Abstract
This study aimed to identify the inflammation-associated 7/4-antigen, which is highly expressed on neutrophils, inflammatory monocytes, some activated macrophages, as well as on bone marrow myeloid-restricted progenitors. The high expression on inflammatory cells is suggestive of a role in inflammation and makes the 7/4-antigen a potential target for the manipulation of inflammatory cells. Consistent with this, the 7/4-antibody mediates specific depletion of 7/4-expressing neutrophils and monocytes. We have identified the 7/4-antigen as a 25- to 30-kDa GPI-anchored glycoprotein synonymous with the Ly-6B.2 alloantigen. We characterized the expression of Ly-6B during the inflammatory reaction induced by zymosan. During the later stages of an experimental, acute, self-resolving inflammatory response, we found that Ly-6B is differentially expressed on macrophages. Ly-6B-expressing macrophages also express more MHCII, CIITA, CCR2, Ly-6C, and CD62L than the Ly-6B-negative macrophages, which in turn, express more of the resident tissue macrophage marker SIGN-R1 and higher CD11b and F4/80. Ly-6B-expressing macrophages incorporate more BrdU than their Ly-6B-negative contemporaries when fed during the resolution phase of the acute inflammatory response. Thus, Ly-6B expression on mature macrophages defines a subset of recently generated inflammatory macrophages that retain monocytic markers and is hence a surrogate marker of macrophage turnover in inflammatory lesions. The definition of the 7/4:Ly-6B antigen will allow further characterization and specific modulation of Ly-6B-expressing cells in vivo.
Keywords: monocyte, surface antigen, inflammation, u-PAR
Introduction
Approximately 25 years ago, the reportedly “neutrophil- specific” rat mAb 7/4 was produced by immunization with nonadherent mouse bone marrow leukocytes [1]. It was noted that the 7/4-antigen was polymorphic and its expression controlled by a single dominant gene [1]. The 7/4-antigen was subsequently shown to be detectable on some populations of activated macrophages, such as those present during malaria and BCG infections [2, 3]. More recently, it has been appreciated that the 7/4-antigen is expressed on murine monocytes and seemingly lost during differentiation to macrophages in inflammatory lesions [4, 5], as it is during differentiation of bone marrow cultures in M-CSF [1, 6]. However, the 7/4-antigen is not expressed by mature tissue resident macrophages, and it has become extremely useful for the differential identification of recruited inflammatory myeloid cells in experimental models [4, 5, 7]. Its high and restricted expression on inflammatory cells is indicative of a role in inflammation and makes the 7/4-antigen a potential target for the manipulation of inflammatory cells. However, the identity of the 7/4-antigen has remained elusive.
The murine Ly-6/u-PAR family on mouse chromosome 15 [8] was classified initially, according to its reactivity with specific antibodies, against Ly-6A–E [9,10,11]. Following the same nomenclature, new members fitting into the mouse Ly-6 family have been identified on chromosome 15, including the myeloid cell-expressed Ly-6G [12] and Ly-6I/M molecules [13, 14]. Like the 7/4-antigen, the Ly-6 genes are polymorphic. Two murine Ly-6 haplotypes have been defined based on the use of anti-Ly-6-sera. A2G, A/Sn, ASW, Balb/c, C3H/HEH, and CBA mouse strains were classified as Ly-6.1, whereas 129J, AKR, C57BL/6, C57BL/10, C58, DBA, and SJL were grouped as Ly-6.2 [9, 11]. NZB and New Zealand White mice were unusual in being Ly-6.1 at all test alloantigens except Ly-6B, where they were recorded as Ly-6B.2 [9].
Collectively, with particular regard to Ly-6C, Ly-6G, and a “common epitope” on these molecules recognized as “Gr-1”, these markers have been extremely useful for the characterization of myeloid cells from the bone marrow to inflammatory settings [4, 5, 12, 15, 16], including the subsetting and identification of monocyte populations [4, 5, 12, 15], and for the definition of plasmacytoid dendritic cells [17]. Gr-1 has also been used extensively to identify myeloid-derived suppressor cells (with a CD11b+Gr-1+ phenotype), which are cells with mixed myeloid phenotype, depending on the disease setting, with a potent ability to suppress T cell responses [18]. CD11b+Gr-1+-expressing myeloid cells also express the 7/4-antigen during steady-state, inflammation, and tumor challenge [5]. However, as mentioned, Gr-1 recognizes two epitopes (Ly-6G and Ly-6C) and can give complex staining patterns that require contextual interpretation, which emphasizes the need for more specific antibodies that can perform more discrete functions, such as targeting of more select myeloid subsets in vivo.
Because of the potential for specific manipulation of inflammatory myeloid cells in vivo, we adopted proteomic, genomic, and candidate techniques to identify the 7/4-antigen. We demonstrate that the 7/4-antigen is synonymous with the Ly-6B.2 alloantigen, a previously uncharacterized member of the murine Ly-6 family, closely related to Ly-6G and Ly-6C. The identification of the Ly-6B gene in mice and its specific recognition by a mAb make it a powerful, alternative target to modulate the activity of specific myeloid subsets in vivo.
MATERIALS AND METHODS
Mice and general reagents
Balb/c, CBA, 129, and C57BL/6 mice and a Balb/c.7/4+ congenic mouse strain created by six generations of backcross of the C57BL/6 7/4+ phenotype onto the Balb/c background were maintained within our institutes and handled in accordance with institutional and UK Home Office guidelines. NZB/OlaHsd mice were purchased from Harlan (UK). The 7/4-antibody as well as biotin-labeled CD204/scavenger receptor A (clone 2F8) and CD11b (clone 5C6) were produced in our laboratory. Anti-mouse IgG-PE, Gr-1-PE, CD93-PE, DX5-PE, CD200R-PE, CD11b-PerCP-Cy5.5, streptavidin-PerCP-Cy5.5, CD3-biotin, CD86-biotin, CD9-biotin, and MHCII-biotin (clone 2G9) were purchased from BD PharMingen (San Diego, CA, USA). F4/80-APC, CD11c-biotin (clone NH18), Ly-6C-AlexaFluor®488, 7/4- AlexaFluor®647, and CD68-AlexaFluor®647 were from Serotec (UK). Ly-6A.2-FITC was from BioLegend (San Diego, CA, USA), and SK38.86 ascites fluid was a kind gift from Prof. Ulrich Hämmerling (Memorial Sloan-Kettering Cancer Center, New York, NY, USA). Percoll was obtained from Pharmacia (Uppsala, Sweden). PI-PLC was obtained from Sigma (Poole, UK).
Subcellular fractionation
Mice were injected with 1 ml 4% (w/v) thioglycollate broth and after 18 h peritoneal lavages, which are highly enriched in neutrophils, were collected in PBS containing 5 mM EDTA. Subcellular fractionation of peritoneal cells was performed by nitrogen cavitation and a Percoll density gradient according to Kjeldsen et al. [19]. Briefly, cells were spun down and resuspended in Krebs-Ringer bicarbonate buffer with glucose (Sigma) and 0.5 mM PMSF (Sigma). After 5 min incubation, cells were spun down, resuspended in disruption buffer (100 mM KCl, 3 mM NaCl, 1 mM ATP Na2, 3.5 mM MgCl2, 10 mM PIPES, and 0.5 mM PMSF), and pressurized under nitrogen for 5 min at 380 pounds per square inch in a nitrogen bomb. The cavitate was collected into EGTA (1.5 mM in 10 ml cell suspension) and spun down at 400 g for 15 min to pellet intact cells and nuclei. Supernatant was collected and transferred slowly over a one-layer Percoll gradient (1 ml 1.12 g/ml Percoll under 20 ml 1.065 g/ml Percoll), followed by centrifugation at 37,000 g for 30 min. After centrifugation, the middle fraction (membranes) was collected and the Percoll removed by centrifugation at 100,000 g for 45 min. The top fraction (cytoplasm) and the pellet containing nuclei were kept for further analysis. Assessment of protein concentration for all of the fractions was performed by the bicinchoninic acid method (Pierce, Rockford, IL, USA).
