampler 96-well mode. The single-sample mode permits very large sample sizes to be analyzed, which is a tremendous advantage for assaying highly variable or subtle phenotypes. However, because samples must be loaded one at a time into the sample chamber, the throughput of this mode is slow and labor-intensive and best suited to small-scale screens. The autosampler mode, enabled by the ReFLX adapter system, allows rapid analysis of liquid-based samples from 96-well plates, which provides tremendous sample throughput. However, the small volumes of 96-well assays limit the number of events per well to sample sizes much smaller than those obtained in the single-sample mode, making the autosampler mode well suited to large-scale genome-wide RNA interference (RNAi) or drug screens that utilize phenotypes of low variability. In the single-sample mode, each file contains the data from one sample. In the autosampler 96-well mode, each file contains the data from every well within a 96-well plate, classified according to well address. In both cases, the time required to filter, extract, and normalize the data; graph the summary results of the screen; compare results among plates; and statistically identify hits is a major rate-limiting step in the screening pipeline. Tools that facilitate the analysis of such large-scale data sets would tremendously advance the throughput capability of COPAS-based assays. Such tools are currently unavailable.
Many different software environments are suitable for the analysis of large-scale COPAS data sets, including R, SAS, and Visual Basic. Another program suitable for such analyses is MATLAB (MathWorks, Natick, MA, USA). MATLAB is a computer interface program specifically designed for analysis of matrix-based data sets, which is typically applied to the automation and standardization of image analysis routines. However, MATLAB can just as easily be applied to analyze any type of numerical data presented in a matrix format. Since the COPAS data file structure is a standardized 26 × n matrix worksheet (where n is the number of events sorted), we reasoned that COPAS-generated data could be analyzed in the MATLAB environment. While analysis of COPAS data is possible in other programming environments, such as Microsoft Excel and Visual Basic, MATLAB offers several significant advantages for COPAS data analyses. First, MATLAB is an interpreted language, making it very easy to learn, use, and modify. It is compatible with many different operating systems (Windows, Linux, Macintosh, etc.) and is therefore accessible to almost all users, regardless of platform. Second, MATLAB can receive user input through custom graphical user interfaces (GUIs); end-users need not have any experience with MATLAB to execute prewritten MATLAB functions. Third, MATLAB provides access to a library of common data handling methods, graphical representations, and statistical tools that can be visualized in highly flexible ways using plotting and imaging commands integrated within the MATLAB program. Such commands must often be written de novo in other programming languages. Since MATLAB is written for science and engineering applications, this library is tailored for analysis of scientific data. Finally, MATLAB is widely used throughout the biomedical research community, providing access to a strong user base for teaching, implementation, and code sharing. These advantages strongly support the use of MATLAB as the software of choice for analysis of COPAS data sets.
Herein, we describe a suite of MATLAB algorithms—COPAquant, COPAmulti, and COPAcompare—which extract, filter, normalize, graph, statistically analyze, and compare intra- and interplate values from COPAS Biosort data files acquired with the Advanced Acquisition Software Package (Union Biometrica). COPAquant analyzes data generated in the single-sample mode, whereas COPAmulti and COPAcompare analyze data obtained in the 96-well autosampling ReFLX mode. Automation of this step within the context of a high-throughput RNAi screen allowed us to rapidly move from secondary validations to hit identification. Although we have used it primarily for screens in C. elegans, the standard file format of COPAS data files, our simple GUI for multiwell plate analyses, and the freely available nature of the algorithms make it widely useful for analysis of any type of COPAS-generated data.
Materials and methods
Strains
The C. elegans strain TJ375 (hsp-16.2p::GFP) was used in this study and was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN, USA). RNAi was conducted as described (7). Worms were dispensed to wells as L1s and given 4 days to grow to adulthood at 16°C. Worms were visually screened for basal GFP fluorescence, heat-shocked at 35°C for 3 h, allowed to recover at 16°C for 3 h, and then visually screened again for wells whose RNAi treatment prevented activation of the heat-shock promoter. Clones identified as hits from the primary screen were rescreened in quadruplicate and compared with an empty vector control by quantitative analysis on the COPAS. Hits were considered verified if their normalized values were ≤60% of the empty vector.
