Abstract
Epigenetic mechanisms have been implicated in syndromes associated with mental impairment but little is known about the role of epigenetics in determining the normal variation in human intelligence. We measured polymorphisms in four DNA methyltransferases (DNMT1, DNMT3A, DNMT3B and DNMT3L) involved in epigenetic marking and related these to childhood and adult general intelligence in a population (n = 1542) consisting of two Scottish cohorts born in 1936 and residing in Lothian (n = 1075) or Aberdeen (n = 467). All subjects had taken the same test of intelligence at age 11yrs. The Lothian cohort took the test again at age 70yrs. The minor T allele of DNMT3L SNP 11330C>T (rs7354779) allele was associated with a higher standardised childhood intelligence score; greatest effect in the dominant analysis but also significant in the additive model (coefficient = 1.40additive; 95%CI 0.22,2.59; p = 0.020 and 1.99dominant; 95%CI 0.55,3.43; p = 0.007). The DNMT3L C allele was associated with an increased risk of being below average intelligence (OR 1.25additive; 95%CI 1.05,1.51; p = 0.011 and OR 1.37dominant; 95%CI 1.11,1.68; p = 0.003), and being in the lowest 40th (padditive = 0.009; pdominant = 0.002) and lowest 30th (padditive = 0.004; pdominant = 0.002) centiles for intelligence. After Bonferroni correction for the number variants tested the link between DNMT3L 11330C>T and childhood intelligence remained significant by linear regression and centile analysis; only the additive regression model was borderline significant. Adult intelligence was similarly linked to the DNMT3L variant but this analysis was limited by the numbers studied and nature of the test and the association was not significant after Bonferroni correction. We believe that the role of epigenetics in the normal variation in human intelligence merits further study and that this novel finding should be tested in other cohorts.
Introduction
Intelligence is a general mental capability which encompasses the ability to reason, plan, solve problems, think abstractly, learn quickly and make sense of our surroundings [1]. Human intelligence is characterised by a high level of heritability [1], [2] but known genetic effects can account for very little of this [1] and it has been suggested that the effect of individual genes may be much smaller than previously assumed [3]. There is growing interest in the potential for epigenetics to influence cognition [4]–[7]. Epigenetic status has recently been defined as “a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence.” [8]. Such epigenetic ‘heritability’ may occur through either mitosis or meiosis and therefore has the potential to explain at least part of the high heritability of intelligence. Epigenetic mechanisms have been implicated in many syndromes associated with mental impairment; Autism (MIM209850), Rett (MIM312750), Immunodeficiency-Centromeric Instability- Facial Abnormalities (ICF) (MIM242860), Prader-Willi (MIM176270), Angelman (MIM105830), Fragile X (MIM300624), Rubinstein-Tabi (MIM180849) [4]–[6], [9]–[13] but little is known about the role of epigenetics in determining the normal variation in human cognitive abilities.
The role of epigenetics in human complex traits such as intelligence is difficult to study for a number of reasons. Epigenetic status can be influenced by factors such as diet [14] and alcohol [15] therefore, depending on the epigenetic mark of interest, there is a danger of reverse causality, where lifestyle choices linked to intelligence may influence epigenetic status. A further difficulty is that the epigenetic status of many genes is tissue specific therefore the epigenome of easily accessible tissues (e.g. blood or buccal cells) may not always reflect the epigenome of the functional organ of interest (e.g. the brain). Perhaps paradoxically, evidence for the involvement of epigenetic processes in intelligence may be obtained from genetic association studies. The goal of genetic association studies is to identify patterns of polymorphisms that vary systematically between individuals with different phenotypes and could therefore represent the effects of risk-enhancing or protective alleles [16]. Association is assumed to arise because the function of the gene has been altered in some way by the measured variant - or another in linkage disequilibrium with it - and that the change in gene function influences the phenotype. Such an approach can help to identify specific biological processes underpinning traits that may otherwise be difficult to study in humans. Analysis of genetic variants within the genes of the epigenetic pathway has been carried out to look for evidence of the involvement of epigenetic processes in the aetiology of diseases such as cancer [17]. We have used the same approach here in relation to human intelligence.
