Abstract
P68 (DDX5) and p72 (DDX17) are members of the DEAD-box RNA helicase family. They can unwind double-stranded RNA and also contribute to the remodeling of ribonucleoprotein complexes. These activities of p68/p72 are required for efficient RNA splicing and microRNA processing. In addition, p68/p72 perform functions that are independent of their enzymatic activity. This is especially common to their role in gene regulation, where p68/p72 coactivate various transcription factors, including the tumor suppressor p53, estrogen receptor α and β-catenin. P68/p72 are posttranslationally modified by SUMO attachment and phosphorylation that regulate their coactivation potential, binding to known interactants or protein stability. Knock-out mouse models revealed that both DDX5 and DDX17 are essential genes during development. Furthermore, together with their ability to stimulate cell proliferation and prevent apoptosis, the reported overexpression of p68/p72 in three of the major human cancers (colon, breast, prostate) strongly suggests that p68/p72 promote tumorigenesis and might even represent proto-oncoproteins. If so, their inhibition holds promise as a novel way to contain or cure various carcinomas
Keywords: Cancer, DDX5, DDX17, DEAD-box, p68 RNA helicase, p72 RNA helicase
Introduction
RNA, for instance in various viruses, can exist in double-stranded form, but single-stranded RNA is also able to form double-stranded regions by engaging in intra- and intermolecular interactions. Examples are the secondary structures of tRNAs or rRNAs within ribosomes. Also, RNA base-pairing is important during transcript splicing and RNA interference, indicating that unwinding as well as forming of double-stranded RNA is involved in numerous cellular processes. RNA helicases support, or are even indispensable for unwinding RNA [1]. Moreover, RNA helicases are capable of disrupting RNA-protein interactions and are thus crucial for the remodeling of many ribonucleoproteins [2, 3].
The largest family of RNA helicases, comprising 38 members in humans, are the DEAD-box (DDX) proteins, which are named after a conserved signature amino acid sequence (Asp-Glu-Ala-Asp, or D-E-A-D). DDX proteins hydrolyze ATP, which is often stimulated by the presence of double- or single-stranded RNA. However, only in a minority of cases has it been demonstrated that a DDX protein is a true RNA helicase [4]. Furthermore, the yeast Dbp9p DEAD-box protein was reported to exhibit DNA helicase activity, suggesting that DDX proteins may not always exclusively act on RNA [5]. This review will focus on two paralogous DEAD-box RNA helicases, p68 (DDX5) and p72 (DDX17), highlighting their function in normal cells and their potential role as tumor promoters.
Identification, structure and expression of p68/ p72 RNA helicases
Three decades ago, an antibody directed against the simian virus 40 large T oncoprotein was shown to cross-react with a 68 kDa cellular protein that resides within the nucleus [6]. This antibody was then utilized to screen a cDNA expression library and the respective DDX5 gene cloned [7]. Sequence analysis revealed homology to the eukaryotic translation initiation factor eIF4A, the first protein identified to unwind RNA in an ATP-dependent manner. Thus, it was no surprise that the p68 protein was subsequently shown to be an RNA-dependent ATPase and a helicase capable of unwinding RNA [8, 9]. Thereafter, the DDX17 gene was cloned and shown to encode for two proteins, p72 and p82 RNA helicase. The larger isoform is due to alternative translation initiating at a non-AUG start codon preceding the AUG codon that defines the translation start site for p72 RNA helicase in the DDX17 mRNA [10, 11]. Western blotting of breast and colon cancer cell lines indicated that p72 and p82 are generally expressed at similar levels [12-14]. Biochemical studies suggest that p72 and p82 have nearly identical properties [11], but this does not exclude that their physiological functions could differ. Moreover, p72/p82 and p68 RNA helicase can form both homo- as well as heterodimers [15], implicating that these paralogous proteins have overlapping functions.
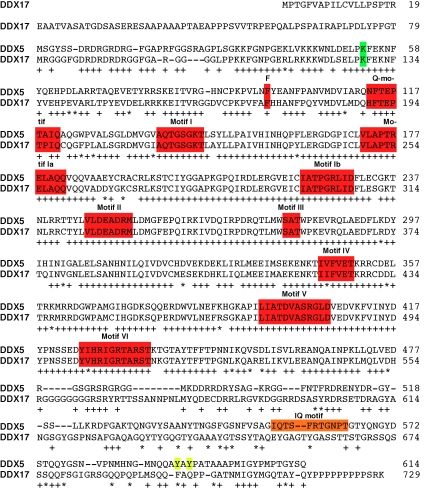
Amino acid alignment exposes that 439 (71.5%) out of the 614 amino acids within p68 RNA helicase are identical and 478 (77.9%) similar to those in p72/p82 (Figure 1). This homology is even higher within the helicase domain consisting of p68 amino acids 96-436: 298 (87.4%) amino acids are identical, and 313 (91.8%) are similar to the DDX17 gene products. In contrast, the N-terminus (amino acids 1-95 of p68) and C-terminus (amino acids 437-614 of p68) are only 71.6% and 54.5% similar to the respective domains in p72/p82, suggesting that DDX5 and DDX17 gene products may perform distinct functions.
Figure 1.
Protein sequence alignment of p68 RNA helicase (DDX5) with DDX17. The 729 amino acid long DDX17 protein corresponds to p82 RNA helicase, whereas DDX17 amino acids 80-729 represent p72 RNA helicase. DDX5 amino acids were derived from NCBI reference sequence NP_004387.1 and DDX17 amino acids from NP_006377.2. Identical amino acids are highlighted by “+” and similar ones by “*”. Nine motifs characteristic for DEAD-box RNA helicases are boxed in red, as is the conserved phenylalanine preceding the Q-motif. Motifs I and II are also known as Walker A and B motifs, respectively. The center of motif II consists of the amino acid sequence Asp-Glu -Ala-Asp, after which the DEAD-box family of RNA helicases is named. Sumoylation and tyrosine phosphorylation sites are highlighted in green and yellow, respectively, and the IQ motif in orange.
