Abstract
Addition of polypeptides belonging to the ubiquitin family to selected lysines residues is a widespread post-translation modification (PTM) that controls many fundamental aspects of cell's life. Specific alterations in the normal turnover of this PTM are frequently observed in tumors. The conjugation/deconjugation cycle of ubiquitin (Ub) or ubiquitin-like (Ubl) proteins influences the activities of oncogenes and tumor suppressor genes. Two families of enzymes work in antagonizing manner to add or remove Ub and Ubl-proteins on target proteins: the E3 ligases and the isopeptidases. These enzymes are the subjects of fervent research with the ambition to comprehend their regulation, their mechanisms of action, their involvement in human diseases, and to develop specific inhibitors for therapeutic intervention. Here we will discuss of isopeptidases, the deconjugating enzymes, with particular emphasis on the proapoptotic activities of the relative inhibitors identified so far.
Keywords: Apoptosis, cancer, proteasome, isopeptidase, DUBs, necrosis, caspase, proteasome, USP
Introduction
Cells have evolved a spectacular array of strategies to modulate protein functions in response to environmental stimuli. In multicellular organisms post-translation modifications (PTMs) are well suited to mach rapid cellular responses to new and unexpected needs of the organism. Ubiquitination is a PTM, which consists in the covalent addition of ubiquitin (Ub), a 76-residue polypeptide, to lysine residues of specific target proteins [1]. The carboxyl group of the C-terminal glycine of ubiquitin forms an isopeptide bond with the ε-aminogroup of lysines present on the target protein [2]. Many recent excellent reviews have discussed the complexity of protein ubiquitination [2-4]. This complexity renders the protein-ubiquitination system (UPS) the fore-most flexible PTM.
There are two main aspects at the origin of the UPS complexity. First, the Ub linkage is subjected to multiple options such as: mono-ubiquitination, poly-ubiquitination on different lysines of the Ub itself (K6, K11, K29, K48 and K63) or on different lysines of the target protein (poly-mono-ubiquitination) and also the aminoterminal ubiquitination [5]. Second, Ub belongs to a protein family, characterized by 14 members (including 3 putative) and classified as ubiquitin-like (Ubl) proteins. Ubl-proteins share structure, but not sequence, similarities with ubiquitin. Differentially from Ub, Ubl-proteins have only regulative but not degradative activities towards their targets [4].
The spectacular collection of options available to cells to modify Lys residues is reflected in a vast assortment of effects on the target proteins, as we begin to comprehend in the case of Ub. Through poly-ubiquitinations, mono-ubiquitination, poly-mono-ubiquitinations this PTM can govern: the proteasomal-mediated degradation of proteins, their assembly into signaling complexes or their localization into specific subcellular compartments. Not surprisingly the Ub and Ubl-proteins are pivotal for several cellular processes, including: cell cycle, apoptosis, DNA repair, membrane trafficking, autophagy, inflammatory response, ribosomal protein synthesis and both the innate and adaptive immune responses [6-8].
Proteins regulated by Ub or Ubl are in general selectively modified by the coordinate action of three Ub-ligase or Ubl-ligases known as the E1, E2 and E3 enzymes. E1 and E2 are responsible for activating the ubiquitin molecule for conjugation, whereas E3 acts as matchmakers between the activated Ub-E2 intermediate and substrate proteins [1-3, 8].
Over the past decade this complex molecular machinery has attracted much attention, not only among molecular and cellular biologists, but also among pharmacologists and oncologists. The protease activity and the unquestion-able involvement of many Ub-targets in the control of cell proliferation inspired the searching for specific inhibitors of the ubiquitin-proteasome system (UPS), to be used in clinic. The approval of Bortezomib/Velcade/PS-341 for the treatment of multiple myeloma and several ongoing clinical trials using bortezomib or other, more recently developed UPS inhibitors, have proved the importance of the UPS as drug-target for anti-neoplastic therapies [9, 10].
DUBs and other isopeptidases
As for other PTMs, such as phosphorylation or acetylation, conjugation of Ub or Ubl-proteins to protein substrates is a reversible process. Isopeptidases, a heterogeneous family of proteolytic enzymes, are involved in this task. The isopeptidases family includes deubiquitinating enzymes (deubiquitinases or DUBs), which in principle should be specifically devoted to the rupture of Ub linkages and other proteolytic enzymes, which are dedicated to deconjugate the Ubl-proteins [4, 11]. Generally, they can be viewed as E3 ligase antagonists. Genomic studies have identified 79 human genes encoding for functionally putative DUBs [12].
From a structural point of view isopeptidases can be grouped into five distinct subfamilies [4, 11]. Four of them are cysteine-proteases subdivided into (i) the Ub-C-terminal hydrolases (UCH), (ii) the ubiquitin specific protease (UBP/ USP), (iii) the ovarian tumor-related (OTU), and (iv) the Machado-Josephin domain (MJD). The last group includes (v) the JAMM, Zn-metalloproteases (AB1/MPN/Mov34 metalloenzyme). In addition, there are many UBL-isopeptidases that do not fully fit within these categories, but nevertheless they are interesting therapeutic targets.
Isopeptidases deconjugation activity can exert diverse outcomes on the substrates [4, 11, 13]. First they can endorse the processing/ maturation of the Ub and Ubl precursors, a necessary step for their subsequent conjugation to the targets. Second, they can antagonize the E3 ligase activities, thus operating as negative regulators of the PTM. Third, DUBs, in particular, can support the degradative phase. Substrate degradation is coupled to its deubiquitination, which is required both for efficient substrate degradation and for recycling the Ub the pool [6].
Curiously, interference with this last function is the manifested consequences of treating cells with non-selective isopeptidase inhibitors (N-SIIs). Accumulation of poly-ubiquitinated proteins in the presence of an active catalytic chamber of the proteasome, is the distinguished trait of these inhibitors [14-16]. Accumulation of poly-ubiquitinated proteins is also elicited by bortezomib, but in this case the catalytic chamber of the proteasome is to be inhibited [10].