DNA methylation is probably the most commonly studied epigenetic phenomenon. We investigated the association between variants in genes involved in the epigenetic marking of DNA by methylation and childhood and adult general intelligence in population based samples with an unusually valuable phenotype: a large number of subjects who had taken the same validated general, IQ-type mental test at age 11. A sub group of these took the same test again almost 60 years later. Polymorphisms were measured in the four DNA methyltransferases: DNMT1 (MIM126375); DNMT3A (MIM602769); DNMT3B (MIM602900); DNMT3L (MIM606588). Genetic variation in all four DNMTs was studied in order to provide additional information on the nature of the epigenetic process which may influence human intelligence. The main function of DNMT1 is to ensure the propagation of existing methylation patterns, whilst DNMT3A and DNMT3B are primarily required for de novo methylation, with DNMT3L acting as an essential cofactor, particularly in the establishment of methylation imprints in the gametes [18]–[22].
Materials and Methods
Subjects
The Lothian and Aberdeen Birth Cohorts 1936 (LBC1936 and ABC1936) comprise surviving participants of the Scottish Mental Survey 1947 (SMS1947) who now live in the Lothian (Edinburgh and its surroundings) and Aberdeen areas of Scotland, respectively.
Ethics statement
Ethics permission for the study was obtained from the Multi-Centre Research Ethics Committee for Scotland (MREC/01/0/56) and from Lothian Research Ethics Committee (LREC/2003/2/29) and Grampian Research Ethics Committee (LREC/01/0299). The research was conducted in compliance with the Helsinki Declaration. All subjects gave written, informed consent.
Mental test
On June 4th 1947 almost all people born in 1936 and attending school in Scotland were tested on a valid general cognitive ability test [23]. The mental test was a version of the Moray House Test (MHT) No. 12, which was concurrently validated against the Terman-Merrill revision of the Binet Scales with a coefficient of approximately 0.8. The test was re-administered to the LBC1936 when participants were seen again at age 70 (IQR = 1.3) yrs, using the same instructions and the same 45-minute time limit. Only two small changes were made to items whose content had become archaic. The test is often referred to as the ‘Verbal Test’ or as a ‘verbal reasoning’ test. However, the test has items of a variety of types: following directions (14 items), same-opposites (11), word classification (10), analogies (8), practical items (6), reasoning (5), proverbs (4), arithmetic (4), spatial items (4), mixed sentences (3), cipher decoding (2), and other items (4). The maximum possible score in the MHT was 76.
Variant selection and genotyping
Blood samples were taken for DNA extraction from white blood cells and the samples stored as described elsewhere [24], [25]. The concentration of DNA was determined by RNaseP assay (Applied Biosystems, Warrington, UK) before genotyping. Genotypes were detected by allelic discrimination assay using TaqMan® MGB probes labelled with 6-FAM™ and VIC® on a 7500 Fast real-time PCR system (Applied Biosystems, Warrington, UK). The gene variants studied were: DNMT1 (MIM126375 19p13.3–p13.2; 21220C>T; rs2114724); DNMT3A (MIM 602769; 2p23; 28510C>T; rs734693); DNMT3B (MIM602900; 20q11.2; 46359C>T; rs2424913); DNMT3L (MIM606588; 21q22.3; 11330C>T; rs7354779). A second variant in DNMT1 (35433A>G; rs2162560) was measured in the ABC1936 cohort but this was found to be highly reciprocally correlated with the DNMT1 21220C>T variant (coefficient = −0.85; p<0.001) and therefore was not measured in the larger LBC1936 and dropped from the study. We used the candidate polymorphism approach which focuses on individual polymorphisms that are suspected of being implicated in biological function [16]. This hypothesis directed approach has the advantage of minimising the number of variants studied and hence reducing the risk of false positive results. Very rare variants are of limited value in association studies as they are only relevant to a small proportion of the population and their effects are difficult to detect in practice therefore we selected only variants where the frequency of the homozygous minor allele was >5%. The DNMT3L 11330C>T variant is a non-synonymous polymorphism resulting in the amino acid change Gly278Arg within the C-terminal portion of DNMT3L. Such non-synonymous variations in the functional domains of the DNMTs are unusual and no non-synonymous polymorphisms have been detected in the catalytic domains of DNMT3A/B or DNMT1 in a European population [26]. Variants in these genes were selected for study primarily on the basis of whether they have been related to a phenotype. Such associations are assumed to arise as a result of non-coding effects [27] or because the variants are in linkage disequilibrium with functional variant(s) within the gene. The DNMT3B variant is located in the promoter region and has been associated with the risk of cancer [28]. For the DNMT1, 3A and 3L variants we have observed phenotypic associations in other cohorts (manuscripts in preparation). All available DNA samples from the cohorts were measured. Only samples in which a valid genotype was not able to be measured were excluded from the data analysis. Genotyping was carried out by laboratory staff blind to the MHT score results.
