Abstract
Background
The human DHRS4 gene cluster consists of three genes, DHRS4, DHRS4L2 and DHRS4L1. Among them, DHRS4 encodes NADP(H)-dependent retinol dehydrogenase/reductase. In a previous study, we investigated the alternative splicing of DHRS4 and DHRS4L2. DHRS4L1 was added to the gene cluster recently, but little is known about its structure and expression. To reveal the regulatory mechanism of the DHRS4 gene cluster expression, we studied the structure and transcription of DHRS4L1 in the context of the transcriptional behaviors of the human DHRS4 gene cluster. Based on the results of bioinformatics analysis, we propose a possible mechanism for the transcriptional regulation of the human DHRS4 gene cluster.
Results
The homologous comparison analysis suggests that DHRS4, DHRS4L2 and DHRS4L1 are three homologous genes in human. DHRS4L1 and DHRS4L2 are paralogues of DHRS4, and DHRS4L2 is the most recent member of the DHRS4 gene cluster. In the minus strand of the human DHRS4 gene cluster, a gene transcribed in an antisense direction was found containing a 5' sequence overlapping the region of exon 1 and promoter of DHRS4. By cloning the full length of RNA variants through 5'RACE and 3'RACE, we identified two transcription start sites, within exon a2 and exon 1, of this newly named gene DHRS4L1 using neuroblastoma cell line BE(2)-M17. Analysis of exon composition in the transcripts of DHRS4 gene cluster revealed that exon 1 was absent in all the transcripts initiated from exon a1 of DHRS4L2 and exon a2 of DHRS4L1.
Conclusions
Alternatively spliced RNA variants are prevalent in the human DHRS4 gene cluster. Based on the analysis of gene transcripts and bioinformatic prediction, we propose here that antisense transcription may be involved in the transcriptional initiation regulation of DHRS4 and in the posttranscriptional splicing of DHRS4L2 and DRHS4L1 for the homologous identity of DHRS4 gene cluster. Beside the alternative transcriptional start sites, the antisense RNA is novel possible factor serving to remove exon 1 from the transcripts initiated from exon a1 and exon a2.
Background
DHRS4, the fourth member of the dehydrogenase/reductase (SDR) family, is a gene encoding NADP(H)-dependent retinol dehydrogenase/reductase (NRDR). Prior to December of 2008, an examination of the DHRS4 gene on chromosome 14q11.2 in GenBank revealed that the human DHRS4 gene cluster consisted of two genes, DHRS4 and DHRS4L2. In 1997, we identified the NRDR proteins from rabbit liver tissues, and found that this protein had strong retinol oxidation and retinal reduction activities and was a crucial enzyme in the metabolism and synthesis of retinoic acid [1], an important intracellular signaling molecule involved in the regulation of cell growth, differentiation of embryonic cells, and the regulation of immune functions. The presence of NRDR in other species was confirmed recently and the enzyme was shown to have similar carbonyl reductase activity but with different substrates [2-4]. In previous studies, we also found the alternative spliced RNAs of DHRS4 and DHRS4L2 [5-7], and identified exon a1 both as a novel exon contained in the RNA variants of DHRS4L2 and an alternative transcription start site (TSS) for DHRS4L2 [7]. Exon a1 matches with a special intergene sequence, approximate 19 kb upstream of the first exon of DHRS4L2 and 559 bp downstream of the last exon of DHRS4 in the genomic DNA.
Due to an information update of GenBank in December of 2008, the gene LOC728635, located downstream of DHRS4 and DHRS4L2 and similar to the peroxisomal short-chain alcohol dehydrogenase, was renamed DHRS4L1. The human DHRS4 gene cluster is now composed of DHRS4, DHRS4L2 and DHRS4L1. To investigate the transcription of DHRS4L1 and the transcriptional regulation of DHRS4 gene cluster, we cloned the full length RNA variants of DHRS4L1 in this study, and found that DHRS4L1 initiated its transcription from two TSS, exon a2 and exon 1. Exon a2 matches with the intergene sequence between DHRS4L2 and DHRS4L1, and is homologous to exon a1 located upstream of DHRS4L2. Through the analysis of the gene structures and transcriptional behaviors of the DHRS4 gene cluster, we propose a possible mechanism involving the antisense transcripts from C14orf167 and the alternative TSS for the transcriptional regulation of the human DHRS4 gene cluster.
