Abstract
Malaria is endemic in the Cukurova region while it is sporadic in other regions of Turkey. Therefore, the laboratory and clinical diagnosis of malaria is important for the treatment of malaria. In this study, 92 blood samples that were taken from the suspected malaria patients for routine diagnosis in a period of 10 years between 1999 and 2009 were analyzed. All of these blood samples were examined by microscopic examinations using Giemsa-stained thick blood films, nested PCR, and real-time PCR. The sensitivity-specificity and positive-negative predictive values for these diagnostic tests were then calculated. It was found that the positive predictive values of microscopic examination of thick blood films, nested PCR, and real-time PCR were 47.8%, 56.5%, and 60.9% for malaria, respectively. The real-time PCR was found to have a specificity of 75% and sensitivity of 100%, while specificity and sensitivity of nested PCR was found 81.2% and 97.7% according to the microscopic examination of thick blood films, respectively.
Keywords: Plasmodium vivax, nested PCR, real-time PCR, malaria
INTRODUCTION
Plasmodium vivax is the most widely distributed malaria species particularly in the Cukurova region in southern parts of Turkey [1]. P. vivax infections are rarely fatal but it can lead to a very debilitating illness [2]. Its clinical features are not enough for an accurate diagnosis, since its clinical features are shared by a host of other occurring infections, such as visceral leishmaniasis, typhoid, and tuberculosis [3]. Therefore, laboratory methods are significant for the diagnosis of P. vivax infections [4]. The laboratory diagnosis of P. vivax infections usually relies on the microscopic examination of Giemsa-stained thick and thin blood films, immunochromatographic tests, and serological methods [5-7]. These methods provide a cost effective, rapid diagnostic tool which can easily be applied in the field [8]. Nonetheless, these methods are prone to misdiagnosis especially in the cases of mixed infections and low level parasitemia [5,9-11].
Molecular techniques had been developed recently, such as real-time PCR and nested PCR, for the diagnosis of malaria. The real-time PCR produce rapid results with very low contamination risks, a high sensitivity and specificity, and the possibility of quantification [12-14]. Nested PCR based methods, on the other hand, are also sensitive and specific but are labor intensive [15].
In this study, 92 malaria-suspected cases were analyzed by conventional methods including microscopic examination of thick blood films, nested PCR, and TaqMan-based real-time PCR qualitative assay, in Cukurova region belonging to the southern part of Turkey. The genus-specific nested PCR was carried out to confirm that the samples were truly positive for malaria and the patients were actually infected with Plasmodium spp. On the other hand, these cases were identified by species-specific TaqMan real-time PCR and nested PCR.
MATERIALS AND METHODS
Blood samples and laboratory methods
After giving informed consent, 92 blood samples were taken from suspected malaria patients coming for the routine diagnosis in the Parasitology Department, Faculty of Medicine, Cukurova University in Turkey for a period of 10 years between 1999 and 2009. Thick blood films were prepared from all blood samples. The smears were examined by 2 parasitologists and were considered negative if no parasites were seen under × 100 objective.
Nested PCR
The genomic DNA was extracted from all of the blood samples using Agencourt Genfind v2 DNA isolation kit (Beckman Coulter, Beverly, Massachusetts, USA) according to the manufacturers' protocol.
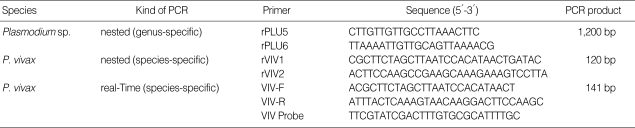
This DNA templates were used for the amplification with thermal cycler (Sensoquest, Göttingen, Germany) using a genusspecific primer set (rPLU5 and rPLU6; Table 1) as described by Perandin et al. [16]. In order to assess P. vivax, a species-specific primer (rVIV1 and rVIV2; Table 1) set was used [16]. All of the PCR reaction mixtures (100 µl of total volume) consisted of 250 mM deoxynucleoside triphosphate (Fermantas RO191), 10 × PCR buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.3], 20 mM MgCl2), 2 U/µl Taq DNA polymerase (Fermantas SB38) and 5 pmol of each primer set. The thermal cycling began with denaturation at 95℃ for 5 min, followed by 25 cycles of 94℃ for 30 sec, 58℃ for 2 min, and 72℃ for 2 min, with final incubation at 72℃ for 5 min. Five-µl aliquots of the product were used for the second amplification cycle (nested 2) using the same PCR conditions and thermal cycle except the use of 30 cycles instead of 25. PCR products were analyzed on agarose gels of 2.5% by electrophoresis at 100 V in 1 × Tris-Boric-EDTA buffer (0.04 M Tris-boric and 1 mM EDTA pH 8.0) and then visualized by UV light after being stained with ethidium bromide.