COPAS Biosort
A COPAS Biosort with Advanced Acquisition Software Version 5.2.69 was utilized. Systems without Advanced Acquisition Software or earlier versions of the COPAS software that do not output data in 26-column format are not compatible with the software as written. Young adult animals fed either empty vector RNAi or gene-specific RNAi were sorted through the COPAS for quantification of GFP fluorescence. Worms were washed from plates with 5–10 mL deionized water, placed in the COPAS sample cup, and analyzed in the single-sample format. COPAS settings were as follows: gain ext, 1; green, 5; yellow, 1; red, 1; threshold signal, 30; TOF minimum, 1; photomultiplier tube (PMT) settings control green, 600; yellow, 0; red, 0. Worms were gated based on TOF to select for adults, and MATLAB analysis was performed specifically on this gated population. Although we prefiltered our data during screening, COPAquant allows users to filter raw data files based on gating status (gated, nongated, or all data). COPAmulti also filters based on gating status and will additionally filter on any COPAS-measured parameter [TOF, EXT, fluorescent channel 1 (Ch1), fluorescent channel 2 (Ch2), or fluorescent channel 3 (Ch3)].
MATLAB
MATLAB version 7.0.1.24704 was used in the creation of this program. MATLAB M-files for COPAquant, COPAmulti, and COPAcompare, as well as sample data files and instructional documentation are freely available through our web site (www.med.upenn.edu/lamitinalab/index.shtml).
Statistics Bar graphs indicate mean values ± sd. In COPAmulti, we implement the mean ± k sd method for hit identification by calculating the plate mean ± plate sd and then determining which wells exceed this minimum sd threshold. The median absolute deviation (MAD) test was conducted using the MAD function in the MATLAB library. Multiple comparison t-tests were conducted using the t-test function in the MATLAB library. It should be noted that user-defined P values must be corrected for multiple comparisons by dividing the selected P value by the number of samples analyzed (Bonferroni correction). A table of standard P values and Bonferroni-corrected P values for 96-well plate samples can be found in Table 1.
Table 1.
Conversion of standard P values to Bonferroni-corrected P values
| P value | Bonferroni-corrected P value (for 96 samples)a |
|---|---|
| 0.050 | 0.000521 |
| 0.025 | 0.000260 |
| 0.010 | 0.000104 |
| 0.005 | 0.000052 |
| 0.001 | 0.000010 |
To compute corrected P value for sample sizes other than 96, divide the desired P value by the number of samples being analyzed.
Results and discussion
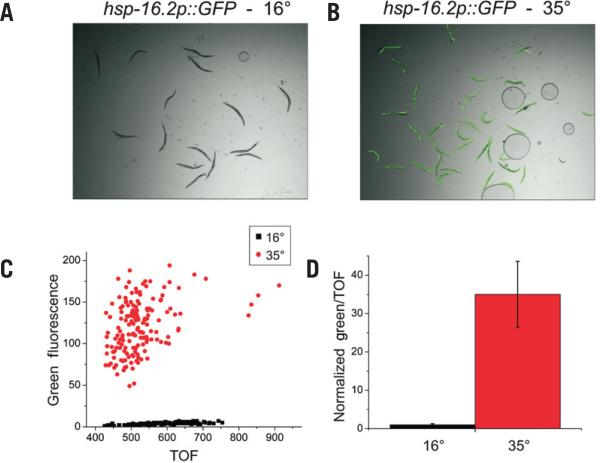
Many RNAi screens performed in C. elegans are based on the in vivo expression of GFP reporters. One such screen under investigation in our laboratory involves the temperature-dependent regulation of an hsp-16p::GFP reporter. In this strain, GFP expression within young adult hermaphrodites (TOF = 400–1000) is negligible under basal conditions (Figure 1A), but is highly induced in almost all cells after a brief heat shock and recovery period (Figure 1B) (see the “Materials and methods” section for a more detailed description of the experiment). Quantification of this induction among young adult animals revealed a wide distribution of GFP expression levels between individuals (Figure 1C), as has been previously reported (9,10). However, the population means accurately reflect the behavior of the transgene (Figure 1D). In order to identify regulators of the heat-shock response pathway in C. elegans, we conducted a genome-wide RNAi screen for suppressors and enhancers of heat shock– dependent hsp-16p::GFP expression (Morton and Lamitina, unpublished data). GFP reporter expression was initially quantified by visual inspection. During the secondary validation screen, RNAi treatments were quantified using the COPAS Biosort in the single-sample mode of screening.