PNGase F treatment
Membranes obtained from subcellular fractionation (200 μg protein content) were resuspended in glycoprotein denaturing buffer (New England BioLabs, Beverly, MA, USA; 0.5% SDS and 1% β-ME) and boiled for 10 min. G7 buffer (New England BioLabs; 50 mM sodium phospate, pH 7.5), 10% Nonidet P-40, and PNGase (New England BioLabs) were added, and the reaction was incubated at 37°C for 1 h. Separation of PNGase-treated and untreated membranes was visualized by SDS-PAGE and by Western blot with the 7/4-antibody.
SDS-PAGE and Western blot
Intact cells or membranes were lysed in Laemmli sample buffer (4% SDS, 20% glycerol, 0.12 M Tris-HCl, pH 6.8) and boiled for 5 min. Equal amounts of protein were separated by SDS-PAGE. For 7/4 or Gr-1 Western blots, nitrocellulose membranes were blocked in 5% milk in PBS for 1 h at room temperature. 7/4- or Gr-1 antibodies were diluted at 10 μg/ml in 5% milk (in PBS) and incubated overnight at 4°C. After washes with PBS-Tween (0.1% Tween-20), antibody binding was detected with anti-rat peroxidase-conjugated antibody (Jackson Laboratory, Bar Harbor, ME, USA ) and ECL (Amersham, UK).
PI-PLC treatment
Mouse bone marrow was isolated as described previously [20] and resuspended in PBS at 2 × 107 cells/ml, and 2 × 106 cells were incubated for 30 min at 4°C or 37°C, with or without PI-PLC (Sigma) in the presence of 150 mM NaCl and 10 mM Tris (pH 7.4). Cells were stained and analyzed by flow cytomtery for expression of markers as detailed below but gating on bone marrow monocytes as F4/80+, CD11b+, SSClow cells and neutrophils as F4/80−, CD11b+, SSChigh cells.
Flow cytometric analysis
Cells (bone marrow-derived or stable cell lines) were incubated in blocking buffer (PBS containing 5% heat-inactivated rabbit serum, 0.5% BSA, 5 mM EDTA, 2 mM NaN3, 10 μg/ml 2.4G2) for 1 h at 4°C. FITC-labeled antibodies (7/4, Ly-6A.2) and Gr-1-PE were added at 10 μg/ml in a final volume of 100 μl washing buffer (PBS containing 0.5% BSA, 5 mM EDTA, and 2 mM NaN3) and incubated for 1 h at 4°C. SK38.86 ascites were used undiluted and incubated as the other antibodies. Cells incubated with FITC- or PE-labeled antibodies were washed three times with washing buffer and resuspended in 1% formaldehyde (in PBS). Cells incubated with SK38.86 were washed twice with washing buffer, and anti-mouse-PE was added for a further 1 h at 4°C. After this time, cells were treated as those incubated with fluorescent antibodies. Data were acquired on a FACSCalibur (Becton Dickinson, San Diego, CA, USA) or CyAn ADP analyzer (Beckman-Coulter, Fullerton, CA, USA), and analysis was performed using FlowJo (Tree Star, Inc., Ashland, OR, USA) or Summit (Beckman-Coulter).
Blood from C57BL/6 mice was obtained by cardiac puncture with EDTA used as an anticoagulant. RBCs were lysed with ACK buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA), and only leukocytes were used for staining with CD11b-PerCP-Cy5.5, F4/80-APC, 7/4-PE, and Ly-6C-FITC or DX5-PE (BD Biosciences, San Jose, CA, USA) and 7/4-Alexa647 and CD3-biotin with streptavidin-PerCP-Cy5.5. Data were acquired and analyzed as indicated above.
Proteomics and ELISA
Thioglycollate-induced neutrophils from C57BL/6 mice were lysed using a SDS-based buffer, and protein extraction was carried out in 50% v/v acetonitrile and 0.1% trifluoroacetic acid buffer for proteomic analysis. Proteins were separated by a ProteomeLab™ PF 2D protein fractionation system (Beckman-Coulter), which combines chromatofocusing (separation based on pI) followed by nonporous reverse-phase chromatography (separation based on protein size). More than 700 liquid fractions were collected and tested by ELISA for the presence of the 7/4-antigen by standard protocols.
Generation of stable cell lines expressing Ly-6 proteins
RNA was isolated from bone marrow cells of Balb/c and C57BL/6, according to standard conditions of the RNAeasy spin protocol (Qiagen, Valencia, CA, USA), and cDNA synthesis was performed using the Advantage RT-PCR kit (BD Biosciences). The Ly-6-degenerate primers, GGACCACCATGGACAVTWSTCAYRYKRYAARGWCCTG (5′) and GGGTCASAKMARGGTCTGYAGRWNGACYGA (3′), were designed based on sequence similarities among Ly-6a, Ly-6c, Ly-6g, Ly-6e, and Ly-6i. Ly-6i was amplified using the following specific primers: GGGGAATTCCACCATGGACACTTCTCACGCGAT (5′) and GGGGAATTCTCACATCAGGGTCTGCAGGAGG (3′). PCR products were cloned into the pCR3.1 vector (Invitrogen Life Technologies, Carlsbad, CA, USA) and sequenced (Sir William Dunn School of Pathology, Sequencing Service, Oxford, UK). Ly-6 proteins were subcloned further into the retroviral vector pFB(neo) (Stratagene, La Jolla, CA, USA) and transfected into the human embryo kidney 293T-based Phoenix ecotropic packaging cells using FuGene 6 (Roche, Indianapolis, IN, USA). Retroviral supernatants were collected after 48 h and used to transduce NIH3T3, RAW264.7, and Ba/F3 cells. Stable cell lines were selected and maintained in 0.6 mg/ml geneticin (Sigma).
Immunofluorescence
NIH3T3 cells were plated at a density of 1 × 105 cells on glass coverslips and incubated overnight at 37°C. The next day, cells were washed twice with PBS and fixed with 2% paraformaldehyde (in PBS) for 10 min at room temperature. Alternatively, Ba/F3 cells were cytospun onto glass slides prior to fixation as above. After three washes with washing buffer (PBS containing 0.5% BSA and 2 mM NaN3), cells were incubated in blocking buffer (PBS containing 5% heat-inactivated rabbit serum, 0.5% BSA, and 2 mM NaN3). 7/4- and rat-IgG2a control antibodies were diluted in washing buffer at 10 μg/ml for 1 h at room temperature. Cells were washed twice with washing buffer, incubated with anti-rat-Alexa488 (1:200) for 1 h at room temperature, and washed three more times. Coverslips were mounted on a slide using fluorescent mounting media (Dako, Denmark) and analyzed by fluorescence microscopy.
Zymosan-induced peritonitis
Zymosan was injected i.p. (2×107 particles in 100 μl PBS) into female C57BL/6 mice (6–8 weeks old), and peritoneal lavages were collected after 4, 18, 72, and 168 h. Resident cells were harvested from uninjected mice. Cells were stained with a range of antibodies and processed and analyzed by flow cytomtery as indicated above.
Antibody-mediated cell depletion
Mice were injected i.p. with 100 μg antibody and were bled by cardiac puncture after 16 h. Blood cells were immunostained (as above), and RBCs were removed by washing with ACK lysis buffer prior to flow cytometric analysis.