Historically much attention has been focused on the role of E3-ligases in the control of protein turnover. Only recently the interest has been manifested towards enzymes that reverse the process. The sprouting of this awareness is witnessed by the rising number of excellent reviews published last year on isopeptidases [13, 17-19].
Isopeptidases and cancer
Isopetidases control the activities of several proteins located at vital nodes of various signaling pathways (Table 1). Oncogenes and tumor suppressor genes can be modulated by cycles of Ub or Ubl addiction/removal [17]. The p53 is emblematic, being conjugated with either, Ub and Ubl proteins [20, 21]. Hence, the discovery of mutations in certain DUBs in cancer was not unexpected. Germline mutations in the CYLD gene, which encodes for a DUB that removes 63 -linked Ub-chains, are responsible for three different syndromes, which share the predisposition for developing multiple skin tumors of the head and neck [22-24]. Translocations of USP6 gene have recently been found to be causative in most aneurysmal bone cysts, that are benign tumors [25]. Inactivating somatic mutations of TNFAIP3/A20 in different lymphomas have been recently reported. A20, which contains deubiquitinase and E3 ligase domains, negatively regulates the NF-kB signalling pathway [26]. On the opposite, some isopeptidases show an oncogenic behaviour and are overexpressed in certain tumors [27].
Table 1.
Structures of the principal non-selective isopeptidase inhibitors (N-SIIs)
| Names | Process/Mechanism | Comments |
|---|---|---|
| USP1 | DNA repair, UAF1 (USP1-associated factor 1) | Deubiquitinates and inactivates PCNA. Implications in Fanconi's anemia-related cancer [82]. |
| USP2 | Cell proliferation. Implications in prostate cancer | Stabilization of FAS (Fatty Acid Synthase). Deubiquitinates and stabilizes MDM2 [83, 84]. |
| USP3 | Cell cycle and genomic stability | Regulator of H2A/H2B ubiquitination and involvement in the response to DNA double-strand breaks [85]. |
| USP4 | Cell proliferation differentiation. Implications in prostate cancer | Influence on Wnt signalling acting as a suppressor of β -catenin dependent transcription [86]. |
| USP5 | p53 pathway | p53 stabilization by USP5 knockdown, accumulation of unanchored polyubiquitin [87]. |
| USP7 | p53 and AKT pathways (DNA damage, oxidative stress) | Deubiquitinates and affects stability of both p53 and MDM2 [70-73]. |
| USP8 | Endocytic trafficking and receptor Internalization | Regulates abundance of EGFR and several receptor tyrosine kinases (RTKs) [76, 88, 89]. |
| AMSH | Endocytic trafficking and receptor degra- dation | Impaired AMSH reduces EGFR degradation without affecting internalization [90]. |
| USP9X | Cell cycle, chromosome partition, apoptosis,TGF-β and oxidative stress signaling | Promiscuous, multiple targets [80, 91-94]. |
| USP10 | Post-endocytic trafficking/p53 pathway | Deubiquitinates p53, reversing Mdm2 induced nuclear export and degradation [95]. |
| USP11 | DNA damage/ NF-kB pathway | Component of the HR double-strand break repair pathway. Negatively regulates NF-kB pathway by targeting IkBα [96, 97]. |
| USP14 | Proteasome/protein degradation | Proteasome associated DUB, implicated in colorectal cancer [98]. |
| USP16 | Cell cycle and chromatin regulation | Deubiquitinates histone H2A. Its downregulation promotes mitotic phase defects in HeLa cells [99]. |
| USP17 | Cell cycle, Ras pathway | USP17 expression blocks Ras membrane localization and inhibits downstream kinases MEK/ERK phos-phorylation [100]. |
| USP18 | Negative regulator of the interferon response | USP18 downregulation augments interferon induced TRAIL expression and apoptosis [81]. Regulates EGFR protein synthesis [101]. |
| USP20 | Receptors recycling, HIF1 regulation, motility | Substrate of von Hippel-Lindau tumor suppressor. Beta2 adrenergic receptor recycling. Deubiquitinates and stabilizes HIF1-alpha [102, 103, 104]. |
| USP21 | Chromatin regulation, NF-kB pathway | Deubiquitinates chromatin bound H2A. Inhibition of TNF-induced NF-kB activation through RIP1 deubiquitination [105, 106]. |
| USP22 | Chromatin modification | Subunit of the hSAGA transcriptional cofactor complex. Deubiquinates histones H2A/H2B [107]. |
| USP28 | DNA damage/cell proliferation | Stabilizes MYC proto-oncogene. Role in the Chk2-p53-PUMA pathway [108, 109]. |
| USP33 | Receptors recycling/Motility | Substrate of von Hippel-Lindau tumor suppressor. Beta2 adrenergic receptor recycling [102, 103]. |
| USP39 | Chromosome segregation, mitotic spindle checkpoint regulation | Involved in splicing of Aurora B and other mRNAs essential for spindle checkpoint function [110]. |
| USP44 | Chromosome segregation, mitotic spindle checkpoint regulation | Deubiquitinates Cdc20 stabilizing the APC-inhibitory complex Mad2-Cdc20 preventing the premature activation of APC itself [111]. |
| CLYD | NF-kB pathway | Inhibits NF-kB signaling by removing K63-liked chains on NEMO, TRAF-2 and TRAF-6 [22, 23, 24, 112]. |
| A20 | NF-kB pathway | Replaces the K63 chains of the NF-kB signaling intermediates TRAF6, RIP1, RIP2 and IKK with proteasome-targeting K48 chains [113]. |
| UCHL1 | Cell cycle and mitosis | Involved in Parkinson's and Alzheimer's disease and cancer [65, 66, 67]. |
Recent excellent reviews have discussed these aspects in detail [19]. Hence we refer the readers to these reviews.