Figure 1. OPAS quantification of a heat shock–inducible GFP reporter.
(A) Photomicrographs of hsp-16p::GFP at 16°C or (B) after 3 h of heat-shock at 35°C and 3 h of recovery at 16°C. (C) Values of TOF and green fluorescence were recorded for each individual adult worm using the COPAS Biosort. (D) The reporter expression in each population was summarized by mean ± sd GFP expression normalized to the TOF and displayed here as the fold-change increase of heat-shocked worms over non–heat-shocked worms. n = 149 for each.
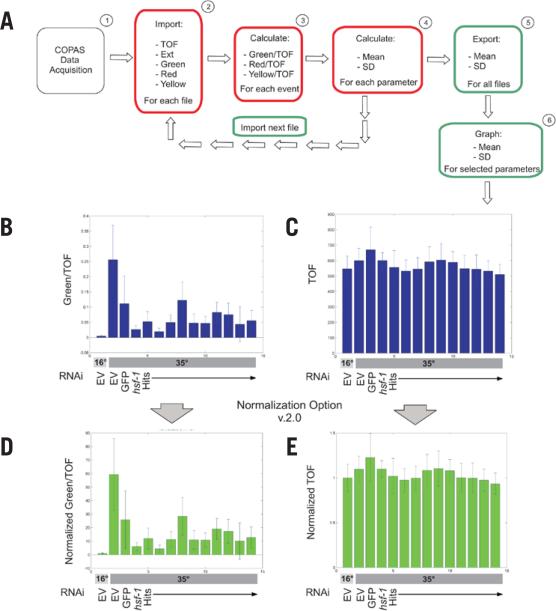
To facilitate analysis of the numerous COPAS data files generated by our RNAi screen, we wrote an algorithm, using the programming platform MATLAB, to automatically extract desired values from COPAS *.txt data files (one file per RNAi condition) (Table 2) . The COPAS exports data in a 26-column format, in which each row represents data from a single worm. The basic function of our COPAquant algorithm, COPASFun, imports numerical values from a COPAS data file. After data import, COPAquant queries the user as to whether the data to be analyzed should be filtered based on gating criteria, which are a unique combination of COPAS parameters (TOF, EXT, Ch1, Ch2, and Ch3) that are user-defined during data acquisition. COPAquant can be instructed to analyze gated data only, nongated data only, or all data. Using our hsp-16p::GFP screen data as an example, we chose to extract gated values for TOF, EXT, and fluorescence values for each of the three fluorescent channels. Because COPAS-measured GFP fluorescence is related to object size (unpublished data), COPASFun can correct for this bias by normalizing to the object TOF, which is a direct measure of object size. These ratio values (Ch1/TOF, Ch2/TOF, Ch3/TOF) are entered into new columns. The resulting columns for our values of interest (TOF; EXT;, Ch1, Ch2, and Ch3; as well as their associated ratios) are then summarized with mean and sd. In the current screen for hsp-16::GFP regulators, meaningful yellow (Ch2) and red (Ch3) data were not obtained, since this strain does not express reporters in either of these fluorescent channels. These statistics, as well as the number of events in the sample (n), are then exported to the function COPASImp (Figure 2A).
Table 2.
Analysis properties of the COPAS MATLAB analysis software
| Program | Purpose | Filtering capability | Parameters analyzed | Data normalization |
|---|---|---|---|---|
| COPAquant | Analysis of single-file data | Gating status | TOF, Ext, Ch1, Ch2, Ch3 | None |
| COPAquant V2 | Analysis of single-file data | Gating status | TOF, Ext, Ch1, Ch2, Ch3 | File 1 |
| COPAmulti | Analysis of 96-well plate data | Gating status, TOF, EXT, Ch1, Ch2, Ch3 | TOF, Ext, Ch1, Ch2, Ch3 | Plate mean |
| COPAmulti V2 | Analysis of 96-well plate data | Gating status, TOF, EXT, Ch1, Ch2, Ch3 | TOF, Ext, Ch1, Ch2, Ch3 | User-selected well(s) |
| COPAcompare | Pair-wise comparison of replicate plates | Gating status, TOF, EXT, Ch1, Ch2, Ch3 | TOF, Ext, Ch1, Ch2, Ch3 | Plate mean |
| COPAcompare V2 | Pair-wise comparison of replicate plates | Gating status, TOF, EXT, Ch1, Ch2, Ch3 | TOF, Ext, Ch1, Ch2, Ch3 | User-selected well(s) |
Figure 2. Data analysis flowchart for COPAquant analysis of single-sample mode data.