Like other RNA helicases, p68 and p72/p82 possess eight conserved motifs (see Figure 1) within their catalytic core [1]. In addition, a Q-motif that is specific for DEAD-box RNA helicases is present at the N-terminus of the catalytic core and is preceded by an aromatic amino acid located 17 residues upstream of the Q-motif. This conserved aromatic amino acid and the Q-motif are regarded as an adenine recognition motif, which is important for binding of RNA substrates, ATP hydrolysis and helicase activity [16, 17]. Motifs I and II (also called Walker A and B motif, respectively) are found in many NTPases and bind ATP as well as associated Mg2+. These domains are indispensable for ATP hydrolysis and thus also helicase activity. Further, motifs III and VI are thought to interact with the γ-phosphate of ATP and couple ATP hydrolysis with helicase activity, whereas motifs Ia, Ib, IV and V are engaged in binding of the RNA substrate [18].
Originally, both p68 and p72 RNA helicase were described as nuclear proteins [6, 9, 10]. However, more detailed biochemical and immunohistochemical analyses revealed that significant amounts of p68 RNA helicase are also present in the cytoplasm of various cell lines, whereas little, if any, p72 is localized within the cytoplasm [13, 19, 20]. Recently, it was found that p68 is indeed shuttling between the cell nucleus and the cytoplasm, which involves two nuclear localization and two nuclear export signals; however, it remains unknown how this shuttling is regulated [21]. Moreover, the intranuclear distribution of p68 is cell cycle-dependent: whereas p68 is excluded from nucleoli during interphase, it appears in nascent nucleoli during telophase [22, 23]. Again, it is unknown how this is regulated and if this bears any physiological relevance. Furthermore, mass spectrometry of purified HeLa nucleoli suggests that p72 is a nucleolar protein similar to p68 RNA helicase, but it was not studied if this is limited to nucleoli in telophase [24].
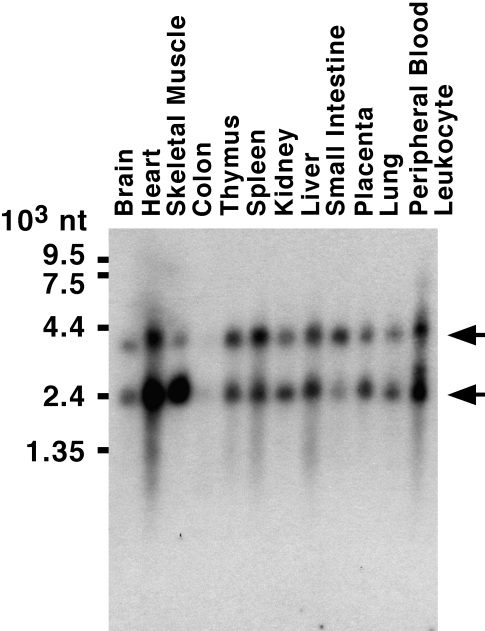
Expression of DDX5 or DDX17 mRNA appears to be ubiquitous, but expression levels vary significantly between different organs [10, 25] (see also Figure 2). Northern blotting reveals two major species of DDX5 mRNA (∼2.4 and ∼4.2 kb), with the longer transcript most likely representing an incompletely spliced mRNA, whose translation is predicted to result in a truncated DDX5 gene product [25]. As such a truncated DDX5 protein has not been observed by Western blotting, it is likely that either this truncated protein is very labile or that the 4.2 kb DDX5 mRNA is inefficiently translated. Similarly, at least two major mRNAs (5.3 and 9.3 kb) are derived from the DDX17 gene [10], indicating that differentially spliced DDX17 mRNAs exist that may give rise to several protein isoforms.
Figure 2.
Human multiple tissue Northern blot containing equal amounts of poly(A)+-RNA in each lane was hybridized with a human DDX5 cDNA probe. Arrows point to major DDX5 mRNAs of 4.2 and 2.4 kb size.
Essential roles in development
The yeast ortholog of p68/p72 is Dbp2p. Although originally reported to be essential for viability, a later study demonstrated that inactivation of the DBP2 gene was not lethal, but rather vastly reduced cell growth and resulted in a cold-sensitive phenotype [22, 26]. Regardless, this was the first hint that p68/p72 RNA helicases perform crucial functions. Moreover, mutations within the Drosophila p68 RNA helicase gene showed phenotypes ranging from sterility to lethality [27], indicating that p68 RNA helicase is essential during development. Indeed, both DDX5 and DDX17 are expressed in chick, frog, mouse and rat embryos [28-30], and individually knocking out DDX5 or DDX17 resulted in lethality in mice [31]. However, there was a distinction between DDX5 and DDX17: whereas DDX5 knockout embryos died at embryonal day 11.5, DDX17 knockout mice survived for up to two days after birth. No gross changes in organogenesis were observed, but both knockout mice displayed malformation of blood vessels.
Analysis of DDX17 -/- mouse embryonic fibroblasts revealed a drastic reduction in cell growth and enhanced apoptosis, and similarly p68 downregulation by RNA interference reduced cell growth and survival [31]. Further, joint downregulation of DDX5 and DDX17 in RKO colon cancer or HeLa cervical carcinoma cells suppressed cell proliferation [13, 32], and likewise knock-down of DDX17, but not DDX5, in MCF-7 breast cancer cells curtailed estrogen-dependent growth [33]. Altogether, these results demonstrate that DDX5 and DDX17 are both essential for development and may have profound effects on cell growth and survival.
Unwinding RNA
P68 and p72 RNA helicase bind to double- and single-stranded RNA, albeit the affinity for the former appears to be much higher. Furthermore, binding to RNA stimulates their ATPase activity, which provides the energy to unwind RNA duplexes in both the 3’→5’ and 5’→3’ direction [34, 35]. In addition, p68 and p72 possess an RNA annealing activity, which, together with their helicase activity, catalyzes the rearrangement of secondary structures in RNA [36]. It may be via such an action that p68/p72 RNA helicases contribute to ribosome biogenesis that requires extensive rRNA remodeling to form mature ribosomes [32], and consistently the yeast ortholog of p68/p72 has been shown to be associated with preribosomal complexes [37].