Results and discussion
Comparative analysis of the DHRS4 gene cluster
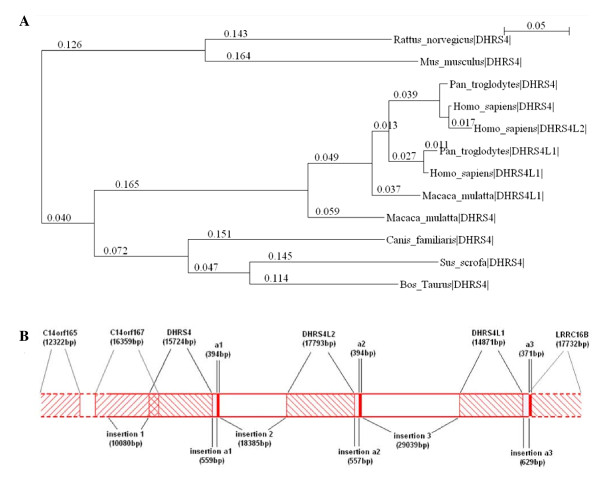
In the three copies of the DHRS4 gene cluster, DHRS4 is common in mammals and aquatic animals, while DHRS4L1 is only found in primates and DHRS4L2 in humans. Humans, therefore, are the only species that contain all three genes in the DHRS4 gene cluster. The phylogenic tree of the DHRS4 gene cluster across different species (Figure 1A) and comparative analysis of the human DHRS4 gene cluster (Table 1 and Table 2) suggest that DHRS4L2 and DHRS4L1 are paralogues of DHRS4, and that DHRS4L2 is the most recent member of the DHRS4 gene cluster. The gene sequence identity between DHRS4L2 and DHRS4L1 is lower than that of DHRS4L2 and DHRS4, indicating that DHRS4L2 was duplicated directly from DHRS4, not from DHRS4L1.
Figure 1.
The homologous comparison of DHRS4 gene cluster. (A) Evolutionary relationships of DHRS4, DHRS4L2 and DHRS4L1 among various species. The phylogeny of DHRS4, DHRS4L2 and DHRS4L1 is shown as a neighbor-joining tree with bootstrap. The scale bar represents 5% sequence divergence. Positions of genes in the corresponding species are shown as vertical hatches. Horizontal bars and the values on them represent the length of the DNA sequence after alignment. (B) The context and composition of the human DHRS4 gene clusters.
Table 1.
Homologous comparison of the human gene DHRS4, DHRS4L2 and DHRS4L1.
| DNA | cDNA | ||||
|---|---|---|---|---|---|
| Insertion* | Gene* | Insertion a | Exon a | ||
| DHRS4 vs DHRS4L2 | 62.0% | 97.5% | 98.7% | 98.9% | 90.9% |
| DHRS4 vs DHRS4L1 | 38.0% | 77.8% | 70.5% | 78.9% | 87.4% |
| DHRS4L2 vs DHRS4L1 | 42.9% | 77.7% | 70.3% | 79.2% | 81.7% |
* Long repeat sequences were removed before comparison.
Table 2.
Exons composition and the homologous relationship of them among the DHRS4 gene cluster.
| * | Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | Exon 8 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| DHRS4 | 161 | 178 | 102 | 71 | 52 | 135 | 56 | 534 | ||
| DHRS4L2 | 258 | 178 | 102 | 71 | 52 | 134 | 56 | 531 | ||
| ** | Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | Exon 8 | Exon 9 | Exon 10 |
| DHRS4L1 | 128 | 178 | 54 | 50 | 7 | 71 | 52 | 135 | 56 | 518 |
* corresponding exon number of DHRS4, DHRS4L2, ** corresponding exon number of DHRS4L1
In our previous study, a novel exon a1 was identified at the 5'end of the DHRS4L2 transcripts, and the sequence of exon a1 matches with the intergene sequence between DHRS4 and DHRS4L2 in genomic DNA [7]. Through sequence BLASTing, two other homologous sequences of exon a1 were found and each was localized downstream of the DHRS4L2 or DHRS4L1 sequences. In this study, we named these three homologous sequence of exon a as exon a1, exon a2 and exon a3, each located downstream of DHRS4, DHRS4L2 and DHRS4L1 respectively. The spaces between an exon a and its upstream gene was named as insertion a1, insertion a2 and insertion a3 while the spaces between exon a and its downstream gene was named as insertion 1, insertion 2 and insertion 3, respectively (Figure 1B). The analysis of the gene structure and homology of the DHRS4 gene cluster suggested that DHRS4 duplication was initiated from the "insertion," leading to the formation of a copy unit, "insertion-DHRS4 gene-insertion a-exon a", which was copied twice in chromosome 14 (14q11.2) with the resultant formation of the DHRS4 gene cluster (Figure 1B). Genome duplications are considered as the results of long-term biological evolution and accounting for the bulkiness and complexity of the human genome [8,9]. Through the comparative analysis, we identified the general structure of DHRS4 gene cluster, and designed the following assays to provide insights into the transcriptional regulation of this gene cluster.