Isopeptidases inhibitors
A complete eradication of the neoplastic cells, after surgical intervention and proapopotic pharmacological strategies, is the best prospect that we can offer to save a patient's life. Unfortunately many incurable tumors show stubborn resistance to die by apoptosis and, as a consequence, an extreme chemoresistance. Generally chemoresistance arises because during tumor progression cancer cells accumulate mutations in critical genes regulating the apoptotic program. Paradoxically, the adverse environment and the host-defense mechanisms, act as a strong selective pressure, which favors the rising of apoptotic resistant clones. Identification and validation of new druggable targets for anti-cancer therapies is an intense area of the bio-medical research, which hopefully will overcome chemoresistance [28, 29].
Due to their protease activity and their involvement in the regulation of important signaling pathways, including apoptosis, isopeptidases are emerging as attractive druggable candidates [10, 19, 30].
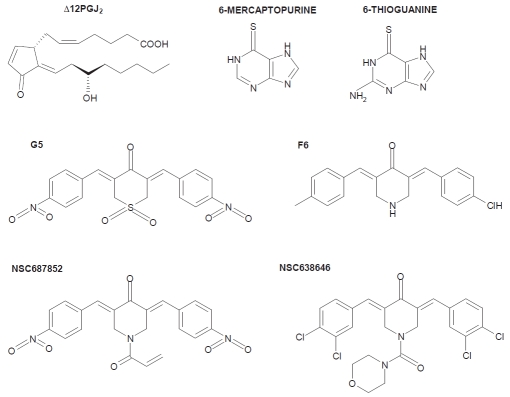
As it often happens in the modern biomedical research, serendipity led to the discovery of the first isopeptidase inhibitors. Studies aimed to explore the proapoptotic functions of certain prostaglandins and high-throughput screening, using chemical libraries of small compounds, aimed to identify new pro-apoptotic drugs, made possible the discovery of the first N-SIIs [14-16] All these compounds share the same pharmacophore, a sterically accessible, cross-conjugated α,β-unsaturated dienone which confers inhibitory activity toward isopeptidases (Figure 1) [31]. When added to cells, N-SIIs elicit the accumulation of poly-ubiquitinated proteins and the induction of classical markers of the cellular response to proteasomal inhibition [14-16].
Figure 1.
Structures of the principal non-selective isopeptidase inhibitors (N-SIIs).
These inhibitors are small molecules susceptible to Michael addition reactions with free sulfhydryl groups of cysteines [32]. Consequently, N-SSIs are irreversible inhibitors. Based on their structural characteristics it is difficult to imagine a strong selectivity towards different isopeptidases. In fact, in vitro studies using recombinant enzymes have demonstrated that N-SIIs can inhibit various isopeptidases such as: UCHs, USP2, USP7 and the Ubl-protease SENP2, with comparable IC50 in the low micromolar range [33]. Hence, compounds containing cross-conjugated α,β-unsaturated dienones can be considered N-SIIs.
In principle, N-SIIs could also attack other proteins or cellular thiols including glutathione (GSH). Although these possibilities cannot be ruled out, we have observed that G5, a compound belonging to this class of inhibitors, when used at a concentration capable of promoting the accumulation of poly-ubiquitinated proteins cannot inhibit caspases, which are cystein proteases [34].
Treatment of cells with GSH depleting agents shows an additive prodeath effect with G5, thus suggesting that glutathione could be involved in its detoxification. However GSH depletion cannot explain N-SIIs pro-apoptotic activities [35].
It is evident that further studies aimed to characterize these inhibitors are needed. In particular it will be important to prove their antitumor activity in vivo, using animal models. Notwith-standing they represent valuable leads for further chemical optimization.
The thiopurines 6-mercaptopurine (6MP) and 6-thioguanine (6TG) probably represent another group of N-SIIs (Figure 1). These compounds are indicated for the treatment of the acute lymphoblastic leukemia and of Crohn's disease, an inflammatory disorder [36]. It has been reported that these compounds can inhibit the SARS-coronavirus (CoV) papain-like protease (PLpro), a cysteine protease with deubiquitinating and deisgylating activity [37]. Deisgylases are involved in the deconjugation of the Ubl-protein ISG15. The expression of this Ubl and of the conjugation/deconjugation system (including the relative E1, E2 and E3 enzymes) is under the control of interferons. Protein isgylation plays important roles in the modulation of the immune response [38].
Structure comparison studies suggest that 6MP and 6TG are also potential inhibitors of USP14 [36]. Hence, these compounds could belong to a new class of N-SIIs.
Death-pathways activated by non-selective isopeptidase inhibitors
Cells can efficiently eliminate themselves through a sophisticated genetic program known as apoptosis. Proteolytic processing of selected cellular proteins, as operated by a unique family of cystein-proteases called caspases, is necessary to induce apoptosis. Two main apoptotic pathways, the extrinsic and the intrinsic, keep in check caspase activation. The extrinsic pathway controls procaspase-8 maturation and it is triggered by the engagement of death receptors placed at the cell surface. By contrast, the intrinsic pathway, which is responsible for procas-pase-9 maturation, is triggered by the release of killer mitochondrial proteins into the cytosol, after MOMP (mitochondrial outer membrane permeabilization) [39].
MOMP is emerging as the point of no return for many death pathways. In the presence of massive MOMP, the cellular demise could take place alternatively, as an effect of the mitochondrial metabolic failure or by means of some still mysterious caspase-independent deaths [40]. MOMP and the release of the mitochondrial proapoptotic factors into the cytosol are under the antagonistic control of the master regulators of the apoptotic process: the members of the Bcl-2 family [41].
