Abstract
Background
In preparation for potential clinical development of Ab-01, an antagonistic antibody directed against the IL21R, studies were undertaken to address translational medicine needs that fall into four categories: 1) development of a pharmacodynamic biomarker assay suitable for use in the clinic, 2) demonstration that Ab-01 has the desired biological activity in vitro and in vivo in cynomolgus monkeys, the preferred safety study species, 3) pre-clinical in vivo proof-of-concept that the assay can be used to detect Ab-01 pharmacodynamic (PD) activity in treated subjects, and 4) comprehensive assessment of the agonistic potential of Ab-01 when cross-linked. This report and a recently published companion report address the first three of these needs. The fourth has been addressed in a separate study.
Methods
Genes that change RNA expression upon ex vivo rhIL21 stimulation of whole blood were identified in human and cynomolgus monkey. The inhibitory effects of exogenously added Ab-01 were measured ex vivo in human and monkey, and the in vivo inhibitory effects of Ab-01 treatment were measured in monkey.
Results
Stimulation of whole human blood for 2 hours with rhIL21 induced robust increases in RNA expression of 6 genes. This response was blocked by Ab-01, indicating that the assay is suitable for measuring Ab-01 activity in blood. rhIL21 induced expression of a similar set of genes in cynomolgus monkey blood. This response was blocked with Ab-01, thus demonstrating that Ab-01 has the desired activity in the species, and that safety studies done in cynomolgus monkeys are relevant. Proof -of-concept for using this assay system to detect PD activity in vivo was generated by measuring the response in monkey blood to ex vivo rhIL21 stimulation before and 5 minutes following in vivo Ab-01 administration.
Conclusions
A robust PD biomarker assay suitable for clinical use has been developed in human whole blood. The successful adaptation of the assay to cynomolgus monkeys has enabled the demonstration of Ab-01 activity both in vitro and in vivo in monkey, thus validating the use of this species in safety studies and establishing proof-of-concept for using this PD assay system to aid in dose selection in clinical studies.
Background
Development of protocols for appropriate dose selection in clinical studies is a clear priority within medical [1] and regulatory [2] communities. The high attrition rate of drugs in development due to toxicity and/or lack of efficacy [3,4] underscores the need for biomarker assays to provide early information on whether the compound being tested does indeed have the expected effect on the targeted pathway. This information can be used to mitigate the risk of entering into lengthy and expensive efficacy studies. To have an impact on clinical development, a robust PD biomarker assay must be developed well in advance of phase I clinical studies. The assay must also function reliably in the population used for phase I studies, which, in the case of compounds directed towards blockade of inflammatory pathways, is often a healthy volunteer population. To develop biomarkers for drugs targeting inflammatory pathways, previous investigators have turned to ex vivo stimulation in whole blood [5,6]. This approach has been particularly useful in the development of p38 MAPK inhibitor compounds [7] in which LPS (lipopolysaccharide)-induced production of inflammatory cytokines can be measured. We followed this basic approach (ex vivo stimulation of whole blood) to develop pharmacodynamic biomarker assays for a candidate therapeutic antibody, Ab-01.
Ab-01, a human antibody generated by phage display, recognizes the high affinity receptor for IL-21, IL21R, blocks IL21-mediated immune activation through antagonist engagement of IL21R and has shown efficacy in a mouse model of lupus [8]. The goal of the biomarker strategy was to provide the means of avoiding toxicity due to unnecessarily high drug levels and lack of efficacy due to ineffective dosing by providing early clinical data on how well the drug hits the target in vivo, and on the best dosing regimen to maintain target engagement/inhibition. A second critical goal while preparing for potential clinical testing was clear demonstration of the desired biological activity in cynomolgus monkeys, the safety study species. In the absence of such data, the relevance of safety studies is uncertain. Therefore, in parallel, we applied our biomarker strategy to cynomolgus monkeys and used it to examine ex vivo and in vivo Ab-01 activity in this species. Here we report the development of PD biomarker assays that measure Ab-01 biological activity in human and cynomolgus monkey samples. In addition we provide pre-clinical proof-of-concept that the assay system can be used to measure PD activity in treated subjects.
Methods
Sample source and human PD biomarker assay development
Pilot studies on whole blood from 12 healthy human donors were performed to identify biomarkers of ex vivo response of blood to stimulation with rhIL21. Human blood samples from healthy volunteers were collected under the Wyeth Human Blood Donor Program - a program approved and administered by Mt Auburn Hospital, Cambridge, MA. Informed consent was obtained from all donors. A total of 7 donors were used for the initial pilot studies used for assay development, and an additional 9 donors were used for the confirmatory experiments reported here. Whole blood samples were collected in BD Vacutainer™ CPT™ cell preparation tubes containing sodium heparin (Catalogue #362753). For all data shown samples were maintained at ambient temperature and were processed within an hour of collection, but additional studies indicated acceptable assay performance in blood that had been stored overnight at room temperature (data not shown).
Protein reagents: rhIL-21, Ab-01, and control antibodies
The protein reagents used in this study - rhIL21 (recombinant human IL21), anti-IL21 receptor antibody Ab-01 (also known as clone VL6 and ATR-107), control antibody human IgG1 α-tetanus triple mutant (IgG1TM, containing the same mutations in the Fc region as Ab-01), were made by the Biological Technologies Department at Wyeth (now Pfizer) Research (Cambridge, MA). Characteristics of rhIL21 are described in Additional file 1. The three mutations common to the Fc portion of Ab01 and IgG1TM reduced their potential effector activity. Antibodies with these mutations had undetectable activity in antibody-dependent cell-mediated cytotoxicity (ADCC) or C1q binding assays [9,10]. An antibody with severely compromised effector function was chosen for development because the therapeutic goal is to block the interaction of IL21 with IL21R, and therefore minimization of effector function is desirable. Endotoxin levels in all proteins reagents were determined to be below 1.0 EU/mg.