Cell sorting
Peritoneal lavages taken 7 days after the i.p. zymosan injection of C57BL/6 female mice were spun down and resuspended in blocking buffer (see Flow cytometric analysis above) at 2 × 107 cells/ml. After 30 min, 7/4-Alexa647 and F4/80-PE were added and incubated for 45 min on ice. Cells were washed twice with PBS containing 0.5% BSA and 5 mM EDTA and resuspended in the same buffer at 1 × 107 cell/ml. Ly-6B+ and Ly-6B− cells were separated by the MoFlo™ cell sorter (Beckman-Coulter), and single preparations were used for RNA isolation.
RT-PCR
Total RNA was extracted using Trizol (Invitrogen Life Technologies), according to the manufacturer’s instructions, and cDNA synthesis was prepared using the RETROscript® kit with an oligo(dT) primer (Ambion, Inc., Austin, TX, USA), as described by the manufacturer. Primers were designed to carry out standard PCR for CCR2 (C-C motif; 5′ TGGCTGT-GTTTGCCTCTCTA; 3′ CCTACAGCGAA-ACAGGGTGT); CIITA (5′ TCTCCAGTGTCCTAATCTAC-CA; 3′ AGATGTGTCCTCTGTCTCATTT); and SIGN-R1 (5′ GCAGGAGAAGATCTA-CCAACA; 3′ AGGCCCGGGCTAGCCTTC-AGTGCATGGGG). β2M (5′ TGACCGGCTTGTATGCTATC; 3′ CAG-TGTCAGCCAGGATATAG) was used as a control housekeeping gene.
BrdU assay
When required, mice were fed BrdU (Sigma) at 0.8 mg/ml in drinking water ad libitum. Cells from peritoneal lavages (106) were blocked and labeled with F4/80-PE and 7/4-AlexaFluor®647 as mentioned above (see Flow cytometric analysis). BrdU detection was performed subsequently using the FITC BrdU flow kit (BD PharMingen).
RESULTS
Distribution of the 7/4-antigen
To identify the main tissue expression of the 7/4-antigen, we obtained samples from C57BL/6 mice including bone marrow, thymus, lung, heart, spleen, lymph nodes, liver, kidney, and small intestine. We observed that the 7/4-antigen was abundant in bone marrow, and considerable expression was also evident in the spleen, lymph nodes, and lung, although the tissues were not perfused prior to analysis. Flow cytometric analysis of collagenase-digested lymph nodes typically identified <0.5% of cells with 7/4 expression and the phenotype of neutrophils or Ly-6C+ monocytes (not shown), and this is consistent with the reduced Western blot signal compared with spleen, where ∼2% of cells are 7/4 high expressors [5, 21]. However, thymus, heart, and liver have low expression levels of the 7/4-antigen (Fig. 1A). This is consistent with the reported expression of the 7/4-antigen on neutrophils and monocytes [1, 4, 5]. We [5] and others [4] have previously reported high 7/4 expression on monocytes, and now, we have confirmed that this expression was restricted to the Ly-6C+ monocyte subset (Fig. 1B). Expression of the 7/4-antigen was also evident on a small subset (15–40%) of CD3−DX5+ NK cells (data not shown).
Figure 1.
Distribution of the 7/4-antigen. (A) Different tissues were obtained and pooled from two C57BL/6 mice and separated by SDS-PAGE. Western blotting with the 7/4-antibody was performed as indicated in Materials and Methods. (B) Peripheral blood leukocytes (129) were labeled with CD11b, F4/80, Ly-6C, and 7/4, as indicated previously. Monocytes were first identified on the FSC/SSC plot and then as CD11b+F4/80+ cells, as described previously [20]. Expression of the 7/4-antigen determined two populations of monocytes (7/4+ and 7/4−), which are comparable with the populations established by the expression of Ly-6C (Ly-6C+and Ly-6C−). Data are derived from blood pooled from four independent animals, and similar results were obtained from three independent experiments with the Gr-1 antibody. (C) Depletion of peripheral myeloid cells by i.p. administration of the 7/4-antibody. Compared with isotype control-injected animals (open symbols), 7/4-injected mice (closed symbols) had substantially reduced numbers of neutrophils (Neu; identified as SSChiCD11b+Ly-6GhiGr-1hi) and Gr-1+ monocytes (Mo; SSClowF4/80+CD11b+Gr-1+) in the periphery. Gr-1− monocytes and eosinophils (Eos; SSCv.hiCD11b+F4/80+Gr-1int) are also shown. Data are expressed as the percentage of peripheral blood leukocytes defined as above, normalized to percentage present in isotype control-injected mice. Each symbol represents a single mouse, and horizontal bars denote means. Data are derived from the larger of two independent experiments and were analyzed by two-way ANOVA with Bonferonni post-tests (*, P<0.05; ***, P<0.001).
7/4-Mediated myeloid cell depletion
As the expression of the 7/4-antigen was relatively high on neutrophils and Gr-1+ monocytes, we tested the capability of the antibody to deplete these cells in vivo. Mice given a single i.p. injection with 100 μg 7/4- or rat IgG2a control antibody 16 h previously were analyzed for the presence of neutrophils and Gr-1+ monocytes in their blood (Fig. 1C). The injection of the 7/4-antibody caused a substantial reduction in the circulating numbers of neutrophils and Gr-1+ monocytes compared with the control antibody-injected mice (Fig. 1C).
7/4-Antigen is a low molecular weight polymorphic antigen
Western blot analysis of SDS-PAGE-resolved bone marrow lysates identifies the 7/4-antigen of C57BL/6 mice as a low molecular weight protein. Under nonreducing conditions, the 7/4-antigen has a size range between 15 and 35 kDa, whereas under reducing conditions, two main bands of ∼25 kDa and >15 kDa are evident (Fig. 2A). The antigen is absent on bone marrow lysates from Balb/c mice treated under the same conditions (Fig. 2A), which is consistent with the original reported polymorphism [1].
Figure 2.
7/4-Antigen is a low molecular weight N-glycosylated polymorphic protein with a GPI anchor. (A) Bone marrow lysates from C57BL/6 and Balb/c mice were separated by SDS-PAGE under nonreducing and reducing conditions. Western blotting with the 7/4-antibody was performed as indicated above. (B) Subcellular fractionation of the nitrogen-cavitated, thioglycollate-elicited, enriched neutrophil population resulted in the separation of membrane, cytoplasm, and nucleus fractions. Each fraction, as well as total lysate, was separated by SDS-PAGE, followed by Western blotting with the 7/4-antibody or Gr-1 as indicated previously. (C) Thioglycollate-elicited, neutrophil-rich membranes obtained from subcellular fractionation were treated with or without PNGase and Western blotted with the 7/4-antibody. (D) Monocytes and neutrophils were treated with PI-PLC at 4°C or 37°C and then labeled with 7/4, Gr-1, or isotype control antibodies for flow cytometric analysis. All experiments were conducted at least twice.
7/4-Antigen is a highly N-glycosylated membrane protein
To determine the location of the 7/4-antigen in the cell, we performed subcellular fractionation of neutrophils as indicated (see Materials and Methods) and tested 7/4 binding by Western blot. The 7/4-antigen was enriched in the membrane fraction, and cytoplasm and nucleus were deficient in its expression (Fig. 2B). A similar expression pattern was observed when the same fractions were probed with Gr-1 antibody (Fig. 2B), which recognizes Ly-6C and Ly-6G, two members of the Ly-6 family.
We treated the membranes obtained from subcellular fractionation with PNGase as indicated above (see Materials and Methods) and assessed 7/4 binding by Western blot to determine whether the 7/4-antigen was an N-linked glyco- protein. Before treatment with PNGase, the 7/4-antigen had a molecular weight of ∼25 kDa, which reduced to ∼15 kDa after PNGase treatment (Fig. 2C).