N-SIIs are potent inducers of apoptosis in various cancer cell lines [14-16, 31, 35]. This apoptotic response, as suspected, shares many traits with apoptosis induced by bortezomib and proteasome inhibitors, including p53 independence, induction of ER stress, activation of the extrinsic pathway, eliciting of MOMP [14, 15, 34, 42]. Interestingly, both bortezomib and N-SIIs can efficiently activate effector caspases in the presence of a defective apoptosome [15, 43]. It seems that a double effect of these in-hibitors, on the extrinsic pathway on one side and, on the IAP antagonist Smac, by stabilization of its cytosolic mitochondrial released form, on the other side, allows the activation of caspase-3/-7 also in absence of active caspase-9 [43, 44].
The mechanism used by UPS inhibitors in general and by N-SIIs in particular to sustain the extrinsic pathway is not clear. UPS inhibitors have several possibilities of interference at different levels of the signaling cascade. The caspase-8 substrate Bid once cleaved can be targeted to proteasome [45]. FLIP, which different isoforms display antagonistic effects on cas-pase-8 activation, can also be regulated by the UPS [46-49]. Caspase-8 activation itself is tunable by poly-ubiquitination. The CUL3 ubiquitin-ligase complex mediates caspase-8 poly-ubiquitination. Next, the ubiquitin-binding protein p62/sequestosome-1 promotes aggregation of CUL3-modified caspase-8, which drives enzyme activation [50]. Since caspase-8 poly-ubiquitination can be reversed by the DUBs A20, this represents another site of intervention for the N-SIIs.
A further possibility of interference for N-SIIs is at the level of membrane trafficking. Localization in submembraneous compartments and vesicle trafficking can influence the activity of death receptors and of their ligands [51]. It is well known that membrane trafficking is modulated by addition of Ub and Ubl-proteins and that the action of specific isopeptidases elicits a tight control over this process [17, 18]. Here again, a possible effect of the N-SIIs on the extrinsic pathway can be envisaged.
Certain tumors become treacherously resistant to chemotherapy-induced apoptosis. Interestingly, N-SIIs, in cells resistant to apoptosis, can activate a peculiar necrotic response, which depends on the reorganization of the actin cytoskeleton [34]. The effect on actin cytoskeleton can be observed at nanomolars concentration of the N-SII G5. In glioblastoma cells the effect on actin cytoskeleton is coupled to a drastic reduction of cell motility, thus suggesting another possible therapeutic window [35]. N-SII can affect not only cancer cells survival but also their invasive and metastatic properties.
The dramatic and variegated effects of N-SIIs on cell survival are probably a consequence of the simultaneous alterations of multiple cellular functions, normally controlled by different isopeptidases, which renders the stress unmanageable for the cells. In fact, microarray analysis of cells treated with prostaglandin J2 (PGJ2) that contains α,β-unsaturated carbonyl groups, revealed the up-regulation of genes of the heat shock and stress response, including proteasomal components, genes controlling protein folding, detoxification and cysteine metabolism [52]. Perturbation of mitochondrial function with a rapid drop of the mitochondrial membrane potential and lysosomal membrane permeabilization [16, 34, 52], although probably late events of the cellular response to N-SIIs are important markers that further prove the induction of unmanageable cellular stress.
Specific isopeptidases inhibitors
The development of more specific isopeptidase inhibitors could enormously improve the opportunities for pharmacologic interventions. The vast majority of isopeptidases are cystein-proteases. As recently highlighted by Daviet and Colland, it is not simple to develop cysteine-proteases inhibitors exhibiting the mandatory clinical properties [30, 53]. More selective inhibitors should minimize toxicities, which is an important limit for clinical applications for these inhibitors. Unfortunately specificity cannot be an easy task. We are dealing with enzyme families that include many different members in which catalyitic cores show extensive homologies [53].
The catalytic core depends on two or three crucial amino acid residues, constituting a catalytic diad or triad, respectively [11, 17, 30, 53]. All isopeptidases with cystein-protease activity utilize a similar catalytic mechanism, which has been described in detail with studies on the papain family of proteases. A His side chain residue, which is positioned next to the catalytic Cys lowers the pKa of Cys, thus enabling the nucleophilic attack on isopeptides linkages [54].
Another important aspect that must be kept in mind is that DUBs catalytic site can be under conformational regulation. Structural studies have revealed that USPs exhibit a conserved three-domains architecture, comprising Fingers, Palm, and Thumb. Fingers specifically manage the interaction with Ub, whereas the catalytic centre lies at the interface between the Palm and Thumb sub-domains [55]. Some isopeptidases are constitutively active, whereas in others substrate binding modulates the enzymatic activity. In the absence of the substrate (without Ub binding) the catalytic domain is in an inactive conformation. Substrate binding induces an allosteric change that activates the catalytic domain [17, 55-58]. In USP7/HAUSP (Herpesvirus associated USP) for example Ub binding elicits the alignment of the catalytic Cys to the His residue [55, 56]. In contrast, the catalytic site of USP14 is perfectly aligned also in the absence of the substrate. Inhibition is obtained by steric hindrance of the active site, as operated by the ubiquitin-binding surface loops. Ubiquitin binding leads to translocation of these loops, allowing access of Ub substrates to the active site [17, 57, 59].
A promising approach to identify specific isopep-tidase is the development of high-throughput screening (HTS) of small compounds libraries. In order to augment specificity and potency, HTS are followed by chemical modification of the most promising hits. A limit of the HTS is the availability of simple and specific assays to measure isopeptidase activities. This weakness is testified by the sprouting of new assays, as described in the recent literature [60-63]. Although the sensitivity and specificity of the assays could be further improved, several research groups have already utilized HTS to isolate isopeptidase inhibitors.