(A) Data flow is diagrammed for extraction of mean and sd of particular parameters from COPAS files. Red boxes represent tasks completed by the function COPASFun, while green boxes represent COPASImp tasks. (B) MATLAB was used to quantify fluorescence in an RNAi screen for suppressors of hsp-16p::GFP expression after heat-shock (35°C). Empty vector (EV) RNAi represents the negative control before and after heat shock. GFP and hsf-1 RNAi represent the positive controls for clones that decrease expression. HSF-1 is a transcription factor that promotes hsp-16.2 expression. Hits are RNAi clones identified as repressing reporter expression in our screen. Values were normalized to TOF. (C) TOF values for the same samples as in panel B were graphed. (D and E) COPASFun version 2.0 normalizes each event value to the mean of the 16°C EV control and returns the new means and standard deviations. Shown are the normalized graphs for the data in panels B and C. For all conditions, n ≥ 41.
The COPASImp function sends multiple COPAS *.txt files to COPASFun for analysis (Figure 2A). Once the MATLAB directory is set to the appropriate folder, COPASImp recognizes and reads all *.txt files within the folder (Figure 2A). Once all the files in the folder have been analyzed, the results are presented in a table titled Results (which is automatically saved as the tab-delimited text file Results.txt for analysis outside of MATLAB) as well as in a structure labeled ImStruc (in which each cell contains the results for one sample). Following analysis, COPASImp queries the user as to which parameter should be represented in graphical format. The user-selected parameter is then plotted and displayed (Figure 2B).
In addition to the form of normalization discussed above, COPAquant V2 will also normalize all samples to a negative control sample to produce a relative fold-change value (Table 2). The program presents data in both the raw form (Figure 2, B and C) and in various normalized forms (Figure 2, D and E), using the lowest numbered file as the negative control reference. The mean of the reference sample is calculated for each parameter, and each event within subsequent samples is divided by this value, creating a new, normalized column of values. The means of the normalized values, as well as their sd values, are exported back to COPASImp (Figure 2, D and E).
Using COPAquant, we dramatically enhanced the rate of data analysis in our screen for regulators of hsp-16p::GFP expression using the single-sample mode of COPAS screening. We were able to rapidly identify hits that affect GFP expression but not worm growth by analyzing both normalized GFP, as well as normalized TOF values (i.e., normalized to the negative control sample—empty vector RNAi in this case). Prior to implementation of COPAquant, the time required for manual analysis of a single day's worth of COPAS data obtained using the single-sample acquisition mode frequently exceeded 8 h. Using COPAquant, data from 1 day of sorting are now analyzed, normalized, and graphed within 10 s, which represents a ~3000-fold increase in data analysis efficiency.