Statistics
Medians are presented with inter-quartile ranges (IQR). The distributions of MHT scores by cohort are presented in kernel density plots. The Moray House Test score percentiles were normalised by transformation to an IQ type scale of mean 100 and standard deviation 15 using the invnorm function in STATA. This transformation facilitates the use of parametric tests and allows the magnitude of any genetic effect to be presented on a commonly understood scale in the field of cognition. Statistical analysis was carried out using STATA/SE version 11 (Stata Corp, College Station, Texas, USA) for both additive (CC vs CT vs TT) and dominant (CC vs CT/TT) models. Associations between genotype and intelligence were evaluated using various statistical tests; linear regression, logistic regression, Chi2. Genotype-sex interactions were assessed by Hosmer and Lemeshow likelihood ratio test. Regression coefficients (un-standardised) are presented with 95% confidence intervals and p values.
Results
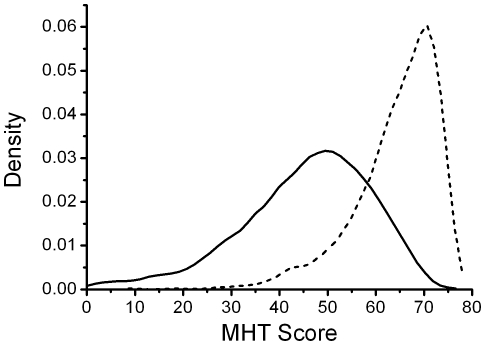
The study population consisted of 50% males and 50% females. The median MHT score at age 11yrs in the entire sample was 47 (IQR = 17); 42 (IQR = 18) for ABC1936 and 50 (IQR = 15) for LBC1936. At age 70yrs the median MHT score was 66 (IQR = 11). The population distributions of childhood, and adult scores are shown in Figure 1 . The scores were standardised to an IQ type scale (mean of 100, standard deviation of 15) for regression analysis; the levels of significance were similar after using other transformations to produce normally distributed MHT data (e.g. squaring in the case of childhood MHT score). The genotype and allele frequencies for the DNMT variants are shown in Table 1 . All the variants studied were in Hardy-Weinberg equilibrium in the combined study population and the individual cohorts (tested by Chi2). The results of linear regression analysis of DNMT genotype on childhood and adult MHT scores are presented in Table 2 . Of the four gene variants studied only DNMT3L was liked to intelligence. The minor allele was associated with a higher level of childhood intelligence in the additive model (coefficient = 1.40; 95%CI 0.22,2.59; p = 0.020). The DNMT3L effect size was larger, and the level of significance greater, in the dominant model which compares carriers of the minor allele (CT/TT) with the homozygote CC (coefficient = 1.99; 95%CI 0.55,3.43; p = 0.007). The DNMT3L genotype accounted for around 0.5% of the variance in intelligence, with the common homozygote (DNMT3L 11330CC) being associated with an approximately 2 point reduction in standardised intelligence. There was no evidence of genotype-sex interaction in relation to childhood intelligence score.
Figure 1. Kernel density plots of childhood Moray House test score in childhood (age 11yrs - - - -) and adulthood (age 70yrs ——) Moray House Test score.