7/4-Antigen is a GPI-anchored protein
To establish whether the 7/4-antigen is bound to the membrane through a GPI anchor, we treated primary bone marrow cells with PI-PLC. By flow cytometric analysis, we observed that PI-PLC-treated neutrophils did not release the 7/4-antigen, whereas monocytes lost the 7/4-antigen after being treated with PI-PLC (Fig. 2D). A similar behavior was seen when we used the Gr-1 antibody, which detected Ly-6C/Ly-6G on PI-PLC-treated neutrophils but not on PI-PLC-treated monocytes (Fig. 2D). This behavior of the Gr-1 antigen is consistent with previous reports regarding the GPI anchor of Ly-6C [22].
7/4-Antigen is genetically linked to the chromosome 15-Ly-6 family
The gene encoding the 7/4-antigen is inherited, according to Mendelian principles of a single gene trait, so to genetically map the gene, 7/4 expression was backcrossed onto the Balb/c 7/4− background. Given the now-apparent, striking similarity between the 7/4-antigen and members of the murine Ly-6-gene family (many of which are small, polymorphic, often acidic, GPI-anchored membrane proteins and in the case of Ly-6C, Ly-6G, and Ly-6I, have similar expression patterns; see Table 1), we investigated the association of the 7/4-antigen with the Ly-6-gene complex on chromosome 15. 7/4+ Congenic Balb/c mice (generated by six generations of backcross) not only expressed the 7/4-antigen on their neutrophils and monocytes but also expressed the C57BL/6-derived Ly-6A.2 allele on their peripheral blood lymphocytes, consistent with a genetic linkage between these two loci (Fig. 3A).
TABLE 1.
Physiochemical Characteristics of Some Ly-6 Family Members Assessed by Prediction Softwarea
| Protein | MW (kDa) | pI | N-Glycosylation | GPI anchor |
|---|---|---|---|---|
| Ly-6A | 14.4 | 4.5 | No | Yes |
| Ly-6C | 14.1 | 6.5 | No | Yes |
| Ly-6G | 11.8 | 4.5 | Yes | Yes |
| Ly-6I | 14.7 | 5.1 | Yes | Yes |
| I830127L07Rik | 14.1 | 5.0 | Yes | Yes |
Provided by the Expasy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB).
Figure 3.
7/4-Antigen is genetically linked to the chromosome 15 Ly-6 family. (A) A congenic Balb/c.7/4+ mouse generated by six generations of backcrossing exhibited expression of the 7/4-antigen on neutrophils in peripheral blood, and this was compared with the expression of Ly-6A.2 on lymphocytes of C57BL/6, Balb/c, and the congenic 7/4+ mouse. Staining in the Balb/c.7/4+ mice is representative of six independent animals. (B) Flow cytometric analysis to assess the expression of the 7/4-antigen on neutrophils derived from C57BL/6 and NZB mice. (C) Spleen lysates from various mouse strains including C57BL/6, 129, NZB, CBA, and Balb/c were separated by SDS-PAGE and Western blotted with the 7/4-antibody. Analysis of NZB mice is representative of two independent animals.
As mentioned, the Ly-6 genes are polymorphic and belong to two primary haplotypes (Ly-6.1 and Ly-6.2). The fundamental difference between the polymorphic expression of the 7/4-antigen and members of the Ly-6 family is that the NZB mouse strain is 7/4+ [1] but Ly-6.1 in regard to most of the Ly-6 family except Ly-6B. However, all other 7/4+ mouse strains are Ly-6.2. NZB mice were reinvestigated for 7/4 expression (Fig. 3). Flow cytometric analysis of peripheral blood confirmed expression of the 7/4-antigen, but it was significantly lower than in C57BL/6 mice (Fig. 3B). By Western blot analysis of spleen lysates from 7/4+ strains (C57BL/6, 129, and NZB) and 7/4− strains (CBA and Balb/c), it was clear that NZB expressed an intermediate amount of protein (Fig. 3C).
Identification of the gene encoding the 7/4-antigen: synonymity with Ly-6B.2
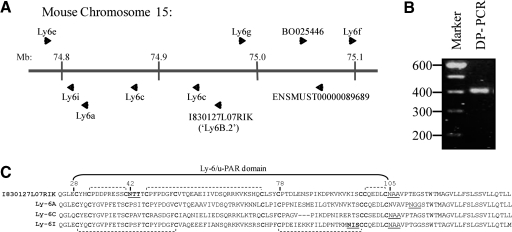
The extended chromosome 15 Ly-6 gene family contains several candidate genes for the 7/4-antigen, which encode small, acidic, GPI-anchored, N-glycosylated proteins. As a consequence of the similarity of protein expression between the 7/4-antigen and Ly-6A, Ly-6E, Ly-6C, Ly-6G, and Ly-6I (shown schematically in their genomic location in Fig. 4A) and the primary sequence similarity between these family members, degenerate PCR primers were designed, which would amplify all of these and hence, additional unknown, similar genes. Degenerate RT-PCR of neutrophil-enriched inflammatory cell peritoneal exudates from C57BL/6 and Balb/c mice generated a single PCR product consistent with the anticipated size of the Ly-6 family members, and the product from C57BL/6 mice was used to clone Ly-6-related, expressed sequences within the RNA preparations (Fig. 4B).
Figure 4.
Genomic location of the Ly-6-family members and protein alignment. (A) Distribution of the Ly-6 genes on the chromosome 15 according to ENSEMBL. A new identified member called I830127L07Rik is located between the Ly6g and Ly6c genes. (B) A single degenerate RT-PCR product of the expected size was obtained after isolating RNA from inflammatory-peritoneal neutrophils and amplification of Ly-6-related, expressed sequences using degenerate primers (DP). (C) Comparison of the primary amino acid sequence of I830127L07Rik to Ly-6A, Ly-6C, and Ly-6I. All of these proteins contain the Ly-6/u-PAR domain with 10 conserved cysteines (bold) linked by disulfide bonds (indicated by dashed brackets). N-Glycosylation was assessed by NetNGlyc, and NXS/T sequons were found in I830127L07Rik and Ly-6I (bold and underlined). Prediction of GPI anchor sites was performed with big PI predictor [23], and these sites are underlined. mb, Megabase.
Sequencing of the clones identified sequences derived from Ly6a, Ly6c, Ly6g, and an uncharacterized member of the family, whose partial sequence was contained in the expressed sequence tag I830127L07Rik (ENSEMBL Gene ID: ENSMUSG00000075601). This sequence obtained for I830127L07Rik also contained the sequence derived from first coding exon identifying the first 24 aa of the protein (MDSSHATRSCVLIFFVALLCAERA). Alignments of the primary protein sequence for Ly-6A, Ly-6C, Ly-6I, and I8302127L07Rik are shown in Figure 4C. The human sequence ENSP00000366895 (ENSEMBL) is the closest match to mouse I830127L07Rik. The gene is located on chromosome 8 in the same region where other human Ly-6 family members (prostate stem cell antigen, LY6K, LY6E, and LY6H) are placed. Further studies are necessary to establish the cellular distribution and function of this Ly-6 protein.
Kimura et al.[9, 10] described an antibody against an antigen called Ly-6B, which although not used extensively, recognized a protein on granulocytes and bone marrow cells of Ly-6.2 mouse strains. Independently, a similar antigen was studied by Hibbs et al. [24] using an antibody called Gm-1.2. Similar to 7/4, these antibodies have intermediate reactivity with cells from NZB mice [9], which have normally been described as a Ly-6.1 strain, indicating that the 7/4-antigen and Ly-6B.2 may be synonymous.