Ex vivo treatment of human blood
Human blood was distributed (1 mL/aliquot) into screw cap cryovials (Nunc, Cat# 375353). All treatments were run in duplicate. rhIL21 (produced from Chinese hamster ovary cells at Wyeth, now Pfizer) was added in volumes ranging from 3 μL to 10 μL to achieve the indicated concentration. A similar volume of PBS was added to unstimulated control samples. Samples were incubated at 37°C for the indicated duration while mixing continuously at approximately 15 revolutions per minute using a Rotamix rotating mixer (ATR Inc, Laurel, MD). To investigate Ab-01-mediated inhibition of rhIL21 response, Ab-01 was added to blood prior to addition of rhIL21. During the assay development phase of the work, Ab-01 was added immediately prior to addition of rhIL21, and total inhibition of the response was observed (data not shown). Since manipulation of samples immediately upon collection would not have been practical in the setting of a clinical study, the final assay protocol included a two hour incubation period in the presence or absence of Ab-01. This protocol mimicked the conditions of the intended clinical use of the assay, since blood from Ab-01 treated subjects (containing Ab-01) would have to placed in a queue in a laboratory prior to addition of rhIL21. The experiments with human blood reported here included a 2 hour incubation at 37°C prior to the addition of rhIL21. Human blood (in 1 mL aliquots) was pre-incubated for 2 hours with Ab-01 or IgG1TM control immunoglobulin at increasing concentrations followed by the addition of rhIL21(10 ng/mL) and subsequent 2 hour incubation.
RNA isolation
Aliquots of blood (0.5 mL) were removed following treatments and added to 2.0 mL microtubes (Axygen Scientific, Union City, CA) containing 1.3 mLs of RNAlater® (Applied Biosystems/Ambion, Austin, TX, Catalogue #AM1928), and mixed thoroughly by 5 complete inversions. Samples were stored at ambient temperature overnight and then frozen at -80°C pending RNA purification. RNA was isolated using the Human RiboPure™-Blood Kit (Applied Biosystems/Ambion Austin TX, Catalogue #AM1928) following the manufacturer's protocol. The Human RiboPure™ RNA isolation procedure involves cell lysis in a guanidinium-based solution and initial purification of the RNA by phenol/chloroform extraction followed by final RNA purification by solid-phase extraction on a glass-fiber filter. The residual genomic DNA was removed according to the manufacturer's instructions by DNAse treatment using the DNA-free™ reagents provided in the kit.
RNA quantity was determined by absorbance at 260 nm with a NanoDrop 1000 (NanoDrop, Wilmington, DE). RNA quality was evaluated using a 2100 Bioanalyzer (Agilent, Palo Alto, CA, Agilent 2100 expert software version B.02.05.SI360), and all samples had RIN (RNA integrity number) [11] >6.6, and all but 2 had RIN values >7.0. Samples were stored at -80°C until cDNA synthesis was performed.
Measurement of gene expression levels using real time RT-PCR
Based on results from the pilot studies (data not shown), assays for gene transcripts with potential as biomarkers were selected for inclusion on a custom TaqMan Low Density Array (TLDA) purchased from Applied Biosystems (ABI) Foster City, CA. This TLDA contained a total of 24 assays measuring 19 potential biomarkers and 5 endogenous controls (Table 1). Two independent measurements of each transcript were obtained from each sample. Following the manufacturer's instruction, 400 ng of total RNA were used to generate cDNA in 40 μL reaction volume in a DNA Engine Peltier Thermal Cycler (MJ Research, GMI Inc., Ramsey, MN) using a High Capacity cDNA Reverse Transcription Kit (ABI, #4368814) with addition of RNase Inhibitor at 50 U/sample (ABI, #N808-0119). Reaction conditions were: 25°C for 10 minutes, 37°C for 2 hours, 85°C 5 seconds and then hold at 4°C. If TLDA amplification reactions were not performed on the same day as cDNA synthesis, the cDNA samples were stored at -20°C. The amount of cDNA to be loaded on the TLDA was determined empirically by titration in a pilot study. Results showed that the amount of cDNA produced from 200 ng of starting RNA yielded values above the lower detection limit for all but two of the candidate biomarkers, and 200 ng (equivalent) was used in all subsequent experiments. The cDNA product (in 20 μl volume) was diluted by addition of 30 μl DEPC water and mixed with 50 μl TaqMan® Universal 2 × PCR Master Mix (ABI, #4304437) for a final volume of 100 μl, and added to each TLDA port. Assay was performed on an ABI PRISM 7900 Sequence Detector (Sequence Detector Software v2.2.2) using universal thermal cycling conditions of 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data output was generated from ABI's SDS 2.2.2 software that determines CT (threshold cycle) values from the PCR amplification plot.
Table 1.