Table 1. DNMT variant genotype and allele frequencies in ABC1936 and LBC1936.
| Gene (variant) | ||||
| DNMT(21220C>T) | DNMT3A(28510C>T) | DNMT3B(46359C>T) | DNMT3L(11330C>T) | |
| Homozygote common | 350 (22.8) | 898 (58.3) | 481 (31.2) | 813 (53.8) |
| Heterozygote | 795 (51.7) | 544 (35.3) | 753 (48.9) | 607 (40.2) |
| Homozygote minor | 393 (25.6) | 99 (6.4) | 307 (19.9) | 92 (6.1) |
| Total | 1538 (100) | 1541 (100) | 1541 (100) | 1512 (100) |
| Common allele frequency | 0.49 | 0.76 | 0.56 | 0.74 |
| Minor allele frequency | 0.51 | 0.24 | 0.44 | 0.26 |
Genotype counts are presented with percentages in brackets.
Table 2. Relationship between DNMT variants and standardised1 Moray House Test (MHT) score.
| Gene (variant) | ||||||||
| DNMT1(21220C>T) | DNMT3A(28510C>T) | DNMT3B(46359C>T) | DNMT3L(11330C>T) | |||||
| Comparison | Regression coefficient(95% CI) | p value | Regression coefficient(95% CI) | p value | Regression coefficient(95% CI) | p value | Regression coefficient(95% CI) | p value |
| Childhood intelligence | ||||||||
| Additive model2 | 0·73(−0·30,1·76) | 0·167 | 0·88(−0·28,2·04) | 0·135 | 0·78(−0·24,1·80) | 0·132 | 1·40(0·22,2·59) | 0·020 |
| Dominant model3 | 0·76(−0·95,2·46) | 0·385 | 0·73(−0·72,2·17) | 0·325 | −1.31(−0·24,2·86) | 0·097 | 1·99(0·55,3·43) | 0·007 |
| Adult intelligence | ||||||||
| Additive model2 | −0.11(−1·40,1·17) | 0·863 | 1.11(−0·33,2·54) | 0·130 | −0·03(−1·30,1·25) | 0·969 | 1·60(0·14,3·05) | 0·032 |
| Dominant model3 | 0·10(−2·07,2·26) | 0·929 | 0·89(−0·91,2·69) | 0·330 | 0·21(−1·73,2·15) | 0·830 | 2·25(0·46,4·05) | 0·014 |
Transformed to normalised IQ type scale of mean 100 and standard deviation 15.
TT>CT>CC.
CT/TT>CC.
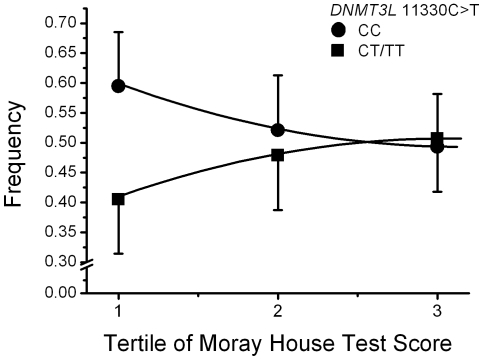
The change in genotype frequency with tertile of intelligence is illustrated in Figure 2 for the dominant model (p = 0.005 by chi2). The DNMT3L change in CC frequency with intelligence was more pronounced at lower levels of intelligence. Logistic regression analysis of the effect of the CC variant on the likelihood of falling in the lowest intelligence group is presented for a range of centiles in Table 3. The DNMT3L 11330 C allele was associated with a higher risk of being below average intelligence – the lowest 50th centile – in both the additive (OR 1.25; 95%CI 1.05,1.51; p = 0.011) and dominant (OR 1.37; 95%CI 1.11,1.68; p = 0.003) models. The effects sizes and levels of significance were similar when considering the lowest 40th centile (OR 1.27additive; 95%CI 1.06,1.51; p = 0.009 and OR 1.40dominant; 95%CI 1.23,1.73; p = 0.002) or the lowest 30th centile (OR 1.34additive; 95%CI 1.10,1.63; p = 0.004 and OR 1.44dominant; 95%CI 1.14,1.82; p = 0.002). For the 20th centile the odds ratios were similarly positive but with one of the comparison groups consisting of only 20% of the data the number of observations was too small to demonstrate significance.
Figure 2. DNMT3L 11330C>T genotype frequencies – CC and T allele carriers (CT/TT) – by MHT score tertile.