RNA splicing involves the association and dissociation of the pre-mRNA with snRNAs, which may be facilitated by RNA helicases. Indeed, mass spectrometric analysis of the spliceosome identified p68 RNA helicase to be an associated protein [38]. Thereafter, it was demonstrated that both p68 and p72 specifically interact with the U1 small nuclear ribonucleoprotein particle that recognizes the 5’ splice site [35, 39, 40]. In fact, p68 molecules devoid of ATPase or RNA helicase activity inhibited the dissociation of U1 from the 5’ splice site, and accordingly down-regulation of DDX5 resulted in the accumulation of unspliced RNA [41]. Moreover, p68 and p72 RNA helicase have been shown to affect alternative splicing [42, 43]. Altogether, these results suggest that p68 and p72 RNA helicase are crucial factors required for efficient RNA splicing.
Another function of p68 and p72/p82 relates to the processing of microRNA (miRNA). The primary miRNA transcript (pri-miRNA) is processed in the cell nucleus by the Drosha complex into pre-miRNA, a hairpin intermediate. Mass spectrometry of the Drosha complex revealed that p68 and p72 are part of this complex [44]. Moreover, knock-down of either DDX5 or DDX17 resulted in impaired processing of selected primiRNAs as well as of the 5.8S rRNA precursor [31]. The fact that ATPase activity is required for p72 to potentiate pri-miRNA processing suggests that p68/p72 unwind RNA to make it accessible for cleavage in the Drosha complex [31]. Another function of p68/p72 in the Drosha complex is based upon their ability to bind Smad proteins and the tumor suppressor p53 [45, 46]. When recruited into the Drosha complex by association with p68 RNA helicase, Smad proteins as well as p53 stimulate the maturation of selected miRNAs [47, 48]. Finally, it has been shown that p68 RNA helicase unwinds the let-7 miRNA duplex in vitro [49]. This suggests that p68 RNA helicase may help loading miRNAs into the silencing complex. In fact, this may not be limited to miRNAs, but also extend to siRNAs [50].
Many other processes involving RNA may require helicase activity and thus p68/p72 RNA helicases. For instance, the yeast and Drosophila orthologs of p68/p72 affect nonsense-mediated mRNA decay and RNA release from chromatin, respectively [27, 51]. Or the zinc-finger antiviral protein, which binds to viral mRNAs and targets them for degradation, interacts with p72 RNA helicase, whose enzymatic activity is thought to restructure viral RNA and thereby makes it more amenable for degradation [52]. On the other hand, cellular p68 is utilized by the hepatitis C virus RNA-dependent RNA polymerase to facilitate the production of negative-strand from positive-strand viral RNA. Of note, the RNA-dependent RNA polymerase binds to p68 RNA helicase and thereby leads to its relocalization from the nucleus into the cytoplasm [53]. Thus, it is likely that also other RNA viruses highjack p68/p72 to promote their replication. And indeed, the influenza virus RNA polymerase complex and the SARS (severe acute respiratory syndrome) coronavirus helicase have been shown to bind to p68 RNA helicase, and DDX5 downregulation led to impaired SARS coronavirus replication [54, 55].
Roles in gene transcription
The transcription of chromosomal genes can be roughly divided into three parts: initiation, elongation and termination. At present, it is unclear whether p68/p72 participate in the latter processes, but ample of evidence suggests that they are involved in the initiation of gene transcription. Consistently, p68/p72 interact with RNA polymerase II and ubiquitous transcriptional cofactors such as CBP, p300 and PCAF [12, 19]. CBP and p300 are two homologous acetyl-transferases that acetylate lysine residues in histones and many transcription factors [56, 57], and PCAF is another acetyltransferase that forms complexes with CBP/p300 [58, 59]. Interestingly, p68/p72 also bind to histone deacetylase (HDAC) 1, 2 and 3 [14, 60] that can antagonize the action of CBP/p300 and PCAF. Which of these interactions prevails (acetyltransferases versus HDACs) may determine whether p68/p72 activate or repress gene transcription.
The first time that p68/p72 were shown to function as transcriptional coactivators was in estrogen-dependent transcription. P68/p72 bind directly to the estrogen receptor α (ERα) [61, 62] and are one of the earliest proteins being recruited to the ERα-regulated pS2 gene promoter after estrogen stimulation [63]. Notably, p68/p72 molecules mutated in their helicase domain were as able to stimulate estrogen-dependent transcription as respective wildtype molecules [33, 61], indicating that p68/p72 do not act as RNA helicases in stimulating ERα-dependent transcription. Similarly, p68/p72 appear to bind to the androgen receptor, but only p68 RNA helicase is capable of stimulating androgen-dependent transcription, which again did not require its catalytic activity [64, 65]. Also, when functioning as a coactivator for the p53 tumor suppressor, Runx2 or MyoD, RNA helicase activity of p68/p72 was not essential [12, 46, 66, 67].
In contrast, RNA helicase activity is required for p68/p72 to activate cyclin D1 transcription [68]. In this case, p68/p72 mediate their effect through binding to β-catenin [13, 20]. Interestingly, there seem to be two modes of how these proteins affect β-catenin: (i) p68, upon platelet-derived growth factor (PDGF) stimulated phosphorylation on Y593, facilitates the translocation of β-catenin from the cytoplasm into the nucleus [20], although this finding is controversial [69]. Notably, p72 is not predicted to act in this way, since it lacks a phosphorylation site that is homologous to Y593 (see Figure 1). (ii) p68/p72 function as transcriptional coactivators of β-catenin in the cell nucleus, and consistently interact with the gene promoters of the β-catenin targets, cyclin D1 and c-Myc [13, 68].