Assays used to measure human genes on custom TaqMan low density array for human studies
| Gene | Gene Description | Assay ID (ABI) |
|---|---|---|
| 18S* | Eukaryotic 18S rRNA | Hs99999901_s1 |
| CCL3 | chemokine (C-C motif ligand | Hs00234142_m1 |
| CD19 | CD19 | Hs00174333_m |
| CXCL10 | chemokine (C-X-C motif ligand 10) | Hs00171042_m1 |
| CXCL11 | chemokine (C-X-C motif ligand 11) | Hs00171138_m1 |
| GNLY | Granulysin | Hs00246266_m1 |
| GAPDH* | glyceraldehyde 3 phosphate dehydrogenase | Hs99999905_m1 |
| GUSB* | glucuronidase, beta | Hs99999908_m1 |
| GZMB | Granzyme B (cytotoxic T lymphocyte-associated serine esterase 1) | Hs00188051_m1 |
| ICAM1 | intercellular adhesion molecule 1 (CD54) | Hs00164932_m1 |
| IFNg | interferon, gamma | Hs00174143_m1 |
| IL10 | interleukin 10 | Hs00174086_m1 |
| IL12A | interleukin 12A (natural killer cell stimulatory factor 1 | Hs00168405_m1 |
| IL1b | interleukin 1, beta | Hs00174097_m1 |
| IL21R | interleukin 21 receptor | Hs00222310_m1 |
| IL2RA | interleukin 2 receptor, alpha, CD25 | Hs00166229_m1 |
| IL6 | interleukin 6 | Hs00174131_m1 |
| IL8 | interleukin 8 | Hs00174103_m1 |
| PGK1* | phosphoglycerate kinase | Hs99999906_m1 |
| PRF1 | perforin 1 (pore forming protein) | Hs00169473_m1 |
| STAT3 | signal transducer and activator of transcription 3 | Hs00234174_m1 |
| TBX21 | T box 21 | Hs00203436_m1 |
| TNF | tumor necrosis factor (TNF superfamily, member 2) | Hs00174128_m1 |
| ZNF592* | zinc finger protein 592 | Hs00206029_m1 |
*Gene used as endogenous normalizer
Description of calibrators and normalization of results using endogenous control genes
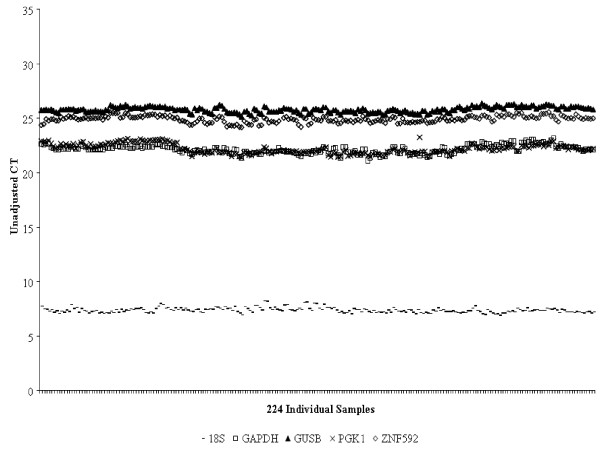
Calibrator samples functioned as the common comparator for RQ (Relative Quantification of RNA expression) calculations. The average CT values for all genes in the unstimulated samples from the first 5 donors following 2 hour incubation served as calibrator for experiments used to determine optimal rhIL21 dose and time course. Similarly, the average CT of the unstimulated samples from the second set of 4 donors was used as calibrator for the experiments related to titration of Ab-01 activity. Since very small differences in the amount of RNA used in the amplification reaction can result in significant differences in CT values, the procedures for normalization of RNA amounts in starting reactions are described in detail here. The genes chosen as normalizer genes were 18S, GAPDH, GUSB, PGK, and ZNF592. (ZNF592 was identified from a large GeneChip database as expressed at very consistent levels in human peripheral blood mononuclear cells, A. Hill and M. O'Toole, unpublished observations). The appropriateness of the 5 genes chosen as normalizers is demonstrated by the consistent expression among samples of each of the 5, Figure 1. RQ values were calculated using the delta delta CT method [12]. CT values of the endogenous controls were averaged for each sample. This average value was used to normalize RNA levels between samples. Expression levels of test genes were calculated as CT of gene - CT of the average of endogenous controls for that sample (delta CT). Gene expression values were calculated as: delta CT of gene minus delta CT of calibrator (delta delta CT) Data were then expressed as RQ (fold change over calibrator, 2E-delta delta CT or 2-ΔΔCT).
Figure 1.
Expression levels of normalizer genes. The unadjusted CT values for 5 genes used as endogenous normalizers are shown and reveal very similar levels of expression in all study samples.
Cynomolgus monkey PD biomarker assay development animals and sample collection
Adult cynomolgus monkeys (Macaca fascicularis; Charles River BRF, Inc, Houston, TX) weighing 6 to 9 kg were singly or pair housed and cared for according to the American Association for Accreditation of Laboratory Animal Care guidelines. The Wyeth Institutional Animal Care and Use Committee approved all aspects of this study. Under ketamine sedation (Ketaset, Fort Dodge Laboratories Inc., Fort Dodge, IA, 10 mg/kg IM), the femoral area was cleaned with povidone-iodine (Betadine; Purdue Frederick Co, Norwalk, CT) preparation solution followed by alcohol. Blood was drawn into Vacutainer CPT mononuclear cell preparation tubes (Catalogue #362761, BD, Franklin Lakes, NJ).
Ex vivo treatment of monkey blood
rhIL-21 was added to aliquoted blood on the same day that the blood was drawn. When samples were treated with both antibody and rhIL21, the antibody was added and mixed thoroughly immediately prior to rhIL21 addition. Samples were then incubated at 37°C for 4 hours. Aliquots (0.5 mL) were removed and added to 2.0 mL microtubes (Axygen Scientific, #10011-744) containing 1.3 mLs of RNAlater® supplied with the Human RiboPure™-Blood Kit and mixed thoroughly by 5 complete inversions. Samples were stored at ambient temperature overnight and then frozen at -80°C pending RNA purification. This report and the report by Vugmeyster et al. [13] document the ex vivo response to rhIL21 stimulation in a combined total of 47 monkeys.
Measurement of Ab-01 PD activity in monkeys dosed with Ab-01
Antibody was administered by means of bolus intravenous (i.v.) infusion via saphenous vein catheter (22G 1" Surflo, Terumo Co). Groups of animals were administered IgG1TM control antibody (n = 3), or Ab-01 (n = 3) at a dose of 10 mg/kg. Blood samples were drawn prior to antibody administration and 5 minutes post dosing.
RNA isolation, description of custom TLDA and assay of RNA concentration for monkey studies
RNA isolation was performed as described above for the human blood assay. The pilot work for the assay was performed on the Human Immune TLDA (ABI), which contains assays measuring the levels of 96 different transcripts. Any assay that detected an IL21 response in human and/or monkey blood was selected for inclusion on a custom TLDA designed for the monkey studies. If the assay for the human gene was capable of measuring the monkey transcript, the human assay was retained on the custom TLDA for monkey studies. For genes that responded to IL21 in humans but were not detectable in the monkey using primers and probes designed for the human sequence, primers and probes designed to detect rhesus genes were used for the custom TLDA because the assay for cynomolgus monkeys were not, in general, available as predesigned Gene Expression Assays from ABI. All TaqMan assays included on the custom TLDA for monkey studies were among the "inventoried" assays available from ABI, and are described in Table 2. cDNA synthesis, preparation of samples for TLDA assay and measurements of RNA concentration were performed as described for the human assay.