Table 3. DNMT3L 11330 genotype and risk of being in the lowest Moray House Test score centile.
| Additive model1 | Dominant model2 | |||
| Lowest centile | Odds ratio(95% CI) | p value | Odds Ratio(95% CI) | p value |
| 50th centile | 1.25(1.05,1.51) | 0.011 | 1.37(1.11,1.68) | 0.003 |
| 40th centile | 1.27(1.06,1.51) | 0.009 | 1.40(1.23,1.73) | 0.002 |
| 30th centile | 1.34(1.10,1.63) | 0.004 | 1.44(1.14,1.82) | 0.002 |
| 20th centile | 1.20(0.96,1.51) | 0.114 | 1.23(0.93,1.62) | 0.140 |
DNMT3L 11330 CC>CT>TT.
DNMT3L 11330 CC>CT/TT.
Adult intelligence was also related to DNMT3L genotypes ( Table 2 ). The minor allele was associated with a higher level of adult intelligence in both the additive (coefficient 1.60; 95%CI 0.14,3.05; p = 0.032) and dominant (coefficient 2.25; 95%CI 0.46,4.05; p = 0.014) models. There was no evidence of genotype-sex interaction in relation to adult intelligence score. Although the level of significance was lower than for the childhood association with DNMT3L genotype it should be noted that this analysis was based only on LBC1936 therefore the numbers were significantly reduced. Interpretation of the adult data is also complicated by the fact that around 95% of the adults improved on their childhood intelligence score with a median improvement of 15 (IQR = 10) score points. This means that the most able children are likely to be limited by the nature of the test in adult life, resulting in a reduction in the power of the test to discriminate intelligence at the highest levels (ceiling effect) and a skewed distribution of scores ( Figure 1 ). There is no way of knowing from the distribution alone which adults would be affected in this way therefore no single cut-off value can be derived. However, we can say that the relationship between adult intelligence and DNMT3L genotype remained significant, by both the additive and dominant models, following sequential exclusion of the highest MHT scores over a large span of the data; from the maximum possible score of 76 down to a score of 67 using the additive model and down to a score of 59 using the dominant model. At lower cut-off values the amount of data on which the analysis was based was too low to provide a meaningful test.
We tested here for the influence of 4 independent gene variants on intelligence. After Bonferroni correction for the number variants tested the minor T allele remained associated with a higher level of childhood intelligence (homozygote CC associated with a lower level of intelligence) by linear regression (p = 0.028). The chi2 analysis of the change in minor allele frequency with childhood intelligence tertile remained significant (p for trend = 0.020). The risk of being in the lowest intelligence centile associated conferred by the CC genotype also remained significant across the same range of values for both the additive and dominant models; below average intelligence (p = 0.044additive, p = 0.012dominant);lowest 40th centile (p = 0.036additive, p = 0.008dominant); lowest 30th centile (p = 0.016additive, p = 0.008dominant). After Bonferroni correction the minor T allele association with adult intelligence by linear regression was only approaching significance (p = 0.056).
Discussion
We observed a significant association between the DNMT3L 11330C>T variant and childhood intelligence in a study population made up of two large birth cohorts born in Scotland in 1936. Adult intelligence was also related to the same DNMT3L genotype but it should be noted that the test of intelligence, designed for children, may not have been sufficiently challenging to discriminate adult performance at the highest levels of intelligence and the number of data points on which the analysis was based (LBC1936 only) was less than for the childhood data. The relationship between DNMT3L and adult intelligence was only approaching statistical significance after Bonferroni adjustment therefore the primary finding here is in relation to childhood intelligence.