The steroid receptor coactivator (SRA) gene encodes for several alternatively spliced mRNAs, which give rise to two different gene products: a non-coding RNA molecule and the SRA protein, SRAP [70-72]. While it is still unresolved which functions SRAP exerts, it is evident that the SRA RNA is a coactivator of steroid hormone receptors [70]. Moreover, p68/p72 bind to the SRA RNA molecule as well as the p160 family of steroid hormone coactivators, thereby enhancing ERα- dependent gene transcription [62]. In addition, p68/p72 cooperate with the SRA RNA in stimulating the transcription factor MyoD [67]. Since no helicase activity of p68/p72 is required for ERα and MyoD coactivation, p68/ p72 are not thought to remodel the exquisite secondary structure of the SRA RNA that is important for its function [73]. Rather, p68/p72 may help to recruit the SRA RNA into transcriptional complexes by virtue of their RNA binding activity. It remains to be determined whether SRA recruitment is a general mechanism of how p68/p72 stimulate gene transcription or if this is limited to ERα- and MyoD-dependent transcription.
Regulation by posttranslational modification
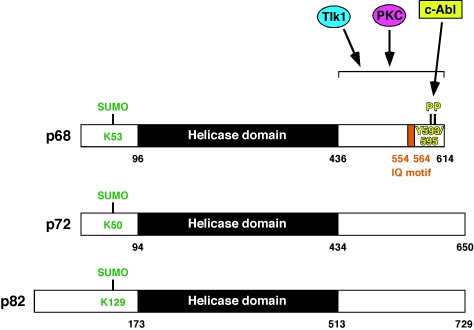
The first demonstration that p68 RNA helicase can be posttranslationally modified was its in vitro phosphorylation by PKC, a protein kinase that stably binds to p68 RNA helicase [74, 75]. Although the phosphorylation site was not mapped, it is likely to occur within the IQ motif (see Figures 1 and 3), a sequence that is involved in calmodulin binding, has a (I/L/V) QXXXRXXXX(R/K) consensus and often contains a PKC or PKA site [76]. Notably, the IQ motif found in p68 is absent in p72/p82 (see Figure 1). It was shown that both PKC-mediated phos-phorlyation and calmodulin binding of p68 RNA helicase inhibit its ATPase activity [74]. Moreover, PKC inhibits the ability of the p68 C-terminal region to bind to single-stranded RNA [77]. A similar phenomenon was observed upon phosphorylation by the serine/threonine kinase Tlk1 [78], suggesting that PKC and Tlk1 may target the same phosphorylation site(s) in p68 RNA helicase.
Figure 3.
Reported sumoylation and phosphorylation sites in p68, p72 or p82 RNA helicase
PDGF as well as tumor necrosis factor-α induce the phosphorylation of p68 on tyrosine [79, 80]. Furthermore, tyrosine phosphorylation of p68 appears to be prominent in cancer cell lines, but not in non-tumorigenic ones, suggesting that tyrosine phosphorylation of p68 may be a marker for tumor cells [80]. However, a systematic analysis of human normal and tumor tissue specimens is needed to validate this assumption. Subsequently, it was shown that PDGF stimulation of colon cancer cells leads to the activation of c-Abl, a non-receptor tyrosine kinase and proto-oncogene [81], which then phosphorylates p68 RNA helicase on Y593. This phosphorylation was reported to be required for efficient interaction with β-catenin, its translocation from the cytoplasm into the cell nucleus and the induction of epithelial-mesenchymal transition [20]. Interestingly, Y593 phosphorylation is also needed for p68 RNA helicase to coactivate androgen-dependent transcription in LNCaP prostate cancer cells [64]. In T98G glioblastoma cells, an autocrine PDGF-loop results into additional phosphorylation of p68 on Y595, and phosphorylation on both Y593 and Y595 confers resistance to TRAIL-induced apoptosis [82]. Figure 1 shows that p72/p82 do not possess any tyrosine residues that are homologous to Y593 and Y595 in p68 RNA helicase, implicating that p68, but not p72/p82, is a downstream effector of PDGF or tumor necrosis factor-α. Finally, p38 MAP kinase phosphorylates p68 RNA helicase, but neither the site(s) of phosphorylation nor the biological significance of this phosphorylation event is known [79].
A second type of posttranslational modification is ubiquitylation. Both p68 and p72 are ubiquitylated in vivo, but whereas p68 shows a high degree of poly-ubiquitylation, p72 is predominantly mono-ubiquitylated [14, 83]. Possibly because of this difference in poly-ubiquitylation that normally targets proteins for degradation, the half-life of p72 is much longer compared to p68 RNA helicase (68 h versus 20 h in 293T cells; ref. [14]). However, up to now the lysine residue(s) becoming ubiquitylated have not been uncovered.
In contrast, one conserved lysine residue in the N-terminus of p68 and p72 RNA helicase (K53 and K50, respectively; see Figures 1 and 3) has been identified to be a target for sumoylation [14, 84]. This conserved lysine residue conforms to the consensus sequence for sumoylation ΨKXE, where Ψ is a large aliphatic amino acid [85]. Sumoylation of p68/p72 has a variety of consequences: (i) It doubles protein stability of p68, whereas the protein half-life of p72 is only slightly increased upon sumoylation. (ii) It enhances the interaction of p68 and p72 with HDAC1, but has no effect on their interaction with HDAC2, HDAC3, p300 or ERα. (iii) It affects the coactivation potential of p68/p72 [14, 84]. However, the impact of sumoylation on the coactivation potential differs dependent on the promoter studied for p68: whereas its K53R mutant was slightly more potent to activate an artificial p53-responsive promoter, it was as potent as wild-type p68 in stimulating p53-dependent MDM2 transcription and drastically less able to stimulate ERα-dependent transcription. In contrast, mutation of the p72 sumoylation site resulted in enhanced ERα-dependent transcription as well as p53-mediated MDM2 upregulation, clearly showing that sumoylation affects p68 and p72 differently. Increased recruitment of HDAC1 to gene promoters normally leads to a reduction of gene transcription. Accordingly, this could explain why p72-K50R, which binds less efficiently to HDAC1 than wild-type p72, is transcriptionally more potent than wild-type p72. And similarly, this provides a plausible explanation why p68-K53R activates an artificial p53-responsive promoter more than wild-type p68, but it does not rationalize why p68-K53R coactivates ERα less efficiently than wild-type p68. However, it has not been resolved if changed HDAC1-interaction is indeed an underlying cause for the differences in the transactivation potential of sumoylated versus non-sumoylated p68/p72.