The cloned Ly-6 sequences were subcloned into a retroviral vector (pFBNeo; Stratagene) to generate stable cell lines. By Western blot, the 7/4-antibody recognized an ∼30-KDa protein present in the lysates of NIH3T3 cells transduced with I830127L07Rik but not those transduced with alternate Ly-6 family members (Fig. 5A). To determine if the 7/4- and anti-Ly-6B.2 (clone SK38.86) antibodies recognized the same gene product, the surface expression of the 7/4-antigen and Ly-6B.2 was assessed on RAW264.7 cells that had been transduced with I830127L07Rik or empty vector control (Fig. 5B). Anti-Ly-6B.2 and anti-7/4 bound to the I830127L07Rik-transduced cells, only indicating that these antigens were synonymous. Ly-6B expression by NIH3T3 cells resulted in primarily membrane-based staining (Fig. 5C). Flow cytometric analysis of I830127L07Rik-transduced NIH3T3 with anti-Gr-1 indicated that the antigen was not similar enough in the region of the epitope to that in Ly-6G/C for cross-reactivity (not shown). Further sequence analysis of I830127L07Rik (now referred to as Ly-6b) from Balb/c mice confirmed the presence of one base change in Balb/c mice, which encodes a premature stop codon in the third exon instead of a glutamine (Q73Stop). This indicates that in Balb/c mice, a truncated protein would be encoded, explaining the 7/4-deficient phenotype associated with these mice (shown schematically in Fig. 5D). Attempts to amplify the same region of the NZB Ly-6G gene with a primer pair that works with 129 and Balb/c mice failed, suggestive of distinct polymorphic variation, which was not investigated further (data not shown).
Figure 5.
7/4-Antigen is synonymous with Ly-6B.2. (A) NIH3T3-stable cell lines expressing the indicated Ly-6 proteins were analyzed by Western blot with the 7/4-antibody, which only recognizes the protein encoded by the complete I830127L07Rik sequence. (B) RAW264.7 cells expressing the I830127L07Rik sequence were labeled with the 7/4-antibody or SK38.86 ascites (black line) and empty vector-transduced control (filled histogram). Analysis was performed by flow cytometry. Data confirming flow cytometric recognition of I830127L07Rik by the 7/4-antibody was obtained twice with RAW264.7 cells and once with NIH3T3 cells. (C) NIH3T3 expressing Ly-6B.2 were cultured on coverslips and stained with FITC-labeled 7/4-antibody or IgG2a (isotype control). Visualization of Ly-6B.2 at the plasma membrane was done by fluorescence microscopy. Similar results were obtained in a second experiment using independently generated, retrovirally transduced Ba/F3 cells (not shown). (D) Schematic representation of the three exons coding for Ly-6B. The third exon on the sequence of Balb/c mice (Ly-6.1) holds a premature stop codon.
Expression of Ly-6B during zymosan-induced peritoneal inflammation
Using i.p. zymosan as an experimental model of inflammation, we determined the expression of Ly-6B on myeloid cells during the inflammatory reaction. Ly-6B is not expressed on peritoneal resident macrophages (F4/80highCD11bhigh), as we have demonstrated previously [5]. During the first hours of inflammation (4 h), Ly-6B is expressed on inflammatory monocytes (F4/80lowCD11blow) recruited to the peritoneal cavity (Fig. 6). After 18 h, Ly-6B is expressed on inflammatory monocytes and macrophages with increasing F4/80 and CD11b staining compared with the inflammatory monocytes present at the earlier time-point but with reducing Ly-6B expression. After 72 h, Ly-6B was evident on a novel F4/80intCD11bint subset of macrophages (72 and 168 h) when the inflammatory reaction was largely resolved (Fig. 6). A small number of F4/80highCD11bhigh macrophages expressed Ly-6B by arbitrary gating and quantitation (Fig. 6B), but this most likely reflects overlap of the distinct Ly-6B+ populations shown in Figure 6A with largely Ly-6B−F4/80highCD11bhigh “resident-like” macrophages, which have returned after the inflammatory insult. In six independent experiments (Day 7), the percentage of “mature” F4/80high macrophages expressing Ly-6B was 15.30 ± 4.64 (mean±sem).
Figure 6.
Expression of Ly-6B during zymosan-induced peritoneal inflammation. (A) C57BL/6 mice were injected with 2 × 107 zymosan particles, and peritoneal lavages were collected after 4, 18, 72, and 168 h. Cells were stained with F4/80, CD11b, and Ly-6B as indicated previously. The first panel shows the expression of F4/80 and CD11b at different time-points. At 4 h, residual resident macrophages (F4/80highCD11bhigh) and inflammatory monocytes (F4/80lowCD11blow) are readily identified. By 18 h, resident macrophages (F4/80highCD11bhigh) and inflammatory monocyte/macrophage (F4/80lowCD11blow) are evident. The F4/80 and CD11b profiles of these two populations “merge” during the 72- and 168-h resolution phases, and they are separated arbitrarily into three populations. The additional population with intermediate expression of F4/80 and CD11b (F4/80intCD11bint) emerges at 72 h and remains present at 168 h. Eosinophils (E) and neutrophils (N) are identified as indicated. Based on the F4/80:CD11bhigh,int,low gates, the second row of panels illustrates the changes of Ly-6B expression through the inflammatory response compared with F4/80. The novel Ly-6B+F4/80int “resolution phase” population is indicated. The third row of panels displays the expression of F4/80 associated with size of the cells (FSC area). (B) The expression of Ly-6B on the cells within the arbitrary F4/80highCD11bhigh, F4/80intCD11bint, and F4/80lowCD11blow gates indicated in A is represented as the mean ± sem of absolute cell counts from independent animals (three mice/group). Total counts for each population are shown with those that are Ly-6B+, indicated with shaded areas of bars. These data are consistent with prior publications at 4 and 18 h [4, 5, 7] and representative of two independent repeats, and the separation of macrophages into Ly-6B+ and Ly-6B− subsets at Days 3 and 7 is representative of two and seven independent experiments, respectively.
Ly-6B− and Ly-6B+ cells display different phenotypes
To gain insight into the nature of the heterogeneity of Ly-6B− and Ly-6B+ cells, we investigated whether Ly-6B+ and Ly-6B− cells also differed in the expression of other surface antigens. After a large screen of markers, we identified notably higher or consistent slightly higher expression of CD86, CD93, CD200R, CD9, CD11b, and CD68 by Ly-6B− macrophages when compared with Ly-6B+ macrophages. Interestingly, the Ly-6B+ macrophages expressed significantly more MHCII than the Ly-6B− cells, although neither cell type expressed significant amounts of CD11c (Fig. 7A). Expression of Ly-6C was also higher on Ly-6B+ cells, albeit very low when compared with peripheral blood monocytes (cf., Fig. 1A). We sorted these two cell populations (Fig. 7B) and isolated RNA from preparations with >90% purity. RT-PCR analysis revealed that Ly-6B+ macrophages harbor higher mRNA levels for CCR2, CD62L, and CIITA than the Ly-6B− macrophages (Fig. 7C). In contrast, Ly-6B− cells have higher expression of the resident cell-associated antigen SIGN-R1 [25], whose mRNA is substantially lower in Ly-6B+ cells (Fig. 7C).
Figure 7.
Ly-6B− and Ly-6B+ cells display different phenotypes. C57BL/6 mice were injected with 2 × 107 zymosan particles, and peritoneal lavages were collected after 7 days (168 h). Cells were stained with the indicated markers and analyzed by flow cytometry. Data are compiled from representative plots obtained with three independent mice or pooled cells from three mice from two independent experiments. (B) Cells were sorted by MoFlo for RNA isolation, as indicated in Materials and Methods. Plots are representative of three independent cell-sorting experiments with cells pooled from C57BL/6 mice. (C) Semi-quantitative RT-PCR analysis of select transcripts in cells sorted based on Ly-6B expression, as described in Materials and Methods. Similar differences were observed in independent RNA analysis of three separately sorted pairs of samples.