Intelligence is a highly complex phenotype which is the net result of a wide rage of biological processes and the effect of individual polymorphisms on intelligence is thought to be very low [3]. In this study the DNMT3L genotype accounted for around 0.5% of the variance in intelligence, with the common DNMT3L homozygote genotype being associated with an approximately 2 point reduction in standardised intelligence. It should be emphasised that this variant could not be used as any sort of test or predictor of intelligence, at either the group or individual level. The value of this finding is that it may point to specific biological processes which are worthy of further study. Current understanding of the role of DNMT3L points to the process of epigenetic regulation and imprinting – parent of origin specific epigenetic marking – in particular [21], [29], [30]. DNMT3L is known to interact with the histone deacetylases [31] and histone methyltransferases [32] but its primary effect is on de novo DNA methylation [21], [29], [30]. DNMT3L is structurally similar to the other methyltransferases. It is essential for de novo methylation but its mode of action primarily involves interaction with the other methyltransferases as it does not have methyltransferase activity itself [18]–[22]. Studies in animals have demonstrated that the progeny of Dnmt3L knockouts exhibit loss of imprinting, stochastic imprinting and biallelic expression of imprinted genes [33], [34]. The DNMT3L 11330C>T variant alters the amino acids sequence of the C-terminal portion of DNMT3L which interacts with the active catalytic methyltransferase domain of DNMT3A and DNMT3B [19]. A recent study reported differences in the methylation level of some genes in association with the DNMT3L 11330C>T variant [26]. These differences were not significant after adjustment for multiple testing but it is unlikely that this study was sufficiently powered for the very large number of tests carried out. The imprint is set during reproduction and, in a human study analogous to the experimental animal knockouts, we measured the effect of maternal DNMT3L 11330C>T genotype on the methylation status of the imprinted gene IGF2 in newborn cord blood (manuscript in preparation). The minor DNMT3L allele, associated here with a higher level of adult intelligence, was associated with a significantly lower level of methylation in IGF2 using the same statistical model (p = 0.035, n = 840).
A mechanism operating through epigenetic regulation, and imprinting in particular, would be consistent with many of the characteristics of human intelligence. Although known imprinted genes make up only around 1% of all genes in humans they primarily affect brain function and behaviour and pre-natal growth [5], [35]. These two key effects of the imprinted genes are consistent with the epidemiological link between IQ and birth weight [36]. The differential maternal and paternal inheritance patterns of many mental disorders would also be consistent with an imprinting mechanism [37], [38]. Direct evidence of an imprinting link to intelligence comes from disorders of imprinting, such as Prader-Willi syndrome (MIM176270) and Angelman syndrome (MIM105830), which are associated with a reduction in IQ [12]. Other mental impairment syndromes have been linked to imprinting changes or genetic polymorphisms relevant to epigenetics [9], [11], [13], [39], [40]. Evidence in support of a role for imprinting in neural function and cognition continues to grow [5], [41]–[47] but direct evidence in humans is difficult to obtain because of the difficulty of studying the brain directly. A link between intelligence and a genetic variant within a gene which is critical to imprinting is therefore a useful piece of additional evidence.
The potential involvement of epigenetics, and imprinting in particular, raises the intriguing possibility that even the heritable component of intelligence could be modifiable by factors such as diet during early development. The ultimate methyl donor for epigenetic-methylation reactions is the folate-methylation cycle and feeding pregnant dams diets deficient in methyl donors results in altered epigenetic regulation of specific genes in the offspring; e.g. axin fused [48] and the Agouti gene which is under imprinting control [49], [50]. Variation in the expression and epigenetic marking of the imprinted genes is also seen in humans [35], [51], [52] and human twin studies have demonstrated the heritability of imprinted gene methylation [53], [54]. We speculate that these two properties of imprinting – heritability and plasticity – could potentially explain the apparent paradox of the high level of IQ heritability [2] alongside the steady rise in IQ test scores from one generation to the next in the so called “Flynn effect” [55], [56].
The association between genetic variation in DNMT3L and childhood intelligence reported here must be considered an initial and as yet un-replicated finding, with the priority being to replicate the association in other populations. An association between DNMT3L and intelligence would be consistent with the critical role of DNMT3L in imprinting and the evidence linking imprinting to cognitive function but more work is needed to determine which function of DNMT3L is influenced by the 11330C>T variant and to investigate how this process might influence human intelligence.