The cancer connection
It is now well established that p68/p72 promote cell proliferation and survival [13, 31-33, 68, 82, 86]. In addition, DDX5 overexpression protects lung carcinoma cells from the topoisomerase-1 poison, camptothecin, which is often employed in cancer therapy [87], and p68 RNA helicase facilitates epithelial-mesenchymal transition [20], a process that is not only important during embryogenesis but also for invasion and metastasis of tumors [88]. Moreover, the DDX5 gene passed three litmus tests for a proto-oncogene: its ectopic expression in NIH3T3 and NC3H10 fibroblasts resulted in the formation of foci on top of a monolayer of cells, caused anchorage-independent growth in soft agar, and induced tumor formation in nude mice [89]. Provided that these latter experiments can be independently confirmed, these data altogether point strongly at a tumor promoting activity of p68 and p72.
Consistent with a possible role as protooncoproteins, both p68 and p72 were found to be overexpressed in colorectal tumors [13, 83]. Immunohistochemical staining clearly showed that p68/p72 expression increased with the progression of the disease from hyperplastic polyps to adenomas and adenocarcinomas, the latter ones overexpressing p68/p72 in ≥90% of all cases. How could this overexpression contribute to colorectal tumor formation? A key event during colon carcinogenesis is the aberrant activation of β-catenin [90, 91], and both p68 and p72 RNA helicase bind to β-catenin and stimulate β-catenin-dependent transcription of target genes such as c-Myc, cyclin D1 and c-jun. Furthermore, downregulation of DDX5/DDX17 in colorectal cancer cells reduced proliferation and tumor formation in nude mice [13, 20, 68]. Collectively, these data suggest that p68/p72 overexpression might be an underlying cause of colon cancer formation by augmenting β-catenin.
Similarly, p68 and p72 were reported to be overexpressed in 30-58% or 72-76% of breast tumors, respectively [14, 33]. Approximately 70% of human breast tumors are ERα-positive and pharmacological inhibition of ERα is one mainstay of therapy [92, 93]. Since both p68 and p72 are coactivators of ERα, their overex-pression may enhance the oncogenic activities of ERα, providing a plausible mechanism how p68/p72 contribute to breast tumorigenesis. Similarly, p68 was found to be overexpressed in prostate tumors and is capable of coactivating androgen receptor [64], the key villain in prostate tumorigenesis [94], and could thereby facilitate prostate tumorigenesis.
One open question is how do p68/p72 become overexpressed in tumors? Recent analyses suggest that DDX5 mRNA is overexpressed in ovarian cancer or multiple myelomas [95, 96], but this seems not to be true for colon carcinomas [83]. Thus, DDX5 and DDX17 gene expression may be upregulated at the transcriptional level in some tumors, but at the posttranscriptional level in others. Notably, sumoylation of p68 or p72 strongly or moderately, respectively, stabilizes these proteins [14]. Further, SUMO1, the SUMO conjugating enzyme Ubc9 and the SUMO ligase PIAS3 appear to be upregulated in breast cancer cells, whereas the SUMO protease SENP6 is downregulated [14, 97], indicating that posttranslational modification by SUMO is generally enhanced in breast tumors. This could, at least in part, explain why p68 and p72 become overexpressed in these tumors. However, more research is needed to elucidate the mechanisms of p68/p72 overexpression in various tumors. As a caveat, not all tumors will show enhanced p68/p72 levels, one example being the apparent downregulation of DDX17 mRNA in meibomian cell carcinoma [98]. And finally, genetic studies are required to prove that p68/p72 are indeed tumor promoters or even true oncoproteins. To this end, transgenic or conditional knockout mouse models should be established and analyzed whether overexpression enhances and lack of p68/p72 suppresses spontaneous as well as chemically or genetically induced tumorigenesis.
Conclusion
The RNA helicases p68 and p72 perform a plethora of cellular functions and are essential for development. In most cases, these two proteins require enzymatic activity to perform their functions and thus behave as true helicases. But their role in transcription is often independent of their RNA helicase activity, suggesting that p68/p72 function similar to other transcriptional cofactors that recruit further proteins to gene promoters or establish the contact with the RNA polymerase II holoenzyme. Many lines of evidence strongly suggest, but do not yet prove, that p68/p72 overexpression promotes tumorigenesis. Notably, p68/p72 may also be involved in other diseases, including obesity [99-101], development of brain defects in Down syndrome [102] or hepatic fibrosis [103, 104].
The facts that p68/p72 are overexpressed in major carcinomas (breast, prostate, colon) and are enzymes makes them attractive drug targets. Small molecules could be developed that fit into their catalytic center and block ATPase and/or helicase activity. If the structures of p68/p72 would be known, rational drug design could be of great help to develop such inhibitors. Unfortunately, the structures of p68/p72 can currently only be modeled from known structures of other RNA helicases [18], although attempts are underway to elucidate the structure of p68 RNA helicase [105]. Finally, if the main oncogenic functions of p68/p72 are independent of their enzymatic activity, other means than blocking the catalytic center of p68/p72 must be found to suppress their harmful action in tumors.
Acknowledgments
Research on RNA helicases in the author’s laboratory was supported by a grant from the Department of Defense Breast Cancer Research Program (W81XWH-06-1-0492).