Table 2.
Assays used to measure monkey genes on custom TaqMan low density array for monkey studies
| Gene ID | Use | Species | Assay ID (ABI) |
|---|---|---|---|
| 18S | Manufacturing QC | Human | Hs99999901_s1 |
| CD19 | Effects of IL21/Ab-01 | Human | Hs00174333_m1 |
| CSF1 | Effects of IL21/Ab-01 | Human | Hs00174164_m1 |
| GUSb | Normalizer | Human | Hs99999908_m1 |
| GZMB | Effects of IL21/Ab-01 | Rhesus | Rh02621701_m |
| ICOS | Effects of IL21/Ab-01 | Rhesus | Rh02621771_m1 |
| IFNγ | Effects of IL21/Ab-01 | Rhesus | Rh02621721_m1 |
| IL10 | Effects of IL21/Ab-01 | Human | Hs00174086_m1 |
| IL12B | Effects of IL21/Ab-01 | Human | Hs00233688_m1 |
| IL21R | Effects of IL21/Ab-01 | Human | Hs00174086_m |
| IL2RA | Effects of IL21/Ab-01 | Human | Hs00166229_m1 |
| IL6 | Effects of IL21/Ab-01 | Rhesus | Rh02621719_u1 |
| IL7 | Effects of IL21/Ab-01 | Human | Hs00174202_m1 |
| IL8 | Effects of IL21/Ab-01 | Human | Hs00174103_m1 |
| PGK1 | Normalizer | Human | Hs99999906_m1 |
| PRF1 | Effects of IL21/Ab-01 | Human | Hs00169473_m1 |
| STAT3 | Effects of IL21/Ab-01 | Human | Hs00234174_m1 |
| TBX2 | Effects of IL21/Ab-01 | Human | Hs00203436_m1 |
| TNF | Effects of IL21/Ab-01 | Human | Hs00174128_m1 |
| ZNF592 | Normalizer | Human | Hs00206029_m1 |
In addition to the assays listed, assays for CCL19, CSF2, IL-17, and REN were run but found to be unreliable in these cynomolgus monkey samples and are not shown.
Results
Time course and dose response of ex vivo response of human whole blood to rhIL21
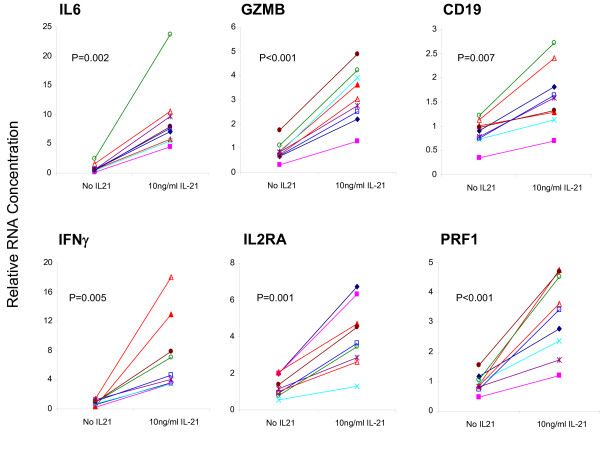
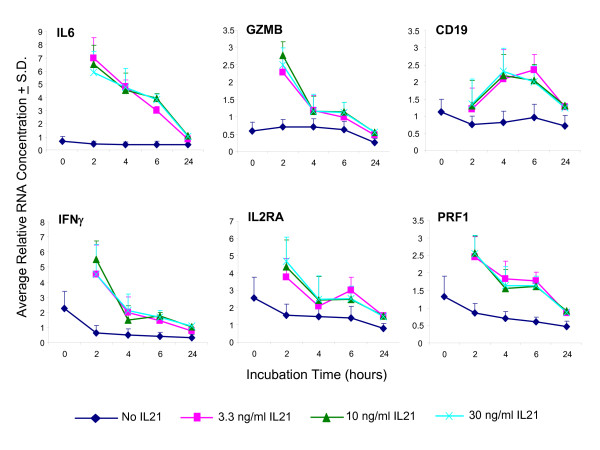
Whole blood samples from 5 healthy human donors were incubated in the presence of 3.3, 10 or 30 ng/mL of rhIL21 for 2, 4, 6 or 24 hours. Consistent with prior pilot exploratory studies performed on 10 human blood donors, the most significant and robust rhIL21-dependent signals were obtained for six genes: IL6, IFNγ, IL2RA, GZMB, PRF1, CD19 (Figure 2). These 6 genes were therefore chosen as biomarkers of IL21 activity in whole blood. The optimal signal for all but CD19 was obtained at 2 hours (Figure 3). There was no difference in the response obtained at 3.3, 10 or 30 ng/mL rhIL21. Based on the results obtained with these 5 donors, the assay conditions chosen for subsequent experiments on ex vivo whole blood response to rhIL21 were 2 hour stimulation with 10 ng/mL of rhIL21.
Figure 2.
Whole human blood response to ex vivo stimulation with 10 ng/mL rhIL21 for 2 hours. Data shown are for the 6 genes (of 19 tested) with the most consistent response among the 9 individual donors, and are identified as 6 preferred biomarkers of rhIL21 activity in whole human blood. These 6 genes were also identified as the best indicators of IL21 response in a series of pilot studies conducted with blood from a different group of donors (data not shown).
Figure 3.
Determination of optimal time point for rhIL21-response for the 6 genes identified as preferred biomarkers. This time point experiment was done using 5 of the donors shown in Figure 2. Earlier pilot studies had suggested that stimulation for 30 minutes was significantly sub-optimal. There was no significant difference in response between all doses tested (30, 10, and 3 ng/mL). The lowest dose of rhIL21 that elicits a response has therefore not been determined.