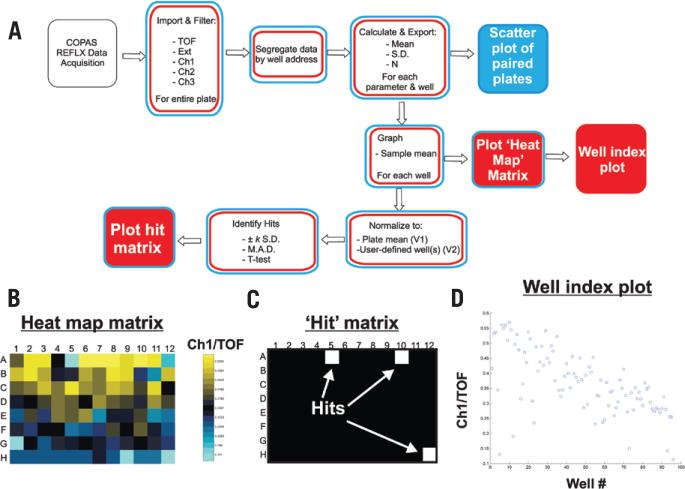
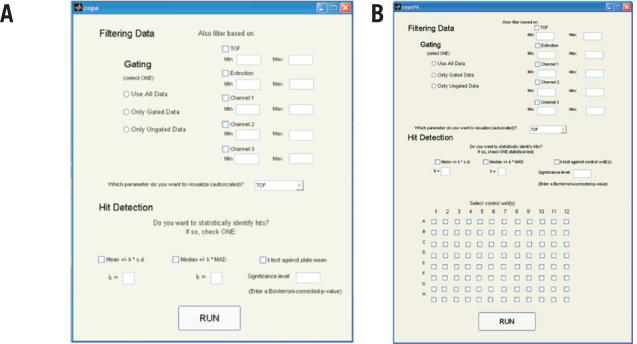
In addition to the single-sample sorting mode described above, some labs also employ an autosampling device called the ReFLX system. ReFLX-equipped COPAS systems sort and quantify events from individual wells of 96-well plates using the optional ReFLX sampler. Data from each well are stored within a single 26-column format file according to their row and column address. To make our MATLAB program applicable to ReFLX screening platforms, we modified our existing single-sample MATLAB code to read ReFLX files. The modified programs, COPAmulti and COPAcompare (Figure 3 and Table 2), read raw *.txt files generated by the ReFLX, filter and extract matrices for each well, and summarize useful parameters. Data from one or more 96-well files (COPAmulti) or a replicate pair of 96-well files (COPAcompare) are analyzed, and the data for each plate is stored in a separate cell of a Results Structure within MATLAB. For each plate analyzed, the raw data (n and well mean ± sd for each of eight different parameters for every well) are exported to a Results Structure, which can be accessed for export to other programs. To make COPAmulti as user-friendly as possible, we implemented a GUI within MATLAB that allows users to define several criteria for data analysis, including filtering cutoffs, the parameter to be utilized for analysis, and statistical criteria and thresholds used to identify hits (Figure 4A). Since these criteria can be adjusted through the GUI and the data are rapidly reanalyzed, the effects of altered filtering and statistical criteria are easily determined.
Figure 3. Data analysis flowchart for COPAmulti and COPAcompare analysis of ReFLX multiwell mode data.
(A) Data flow is diagrammed for extraction of well mean and sd of user-defined parameters from COPAS ReFLX files. Red boxes represent tasks completed by COPAmulti. Blue boxes indicate specific tasks completed by COPAcompare. Solid boxes indicate plots generated by COPAmulti or COPAcompare. (B) Heat map plot for the well means of Ch1/TOF data from a hypothetical 96-well ReFLX file. Note that the coloring is autoscaled according to the specific data for each plate. (C) Hit matrix plot indicating wells that passed a user-defined statistical threshold (in this case, MAD > 3 for Ch1/TOF). Hits are plotted in white, and non-hits are plotted in black. (D) Well index graph plotting the GUI-selected parameter for each well. If multiple 96-well plates are analyzed, all wells from all plates are plotted (i.e., plate 1, wells 1–96; plate 2, wells 97–192; plate 3, wells 193–278; etc.).
Figure 4. Graphical user interface for COPAmulti.
(A) Screen shot of the COPAmulti GUI demonstrating user-configurable parameters for multiwell plate analyses. Ch1, Ch2, and Ch3 refer to the respective fluorescence channel (green, yellow, and red on most, but not all, COPAS systems). The parameter to be analyzed is selected from the drop-down menu in the middle of the GUI. Hit identification is accomplished via selection of one statistical test and associated threshold criteria. (B) Screen shot of the COPAmulti GUI that allows users to select negative control normalization well(s).