References
- 1.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 3.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 4.Linder P. Dead-box proteins: a family affair– active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuma T, Ohtsu M, Utsugi T, Koga S, Okuhara K, Eki T, Fujimori F, Murakami Y. Dbp9p, a member of the DEAD box protein family, exhibits DNA helicase activity. J Biol Chem. 2004;279:20692–20698. doi: 10.1074/jbc.M400231200. [DOI] [PubMed] [Google Scholar]
- 6.Lane DP, Hoeffler WK. SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature. 1980;288:167–170. doi: 10.1038/288167a0. [DOI] [PubMed] [Google Scholar]
- 7.Ford MJ, Anton IA, Lane DP. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature. 1988;332:736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- 8.Hirling H, Scheffner M, Restle T, Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 9.Iggo RD, Lane DP. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamm GM, Nicol SM, Fuller-Pace FV, Lamond AI. p72: a human nuclear DEAD box protein highly related to p68. Nucleic Acids Res. 1996;24:3739–3747. doi: 10.1093/nar/24.19.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlmann-Schiffler H, Rössler OG, Stahl H. The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J Biol Chem. 2002;277:1066–1075. doi: 10.1074/jbc.M107535200. [DOI] [PubMed] [Google Scholar]
- 12.Shin S, Janknecht R. Concerted activation of the Mdm2 promoter by p72 RNA helicase and the coactivators p300 and P/CAF. J Cell Biochem. 2007;101:1252–1265. doi: 10.1002/jcb.21250. [DOI] [PubMed] [Google Scholar]
- 13.Shin S, Rossow KL, Grande JP, Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007;67:7572–7578. doi: 10.1158/0008-5472.CAN-06-4652. [DOI] [PubMed] [Google Scholar]
- 14.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 15.Ogilvie VC, Wilson BJ, Nicol SM, Morrice NA, Saunders LR, Barber GN, Fuller-Pace FV. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 2003;31:1470–1480. doi: 10.1093/nar/gkg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 17.Cordin O, Tanner NK, Doere M, Linder P, Banro-ques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Rossow KL, Janknecht R. Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene. 2003;22:151–156. doi: 10.1038/sj.onc.1206067. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Lin C, Liu ZR. P68 RNA Helicase Mediates PDGF-Induced Epithelial Mesenchymal Transition by Displacing Axin from beta-Catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Gao X, Huang Y, Yang J, Liu ZR. P68 RNA helicase is a nucleocytoplasmic shuttling protein. Cell Res. 2009;19:1388–1400. doi: 10.1038/cr.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iggo RD, Jamieson DJ, MacNeill SA, Southgate J, McPheat J, Lane DP. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol Cell Biol. 1991;11:1326–1333. doi: 10.1128/mcb.11.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicol SM, Causevic M, Prescott AR, Fuller-Pace FV. The nuclear DEAD box RNA helicase p68 interacts with the nucleolar protein fibrillarin and colocalizes specifically in nascent nucleoli during telophase. Exp Cell Res. 2000;257:272–280. doi: 10.1006/excr.2000.4886. [DOI] [PubMed] [Google Scholar]
- 24.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 25.Rössler OG, Hloch P, Schutz N, Weitzenegger T, Stahl H. Structure and expression of the human p68 RNA helicase gene. Nucleic Acids Res. 2000;28:932–939. doi: 10.1093/nar/28.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barta I, Iggo R. Autoregulation of expression of the yeast Dbp2p ‘DEAD-box’ protein is mediated by sequences in the conserved DBP2 intron. EMBO J. 1995;14:3800–3808. doi: 10.1002/j.1460-2075.1995.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buszczak M, Spradling AC. The Drosophila P68 RNA helicase regulates transcriptional deactivation by promoting RNA release from chromatin. Genes Dev. 2006;20:977–989. doi: 10.1101/gad.1396306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson RJ, Hamilton SJ, MacCallum DE, Hall PA, Fuller-Pace FV. Expression of the ‘DEAD box’ RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J Pathol. 1998;184:351–359. doi: 10.1002/(SICI)1096-9896(199804)184:4<351::AID-PATH1235>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 29.Seufert DW, Kos R, Erickson CA, Swalla BJ. p68, a DEAD-box RNA helicase, is expressed in chordate embryo neural and mesodermal tissues. J Exp Zool. 2000;288:193–204. doi: 10.1002/1097-010x(20001015)288:3<193::aid-jez1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.Ip FC, Chung SS, Fu WY, Ip NY. Developmental and tissue-specific expression of DEAD box protein p72. Neuroreport. 2000;11:457–462. doi: 10.1097/00001756-200002280-00006. [DOI] [PubMed] [Google Scholar]
- 31.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, Akimoto C, Yamamoto Y, Katagiri T, Foulds C, Takezawa S, Kitagawa H, Takeyama K, O'Malley BW, Kato S. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 32.Jalal C, Uhlmann-Schiffler H, Stahl H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 2007;35:3590–3601. doi: 10.1093/nar/gkm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wortham NC, Ahamed E, Nicol SM, Thomas RS, Periyasamy M, Jiang J, Ochocka AM, Shousha S, Huson L, Bray SE, Coombes RC, Ali S, Fuller-Pace FV. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene. 2009;28:4053–4064. doi: 10.1038/onc.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277:12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- 35.Lee CG. RH70, a bidirectional RNA helicase, copurifies with U1snRNP. J Biol Chem. 2002;277:39679–39683. doi: 10.1074/jbc.C200337200. [DOI] [PubMed] [Google Scholar]