Ly-6B is expressed on recently generated macrophages
To determine whether Ly-6B is expressed on newly generated macrophages rather than resident peritoneal macrophages, we treated mice with 2 × 107 zymosan particles for 7 days. We fed the mice with 0.8 mg/ml BrdU in drinking water for the last 2 and 4 days prior to Day 7 (three mice/group). Peritoneal lavages were collected on Day 7 to detect BrdU incorporation (see above). Ly-6B+ cells showed higher incorporation of BrdU than Ly-6B− cells after 2 or 4 days BrdU treatment (Fig. 8). This indicates that Ly-6B+ macrophages represent cells recently generated by cell division. Mature, resident peritoneal macrophages have lower rates of proliferation.
Figure 8.
Proliferation of macrophages assessed by BrdU incorporation. C57BL/6 mice were injected with 2 × 107 zymosan particles. Mice were fed with 0.8 mg/ml BrdU in drinking water for 2 and 4 days before the 7-day time-point. A group of mice without BrdU feeding was used as control. Peritoneal lavages were collected after 7 days, and BrdU detection was performed using the BrdU flow kit (BD PharMingen), as indicated in Materials and Methods. (A) Representative flow cytometric analysis of mice showing the incorporation of BrdU after 2 and 4 days by Ly-6B− and Ly-6B+ cells. Gating was determined using mice that had not been fed BrdU. (B) Percentage of BrdU incorporation by Ly-6B− and Ly-6B+ cells. Each symbol represents an individual mouse from the same experiment respresented in A, and background data, from mice that had not been fed BrdU, were subtracted. Data are representative of one and two (2 days) independent experiments, and contemporary experiments were analyzed by two-way ANOVA as indicated. Dashed lines indicate the samples derived from the same mice.
DISCUSSION
In this study, we have found that the 7/4-antigen is synonymous with the previously unidentified Ly-6B.2 alloantigen, named in accordance with the Ly-6 classification, established previously by Kimura et al. [9]. We have identified the gene encoding the antigen and shown that it exhibits particularly high expression on the surface of neutrophils and Ly-678C+ monocytes. This relatively high surface expression renders these cells susceptible to the 7/4-antibody-mediated depletion in vivo and highlights the potential for manipulation of these types of myeloid subsets by targeting through this surface receptor.
Ly-6B is encoded by a gene located on chromosome 15 with other members of the murine Ly-6 family. The receptor itself is a low molecular weight, GPI-anchored, heavily N-glycosylated protein with low pI. We had made extensive attempts to identify the 7/4-antigen from a neutrophil-rich lysate from C57BL/6 mice using proteomic approaches. Separating by chromatofocusing (based on pI), followed by nonporous reverse-phase chromatography (based on protein size), identified the 7/4-antigen as an acidic protein (pI 4.5–5.0; data not shown), but MS analysis of positive fractions did not reveal the presence of likely 7/4-antigen candidates, based on size and physical properties of the proteins identified (data not shown). Repeated attempts to immunoprecipitate the 7/4-antigen in significant enough quantities to permit MS identification also failed as a result of the poor solubility of the protein and very poor yields of 7/4-antigen immunoreactive bands on SDS-PAGE (data not shown).
The 7/4-coding sequence was ultimately identified by a degenerate RT-PCR approach on neutrophil-enriched RNA, which was targeted to a subcluster of Ly-6 family members with similar expression and physical characteristics (Table 1). Its synonymity with Ly-6B.2 was confirmed with the demonstration that a noncommercial, Ly-6B.2-specific mAb (SK38.86) and the 7/4-antibody recognized the same gene product in specific transductants. We therefore concluded that the 7/4-antigen is the Ly-6B.2 protein described earlier by Kimura et al. [9].
Ly-6B is attached to the membrane via a GPI anchor (Fig. 3). PI-PLC treatment, however, did not remove Ly-6B from neutrophils. This behavior has also been observed in other GPI-anchored proteins as a result of the interaction of the GPI anchor with other membrane components or the addition of extra fatty acids, which impair the activity of PI-PLC, to the inositol ring [26]. The role of GPI-anchored proteins in cellular functions has been rather elusive. The presence of GPI anchors plays a significant role in the localization of proteins at plasma membranes. Although Ly-6 proteins have been recognized as differentiation markers (Ly-6A.2 on T cells, Ly-6C on monocytes/macrophages, Ly-6G on neutrophils, and Sca-1 on hematopoietic stem cells amongst other cell types), their physiological relevance is still unknown. Several studies have shown that cross-linking of Ly-6A/E (T cell-activating protein) using mAb induces lymphocyte proliferation subsequent to an increase of IL-2 production [27,28,29], and the presence of the GPI anchor seems to be essential for cell activation [30]. Ligation of other GPI-anchored proteins, including CD14, CD55, CD59, and CD90 (Thy.1), among others, has also been associated with responses such as oxidative burst, increase of cytoplasmic Ca++, and tyrosine phosphorylation [31]. As these proteins lack intracellular domains, it is difficult to explain how they interact with cytoplasmic signaling molecules. It has been suggested that they can interact with Src kinases or G-proteins, which are also anchored into lipid rafts through post-translational modifications such as palmitoylation and prenylation [31]. GPI-anchored proteins could also interact with transmembrane receptors. Some studies have described a physical association between FcγRs and GPI-anchored proteins as well as their crucial cooperation in FcγR effector functions, including FcR γ-chain phosphorylation, TNF-α release, dendritic cell maturation, and antigen presentation [32,33,34]. We carried out cross-linking experiments using the 7/4-antibody on Ly-6B-transduced Ba/F3 cells and in vitro-differentiated neutrophils, the latter using protocols published previously [35], but we were unable to detect an increase in Ca++ levels, specific tyrosine phosphorylation, or a release of reactive oxygen species consequent to respiratory burst (data not shown).
For a long time, the 7/4-antibody has been used as a neutrophil marker [1], and more recently, it is appreciated that it is also expressed by inflammatory monocytes [4, 5], some activated macrophages [2, 3], and bone marrow-restricted progenitors [6]. Here, we also show that the two main populations of monocytes in peripheral blood, Ly-6C− and Ly-6C+, are Ly-6B− and Ly-6B+, respectively (Fig. 1B). Although both major populations of monocytes are associated with recruitment to inflammatory lesions, the Ly-6C+ subset is characterized by higher expression of CCR2 and CD62L [16, 36]. It has been suggested that the Ly-6C+ subset precedes the Ly-6C− subset developmentally [37, 38]. The labeling of Ly-6B on monocytes resembles that of Ly-6C, which could be suggestive of similar gene regulation mechanisms that would not be too surprising, given their close genomic proximity. Like Ly-6C, the role of Ly-6B in homeostasis or infection and inflammation remains to be established.