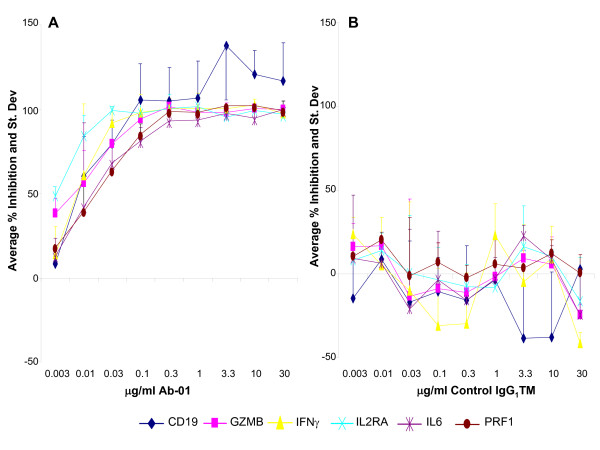
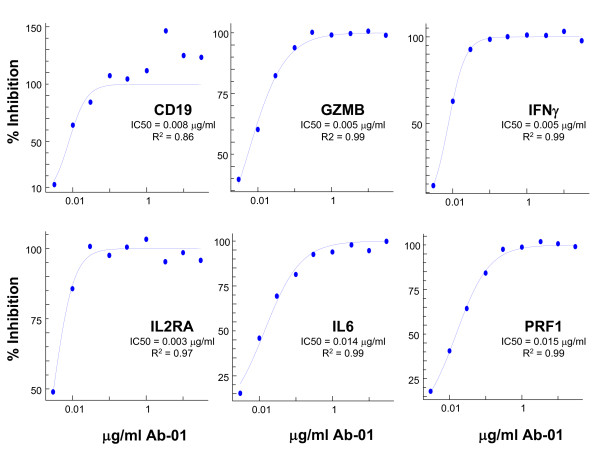
Titration of Ab-01 inhibition of ex vivo response to rhIL21
Samples from 4 individual healthy human donors were pre-incubated for 2 hours at the indicated concentration of Ab-01 or the control IgG1TM prior to addition of 10 ng/mL rhIL21 and 2 hr incubation, and the effect on the 6 biomarkers was then assessed (Figures 4 and 5). For the first 2 donors tested, even the lowest concentration of Ab-01 (0.1 μg/mL, 0.66 nM) resulted in complete inhibition of the rhIL21 response, therefore the two subsequent donors were tested at increasing concentrations of Ab-01 starting at 0.003 μg/mL. Ab-01 inhibited the response of all 6 genes in all 4 donors. IC50 values ranged between 0.003 and 0.015 μg/mL Ab-01(Figure 5). Control IgG1TM had no significant effect on rhIL21 response (Figure4B).
Figure 4.
Average percent inhibition of the expression level of 6 IL21-responsive genes. Percent inhibition values were calculated based on RQ (relative quantification) values of untreated control and rhIL21-treated samples for each of the 4 donors, and subsequently the mean and standard deviation were determined for each gene shown. A: Percent inhibition in presence of Ab-01. B: Percent inhibition in presence of control IgG1TM. Data for the 0.1 μg/mL , 0.3 μg/mL and 1 μg/mL concentrations were generated using 4 donors. Data for the higher and the lower concentrations were generated using 2 donors.
Figure 5.
Inhibition by Ab-01 at the indicated concentration is shown for 6 IL21-responsive genes. IC50 values of inhibition curves shown in Figure 4A were calculated using curve fit (XLfit) program for each of the referred biomarker genes. Values for the 0.1 μg/mL, 0.3 μg/mL and 1 μg/mL concentrations were generated using 4 donors. Data for the higher and the lower concentrations were generated using 2 donors each.
Ab-01 blocks signal transduction through cynomolgus rhIL21R
In order to determine if cynomolgus monkey was a suitable choice for safety studies, we tested whether the activity of rhIL21 on monkey cells was blocked by Ab-01. We therefore first examined the activation effects of ex vivo rhIL21 stimulation on cynomolgus blood cells, and observed very similar results to those observed with human blood. Results for 5 genes most significantly increased in monkey blood stimulated ex vivo with rhIL21 are shown in Table 3. The most robust (largest magnitude change and most consistent change) rhIL21-mediated change in cynomolgus monkeys was observed for IL2RA. All animals tested (n = 48) for effects of rhIL21 on IL2RA expression levels gave a response of >1.5 fold change [13], and therefore this gene was selected as the biomarker for subsequent monkey studies. To determine if Ab-01 had the desired blocking activity of rhIL21/IL21R dependent activation of cynomolgus blood cells, its ability to block rhIL21-dependent IL2RA activation was tested. The IL2RA response was effectively blocked by Ab-01 treatment (Table 4). There was no significant difference between the response to rhIL21 in the presence and absence of control IgG1TM, while the response in the presence of Ab-01 was blocked (P < 0.001). These results show that signal induction through the interaction of cynomolgus IL21R with rhIL21 is inhibited by Ab-01, an antibody to human IL21R. Therefore Ab-01 has the intended activity in cynomolgus monkeys.
Table 3.
rhIL21-responsive genes in whole cynomolgus monkey blood
| Gene | Average Fold Change (with IL21) | SD | P-value paired t-test |
|---|---|---|---|
| IL2RA | 4.1 | 1.76 | < 0.0001 |
| PRF1 | 1.8 | 0.78 | <0.0001 |
| IL21R | 2.3 | 0.63 | <0.0001 |
| GZMB | 1.6 | 0.39 | <0.0001 |
| IL6 | 2.7 | 1.72 | 0.0007 |
rhIL21-mediated average fold increase in RNA expression in blood of 18 individual cynomolgus monkeys stimulated in vitro for 4 hours. Control (no rhIL21) RNA expression values were normalized to 1, and p values were calculated using log2 of the fold change paired with the log2 of 1 (0).
Table 4.