- 36.Rössler OG, Straka A, Stahl H. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 2001;29:2088–2096. doi: 10.1093/nar/29.10.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multiprotein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 39.Liu ZR, Sargueil B, Smith CW. Detection of a novel ATP-dependent cross-linked protein at the 5’ splice site-U1 small nuclear RNA duplex by methylene blue-mediated photocross-linking. Mol Cell Biol. 1998;18:6910–6920. doi: 10.1128/mcb.18.12.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu ZR. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5’ splice site duplex. Mol Cell Biol. 2002;22:5443–5450. doi: 10.1128/MCB.22.15.5443-5450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C, Yang L, Yang JJ, Huang Y, Liu ZR. AT-Pase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol Cell Biol. 2005;25:7484–7493. doi: 10.1128/MCB.25.17.7484-7493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hönig A, Auboeuf D, Parker MM, O'Malley BW, Berget SM. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol Cell Biol. 2002;22:5698–5707. doi: 10.1128/MCB.22.16.5698-5707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guil S, Gattoni R, Carrascal M, Abian J, Stevenin J, Bach-Elias M. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol. 2003;23:2927–2941. doi: 10.1128/MCB.23.8.2927-2941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 45.Warner DR, Bhattacherjee V, Yin X, Singh S, Mukhopadhyay P, Pisano MM, Greene RM. Functional interaction between Smad, CREB binding protein, and p68 RNA helicase. Biochem Biophys Res Commun. 2004;324:70–76. doi: 10.1016/j.bbrc.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, Gregory DJ, Lane DP, Perkins ND, Fuller-Pace FV. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005;24:543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 49.Salzman DW, Shubert-Coleman J, Furneaux H. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem. 2007;282:32773–32779. doi: 10.1074/jbc.M705054200. [DOI] [PubMed] [Google Scholar]
- 50.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bond AT, Mangus DA, He F, Jacobson A. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol Cell Biol. 2001;21:7366–7379. doi: 10.1128/MCB.21.21.7366-7379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen G, Guo X, Lv F, Xu Y, Gao G. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc Natl Acad Sci USA. 2008;105:4352–4357. doi: 10.1073/pnas.0712276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goh PY, Tan YJ, Lim SP, Tan YH, Lim SG, Fuller-Pace F, Hong W. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J Virol. 2004;78:5288–5298. doi: 10.1128/JVI.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jorba N, Juarez S, Torreira E, Gastaminza P, Zamarreno N, Albar JP, Ortin J. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics. 2008;8:2077–2088. doi: 10.1002/pmic.200700508. [DOI] [PubMed] [Google Scholar]
- 55.Chen JY, Chen WN, Poon KM, Zheng BJ, Lin X, Wang YX, Wen YM. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch Virol. 2009;154:507–512. doi: 10.1007/s00705-009-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janknecht R, Hunter T. Versatile molecular glue. Transcriptional control. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 57.Janknecht R. The versatile functions of the transcriptional coactivators p300 and CBP and their roles in disease. Histol Histopathol. 2002;17:657–668. doi: 10.14670/HH-17.657. [DOI] [PubMed] [Google Scholar]
- 58.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 59.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 60.Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S. Purification and identification of p68 RNA helicase acting as a tran-scriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Watanabe M, Yanagisawa J, Kitagawa H, Take-yama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, Masuhiro Y, Kato S. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 64.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, Fuller-Pace FV, Robson CN. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overex-pressed in prostate cancer. Cancer Res. 2008;68:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong HY, Demmers JA, Bezstarosti K, Grootegoed JA, Brinkmann AO. DNA dependent recruitment of DDX17 and other interacting proteins by the human androgen receptor. Biochim Biophys Acta. 2009;1794:193–198. doi: 10.1016/j.bbapap.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Jensen ED, Niu L, Caretti G, Nicol SM, Teplyuk N, Stein GS, Sartorelli V, van Wijnen AJ, Fuller-Pace FV, Westendorf JJ. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. J Cell Biochem. 2008;103:1438–1451. doi: 10.1002/jcb.21526. [DOI] [PubMed] [Google Scholar]
- 67.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA Helicases p68/p72 and the Noncoding RNA SRA Are Co-regulators of MyoD and Skeletal Muscle Differentiation. Dev Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Yang L, Lin C, Zhao S, Wang H, Liu ZR. Phos-phorylation of p68 RNA helicase plays a role in platelet-derived growth factor-induced cell pro-liferation by up-regulating cyclin D1 and c-Myc expression. J Biol Chem. 2007;282:16811–16819. doi: 10.1074/jbc.M610488200. [DOI] [PubMed] [Google Scholar]
- 69.Stucke VM, Gorses D, Hofmann F. DEAD-box RNA helicase p68 is not required for nuclear translocation of beta-catenin in colon cancer cells. Cell Cycle. 2008;7:830–832. doi: 10.4161/cc.7.6.5614. [DOI] [PubMed] [Google Scholar]
- 70.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 71.Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor coactivator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Bio-chem J. 2003;369:163–171. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emberley E, Huang GJ, Hamedani MK, Czosnek A, Ali D, Grolla A, Lu B, Watson PH, Murphy LC, Leygue E. Identification of new human coding steroid receptor RNA activator isoforms. Bio-chem Biophys Res Commun. 2003;301:509–515. doi: 10.1016/s0006-291x(02)03070-x. [DOI] [PubMed] [Google Scholar]
- 73.Lanz RB, Razani B, Goldberg AD, O'Malley BW. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc Natl Acad Sci USA. 2002;99:16081–16086. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buelt MK, Glidden BJ, Storm DR. Regulation of p68 RNA helicase by calmodulin and protein kinase C. J Biol Chem. 1994;269:29367–29370. [PubMed] [Google Scholar]
- 75.Rosenberger U, Lehmann I, Weise C, Franke P, Hucho F, Buchner K. Identification of PSF as a protein kinase Calpha-binding protein in the cell nucleus. J Cell Biochem. 2002;86:394–402. doi: 10.1002/jcb.10233. [DOI] [PubMed] [Google Scholar]
- 76.Bähler M, Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002;513:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 77.Yang L, Yang J, Huang Y, Liu ZR. Phosphorylation of p68 RNA helicase regulates RNA binding by the C-terminal domain of the protein. Bio-chem Biophys Res Commun. 2004;314:622–630. doi: 10.1016/j.bbrc.2003.12.129. [DOI] [PubMed] [Google Scholar]