Transcription of the human DHRS4 gene cluster
The reference sequences of the human DHRS4 and DHRS4L2 cDNA are 1289 bp and 1382 bp respectively, each consisting of 8 exons. DHRS4L1 has a full length of 1249 bp and is composed of 10 exons (Table 2). Based on analysis of the RNA variants of DHRS4 and DHRS4L2 identified in our previous studies and related data from GenBank, we found that the alternative splicing was prevalent in DHRS4 and DHRS4L2 (Table 3), moreover, DHRS4L2 harbored two alternative TSS (exon a1 and exon 1), while DHRS4 initiated its transcription only from one site (exon 1). To explore the transcriptional regulation mechanism of the human DHRS4 gene cluster, we first tested the presence of the DHRS4L1 RNA variants in this study since little was known about the transcription of DHRS4L1.
Table 3.
Alternative splicing variants of the human DHRS4 gene cluster.
| DHRS4 | DHRS4L2 | DHRS4L1 | |||||
|---|---|---|---|---|---|---|---|
| No. | Exon | No. | Exon | No. | Exon | No. | Exon |
| NM_021004 | 1-2-3-4-5-6-7-8 | NM_198083 | 1-2-3-4-5-6-7-8 | NM_0010824 | 1-2-3-4-5-6-7-8-9-10 | ||
| AB045131 | 1-2-3-4-5-6-7-8 | AK301373 | 1-2-6-7 | AY616183 | a1-2-3-7-8 | AA255746 | 8-9-10 |
| AF044127 | 1-2-3-4-5-6-7-8 | BC000663 | 1-2-3-4-5-6-7-8 | AY943857 | a1-2-7-8 | AV762338 | 1-8-9-10 |
| AI087304 | 3-7-8 | BC006125 | 1-2-3-4-5-6-7-8 | CD244539 | a1-2-3-4-5-6 | BC171914 | 1-8-9-10 |
| AK308436 | 1-2-3-7 | BC101812 | 1-2-3-4-5-6-7-8 | DN237879 | a1-2 | BC171918 | 1-8-9-10 |
| AK314448 | 1-2-3-4-5-6-7-8 | BC101814 | 1-2-3-4-5-6-7-8 | DN237881 | a1-2-3-6-7-8 | BX117130 | 1-2 |
| AY071856 | 1-2-3-7-8 | DB449220 | 1-2 | DN237882 | a1-2-3-7-8 | GQ871921 | a2-8-9-10 |
| AY358638 | 1-2-3-4-5-6-7-8 | DN237888 | 2-3-4-5-6-7 | DN237883 | a1-2-3-7-8 | GQ871922 | 1-8-9-10 |
| AY616182 | 1-2-3-5-7-8 | DN237893 | 2-4-5-6-7 | DN237884 | a1-2-3 | GQ871923 | 1-8-9-10 |
| BC003019 | 1-2-3-4-5-6-7-8 | DN237895 | 2-3-5 | DN237885 | a1-2-3-5 | GQ871924 | a2-2-8-9-10 |
| BU529016 | 1-2-3-7-8 | DN237896 | 2-3-7-8 | DN237886 | a1-2-3-4-5-6-7 | GQ871925 | a2-8-9-10 |
| DN237893 | 2-3-4-5-6-7-8 | BQ030242 | 8 | DN237887 | a1-2-3-5 | GQ871926 | 1-9-10 |
| DQ325464 | 1-2-4-5-6-7-8 | DQ088987 | 3-4-5-6-7-8 | DN237890 | a1-2-3-4-5 | GQ871927 | 10 |
| DQ338571 | 1-2-4-5-7-8 | DQ088988 | 3-4-5-6-8 | DN237891 | a1-2-3-5 | ||
| DQ344810 | 1-2-3-4-5-6-7-8 | DN237892 | a1-2-4-5 | ||||
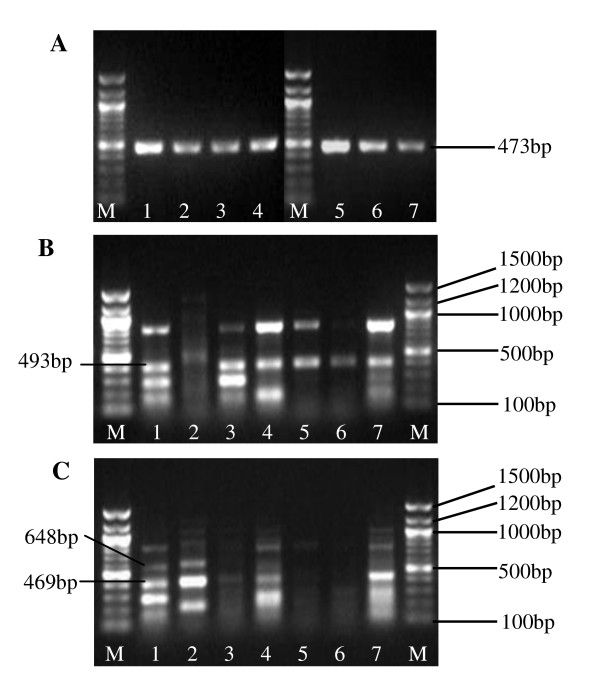