Inhibition by Ab-01 of ex vivo rhIL21-dependent IL2RA expression
| Animal | rHuIL21 alone | rHuIL21 + Control IgG1TM | rHuIL21 + Ab-01 |
|---|---|---|---|
| 1 | 4.7 | 3.5 | 1.2 |
| 2 | 3.8 | 3.8 | 0.9 |
| 3 | 4.1 | 5.1 | 1.4 |
| 4 | 2.8 | 2.8 | 0.9 |
| 5 | 3.3 | 3.2 | 1.3 |
| 6 | 2.8 | 2.9 | 1 |
| 7 | 3.6 | 3.7 | 1.1 |
| 8 | 7.2 | 8.8 | 1.1 |
Each value represents the fold change over the no-treatment group in expression levels of IL2RA in monkey blood samples following 4 hour ex vivo treatment as indicated.
rhIL21-mediated activation is blocked in blood from monkeys dosed with Ab-01
The utility of the ex vivo rhIL21 stimulation assay as a read-out of Ab-01 PD activity in treated individuals was tested by comparing the rhIL21 response in 3 monkeys before and 5 minutes following administration of a 10 mg/kg i.v. dose of Ab-01. The control group consisted of 3 monkeys dosed with 10 mg/kg i.v. dose of IgG1TM. Consistent with the results from the many other monkeys tested, prior to dosing with Ab-01 an increase in IL2RA expression was observed in the blood of all 6 monkeys when stimulated ex vivo with rhIL21 (Table 5). No IL2RA response was observed in blood drawn 5 minutes following i.v. administration of Ab-01 (Table 5). Dosing with control IgG1TM did not affect the subsequent ex vivo response to rhIL21. Additional blood samples were tested at later time points following the single dose of Ab-01. Results showed that as the circulating levels of Ab-01 fell over time, the ex vivo rhIL21-mediated response was restored [13]. These results, together with the pre-dose and 5 minute post-dose data in Table 5 establish that all three monkeys dosed with Ab-01 were responsive to rhIL21 before dosing, did not respond when Ab-01 was present in the blood, and returned to responsiveness when Ab-01 was cleared from circulation.
Table 5.
rhIL21 induced IL2RA expression in whole blood from cynomolgus monkeys dosed with Ab-01
| Animal | Treatment Group | Time point | Ex vivoIL2RA Fold Change Response to IL21 Stimulation | Cpeak |
|---|---|---|---|---|
| 1 | Ab-01 | pre-dose | 2.8 | not applicable |
| 1 | Ab-01 | 5 minute post dose | 0.8 | 200 |
| 2 | Ab-01 | pre-dose | 4.8 | not applicable |
| 2 | Ab-01 | 5 minute post dose | 0.8 | 139 |
| 3 | Ab-01 | pre-dose | 4.2 | not applicable |
| 3 | Ab-01 | 5 minute post dose | 0.9 | 153 |
| 4 | Control IgG1TM | pre-dose | 4.2 | not applicable |
| 4 | Control IgG1TM | 5 minute post dose | 4.3 | not applicable |
| 5 | Control IgG1TM | pre-dose | 2.7 | not applicable |
| 5 | Control IgG1TM | 5 minute post dose | 4.4 | not applicable |
| 6 | Control IgG1TM | pre-dose | 9.1 | not applicable |
| 6 | Control IgG1TM | 5 minute post dose | 7.7 | not applicable |
Relative RNA concentration of IL2RA induced by ex vivo addition of rhIL-21 to whole blood obtained from cynomolgus monkeys before and after treatment with either Ab-01 or control IgG1TM is shown. Pre-dose whole blood sample was taken on day -13 for animal 1, and on day 0 (the day of dosing) for all others. All monkeys were dosed with 10 mg/kg via intravenous (IV) route. The ex vivo response to IL21 was completely blocked in monkeys treated with Ab-01 and was not blocked in control IgG1TM treated monkeys
Discussion
We have developed a human blood biomarker assay that detects the blocking activity of Ab-01, an antibody to IL21R. In parallel we have developed an adaptation of this assay and used it to demonstrate that the IL21-dependent response in cynomolgus monkey blood is blocked both by ex vivo addition of Ab-01 to blood, and by i.v. administration prior to blood collection. These results support the use of cynomolgus monkeys for safety studies by establishing that Ab-01 hits its target in vivo and has the desired biological activity in the species. We have shown that this assay system is well suited to examining the relationship between pharmacokinetics and pharmacodynamics (PK/PD), the intended clinical use of the assay system [13]. A third critical contribution, demonstrating lack of Ab-01 agonistic activity even under circumstances designed to force an agonistic signal is described in Guo et al [14]. The data reported here and in these related reports on Ab-01 are unified by their focus on addressing central challenges of translational medicine - dose selection, elucidation of PK/PD relationships, choice of safety study species, and mitigation of risk of immunotoxicity.
The assay reported here relies on an ex vivo stimulation procedure. It is difficult to develop a robust assay for determining Ab-01 PD activity by relying upon the endogenous levels of IL21 activity on biomarker gene expression in samples upon collection, especially in healthy donors. It is possible that such a strategy could correlate PK parameters and biomarker movement during the treatment period, but it would not show direct linkage between the movement of the biomarker and the engagement of the target. Following the biomarker assay development strategy previously employed by others, we have developed a whole blood ex vivo stimulation assay. Our biomarker discovery and assay development strategies have, from the start, proceeded with the realities of the clinical setting in mind. The volume of blood required is less than 5 mL, well below the limits of a routine blood draw. The live blood sample is subjected to minimal manipulation, consisting of separation into two aliquots, addition of rhIL21 to one of the aliquots, and 2 hour incubation at 37°C with rotation. RNA preserving solution is then added, and the sample frozen. All subsequent procedures can be carried out on batched samples in a validated lab. The assay is sensitive and robust, and has shown consistent performance among healthy human and primate subjects. We have shown that the assay detects Ab-01 in the 100 pM range in humans, well within the range for potential clinical utility.