Acknowledgments
The Lothian work was undertaken within The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The genotype measurements were funded by the Scottish Government Rural and Environment Research and Analysis Directorate (RERAD) and the Alzheimer's Research Trust. PH and GH acknowledge the support of RERAD. The Aberdeen data and sample collection was funded by BBSRC. The Lothian work was undertaken within The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative, with funding from BBSRC, EPSRC, ESRC and MRC. The Lothian data collection was supported by a programme grant from Research Into Ageing, which continues as the Disconnected Mind project funded by Help the Aged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Deary IJ, Spinath FM, Bates TC. Genetics of intelligence. Eur J Hum Genet. 2006;14:690–700. doi: 10.1038/sj.ejhg.5201588. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard TJ., Jr Genetic and environmental influences on adult intelligence and special mental abilities. Hum Biol. 1998;70:257–279. [PubMed] [Google Scholar]
- 3.Butcher LM, Meaburn E, Knight J, Sham PC, Schalkwyk LC, et al. SNPs, microarrays and pooled DNA: identification of four loci associated with mild mental impairment in a sample of 6000 children. Hum Mol Genet. 2005;14:1315–1325. doi: 10.1093/hmg/ddi142. [DOI] [PubMed] [Google Scholar]
- 4.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 6.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 7.Whalley LJ, Dick FD, McNeill G. A life-course approach to the aetiology of late-onset dementias. Lancet Neurol. 2006;5:87–96. doi: 10.1016/S1474-4422(05)70286-6. [DOI] [PubMed] [Google Scholar]
- 8.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes & Development. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amir RE, Van dV, I, Wan M, Tran CQ, Francke U, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 10.Ausio J, Levin DB, De Amorim GV, Bakker S, Macleod PM. Syndromes of disordered chromatin remodeling. Clin Genet. 2003;64:83–95. doi: 10.1034/j.1399-0004.2003.00124.x. [DOI] [PubMed] [Google Scholar]
- 11.Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittington J, Holland A, Webb T, Butler J, Clarke D, et al. Cognitive abilities and genotype in a population-based sample of people with Prader-Willi syndrome. J Intellect Disabil Res. 2004;48:172–187. doi: 10.1111/j.1365-2788.2004.00556.x. [DOI] [PubMed] [Google Scholar]
- 13.Xie ZH, Huang YN, Chen ZX, Riggs AD, Ding JP, et al. Mutations in DNA methyltransferase DNMT3B in ICF syndrome affect its regulation by DNMT3L. Hum Mol Genet. 2006;15:1375–1385. doi: 10.1093/hmg/ddl059. [DOI] [PubMed] [Google Scholar]
- 14.Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 15.Bonsch D, Lenz B, Reulbach U, Kornhuber J, Bleich S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2004;111:1611–1616. doi: 10.1007/s00702-004-0232-x. [DOI] [PubMed] [Google Scholar]
- 16.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 17.Cebrian A, Pharoah PD, Ahmed S, Ropero S, Fraga MF, et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27:1661–1669. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- 18.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 21.Trasler JM. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod Fertil Dev. 2006;18:63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- 22.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scottish Council for Research in Education. The trend of Scottish intelligence: A comparison of the 1947 and 1932 Surveys of the intelligence of eleven-year-old pupils. London: University of London Press; 1949. [Google Scholar]
- 24.Deary IJ, Whiteman MC, Starr JM, Whalley LJ, Fox HC. The impact of childhood intelligence on later life: following up the Scottish mental surveys of 1932 and 1947. J Pers Soc Psychol. 2004;86:130–147. doi: 10.1037/0022-3514.86.1.130. [DOI] [PubMed] [Google Scholar]
- 25.Deary IJ, Gow AJ, Taylor MD, Corley J, Brett C, et al. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr. 2007;7:28. doi: 10.1186/1471-2318-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Maarri O, Kareta MS, Mikeska T, Becker T, az-Lacava A, et al. A systematic search for DNA methyltransferase polymorphisms reveals a rare DNMT3L variant associated with subtelomeric hypomethylation. Hum Mol Genet. 2009;18:1755–1768. doi: 10.1093/hmg/ddp088. [DOI] [PubMed] [Google Scholar]