With this approach O-acyloxime derivatives of isatins have been isolated as inhibitors of UCH-L1, in the in low micromolar range (Table 2). Compounds structure analysis allowed the generation of further isoforms specific, UCH-L1 or UCH-L3 inhibitors, showing increased inhibitor potency and specificity [64]. The mechanism proposed for this class of compounds is a reversible competitive inhibition of the active site. The compounds were also able to inhibit the growth of different tumor cell lines, mimicking the effect of UCH-L1 specific siRNA.
Table 2.
Selective isopeptidase inhibitors
| Compound | Isopeptidase | Activity | Specificity | Mechanism | Cell effects | Ref. |
|---|---|---|---|---|---|---|
| O-acyl oxime derivatives of isatins | UCH-L1 | IC50 0.80-0.94 μM | IC50 for UCH-L3 17-25 μM | Reversible competitive active site directed | Promote proliferation of H1299 and SHSY5Y cells. | [64] |
| 3-Amino-2-keto-7H-thieno[2,3-b] pyridin-6-one derivatives | UCH-L1 | Ki 2.8 μM | Not active for UCHL3, and various cystein proteases | Uncompetitive substrate/enzyme complex directed | Not reported | [68] |
| N,N0-4,40- Biphenyldiylbis(4 Ethylbenzenesulfo namide) | UCH-L1 | IC50 15 μM | Not reported | Active site directed | Not reported | [69] |
| 2-methyl- N -[1-(2-naphthyl) ethyl] benzamide derivative | PLpro | IC50 0.6 μM | IC50>100 for various DUBs and cystein proteases | Reversible competitive directed active site | Antiviral activity in Vero E6 cells, EC50=10-15 μM | [78] |
| Cyanoidenopyrazine derivative | USP7 | IC50 0.42 μM | Partial specificity | Reversible, non competitive | Induction of p53 activity, inhibition of cell proliferation and induction of p53 dep. apoptosis | [74] |
| Not reported HBX 28,231 HBX 28,218 | USP7 | Low μmolar range | No activity against various DUBs and cystein proteases. | Not reported | Induction of p53 activity, inhibition of cell proliferation and induction of p53 dep. Apoptosis. | [30][75] |
| 9-Oxo-9h-indeno [1,2-b]pyrazine-2,3-dicarbonitrile analogues | USP8 | IC50 0.98-0.56 μM | No activity against various DUBs and cystein proteases, partial activity against UCH-L3. | Not reported | Affected viability of cancer cells lines HTC116 and PC-3 IC50 0.5-1.5 μM | [77] |
UCH-L1 is a protease involved in Parkinson's disease (PD) and Alzheimer's disease (AD) as well in promoting cancer cell phenotypes [65-67]. Interestingly, whereas PD and AD are associated with reduced levels of UCH-L1 the tumor progression is dependent on the isopeptidase activity. Compounds inhibiting UCH-L1 should be beneficial for cancer treatment, whereas an enzyme activator could be effective toward PD and AD.
Derivatives of 33-Amino-2-keto-7H-thieno[2,3-b] pyridin-6-one, with a Kiaap of 2.8 µM against UCH-L1 have been recently developed with a similar screening strategy [68]. These inhibitors seem to be specific, since at the same concentration they are non-effective against other DUBs and cystein proteases. Differentially from the previous case the mechanism is a non-competitive inhibition of the Michaelis complex rather than of the free enzyme (Table 2).
Following a different strategy, an in silico virtual drug screening based upon the crystal structure data of UCH-L1, an enzyme potentiator and several inhibitors, the best one with an IC50 of 15 µM, were identified [69]. No data are available about specificity or in vivo effects of these compounds.
HTS approaches, using Ub-AMC (Ub- ubiquitin-7-amido-4-methylcoumarin) as substrate, have been used to isolate specific inhibitors of USP7.
USP7/HAUSP destabilizes p53, by antagonizing the autoubiquitination activity of MDM2 [70, 71]. Silencing of USP7 has p53-dependent growth suppressive effects in tumor cells. USP7 can deubiquinate additional substrates including FOXO4 and PTEN. In both cases USP7 promotes nuclear export and inactivation of the two proteins which leads to the reinforcement of the PI3K/PKB signaling pathway [72, 73].
After the HTS and subsequent optimization, a cyano-indenopyrazine derivative, HBX 41,108 was isolated as USP7 inhibitor [74]. This compound shows an IC50 0.42 µmol/L on Ub-AMC and further analysis suggest for a non-competitive reversible mechanism of inhibition. When tested against other cystein- proteases such as Cathepsin D, L, S or UCH-L1, an inhibitory activity can be measured (IC50>1µmol/L). Curiously, it was less effective in inhibiting the SUMO-protease SENP1 (IC50>10µmol/L). When added to colon cancer cell lines HBX 41,108 can trigger a p53-dependent apoptotic response. After this first discovery, a second-generation of more specific USP7 inhibitors would be on the way [30, 75].
USP8 is a regulator of membrane trafficking and endocytosis and it interacts with the epidermal growth factor receptor (EGFR) [76]. One of the USP7 inhibitor identified in the HTS screening of Colland [74], the 9-oxo-9h-indeno [1,2-b] pyrazine-2,3-dicarbonitrile was active in the low micromolar and submicromolar range against USP7 and USP8 respectively. Chemical modification allowed the development of a derivative without activity toward USP7, but with submicro-molar activity against USP8 [77]. The resulting compound was inactive against other cystein-proteases and DUBs with the exception of a partial activity toward UCH-L3. The inhibitor series were also able to affect cancer cell lines viability.
GRL0617, an inhibitor of the papain-like protease (PLpro), has been recently isolated by means of HTS. GRL0617, after synthetic optimization, shows an IC50 of 0.6 µM for the Plpro activity and an IC50 of 15 µM versus viral replication. Structural studies have evidenced that it binds within the S4-S3 subsites of the enzyme and induces a loop closure that shuts down catalysis at the active site [78]. Interestingly, USP7/HAUSP, USP18, UCH-L1 and UCH-L3 are not inhibited by GRL06017.