We believe that the RNA assay system reported here has significant advantages over a protein secretion assay. First, the read-out of RNA is a more proximal event than protein secretion, and we have found that, in this assay system at least, the RNA signal is more easily and reliably detected than the protein signal [14]. Detection of protein secretion required a much longer incubation period, necessitating a shift from an assay with whole blood to an assay requiring purification and culturing of peripheral blood mononuclear cells. Such sample manipulation steps compromise the ease of adaptation for clinical use and introduce additional sources of variability. Secondly, measurements made on well purified RNA are not subject to the factors (such as for example, specific binding factors, charged proteins, or rheumatoid factor), which can confound ELISA assays performed in human serum or plasma [15]. We have also found that the cost of RNA assay systems compares favorably with that of ELISA systems, especially with considerations of standardization methodology.
Concordance between humans and cynomolgus monkeys with respect to genes activated was observed for IL2RA, GZMB, IL6 and PRF1. IFNγ also increases dramatically in both species, but detection of this increase in cynomolgus monkeys required a protocol modification (purification of PBMCs to increase IFNγ RNA yield) that was not performed for the 18-monkey cohort shown in Table 3. While CD19 is among the genes listed as most significantly increased by rhIL21 in humans, its absence from the list of genes in cynomolgus monkeys does not reflect a difference between the species, but rather the unavailability of a reliable TaqMan assay for CD19 in cynomolgus monkeys. A significant rhIL21-mediated IL21R elevation was observed in both humans and monkeys. The presence of IL21R on the top gene list for monkeys and not for humans reflects a higher relative ranking in monkeys due to the lack of IFNγ and CD19 data in cynomolgus monkeys (for the reason detailed above). We conclude that the whole blood rhIL21 responses of human and monkey are remarkably similar, and our work does not identify differences between them.
The goal of this work was to identify markers to be used to assess the PD activity of Ab-01 in treated subjects. For this reason, the most reliable read-outs detectable in whole blood were selected as the most useful from a clinical operations perspective. IL21 exerts many different effects on a wide variety of lymphoid and non-lymphoid cells including fibroblasts and intestinal epithelial cells [16-18]. The activities of IL21 have been shown to be highly dependent on the lineage of the target cell, the activation state of the target cell and the presence of other co-stimulators and immune modulators [19,20]. Previous studies have shown that IL21 activates both IFNγ and IL2RA in purified NK and purified T cells [21]. CD19 and IL21R were also among the genes that changed significantly upon ex vivo exposure of whole blood to rhIL21, and IL21 has been shown to up regulate both CD19 and IL21R expression in activatec B cells [22]. Therefore the markers identified in this study include genes with previously demonstrated links to activation of the IL21 pathway in a variety of cell types. In light of the well established complexities of IL21 biological activity, it is fortuitous from a clinical development perspective that whole blood provides a useful analytical sample for detecting inhibition by a candidate therapeutic of signal transduction through IL21R.
Conclusions
The PD biomarker assay described here has been developed to facilitate safe and efficient clinical testing by positioning a clinical team to make dose selection decisions based on reliable information on in vivo biological activity. The assay system monitors levels of activity in blood and will indicate when levels are too low to block activity and when levels are significantly higher than they need to be. The basic approach here is also applicable to other therapeutic candidates, especially in indications related to inflammation. The demonstration of the desired biological activity in the safety study species is also a significant contribution in preparing for transition to the clinic.
Competing interests
All authors were employees of Pfizer (formerly Wyeth) at the time this work was performed.
Authors' contributions
MA performed all the studies on human samples reported here, analyzed the data and co-wrote the manuscript. SJ and AAW performed all the pilot studies in humans and monkeys that identified the candidate biomarkers, and they participated in the data analyses. AAH developed the customized Spotfire tool used for data analyses and reviewed statistical analyses. YG participated in the assessment and selection of TaqMan PCR assays for studies in monkeys and participated in the biomarker discovery (pilot) phase of the project. AGB performed all in vivo procedures on monkeys and contributed to the writing of the manuscript. MFS was responsible for interface with the clinical team and for securing and scheduling the resources that will be required upon hand-off for assay validation. SJ, AW, SWA and KA performed the in vitro assays on monkeys treated with Ab-01 and control Ig and analyzed the data, and KA and SA performed the experiments on monkeys dosed in vivo with Ab-01 or IgG1TM. ERL performed data analysis and review and participated in the drafting of the manuscript. DY co-led the Ab-01 team and was responsible for studies related to the characterization of the biological properties of the antibody. LB co-led the Ab-01 team and was responsible for lead antibody selection and development. MO devised the PD assay strategy, directed the biomarker discovery process, supervised assay development and co-wrote the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Description of rhIL21. Sequence and information on preparation and activity of the rhIL21 protein preparation used in these studies is shown.
Contributor Information
Maya Arai, Email: maya.arai@pfizer.com.
Sadhana Jain, Email: sadhana.jain@pfizer.com.
Amy A Weaver, Email: amy.weaver@pfizer.com.
Andrew A Hill, Email: andrew.hilll@pfizer.com.
Yongjing Guo, Email: jane.guo@pfizer.com.
Andrea G Bree, Email: andrea.bree@pfizer.com.
Michael F Smith, Jr, Email: ms6@roche.com.
Scott W Allen, Email: scott.allen@pfizer.com.
Edward R LaVallie, Email: edward.lavallie@pfizer.com.
Deborah Young, Email: deborah.young@pfizer.com.
Laird Bloom, Email: laird.bloom@pfizer.com.
Karissa Adkins, Email: karissa.adkins@pfizer.com.
Margot O'Toole, Email: margot.otoole@pfizer.com.
Acknowledgements
We thank Holly Legault, Leslie Lowe and Vikki Spaulding for exploratory studies related to assay development, Khetemenee Lam and Stephane Olland for providing rhIL21, and Mary Collins, Cheryl Nutter and Davinder Gill for critical review of the manuscript.