Through 5'RACE, 3'RACE and RT-PCR (Figure 2), three full length and four partial RNA sequences of DHRS4L1 were amplified and identified in human neuroblastoma cell line BE(2)-M17. The sequences of these novel transcript variants were submitted to NCBI and recorded as GQ871921, GQ871922, GQ871923, GQ871924, GQ871925, GQ871926 and GQ871927. The analysis of the full length RNA variants indicates that DHRS4L1 starts its transcription from two alternative sites, exon a2 and exon 1. This suggests that the sequence upstream exon a2 has the potential promoter activity, similar to exon a1, to initiate the transcription of DHRS4L1. Exon a3, the homologous sequence of exon a1 downstream of DRHS4 and exon a2 downstream of DHRS4L2, is located downstream of DHRS4L1 and only 629bp away from the last exon of DHRS4L1, while the mRNA sequences in GenBank indicates that exon a3 is within the first exon of the LRRC16B (leucine rich repeat containing 16B) gene not related to DHRS4 gene cluster.
Figure 2.
Transcription of DHRS4L1. Lane 1. BE(2)-M17, 2. SK-SY5Y, 3. SK-N-SH, 4. HeLa, 5. Eca-109, 6. Hep G2, 7. HL-7702, M. marker(100 bp DNA ladder). (A) β-actin shown as a control of RT-PCR. (B) RT-PCR amplifying the exon 1-exon 10 of DHRS4L1. Product of 493 bp is composed of exon 1-part of intron 2-exon 8-exon 9-exon 10. (C) RT-PCR amplifying the exon a2-exon 10 of DHRS4L1. Product of 469 bp is composed of exon a2- exon 8-exon 9-exon 10. Product of 648 bp is composed of exon a2-part of insertion 3-exon 8-exon 9-exon 10.
Alternative splicing patterns often show specificity related to tissue types, development stages and physiological conditions of the cells [10]. It is evident in nerve tissue that alternative splicing is correlated with the complex functions of the brain [11,12]. The results of RT-PCR to amplify the RNAs from exon 1 to exon 10 and exon a2 to exon 10 of DHRS4L1 showed that the DHRS4L1 RNA isoforms existed not only in cell lines from the nervous system, but also in cells from the cervical, hepatic and esophageal tissues, while different first exon (exon a2 and exon 1) may be utilized to initiate the transcription of DHRS4L1 among these cell lines (Figure 2).
Through analyzing the homologous identity of DNA and the exon composition in the transcripts variants of the DHRS4 gene cluster, we found that exon 1 was absent in all the spliced variants of DHRS4L2 and DHRS4L1 initiated from exon a1 and exon a2 respectively (Table 3). To verify if the absence of exon 1 is just one example found accidentally in cloned sequences or true for all the transcripts of DHRS4 cluster, we used forward primer in exon a (completely matching the sequences of exon a1 and exon a2) and reverse primer in exon 1 of DHRS4L2 and DHRS4L1 respectively to perform RT-PCR in the human neuroblastoma cell lines BE(2)-M17 and cervical carcinoma cell line HeLa. No amplifications was observed (data not shown), suggesting that exons a1/a2 and exon 1 were mutually exclusive.