- 78.Kodym R, Henockl C, Furweger C. Identification of the human DEAD-box protein p68 as a sub-strate of Tlk1. Biochem Biophys Res Commun. 2005;333:411–417. doi: 10.1016/j.bbrc.2005.05.136. [DOI] [PubMed] [Google Scholar]
- 79.Yang L, Lin C, Liu ZR. Signaling to the DEAD box –regulation of DEAD-box p68 RNA helicase by protein phosphorylations. Cell Signal. 2005;17:1495–1504. doi: 10.1016/j.cellsig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 80.Yang L, Lin C, Liu ZR. Phosphorylations of DEAD box p68 RNA helicase are associated with cancer development and cell proliferation. Mol Cancer Res. 2005;3:355–363. doi: 10.1158/1541-7786.MCR-05-0022. [DOI] [PubMed] [Google Scholar]
- 81.Lin J, Arlinghaus R. Activated c-Abl tyrosine kinase in malignant solid tumors. Oncogene. 2008;27:4385–4391. doi: 10.1038/onc.2008.86. [DOI] [PubMed] [Google Scholar]
- 82.Yang L, Lin C, Sun SY, Zhao S, Liu ZR. A double tyrosine phosphorylation of P68 RNA helicase confers resistance to TRAIL-induced apoptosis. Oncogene. 2007;26:6082–6092. doi: 10.1038/sj.onc.1210427. [DOI] [PubMed] [Google Scholar]
- 83.Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, Fuller-Pace FV. Overex-pression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene. 2001;20:7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 84.Jacobs AM, Nicol SM, Hislop RG, Jaffray EG, Hay RT, Fuller-Pace FV. SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene. 2007;26:5866–5876. doi: 10.1038/sj.onc.1210387. [DOI] [PubMed] [Google Scholar]
- 85.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 86.Kahlina K, Goren I, Pfeilschifter J, Frank S. p68 DEAD box RNA helicase expression in keratinocytes. Regulation, nucleolar localization, and functional connection to proliferation and vascular endothelial growth factor gene expression. J Biol Chem. 2004;279:44872–44882. doi: 10.1074/jbc.M402467200. [DOI] [PubMed] [Google Scholar]
- 87.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 88.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 89.Wei Y, Hu MH. [The study of P68 RNA helicase on cell transformation] Yi Chuan Xue Bao. 2001;28:991–996. [PubMed] [Google Scholar]
- 90.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 91.de Lau W, Barker N, Clevers H. WNT signaling in the normal intestine and colorectal cancer. Front Biosci. 2007;12:471–491. doi: 10.2741/2076. [DOI] [PubMed] [Google Scholar]
- 92.Normanno N, Di Maio M, De Maio E, De Luca A, de Matteis A, Giordano A, Perrone F. Mechanisms of endocrine resistance and novel therapeutic strategies in breast cancer. Endocr Relat Cancer. 2005;12:721–747. doi: 10.1677/erc.1.00857. [DOI] [PubMed] [Google Scholar]
- 93.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 94.Attard G, Cooper CS, de Bono JS. Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell. 2009;16:458–462. doi: 10.1016/j.ccr.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 95.Stone B, Schummer M, Paley PJ, Thompson L, Stewart J, Ford M, Crawford M, Urban N, O'Briant K, Nelson BH. Serologic analysis of ovarian tumor antigens reveals a bias toward antigens encoded on 17q. Int J Cancer. 2003;104:73–84. doi: 10.1002/ijc.10900. [DOI] [PubMed] [Google Scholar]
- 96.Felix RS, Colleoni GW, Caballero OL, Yamamoto M, Almeida MS, Andrade VC, Chauffaille Mde L, Silva WA, Jr, Begnami MD, Soares FA, Simpson AJ, Zago MA, Vettore AL. SAGE analysis high-lights the importance of p53csv, ddx5, mapkapk2 and ranbp2 to multiple myeloma tumorigenesis. Cancer Lett. 2009;278:41–48. doi: 10.1016/j.canlet.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 97.Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep. 2004;11:1319–1324. [PubMed] [Google Scholar]
- 98.Kumar A, Kumar Dorairaj S, Prabhakaran VC, Prakash DR, Chakraborty S. Identification of genes associated with tumorigenesis of meibomian cell carcinoma by microarray analysis. Genomics. 2007;90:559–566. doi: 10.1016/j.ygeno.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 99.Oishi M, Taniguchi Y, Nishimura K, Yamada T, Sasaki Y. Characterisation of gene expression in bovine adipose tissue before and after fattening. Anim Genet. 2000;31:166–170. doi: 10.1046/j.1365-2052.2000.00626.x. [DOI] [PubMed] [Google Scholar]
- 100.Kitamura A, Nishizuka M, Tominaga K, Tsuchiya T, Nishihara T, Imagawa M. Expression of p68 RNA helicase is closely related to the early stage of adipocyte differentiation of mouse 3T3 -L1 cells. Biochem Biophys Res Commun. 2001;287:435–439. doi: 10.1006/bbrc.2001.5577. [DOI] [PubMed] [Google Scholar]
- 101.Bolduc C, Larose M, Yoshioka M, Ye P, Belleau P, Labrie C, Morissette J, Raymond V, Labrie F, St-Amand J. Effects of dihydrotestosterone on adipose tissue measured by serial analysis of gene expression. J Mol Endocrinol. 2004;33:429–444. doi: 10.1677/jme.1.01503. [DOI] [PubMed] [Google Scholar]
- 102.Kircher SG, Kim SH, Fountoulakis M, Lubec G. Reduced levels of DEAD-box proteins DBP-RB and p72 in fetal Down syndrome brains. Neuro-chem Res. 2002;27:1141–1146. doi: 10.1023/a:1020921324871. [DOI] [PubMed] [Google Scholar]
- 103.Huang H, Shiffman ML, Cheung RC, Layden TJ, Friedman S, Abar OT, Yee L, Chokkalingam AP, Schrodi SJ, Chan J, Catanese JJ, Leong DU, Ross D, Hu X, Monto A, McAllister LB, Broder S, White T, Sninsky JJ, Wright TL. Identification of two gene variants associated with risk of advanced fibrosis in patients with chronic hepatitis C. Gastroenterology. 2006;130:1679–1687. doi: 10.1053/j.gastro.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 104.Guo J, Hong F, Loke J, Yea S, Lim CL, Lee U, Mann DA, Walsh MJ, Sninsky JJ, Friedman SL. A DDX5 S480A polymorphism is associated with increased transcription of fibrogenic genes in hepatic stellate cells. J Biol Chem. 2010;285:5428–5437. doi: 10.1074/jbc.M109.035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi YW, Dutta S, Fielding BC, Tan YJ. Expression, purification and preliminary crystallo-graphic analysis of recombinant human DEAD-box polypeptide 5. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:192–194. doi: 10.1107/S1744309109052956. [DOI] [PMC free article] [PubMed] [Google Scholar]