A patent as been recently filled for compounds, identified in a fluorescence polarization assay, active against USP2 and UCH-L3. The reported IC50 range from 100 nM to 50 μM and from 100 nM to 100 μM for UCH-L3 and USP2 respectively [79].
Conclusions
Isopeptidases in general and deubiquitinases more in specific, exhibit many characteristics of a promising therapeutic target. Different biotech companies are investing on this class of enzymes and specific inhibitors are emerging from different kinds of screening. The searching for specific inhibitors of isopeptidases is a growing business. Perhaps we should ask to ourselves whether in fact highly specific inhibitors would be able to trigger apoptosis in cancer cells [80]. Or whether more broad inhibitors, hitting groups of isopeptidases at the same time, might be favored to kill tumor cells highly resistant to apoptosis.
Recently, we silenced 53 different isopepti-dases using a small siRNA library, in cancer cells. We found that down-regulation of certain isopeptidases synergistically enhanced the pro-apoptotic activity of known anti-tumor drugs. However, the simple down-regulation of a single isopeptidase was insufficient to trigger a robust apoptotic response [81].
A final consideration regards what we know about isopeptidases. For many aspects our knowledge in this field are at an early stage. In order to develop anti-cancer strategies, based on isopeptidase inhibition is of paramount im-portance understanding in detail the role played by individual isopeptidase in cell cycle, apoptosis and growth-related pathways. New pieces of information are accumulating, but further work is waiting for us.
References
- 1.Hershko A, Ciechanover A. The Ubiquitin System. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Eddins MJ. Ubiquitstructures, functions, mechanisms. Biochem Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of Proteins by Ubiquitin and Ubiq-uitin-Like Proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. “Protein Modifications: Beyond the Usual Suspects.”. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finley D. Recognition and Processing of Ubiquitin -Protein Conjugates by the Proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ZJ, Sun LJ. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Kodadek T. No Splicing, No Dicing: Non Proteolytic Roles of the Ubiquitin Proteasome System In Transcription. J Biol Cell. 2010;285:2221–2226. doi: 10.1074/jbc.R109.077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chauhan D, Bianchi G, Anderson KC. Targeting the UPS as therapy in multiple myeloma. BMC Biochem. 2008;1(9 Suppl):S1. doi: 10.1186/1471-2091-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brancolini C. Inhibitors of the Ubiquitin-Proteasome System and the Cell Death Machinery: How Many Pathways are Activated? Curr Mol Pharmacol. 2008;1:24–37. doi: 10.2174/1874467210801010024. [DOI] [PubMed] [Google Scholar]
- 11.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochem Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A Genomic and Functional Inventory of Deubiquitinating Enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullally JE, Moos PJ, Edes K, Fitzpatrick FA, Mullally JE, Moos PJ, Edes K, Fitzpatrick FA. Cyclopentenone prostaglandins of the J series inhibit the ubiquitin isopeptidase activity of the proteasome pathway. J Biol Chem. 2001;276:30366–30373. doi: 10.1074/jbc.M102198200. [DOI] [PubMed] [Google Scholar]
- 15.Aleo E, Henderson CJ, Fontanini A, Solazzo B, Brancolini C. Identification of New Compounds That Trigger Apoptosome-Independent Caspase Activation and Apoptosis. Cancer Res. 2006;66:9235–9244. doi: 10.1158/0008-5472.CAN-06-0702. [DOI] [PubMed] [Google Scholar]
- 16.Berndtsson M, Beaujouin M, Rickardson L, Havelka AM, Larsson R, Westman J, Liaudet-Coopman E, Linder S. Induction of the lysosomal apoptosis pathway by inhibitors of the ubiquitin-proteasome system. Int J Cancer. 2009;124:1463–1469. doi: 10.1002/ijc.24004. [DOI] [PubMed] [Google Scholar]
- 17.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 18.Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 20.Watson IR, Irwin MS. Ubiquitin and ubiquitin-like modifications of the p53 family. Neoplasia. 2006;8:655–666. doi: 10.1593/neo.06439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Diff. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 23.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 24.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira AM, Hsi BL, Weremowicz S, Rosenberg AE, Dal Cin P, Joseph N, Bridge JA, Perez-Atayde AR, Fletcher JA. USP6 (Tre2) fusion oncogenes in aneurysmal bone cyst. Cancer Res. 2004;64:1920–1923. doi: 10.1158/0008-5472.can-03-2827. [DOI] [PubMed] [Google Scholar]
- 26.Shembade N, Ma A, Harhaj EW. Inhibition of NFkappa-B Signaling by A20 Through Disruption of Ubiquitin Enzyme Complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacco JJ, Coulson JM, Clague MJ, Urbé S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabner BA, Roberts TG., Jr Chemotherapy and the war on cancer. Nature Rev Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 29.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 30.Colland F. The therapeutic potential of deubiquitinating enzyme inhibitors. Biochem Soc Trans. 2010;38:137–143. doi: 10.1042/BST0380137. [DOI] [PubMed] [Google Scholar]
- 31.Mullally JE, Fitzpatrick FA. Pharmacophore model for novel inhibitors of ubiquitin isopepti-dases that induce p53-independent cell death. Mol Pharmacol. 2002;62:351–358. doi: 10.1124/mol.62.2.351. [DOI] [PubMed] [Google Scholar]
- 32.Straus DS, Glass CK. Cyclopentenone pros-taglandins: new insights on biological activities and cellular targets. Med Res Rev. 2001;21:185–210. doi: 10.1002/med.1006. [DOI] [PubMed] [Google Scholar]