In humans, majority of genes are alternatively spliced to express multiple proteins [13]. The analysis of the predicted proteins based on different DHRS4 RNA isoforms showed that most of them contained a domain centre of the short-chain dehydrogenase/reductase(ADH-short) family signature, possibly pointing to the functional conservation of the spliced variants. In our previous study, only one protein isoform of DHRS4 was identified and found to be correlated with the tumorigenesis of the cervical epithelia [5], while most of the spliced RNA isoforms of the DHRS4 gene cluster lacked an obvious open reading frame. Moreover, the full length RNA sequences of DHRS4L2 in the previous study [7] and that of DHRS4L1 in present study were investigated using the software RNAStructure4.5. This analysis predicted that low minimum free energy was required for them to form a double-stranded RNA structure. Considering the potential to form a stable secondary RNA structure and the lack of open reading frame, we speculate that most RNA variants of the DHRS4 gene cluster might function as non-coding RNAs to regulate gene expression.
Transcriptional regulation of the DHRS4 gene cluster
Focusing our work on the alternative splicing of the DHRS4 gene cluster and the roles of these alternatively spliced RNAs, we have also been interested in alternative transcriptional regulation of the DHRS4 gene cluster. DHRS4L2 and DHRS4L1 initiate their transcription from two alternative TSS, and exon 1 is absent in all the transcripts initiated from the exons a1 and a2 of DHSR4L2 and DHSR4L1 respectively. Previous studies reported that alternative promoter was accompanied by alternative splicing of the initial exon [14,15]. Analysis of the sequence upward the exon 1 of DHRS4L2 and DHRS4L1 indicates that exon 1 lacks a canonical splice site of AG at its 5' end. It seems that the transcripts initiated from exon a1/a2 is spliced directly to the next available splice acceptor site, resulting in the removal of exon 1.
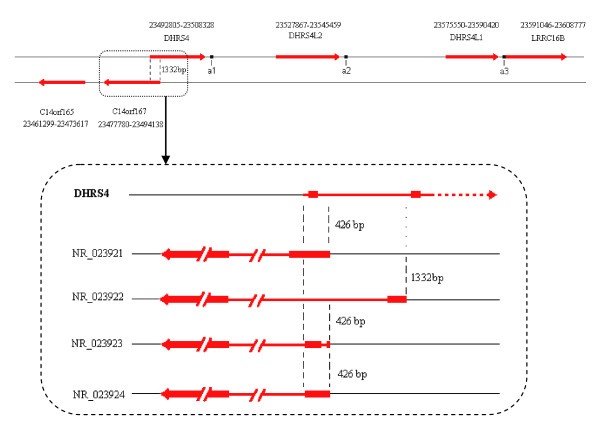
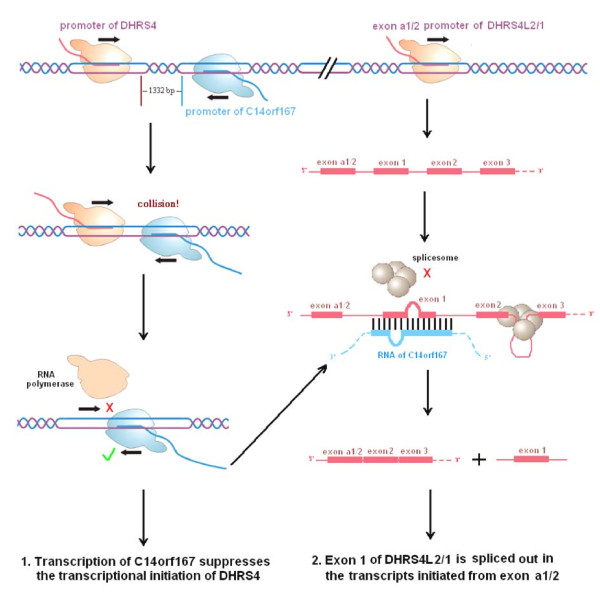
In addition, the homologous comparison of the DHRS4 gene cluster and analysis of the positional relationship of DHRS4 with the antisense gene C14orf167 indicate a possible mechanism for the transcriptional regulation of the DHRS4 gene cluster via an antisense transcribed gene. Firstly, C14orf167 is a typical naturally occurring antisense gene to DHRS4. These two genes have overlapped promoters and 5' 1332 bp coding regions (Figure 3) that belongs to the cis head-to-head bidirectional transcription [16-19]. Given such a positional relationship in the 5' sequence, we propose that DHRS4 and C14orf167 will interfere with each other in transcription initiation (Figure 4). Secondly, antisense RNAs may also participate in the transcriptional regulation of the trans-encoded RNAs transcribed from different loci [20,21]. Previous studies suggest that antisense RNA sequences matching with exon-intron border of the primary transcripts may prevent the binding of the splicesome to the splice site, and consequently affecting the posttranscriptional splicing [22-24]. Due to the homologies within the DHRS4 gene cluster (Table 1 and Table 2), the transcripts of C14orf167 match with exon 1 in the primary transcripts of DHRS4L2 or