- 27.Chamary JV, Hurst LD. The price of silent mutations. Sci Am. 2009;300:46–53. doi: 10.1038/scientificamerican0609-46. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Wang L, Wang LE, Sturgis EE, Wei Q. Polymorphisms of the DNMT3B gene and risk of squamous cell carcinoma of the head and neck: A case-control study. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 31.Aapola U, Liiv I, Peterson P. Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Res. 2002;30:3602–3608. doi: 10.1093/nar/gkf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnaud P, Hata K, Kaneda M, Li E, Sasaki H, et al. Stochastic imprinting in the progeny of Dnmt3L(−/−) females. Hum Mol Genet. 2006;15:589–598. doi: 10.1093/hmg/ddi475. [DOI] [PubMed] [Google Scholar]
- 34.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 35.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 36.Shenkin SD, Starr JM, Deary IJ. Birth weight and cognitive ability in childhood: a systematic review. Psychol Bull. 2004;130:989–1013. doi: 10.1037/0033-2909.130.6.989. [DOI] [PubMed] [Google Scholar]
- 37.Badcock C, Crespi B. Imbalanced genomic imprinting in brain development: an evolutionary basis for the aetiology of autism. J Evol Biol. 2006;19:1007–1032. doi: 10.1111/j.1420-9101.2006.01091.x. [DOI] [PubMed] [Google Scholar]
- 38.Davies W, Isles AR, Wilkinson LS. Imprinted genes and mental dysfunction. Ann Med. 2001;33:428–436. doi: 10.3109/07853890108995956. [DOI] [PubMed] [Google Scholar]
- 39.Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, El-Maarri O, et al. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet. 2003;72:571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Catchpoole D, Lam WW, Valler D, Temple IK, Joyce JA, Reik W, et al. Epigenetic modification and uniparental inheritance of H19 in Beckwith-Wiedemann syndrome. J Med Genet. 1997;34:353–359. doi: 10.1136/jmg.34.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunningham MD, Kassis JA, Pfeifer K. Chromatin modifiers, cognitive disorders, and imprinted genes. Dev Cell. 2010;18:169–170. doi: 10.1016/j.devcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies W, Lynn PM, Relkovic D, Wilkinson LS. Imprinted genes and neuroendocrine function. Front Neuroendocrinol. 2008;29:413–427. doi: 10.1016/j.yfrne.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Davies W, Isles AR, Humby T, Wilkinson LS. What are imprinted genes doing in the brain? Adv Exp Med Biol. 2008;626:62–70. doi: 10.1007/978-0-387-77576-0_5. [DOI] [PubMed] [Google Scholar]
- 44.Goos LM, Silverman I. The inheritance of cognitive skills: does genomic imprinting play a role? J Neurogenet. 2006;20:19–40. doi: 10.1080/01677060600685840. [DOI] [PubMed] [Google Scholar]
- 45.Goos LM, Ragsdale G. Genomic imprinting and human psychology: cognition, behavior and pathology. Adv Exp Med Biol. 2008;626:71–88. doi: 10.1007/978-0-387-77576-0_6. [DOI] [PubMed] [Google Scholar]
- 46.Whittington J, Holland A, Webb T. Relationship between the IQ of people with Prader-Willi syndrome and that of their siblings: evidence for imprinted gene effects. J Intellect Disabil Res. 2009;53:411–418. doi: 10.1111/j.1365-2788.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhao X, Pak C, Smrt RD, Jin P. Epigenetics and Neural developmental disorders: Washington DC, September 18 and 19, 2006. Epigenetics. 2007;2:126–134. doi: 10.4161/epi.2.2.4236. 4236 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, et al. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 49.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 50.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 51.Sakatani T, Wei M, Katoh M, Okita C, Wada D, et al. Epigenetic heterogeneity at imprinted loci in normal populations. Biochem Biophys Res Commun. 2001;283:1124–1130. doi: 10.1006/bbrc.2001.4916. [DOI] [PubMed] [Google Scholar]
- 52.Sandovici I, Leppert M, Hawk PR, Suarez A, Linares Y, et al. Familial aggregation of abnormal methylation of parental alleles at the IGF2/H19 and IGF2R differentially methylated regions. Hum Mol Genet. 2003;12:1569–1578. doi: 10.1093/hmg/ddg167. [DOI] [PubMed] [Google Scholar]
- 53.Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 54.Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 55.Flynn JR. IQ gains, WISC subtests and fluid g: g theory and the relevance of Spearman's hypothesis to race. Novartis Found Symp. 2000;233:202–216. doi: 10.1002/0470870850.ch13. [DOI] [PubMed] [Google Scholar]
- 56.Lynn R, Hampson SL, Mullineux JC. A long-term increase in the fluid intelligence of English children. Nature. 1987;328:797. doi: 10.1038/328797a0. [DOI] [PubMed] [Google Scholar]