We investigated the expression of Ly-6B during the inflammatory response induced by administration of zymosan and thioglycollate. Ly-6B is expressed on neutrophils and monocytes recruited into the peritoneal cavity within a few hours, as has been shown in other studies [1, 5, 39]. The majority of inflammatory monocytes and macrophages present during the acute phase of the response (4 and 18 h) expresses Ly-6B, although the expression of Ly-6B declines as it matures, and F4/80, CD11b, and cell size (FSC) increase (Fig. 6) [4, 5]. As mentioned above (Fig. 6), we identified Ly-6B+ and Ly-6B− macrophages during the resolution phase of a model inflammatory reaction induced with zymosan. At this time, the inflammatory response is largely resolved, and few neutrophils (F4/80−Ly-6B+) and inflammatory monocytes (F4/80lowLy-6B+) can be detected in the lesion (see Fig. 7). Ly-6B− macrophages are comparable with resident peritoneal macrophages in marker expression (data not shown), consistent with their representing the recovery of the resident peritoneal macrophage population after an inflammatory response. The expression of markers such as CD11b, CD86, CD9, CD93, and CD200R is higher in the Ly-6B− rather than Ly-6B+ macrophages. Interestingly, Ly-6B+ macrophages expressed the highest levels of MHCII and Ly-6C and at the mRNA level, CCR2, CD62L, and CIITA. The coexpression of the “Ly-6C+ monocyte marker” Ly-6B with other markers associated with the Ly-6C+ monocyte subset (CCR2 and CD62L) is suggestive of recent arrival and differentiation of these cells from inflammatory Ly-6B+Ly-6C+ monocytes. Expression of MHCII is regulated by the transcriptional coactivator CIITA, whose repression has been associated with the terminal differentiation of B cells into plasma cells and maturation of dendritc cells [40, 41]. We observed that CIITA is expressed at lower levels in Ly-6B− cells (Fig. 7C), and expression of MHCII is notably lower, which could be part of the maturation process of peritoneal “resident” macrophages and again, would be consistent with the hypothesis that Ly-6B-expressing macrophages are recently generated macrophages in the process of differentiating into, in this setting, resident peritoneal macrophages. The suggestion that these Ly-6B+ cells are more mature compared with inflammatory monocytes is supported by their higher expression of F4/80 and CD11b, lower expression of Ly-6C, and increased cell size. The fact that they may be less mature than their Ly-6C− contemporaries is supported by BrdU incorporation experiments, which indicate that the Ly-6B+ cells are generated more recently by cell division than their Ly-6B− contemporaries. It is also supported by the observation that they express less CD11b and F4/80 and are smaller than the Ly-6B− cells but retain expression of monocytic markers (Ly-6C, CCR2, and CD62L). Collectively, this also supports the speculation that they are not derived from Ly-6B− macrophages by marker regulation.
Taken in context with other studies about the distribution of the 7/4-antigen [4, 5] and the biology of the Ly-6C+ monocyte subset, it seems likely that Ly-6B+ monocytes are recruited into the peritoneal cavity, where they lose Ly-6B expression as they differentiate [4, 5]. This differentiation can be into an inflammatory macrophage or dendritic cell phenotype or as a replacement for tissue resident macrophage populations (most likely during the resolution phase of the inflammatory response). Although the origins of the tissue resident macrophages and dendritic cells are poorly characterized and likely to differ under inflammatory circumstances, it seems unlikely that these late inflammatory response Ly-6B+ cells are becoming “resident peritoneal” dendritic cell populations, as the marker phenotype of the two cells is quite different (data not shown and ref. [42]). We speculate that Ly-6B expression by macrophages in inflammatory lesions, such as seen during parasite infection [3] and BCG granulomas [2] or on cells recruited to tumor challenge [5], represents the arrival of recently generated macrophages. As such, Ly-6B expression may serve as a useful surrogate marker for the kinetics of monocyte/macrophage turnover in chronic inflammation, as it highlights recently recruited monocyte-derived inflammatory macrophages and other potentially more obscure sources of macrophages in an inflammatory context. Also of interest is the expression of Ly-6B by cells with the phenotype of myeloid-derived suppressor cells (Gr-1+CD11b+) during steady-state inflammation and tumor challenge [5, 18]. Myeloid-derived suppressor cells are noted for their “immature”-like phenotype [18]. This would be consistent with the suggestion that myeloid-derived suppressor cells are also relatively, recently produced cells retaining Ly-6B (and Gr-1) expression from their neutrophilic/monocytic roots in a way directly analogous to that proposed above for Ly-6B expression in our experimental peritonitis model and with direct correlation to previous malarial and BCG infection models [2, 3]. The ability, by, for example, exploiting Ly-6B expression, to target these relatively immature, neutrophilic- and monocytic-like cells specifically in vivo could represent a valuable, new approach to many aspects of myeloid cell biology.
AUTHORSHIP
M. R. and P. R. T. designed and performed the experiments and co-wrote the manuscript. B. T. and M. S. designed and conducted experiments. S. G. and P. R. T. conceived the project. All authors contributed to the interpretation of data.
ACKNOWLEDGMENTS
This work was funded by the Wellcome Trust (070579) and the Medical Research Council (G0601617). We thank Alexandre Akoulitchev for his support and advice during the proteomics analysis. We also thank the staff of our animal facilities for the care of the animals used in this study.
Footnotes
Abbreviations: APC=allophycocyanin, β2M=β2-microglobulin, BCG=bacillus Calmette-Guerin, CIITA=class II, MHC, transactivator, CD62L=CD62 ligand, FSC=forward-scatter, MS=mass spectromic, NZB=New Zealand Black, pI=isoelectric point, PI-PLC=phosphatidylinositol-specifc phospholipase C, PNGase F=peptide N-glycanase F, SIGN-R1=specific ICAM-3-grabbing nonintegrin-related 1, SSC=side-scatter, u-PAR=urokinase-type plasminogen-activated receptor
References
- Hirsch S, Gordon S. Polymorphic expression of a neutrophil differentiation antigen revealed by monoclonal antibody 7/4. Immunogenetics. 1983;18:229–239. doi: 10.1007/BF00952962. [DOI] [PubMed] [Google Scholar]
- Gordon S, Keshav S, Stein M. BCG-induced granuloma formation in murine tissues. Immunobiology. 1994;191:369–377. doi: 10.1016/S0171-2985(11)80442-0. [DOI] [PubMed] [Google Scholar]
- Lee S H, Crocker P, Gordon S. Macrophage plasma membrane and secretory properties in murine malaria. Effects of Plasmodium yoelii blood-stage infection on macrophages in liver, spleen, and blood. J Exp Med. 1986;163:54–74. doi: 10.1084/jem.163.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R B, Hobbs J A, Mathies M, Hogg N. Rapid recruitment of inflammatory monocytes is independent of neutrophil migration. Blood. 2003;102:328–335. doi: 10.1182/blood-2002-10-3228. [DOI] [PubMed] [Google Scholar]
- Taylor P R, Brown G D, Geldhof A B, Martinez-Pomares L, Gordon S. Pattern recognition receptors and differentiation antigens define murine myeloid cell heterogeneity ex vivo. Eur J Immunol. 2003;33:2090–2097. doi: 10.1002/eji.200324003. [DOI] [PubMed] [Google Scholar]
- Bertoncello I, Bartelmez S H, Bradley T R, Hodgson G S. Changes in cell surface antigen expression during murine bone marrow cell regeneration in vivo and proliferation in vitro. Immunol Cell Biol. 1989;67:127–133. doi: 10.1038/icb.1989.18. [DOI] [PubMed] [Google Scholar]