DHRS4L1, and possibly take part in the splicing out of exon 1 from the transcripts initiated from exon a1/a2 (Figure 4). Although the alternative promoter may affect the splicing of RNA as described above, several subclass models of the splice sites were reported in mammalian pre-mRNAs and they functioned more frequently on the first or second intron than on the other order intron [25-28]. The sequence at the 5'end of DHRS4L2 exon 1 is consistent with the submodel splice acceptor AC, instead of the typical AG sequence. AC functioned as a real splice acceptor site in our previous identified RNA variants (DN237887) of DHRS4L2. If the splice acceptor sequence at the 5'end of exon 1 in the DHRS4 gene cluster is efficient, the masking by antisense RNAs is a likely mechanism to explain the removal of exon 1 from the transcripts initiated from exon a1 or exon a2, although it needs further study to verify.
Figure 3.
The antisense gene C14orf167 and its relationship to DHRS4.
Figure 4.
The effects of antisense transcription from C14orf167 on the transcriptional regulation of the DHRS4 gene cluster. (1) Bidirectional transcription of C14orf167 inhibits the initiation of the DHRS4 transcription through impeding the binding of RNA polymerase II and transcriptional factors to DHRS4. (2) The antisense RNAs from C14orf167 match with exon 1 in the primary transcripts initiated from exon a1/a2, and possibly resulting in the removing of exon 1 in the spliced RNAs. DHRS4L2 and DHRS4L1 are under the same condition to C14orf167 RNAs.
Conclusions
The human DHRS4 gene cluster consists of DHRS4, DHRS4L2 and DHRS4L1. DHRS4L2 and DHRS4L1 initiate their transcription from two transcription start sites. Furthermore, exon 1 is absent in the transcripts initiated from alternative transcription start sites exon a1 of DHRS4L2 or exon a2 of DHRS4L1. Based on the analysis of the sequence relationship between C14orf167 and DHRS4, we propose that transcriptional process of C14orf167 may affect the transcriptional initiation of DHRS4. Given the homologous identities of genes within DHRS4 gene cluster, antisense RNAs of C14orf167 may take part in the posttranscriptional splicing regulation of DHRS4L2 and DHRS4L1 through masking the splicing sites and removing of exon 1 from RNA transcripts. This is a rare example of one antisense transcript regulating the transcription of different gene in cis and trans manner simultaneously, though antisense RNA and bidirectional transcription have been the focus of extensive studies recently.
Methods
Databases and software
The DNA, cDNA and EST sequences of the DHRS4 gene cluster were retrieved from the NCBI GenBank http://www.ncbi.nlm.nih.gov and UCSC Genome Browser http://www.genome.ucsc.edu. Jellyfish3.2 http://www.jellyfishsoftware.com and BLAST online http://blast.ncbi.nlm.nih.gov were employed for analyzing the homologous identity of the human DHRS4, DHRS4L2 and DHRS4L1. The sequences of the DHRS4 gene cluster in different species were aligned with Clustal X1.83 http://www.clustal.org/ and all the positions containing gaps were eliminated. A bootstrapped biological phylogenic tree was constructed using MEGA4 with the neighbor-joining method. The RNA secondary structures were predicted and analyzed with RNAstructure4.5 http://rna.urmc.rochester.edu/rnastructure.html.