- 33.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontanini A, Foti C, Potu H, Crivellato E, Maestro R, Bernardi P, Demarchi F, Brancolini C. The Isopeptidase Inhibitor G5 Triggers a Caspase-independent Necrotic Death in Cells Resistant to Apoptosis: a comparative study with the proteosome inhibitor bortezomib. J Biol Chem. 2009;284:8369–8381. doi: 10.1074/jbc.M806113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foti C, Florean C, Pezzutto A, Roncaglia P, Tomasella A, Gustincich S, Brancolini C. Charac-terization of caspase-dependent and caspase-independent deaths in glioblastoma cells treated with inhibitors of the ubiquitin-proteasome system. Mol Cancer Ther. 2009;8:3140–3150. doi: 10.1158/1535-7163.MCT-09-0431. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Chou CY, Chang GG. Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antivir Chem Chemother. 2009;19:151–156. doi: 10.1177/095632020901900402. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends Biochem Sci. 2007;32:37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 Family Reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demarchi F, Brancolini C. Altering protein turnover in tumor cells: new opportunities for anti-cancer therapies. Drug Resist Updat. 2005;8:359–368. doi: 10.1016/j.drup.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Henderson CJ, Aleo E, Fontanini A, Maestro R, Paroni G, Brancolini C. Caspase activation and apoptosis in response to proteasome inhibitors. Cell Death Differ. 2005;12:1240–1254. doi: 10.1038/sj.cdd.4401729. [DOI] [PubMed] [Google Scholar]
- 44.Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, Mitsiades C, Mitsiades N, Yasui H, Letai A, Ovaa H, Berkers C, Nicholson B, Chao TH, Neuteboom ST, Richardson P, Palladino MA, Anderson KC. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;5:407–419. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J Biol Chem. 2000;275:21648–21652. doi: 10.1074/jbc.M001083200. [DOI] [PubMed] [Google Scholar]
- 46.Kim Y, Suh N, Sporn M, Reed JC. An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAILinduced apoptosis. J Biol Chem. 2002;277:22320–22329. doi: 10.1074/jbc.M202458200. [DOI] [PubMed] [Google Scholar]
- 47.Peter ME. The flip side of FLIP. Biochem J. 2004:e1–e3. doi: 10.1042/BJ20041143. 3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poukkula M, Kaunisto A, Hietakangas V, Denessiouk K, Katajamaki T, Johnson MS, Sistonen L, Eriksson JE. Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J Biol Chem. 2005;280:27345–27355. doi: 10.1074/jbc.M504019200. [DOI] [PubMed] [Google Scholar]
- 49.Sayers TJ, Brooks AD, Koh CY, Ma W, Sek N, Raziuddin A, Blazar BR, Zhang X, Elliott PJ, Murphy WJ. The proteasome inhibitor PS-341 sensitizes neoplastic cells to TRAIL mediated apoptosis by reducing levels of c-FLIP. Blood. 2003;102:303–310. doi: 10.1182/blood-2002-09-2975. [DOI] [PubMed] [Google Scholar]
- 50.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-Based Polyubiquitination and p62-Dependent Aggregation of Caspase-8 Mediate Extrinsic Apoptosis Signaling. Cell. 2009;137:604–606. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 51.Song JH, Tse MC, Bellail A, Phuphanich S, Khuri F, Kneteman NM, Hao C. Lipid rafts and nonrafts mediate tumor necrosis factor related apoptosis inducing ligand induced apoptotic and nonapoptotic signals in non small cell lung carcinoma cells. Cancer Res. 2007;67:6946–6955. doi: 10.1158/0008-5472.CAN-06-3896. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Aris VM, Ogburn KD, Soteropoulos P, Figueiredo-Pereira ME. Prostaglandin J2 alters pro-survival and pro-death gene expression patterns and 26 S proteasome assembly in human neuroblastoma cells. J Biol Chem. 2006;281:21377–21386. doi: 10.1074/jbc.M601201200. [DOI] [PubMed] [Google Scholar]
- 53.Daviet L, Colland F. Targeting ubiquitin specific proteases for drug discovery. Biochimie. 2008;90:270–283. doi: 10.1016/j.biochi.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Storer AC, Menard R. Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 55.Hu M, Li P, Li M, Li W, Yao T, Wu J W, Gu W, Cohen R E, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 56.Reyes Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase. J Biol Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 58.Renatus M, Parrado SG, D'Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, Kroemer M. Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome associated deubiquitinating enzyme USP14. EMBO J. 2005;21:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engels IH, Daguia C, Huynh T, Urbina H, Buddenkotte J, Schumacher A, Caldwell JS, Brinker A. A time-resolved fluorescence resonance energy transfer-based assay for DEN1 peptidase activity. Anal Biochem. 2009;390:85–87. doi: 10.1016/j.ab.2009.03.035. [DOI] [PubMed] [Google Scholar]
- 61.Arnold JJ, Bernal A, Uche U, Sterner DE, Butt TR, Cameron CE, Mattern MR. Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity. Anal Biochem. 2006;350:214–221. doi: 10.1016/j.ab.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang SH, Park JJ, Chung SS, Bang OS, Chung CH. Strategies for assaying deubiquitinating enzymes. Methods Enzymol. 2005;398:500–508. doi: 10.1016/S0076-6879(05)98041-5. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, Yeh LA, Cuny GD, Stein RL, Lansbury PT., Jr Discovery of Inhibitors that Elucidate the Role of UCH-L1 Activity in the H1299 Lung Cancer Cell Line. Chem Biol. 2003;10:837–846. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 66.Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim HJ, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- 67.Rolén U, Freda E, Xie J, Pfirrmann T, Frisan T, Masucci MG. The ubiquitin C-terminal hydrolase UCH-L1 regulates B-cell proliferation and integrin activation. J Cell Mol Med. 2008;13:1666–1678. doi: 10.1111/j.1582-4934.2008.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mermerian AH, Case A, Stein RL, Cuny GD. Structure-activity relationship, kinetic mechanism, and selectivity for a new class of ubiquitin C-terminal hydrolase-L1 (UCH-L1) inhibitors. Bioorganic & Medicinal Chemistry Letters. 2007;17:3729–3732. doi: 10.1016/j.bmcl.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 69.Mitsui T, Hirayama K, Aoki S, Nishikawa K, Uchida K, Matsumoto T, Kabuta T, Wada K. Identification of a novel chemical potentiator and inhibitors of UCH-L1 by in silico drug screening. Neurochemistry International. 2010;56:679–686. doi: 10.1016/j.neuint.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 70.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression:disruption of HAUSP gene stabilizes p53. Nature. 2004;428(1):486. doi: 10.1038/nature02501. following. [DOI] [PubMed] [Google Scholar]