Since ReFLX files offer unique analysis challenges and opportunities not present in single-sample data collection modes, we implemented several additional features common to high-throughput multiwell-based RNAi screening for ReFLX file analysis. First, the mean of a user-selected parameter from each well is plotted in an 8 × 12 matrix heat map that is color-coded by well value (Figure 3B). This visualization strategy is a useful way to compare the data across a plate and often helps in the identification of plate edge effects, a common confounder in high-throughput RNAi screening (11). Second, instead of normalizing to a single negative control sample (as we do for single-sample data analysis), COPAmulti takes advantage of the large number of samples and uses the plate mean (calculated from the median 80% of nonzero value samples to remove effects of outliers) as the negative control value. This approach is a well-accepted data normalization strategy for multiwell plate assays that can be uniformly applied across all plates (11). In addition to this normalization strategy, we also implemented a second approach (COPAmulti V2) that allows users to define the well(s) that contain negative control data through the COPAmulti GUI (Figure 4B). Using these calculated negative control reference values, we implement three common statistical tests for hit identification that have been previously utilized in RNAi screening formats: (i) mean ± k sd; (ii) median ± k MAD; and (iii) the multiple-comparisons t-test with Bonferroni correction. The specific significance test and threshold for each test is set within the user-adjustable GUI. Each test has specific strengths and weaknesses and in some cases may not represent the best statistical approach for data analysis. Nonetheless, these methods are among the most commonly used approaches for analysis of high-throughput RNAi screening data (11), and the best approach is usually to compare results obtained with each statistical method. In general, the mean ± k sd test is the most commonly used hit identification technique for RNAi screening, due to its ease of calculation (12,13). Most screeners utilize a 3-sd cutoff with this approach. However this method is sensitive to outlier data and frequently misses weaker positives. Decreasing the sd cutoff usually increases false positives to an unacceptably high rate. An alternative approach is the median ± k MAD test. Like the mean ± k sd test, MAD is relatively easy to calculate but is much less sensitive to outlier data. MAD also does a good job of identifying weak hits while controlling false positives (14). A shortcoming of MAD is that it is not easily linked to probability distributions and P values. Despite this shortcoming, others have recommended MAD as the method-of-choice for hit selection in high-throughput RNAi screens (14). MAD values of ≥2 are commonly used for hit identification in genome-wide RNAi screens (14). A final common statistical test for RNAi screening is the multiple-comparison t-test. This statistic is easy to calculate (due to the large number of events in each well), but is extremely sensitive to outliers and requires multiple-comparison correction (11). For multiple comparison t-tests, the simplest form of correction is the Bonferroni correction, which scales the desired P value by the number of samples to obtain an equivalent multiple comparison P value. A table of Bonferroni-corrected P values for common thresholds is listed in Table 1. In general, users should analyze their data with each statistical approach and utilize the method or combination of methods that most frequently identifies known positive controls. A major advantage of our software is that it allows users to rapidly adjust and test each of these statistical methods for hit identification through the simple GUI. For users that wish to perform statistical analysis of their data using other approaches, COPAmulti automatically exports both summarized and raw data to delimited text files for further analysis.
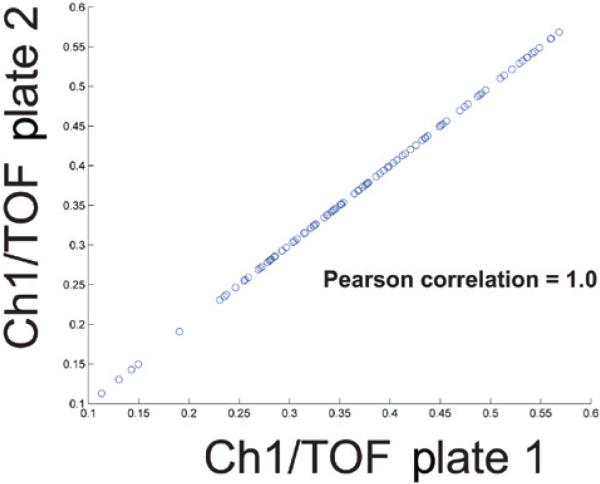
Following statistical analysis, hits meeting user-determined thresholds are binarized in an 8 × 12 matrix, with hits plotted in white and non-hits plotted in black (Figure 3C). We also visualize all data from all plates using a well index plot (Figure 3D). Such plots are useful indicators of screen phenotypic behavior among plates and can help identify plates with phenotypic drift or substantial variance. For example, data in Figure 3 demonstrate lower values toward the end of the plate as compared with the beginning of the plate. Finally, since some users may screen in duplicate, we implemented a separate algorithm, COPAcompare, that allows users to compare results between two plates (Figure 5). COPAcompare plots a user-selected parameter for each well between two user-selected plates. The degree of overall plate-to-plate correlation is determined by calculating the Pearson correlation coefficient (R), where an R value of 1 equals perfect correlation among all wells and -1 equals perfect opposite correlation among all wells.