Cell culture and RNA extraction
Human neuroblastoma cell lines BE(2)-M17, SK-N-SH, SH-SY5Y, human cervical carcinoma cell line HeLa, human esophageal carcinoma cell line Eca-109, human hepatocarcinoma cell line Hep G2 and human hepatocyte cell line HL-7702 were all obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cell lines were maintained in DMEM medium supplemented with 10% (v/v) fetal bovine serum in a humidified 37°C incubator with 5% CO2. Total RNAs of the cell lines were extracted with TRIzol® reagent (Invitrogen) and RNA quality was examined by OD260/OD280 and RNA electrophoresis.
5'RACE, 3'RACE and RT-PCR
The 5'end amplification of DHRS4L1 splicing variants in the human neuroblastoma cell line BE(2)-M17 was carried out with SMARTer™ RACE kit (Clontech), which generated a complete cDNA copy of the original mRNAs with the additional SMARTer sequence at the 5'end by the joint action of the SMARTer II A oligonucleotide and SMARTScribe reverse transcriptase. It was then used to amplify 5'end of DHRS4L1 through 5'RACE touchdown PCR.
The 3'end amplification was carried out using the 3'RACE system (Takara), which depended on the poly A to ligate the adaptor on the 3'end of mRNAs from the neuroblastoma cell line BE(2)-M17 for the reverse transcription into cDNA and subsequent amplification of the 3'end of DHRS4L1 through 3'RACE nest PCR.
RT-PCR of DHRS4L1 in cell lines BE(2)-M17, SK-N-SH, SH-SY5Y, HeLa, Hep G2, Eca-109 and HL-7702 were performed using Platinum Taq DNA Polymerase mixture (Invitrogen) after the RNA samples were reverse transcribed into cDNA with QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's protocol.
All the PCR primers used are listed in Table 4. Amplified products of PCR were electrophoresed in 1.2% agarose gels and visualized with ethidium bromide (Amresco) staining. The bands of PCR product from the neuroblastoma cell line BE(2)-M17 were cut and DNA was purified using the Gel Extraction Kit (Promega). Then the PCR products were cloned into the pGEM-T Easy vector (Promega) for sequence identification in an Applied Biosystems 3100 DNA sequencer.
Table 4.
Primers used to amplify DHRS4L1 mRNA.
| usage | Primer | Sequence (5'→3) |
|---|---|---|
| 5'RACE (touchdown PCR) |
UPA-F | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT (long) CTAATACGACTCACTATAGGGC (short) |
| DHRS4L1 E10-R | GAGCACAGGAAAGACACGATGCCAAGAG | |
| 3'RACE (1st round of nest PCR) |
DHRS4L1 outer F | GGATGGACAAGGAAAAAGAGG |
| Adaptor 3'outer R | TACCGTCGTTCCACTAGTGATTT | |
| 3'RACE (2nd round of nest PCR) |
DHRS4L1 inner F | TTAGGCGAGCCAGAGGATTCTCTT |
| Adaptor 3' inner R | CGCGGATCCTCCACTAGTGATTTCACTATAGG | |
| RT-PCR | DHRS4L1 Ea F | CAAGCCCACCGTGGAGCTCATCTGA |
| DHRS4L1 E10-R | GAGCACAGGAAAGACACGATGCCAAGAG | |
| RT-PCR | DHRS4L1 E1 F | ATGCACAAGGCGCGGCTACGAG |
| DHRS4L1 E10 R | GAGCACAGGAAAGACACGATGCCAAGAG | |
| RT-PCR | β-actin F | AAATCGTGCGTGACATTAA |
| β-actin R | CTCGTCATACTCCTGCTTG |
Abbreviations
DHRS4: dehydrogenase/reductase (SDR family) member 4; DHRS4L1: DHRS4 like 1; DHRS4L2: DHRS4 like 2; NRDR: NADP(H)-dependent retinol dehydrogenase/reductase; RACE: rapid amplification of cDNA ends; RT-PCR: reverse transcription- polymerase chain reaction; TSS: transcription start site.
Authors' contributions
ZJS and QXZ conceived and designed this study. ZJS performed the RACE PCR, RT-PCR, collected and analyzed the bioinformatics data and drafted the manuscript. GFL, XHS, QL, RJW, and HBC participated in the experiment design and the data analysis. XYX and XXS carried out part of the sequence blasting. DYH is responsible for the design, organization and supervision of the whole study. QXZ and DYH provided funding for the research. All authors read and approved the final manuscript.
Contributor Information
Zhong-Jing Su, Email: g_zjsu@stu.edu.cn.