- Taylor P R, Reid D M, Heinsbroek S E, Brown G D, Gordon S, Wong S Y. Dectin-2 is predominantly myeloid restricted and exhibits unique activation-dependent expression on maturing inflammatory monocytes elicited in vivo. Eur J Immunol. 2005;35:2163–2174. doi: 10.1002/eji.200425785. [DOI] [PubMed] [Google Scholar]
- LeClair K P, Rabin M, Nesbitt M N, Pravtcheva D, Ruddle F H, Palfree R G, Bothwell A. Murine Ly-6 multigene family is located on chromosome 15. Proc Natl Acad Sci USA. 1987;84:1638–1642. doi: 10.1073/pnas.84.6.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Tada N, Liu-Lam Y, Hammerling U. Studies of the mouse Ly-6 alloantigen system. II. Complexities of the Ly-6 region. Immunogenetics. 1984;20:47–56. doi: 10.1007/BF00373446. [DOI] [PubMed] [Google Scholar]
- Kimura S, Tada N, Nakayama E, Hammerling U. Studies of the mouse Ly-6 alloantigen system. I. Serological characterization of mouse Ly-6 alloantigen by monoclonal antibodies. Immunogenetics. 1980;11:373–381. doi: 10.1007/BF01567804. [DOI] [PubMed] [Google Scholar]
- Rock K L, Reiser H, Bamezai A, McGrew J, Benacerraf B. The LY-6 locus: a multigene family encoding phosphatidylinositol-anchored membrane proteins concerned with T-cell activation. Immunol Rev. 1989;111:195–224. doi: 10.1111/j.1600-065x.1989.tb00547.x. [DOI] [PubMed] [Google Scholar]
- Fleming TJ, O'hUigin C, Malek T R. Characterization of two novel Ly-6 genes. Protein sequence and potential structural similarity to α-bungarotoxin and other neurotoxins. J Immunol. 1993;150:5379–5390. [PubMed] [Google Scholar]
- Patterson J M, Johnson M H, Zimonjic D B, Graubert T A. Characterization of Ly-6M, a novel member of the Ly-6 family of hematopoietic proteins. Blood. 2000;95:3125–3132. [PubMed] [Google Scholar]
- Pflugh D L, Maher S E, Bothwell A L. Ly-6I, a new member of the murine Ly-6 superfamily with a distinct pattern of expression. J Immunol. 2000;165:313–321. doi: 10.4049/jimmunol.165.1.313. [DOI] [PubMed] [Google Scholar]
- De Bruijn M F, Slieker W A, van der Loo J C, Voerman J S, van Ewijk W, Leenen P J. Distinct mouse bone marrow macrophage precursors identified by differential expression of ER-MP12 and ER-MP20 antigens. Eur J Immunol. 1994;24:2279–2284. doi: 10.1002/eji.1830241003. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Jung S, Littman D R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- Nakano H, Yanagita M, Gunn M D. CD11c(+)B220(+)Gr-1(+) cells in mouse lymph nodes and spleen display characteristics of plasmacytoid dendritic cells. J Exp Med. 2001;194:1171–1178. doi: 10.1084/jem.194.8.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich D I, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeldsen L, Sengelov H, Borregaard N. Subcellular fractionation of human neutrophils on Percoll density gradients. J Immunol Methods. 1999;232:131–143. doi: 10.1016/s0022-1759(99)00171-4. [DOI] [PubMed] [Google Scholar]
- Taylor P R, Brown G D, Reid D M, Willment J A, Martinez-Pomares L, Gordon S, Wong S Y. The β-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol. 2002;169:3876–3882. doi: 10.4049/jimmunol.169.7.3876. [DOI] [PubMed] [Google Scholar]
- Taylor P R, Tsoni S V, Willment J A, Dennehy K M, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown G D. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutila D B, Kurk S, Jutila M A. Differences in the expression of Ly-6C on neutrophils and monocytes following PI-PLC hydrolysis and cellular activation. Immunol Lett. 1994;41:49–57. doi: 10.1016/0165-2478(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- Hibbs M L, Hogarth P M, Scott B M, Harris R A, McKenzie I F. Monoclonal antibody to murine neutrophils: identification of the Gm-2.2 specificity. J Immunol. 1984;133:2619–2623. [PubMed] [Google Scholar]
- Taylor P R, Brown G D, Herre J, Williams D L, Willment J A, Gordon S. The role of SIGNR1 and the β-glucan receptor (dectin-1) in the nonopsonic recognition of yeast by specific macrophages. J Immunol. 2004;172:1157–1162. doi: 10.4049/jimmunol.172.2.1157. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Mayor S. The GPI-anchor and protein sorting. Cell Mol Life Sci. 2001;58:1969–1987. doi: 10.1007/PL00000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S C, Kamdar M M, Bamezai A. Ly-6A.2 expression regulates antigen-specific CD4+ T cell proliferation and cytokine production. J Immunol. 2002;168:118–126. doi: 10.4049/jimmunol.168.1.118. [DOI] [PubMed] [Google Scholar]
- Toulon M, Palfree R G, Palfree S, Dumont F J, Hammerling U. Ly-6 A/E antigen of murine T cells is associated with a distinct pathway of activation. Requirements for interferon and exogenous interleukin 2. Eur J Immunol. 1988;18:937–942. doi: 10.1002/eji.1830180616. [DOI] [PubMed] [Google Scholar]
- Yeh E T, Reiser H, Daley J, Rock K L. Stimulation of T cells via the TAP molecule, a member in a family of activating proteins encoded in the Ly-6 locus. J Immunol. 1987;138:91–97. [PubMed] [Google Scholar]
- Su B, Waneck G L, Flavell R A, Bothwell A L. The glycosyl phosphatidylinositol anchor is critical for Ly-6A/E-mediated T cell activation. J Cell Biol. 1991;112:377–384. doi: 10.1083/jcb.112.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horejsi V, Cebecauer M, Cerny J, Brdicka T, Angelisova P, Drbal K. Signal transduction in leucocytes via GPI-anchored proteins: an experimental artefact or an aspect of immunoreceptor function? Immunol Lett. 1998;63:63–73. doi: 10.1016/s0165-2478(98)00054-6. [DOI] [PubMed] [Google Scholar]
- Chuang F Y, Sassaroli M, Unkeless J C. Convergence of Fc γ receptor IIA and Fc γ receptor IIIB signaling pathways in human neutrophils. J Immunol. 2000;164:350–360. doi: 10.4049/jimmunol.164.1.350. [DOI] [PubMed] [Google Scholar]
- Ding L, Shevach E M. Inhibition of the function of the FcγRIIB by a monoclonal antibody to thymic shared antigen-1, a Ly-6 family antigen. Immunology. 2001;104:28–36. doi: 10.1046/j.0019-2805.2001.01275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazenbos W L, Clausen B E, Takeda J, Kinoshita T. GPI-anchor deficiency in myeloid cells causes impaired FcγR effector functions. Blood. 2004;104:2825–2831. doi: 10.1182/blood-2004-02-0671. [DOI] [PubMed] [Google Scholar]
- Wang G G, Calvo K R, Pasillas M P, Sykes D B, Hacker H, Kamps M P. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- Sunderkotter C, Nikolic T, Dillon M J, Van Rooijen N, Stehling M, Drevets D A, Leenen P J. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172:4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- Varol C, Yona S, Jung S. Origins and tissue-context-dependent fates of blood monocytes. Immunol Cell Biol. 2009;87:30–38. doi: 10.1038/icb.2008.90. [DOI] [PubMed] [Google Scholar]
- Bertoncello I, Bradley T R, Watt S M. An improved negative immunomagnetic selection strategy for the purification of primitive hemopoietic cells from normal bone marrow. Exp Hematol. 1991;19:95–100. [PubMed] [Google Scholar]
- Landmann S, Muhlethaler-Mottet A, Bernasconi L, Suter T, Waldburger J M, Masternak K, Arrighi J F, Hauser C, Fontana A, Reith W. Maturation of dendritic cells is accompanied by rapid transcriptional silencing of class II transactivator (CIITA) expression. J Exp Med. 2001;194:379–391. doi: 10.1084/jem.194.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Waldburger J M, Krawczyk M, Otten L A, Suter T, Fontana A, Acha-Orbea H, Reith W. Mini-review: specificity and expression of CIITA, the master regulator of MHC class II genes. Eur J Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- Dioszeghy V, Rosas M, Maskrey B H, Colmont C, Topley N, Chaitidis P, Kuhn H, Jones S A, Taylor P R, O'Donnell V B. 12/15-Lipoxygenase regulates the inflammatory response to bacterial products in vivo. J Immunol. 2008;181:6514–6524. doi: 10.4049/jimmunol.181.9.6514. [DOI] [PubMed] [Google Scholar]