Figure 5. Two plate comparison using COPAcompare.
Screen shot of the results from a hypothetical COPA-compare two plate comparison. Two identical hypothetical ReFLX files were compared with one another, resulting in a calculated Pearson correlation coefficient of 1. The calculated Pearson Correlation is displayed within the MATLAB command console, as illustrated in our online tutorials (www.med.upenn.edu/lamitinalab/downloads.shtml). Each point on the graph represents a single well, with the x-coordinate representing data from plate 1 and the y-coordinate representing data from plate 2. Overall correlation was determined using the Pearson Correlation function within the MATLAB library.
We developed a suite of MATLAB-based programs to process large COPAS file data sets such as those associated with C. elegans RNAi screens. We implemented one program, COPAquant, for comparisons among data collected in the single-sample format, which is useful for small-scale screens with larger populations. We also implemented two additional programs, COPAmulti and COPAcompare, that use more advanced filtering, analysis, normalization, and statistical analysis of data from 96-well plates obtained using the COPAS ReFLX system. Both programs allow users to rapidly move from raw COPAS data to graphical data representation, replicate plate comparison, and hit identification without extensive knowledge of or experience with the programming environment. Our software greatly simplifies the analysis of COPAS data and fills a major gap in our need for data analysis tools for high-throughput screening using this platform. While we used this program in the validation steps of an RNAi screen for regulators of a heat shock–inducible reporter in C. elegans, the program is customized to the standard data format output by COPAS Biosort instruments and thus can be used in any type of COPAS application, including data obtained from other organisms.
Acknowledgments
This paper was supported by a grant from the National Institutes of Health (NIH; grant no. 1R01AA017580 to T.L.) and an NIH training grant in Cell and Molecular Biology [no. T32 GM-07229 to E.M. (principle investigators Richard Schultz and Marisa Bartolomei)]. The authors wish to thank Hernan Garcia and Michael Springer for helpful training in MATLAB-based analysis, Weon Bae and Rock Pulak for providing sample ReFLX data, and the suggestions of four anonymous reviewers that greatly improved the manuscript. This paper is subject to the NIH Public Access Policy.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Smith MV, Boyd WA, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. A discrete time model for the analysis of medium-throughput C. elegans growth data. PLoS One. 2009;4:e7018. doi: 10.1371/journal.pone.0007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd WA, Smith MV, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. Application of a mathematical model to describe the effects of chlorpyrifos on Caenorhabditis elegans development. PLoS One. 2009;4:e7024. doi: 10.1371/journal.pone.0007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd WA, Smith MV, Kissling GE, Freedman JH. Medium- and high-throughput screening of neurotoxicants using C. elegans. Neurotoxicol. Teratol. 2009;32:68–73. doi: 10.1016/j.ntt.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprando RL, Olejnik N, Cinar HN, Ferguson M. A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a Complex Object Parametric Analyzer and Sorter, and axenic liquid media. Food Chem. Toxicol. 2009;47:722–728. doi: 10.1016/j.fct.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Doitsidou M, Flames N, Lee AC, Boyanov A, Hobert O. Automated screening for mutants affecting dopaminergic-neuron specification in C. elegans. Nat. Methods. 2008;5:869–872. doi: 10.1038/nmeth.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns AR, Kwok TCY, Howard A, Houston E, Johanson K, Chan A, Cutler SR, McCourt P, Roy PJ. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat. Protocols. 2006;1:1906–1914. doi: 10.1038/nprot.2006.283. [DOI] [PubMed] [Google Scholar]
- 7.Lamitina T, Huang CG, Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc. Natl. Acad. Sci. USA. 2006;103:12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, Ewbank JJ. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008;4:e1000105. doi: 10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–242. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 13.DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 14.Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J. Biomol. Screen. 2008;13:149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]