- 71.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 72.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 74.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, Calabrese A, Borg-Capra C, Sippl W, Collura V, Boissy G, Rain JC, Guedat P, Delansorne R, Daviet L. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 75.Lopez R, Collura V, Sippl W, Colland F. Novel specific inhibitors of ubiquitin specific protease 7, the pharmaceutical compositions thereof and their therapeutic applications. Eur Pat. 2009 Application 09305030.0. [Google Scholar]
- 76.Wiliams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 77.Colombo M, Vallese S, Peretto I, Jacq X, Rain JC, Colland F, Guedat P. Synthesis and Biological Evaluation of 9-Oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile Analogues as Potential Inhibitors of Deubiquitinating Enzymes. ChemMed-Chem. 2010;5:552–558. doi: 10.1002/cmdc.200900409. [DOI] [PubMed] [Google Scholar]
- 78.Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Mesecar AD. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008;105:16119–24. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Novartis AG. 2007. WO2007009715;
- 80.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, Yue P, Dornan D, Huang DC, Dixit VM. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 81.Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an Important Regulator of the Susceptibility to IFN-α and Drug-Induced Apoptosis. Cancer Res. 2010;70:1–11. doi: 10.1158/0008-5472.CAN-09-1942. [DOI] [PubMed] [Google Scholar]
- 82.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 83.Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, Lechpammer M, Huesken D, Zimmermann J, Signoretti S, Loda M. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 84.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 86.Zhao B, Schlesiger C, Masucci MG, Lindsten K. The ubiquitin specific protease 4 (Usp4) is a new player in the Wnt signalling pathway. J Cell Mol Med. 2009;13:1886–1895. doi: 10.1111/j.1582-4934.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY -mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Niendorf S, Oksche A, Kisser A, Löhler J, Prinz M, Schorle H, Feller S, Lewitzky M, Horak I, Knobeloch KP. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27:5029–39. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma YM, Boucrot E, Villén J, Affar el B, Gygi SP, Göttlinger HG, Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J Biol Chem. 2007;282:9805–12. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 91.Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 92.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–35. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 93.Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999;4:757–67. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- 94.Nagai H, Noguchi T, Homma K, Katagiri K, Takeda K, Matsuzawa A, Ichijo H. Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol Cell. 2009;36:805–18. doi: 10.1016/j.molcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 95.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 Regulates p53 Localization and Stability by Deubiquitinating p53. Cell. 2010;140:1–13. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wiltshire TD, Lovejoy CA, Wang T, Xia F, O'Connor MJ, Cortez D. Sensitivity to poly (ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J Biol Chem. 2010 doi: 10.1074/jbc.M110.104745. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamaguchi T, Kimura J, Miki Y, Yoshida K. The deubiquitinating enzyme USP11 controls an IkB kinase α (IKKα)-p53 signaling pathway in response to tumor necrosis factor α (TNFα) J Biol Chem. 2007;282:33943–33948. doi: 10.1074/jbc.M706282200. [DOI] [PubMed] [Google Scholar]
- 98.Shinji S, Naito Z, Ishiwata S, Ishiwata T, Tanaka N, Furukawa K, Suzuki H, Seya T, Matsuda A, Katsuta M, Tajiri T. Ubiquitin-specific protease 14 expression in colorectal cancer is associated with liver and lymph node metastases. Oncology Rep. 2006;15:539–543. [PubMed] [Google Scholar]
- 99.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 100.Burrows JF, Kelvin AA, McFarlane C, Burden RE, McGrattan MJ, De la Vega M, Govender U, Quinn DJ, Dib K, Gadina M, Scott CJ, Johnston JA. USP17 regulates Ras activation and cell proliferation by blocking RCE1 activity. J Biol Chem. 2009;284:9587–9595. doi: 10.1074/jbc.M807216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duex JE, Sorkin A. RNA interference screen identifies Usp18 as a regulator of epidermal growth factor receptor synthesis. Mol Biol Cell. 2009;20:1833–1844. doi: 10.1091/mbc.E08-08-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Commun. 2002;294:700–9. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 103.Berthouze M, Venkataramanan V, Li Y, Shenoy SK. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 2009;28:1684–96. doi: 10.1038/emboj.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–8. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T. Deubiquitylation of histone H2A activates transcriptional initiation via transhistone crosstalk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu G, Tan X, Wang H, Sun W, Shi Y, Burlingame S, Gu X, Cao G, Zhang T, Qin J, Yang J. Ubiquitin -specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285:969–78. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycombcatalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7:1522–4. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 109.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 110.Van Leuken RJ, Luna-Vargas MP, Sixma TK, Wolthuis RM, Medema RH. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle. 2008;7:2710–9. doi: 10.4161/cc.7.17.6553. [DOI] [PubMed] [Google Scholar]
- 111.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446(8):76–81. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 112.Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, Jones C, Hansen J, Blair E, Hofmann B, Siebert R, Turner G, Evans DG, Schrander-Stumpel C, Beemer FA, van Den Ouweland A, Halley D, Delpech B, Cleveland MG, Leigh I, Leisti J, Rasmussen S. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–5. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 113.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. Deubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]