Qiao-Xia Zhang, Email: qiaoxiazhang@163.com.
Ge-Fei Liu, Email: gefei.victor@gmail.com.
Xu-Hong Song, Email: songxuhong@stu.edu.cn.
Qi Li, Email: 77liqi@sohu.com.
Rui-Jian Wang, Email: wrjww@163.com.
Hai-Bin Chen, Email: chenhb@stu.edu.cn.
Xiao-Yuan Xu, Email: g_xyxu@stu.edu.cn.
Xu-Xia Sui, Email: suixuxia@126.com.
Dong-Yang Huang, Email: huangdy@stu.edu.cn.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant number 30800639 and 30970626] and the Natural Science Foundation of Guangdong Province [grant number 8251503102000003].
References
- Huang DY, Ichikawa Y. Purification and characterization of a novel cytosolic NADP(H)-dependent retinol oxidoreductase from rabbit liver. Biochim Biophys Acta. 1997;1338:47–59. doi: 10.1016/s0167-4838(96)00183-5. [DOI] [PubMed] [Google Scholar]
- Endo S, Matsunaga T, Nagano M, Abe H, Ishikura S, Imamura Y, Hara A. Characterization of an oligomeric carbonyl reductase of dog liver: its identity with peroxisomal tetrameric carbonyl reductase. Biol Pharm Bull. 2007;30:1787–1791. doi: 10.1248/bpb.30.1787. [DOI] [PubMed] [Google Scholar]
- Endo S, Maeda S, Matsunaga T, Dhagat U, El-Kabbani O, Tanaka N, Nakamura KT, Tajima K, Hara A. Molecular determinants for the stereospecific reduction of 3-ketosteroids and reactivity towards all-trans-retinal of a short-chain dehydrogenase/reductase (DHRS4) Arch Biochem Biophys. 2009;481:183–190. doi: 10.1016/j.abb.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Endo S, Maeda S, Ishikura S, Tajima K, Tanaka N, Nakamura KT, Imamura Y, Hara A. Characterization of human DHRS4: an inducible short-chain dehydrogenase/reductase enzyme with 3beta-hydroxysteroid dehydrogenase activity. Arch Biochem Biophys. 2008;477:339–347. doi: 10.1016/j.abb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Song XH, Liang B, Liu GF, Li R, Xie JP, Du K, Huang DY. Expression of a novel alternatively spliced variant of NADP(H)-dependent retinol dehydrogenase/reductase with deletion of exon 3 in cervical squamous carcinoma. Int J Cancer. 2007;120:1618–1626. doi: 10.1002/ijc.22306. [DOI] [PubMed] [Google Scholar]
- Du K, Liu GF, Xie JP, Song XH, Li R, Liang B, Huang DY. A 27.368 kDa retinal reductase in New Zealand white rabbit liver cytosol encoded by the peroxisomal retinol dehydrogenase-reductase cDNA: purification and characterization of the enzyme. Biochem Cell Biol. 2007;85:209–217. doi: 10.1139/O06-183. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Liu G, Xu X, Song X, Liang B, Li R, Xie J, Du M, Xiao L, Gan X, Huang D. Alternative transcription initiation and splicing variants of the DHRS4 gene cluster. Biosci Rep. 2009;29:47–56. doi: 10.1042/BSR20080040. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/S0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/S0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/S0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer GA, Wu J, Yan P, Plass C, Huang TH, Davuluri RV. Genome-wide analysis of alternative promoters of human genes using a custom promoter tiling array. BMC Genomics. 2008;9:349. doi: 10.1186/1471-2164-9-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan M, Kondo S, Schonbach C, Adachi J, Hume DA, Hayashizaki Y, Gaasterland T. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: A critical look in both directions. Cell Mol Life Sci. 2008;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev SA, Korneeva EI, Lagarkova MA, Kiselev SL, Critchley G, O'Shea M. Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. RNA. 2008;14:2030–2037. doi: 10.1261/rna.1084308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg K, Green P. Differing patterns of selection in alternative and constitutive splice sites. Genome Res. 2007;17:1015–1022. doi: 10.1101/gr.6347907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RC, Incorvaia R, Padgett RA. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1:151–160. doi: 10.1016/S1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- Clark F, Thanaraj TA. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum Mol Genet. 2002;11:451–464. doi: 10.1093/hmg/11.4.451. [DOI] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]