References
- Kuhlmann J, Wensing G. The applications of biomarkers in early clinical drug development to improve decision-making processes. Curr Clin Pharmacol. 2006;1:185–191. doi: 10.2174/157488406776872578. [DOI] [PubMed] [Google Scholar]
- Guidance for Industry Dosage and Administration Section of Labeling for Human Prescription Drug and Biological Products. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryIn formation/Guidances/UCM075066.pdf
- Danhof M, de Lange EC, Della Pasqua OE, Ploeger BA, Voskuyl RA. Mechanism-based pharmacokinetic-pharmacodynamic (PK-PD) modeling in translational drug research. Trends Pharmacol Sci. 2008;29:186–191. doi: 10.1016/j.tips.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Lavallie ER, Dorner AJ, Burczynski ME. Use of ex vivo systems for biomarker discovery. Curr Opin Pharmacol. 2008;8:647–653. doi: 10.1016/j.coph.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Ray CA, Dumaual C, Willey M, Fill J, O'Brien PJ, Gourley I, Devanarayan V, Konrad RJ. Optimization of analytical and pre-analytical variables associated with an ex vivo cytokine secretion assay. J Pharm Biomed Anal. 2006;41:189–195. doi: 10.1016/j.jpba.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Parasrampuria DA, de Boer P, Desai-Krieger D, Chow AT, Jones CR. Single-dose pharmacokinetics and pharmacodynamics of RWJ 67657, a specific p38 mitogen-activated protein kinase inhibitor: a first-in-human study. J Clin Pharmacol. 2003;43:406–413. doi: 10.1177/0091270002250615. [DOI] [PubMed] [Google Scholar]
- Vugmeyster Y, Guay H, Szklut P, Qian MD, Jin M, Widom A, Spaulding V, Bennett F, Lowe L, Andreyeva T, Lowe D, Lane S, Thom G, Valge-Archer V, Gill D, Young D, Bloom L. In vitro potency, pharmacokinetic profiles, and pharmacological activity of optimized anti-IL-21R antibodies in a mouse model of lupus. MAbs. 2010;2:335–346. doi: 10.4161/mabs.2.3.11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black R, Ekman L, Lieberburg I, Grundman M, Callaway J, Gregg KM, Jacobsen JS, Gill D, Tchistiakova L, Windom A. Immunotherapy regimes dependent on APOE status, patent application number 20090155256. 2009. http://appft.uspto.gov
- Kasaian MT, Tan XY, Jin M, Fitz L, Marquette K, Wood N, Cook TA, Lee J, Widom A, Agostinelli R, Bree A, Schlerman FJ, Olland S, Wadanoli M, Sypek J, Gill D, Goldman SJ, Tchistiakova L. Interleukin-13 neutralization by two distinct receptor blocking mechanisms reduces immunoglobulin E responses and lung inflammation in cynomolgus monkeys. J Pharmacol Exp Ther. 2008;325:882–892. doi: 10.1124/jpet.108.136515. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Vugmeyster Y, Allen S, Szklut P, Bree A, Ryan M, Ma M, Spaulding V, Young D, Guay H, Bloom L, Leach MW, O'Toole M, Adkins K. Correlation of pharmacodynamic activity, pharmacokinetics, and anti-product antibody responses to anti-IL-21R antibody therapeutics following IV administration to cynomolgus monkeys. J Transl Med. 2010;8:41. doi: 10.1186/1479-5876-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Hill A, Ramsey R, Immermann F, Corcoran C, Young D, LaVallie E, Pfeifer R, Warner G, Bologna M, Bloom L, O'Toole M. Assessing agonistic potential of a candidate therapeutic anti-IL21R antibody. J Transl Med. 2010;8:50. doi: 10.1186/1479-5876-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole M, Legault H, Ramsey R, Wynn TA, Kasaian MT. A novel and sensitive ELISA reveals that the soluble form of IL-13R-alpha2 is not expressed in plasma of healthy or asthmatic subjects. Clin Exp Allergy. 2008;38:594–601. doi: 10.1111/j.1365-2222.2007.02921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso R, Fina D, Peluso I, Stolfi C, Fantini MC, Gioia V, Caprioli F, Del Vecchio Blanco G, Paoluzi OA, Macdonald TT, Pallone F, Monteleone G. A functional role for interleukin-21 in promoting the synthesis of the T-cell chemoattractant, MIP-3alpha, by gut epithelial cells. Gastroenterology. 2007;132:166–175. doi: 10.1053/j.gastro.2006.09.053. [DOI] [PubMed] [Google Scholar]
- Monteleone G, Caruso R, Fina D, Peluso I, Gioia V, Stolfi C, Fantini MC, Caprioli F, Tersigni R, Alessandroni L, MacDonald TT, Pallone F. Control of matrix metalloproteinase production in human intestinal fibroblasts by interleukin 21. Gut. 2006;55:1774–1780. doi: 10.1136/gut.2006.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungel A, Distler JH, Kurowska-Stolarska M, Seemayer CA, Seibl R, Forster A, Michel BA, Gay RE, Emmrich F, Gay S, Distler O. Expression of interleukin-21 receptor, but not interleukin-21, in synovial fibroblasts and synovial macrophages of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50:1468–1476. doi: 10.1002/art.20218. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Spolski R. Interleukin-21: a modulator of lymphoid proliferation, apoptosis and differentiation. Nat Rev Immunol. 2005;5:688–698. doi: 10.1038/nri1688. [DOI] [PubMed] [Google Scholar]
- Zeng R, Spolski R, Finkelstein SE, Oh S, Kovanen PE, Hinrichs CS, Pise-Masison CA, Radonovich MF, Brady JN, Restifo NP, Berzofsky JA, Leonard WJ. Synergy of IL-21 and IL-15 in regulating CD8 + T cell expansion and function. J Exp Med. 2005;201:139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strengell M, Sareneva T, Foster D, Julkunen I, Matikainen S. IL-21 up-regulates the expression of genes associated with innate immunity and Th1 response. J Immunol. 2002;169:3600–3605. doi: 10.4049/jimmunol.169.7.3600. [DOI] [PubMed] [Google Scholar]
- Ettinger R, Kuchen S, Lipsky PE. The role of IL-21 in regulating B-cell function in health and disease. Immunol Rev. 2008;223:60–86. doi: 10.1111/j.1600-065X.2008.00631.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of rhIL21. Sequence and information on preparation and activity of the rhIL21 protein preparation used in these studies is shown.