Table 1.
Primers and probe for 18S rRNA gene in P. vivax
Real-Time PCR
The real-time PCR assay was performed using species-specific primer (VIV-F and VIV-R) and TaqMan fluorescence-labeled probe (VIV probe) for the detection of P. vivax (Table 1) [16]. The probe was labeled with 3'6-carboxyfluorescein (FAM) and TaqMan fluorescence. The real-time PCR mix comprised of 25 µl of TaqMan 2x universal PCR master mix (Applied Biosystems, Inc., Foster City, California, USA), a 300 nM concentration of P. vivax species-specific primer set, a 200 nM concentration of probe, and 5 µl of genomic DNA samples. The real-time PCR mixtures were preincubated at 50℃ for 2 min followed by denaturation at 95℃ for 10 min and 50 cycles of 95℃ for 15 sec and 60℃ for 1 min using Stratagene Mx3,000 p Real-Time PCR system.
RESULTS
Total 92 blood samples from patients suspected with malaria were examined by the microscopic examination of thick blood films, nested PCR, and real-time PCR. Microscopic examination was positive for 47.8% patients (44/92).
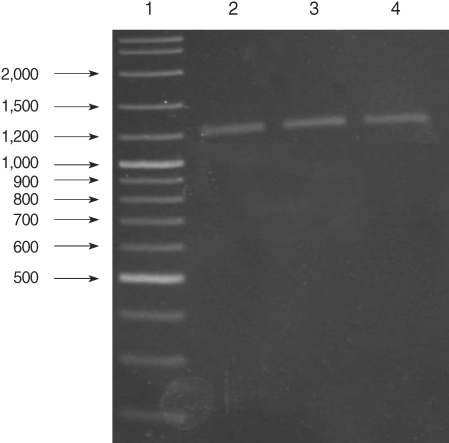
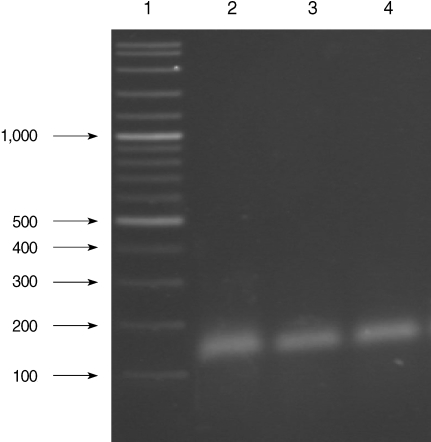
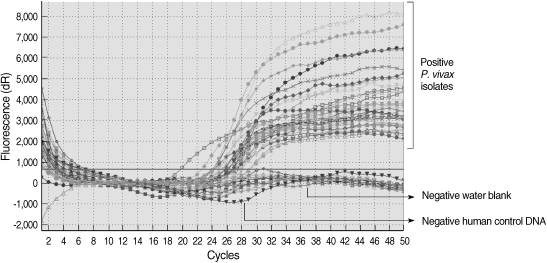
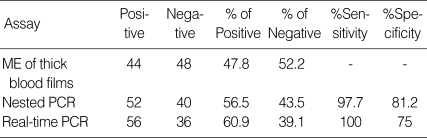
All of the DNA samples were screened for the presence of Plasmodium spp. with the rPLU5 and rPLU6 genus-specific primers; and the PCR products were considered positive when a band of correct size of ~1,200 bp was observed for Plasmodium isolates (Fig. 1). Moreover, all of the DNA samples were screened for the identification of Plasmodium spp. with rVIV1 and rVIV2 species-specific primers for P. vivax. The PCR product sizes of Plasmodium spp. isolates were found to be approximately 120 bp for P. vivax (Fig. 2). All of the samples were identified as containing P. vivax; therefore, the other Plasmodium species were not screened by other species-specific primers. Nested PCR was 56.5% positive (52/92) in all samples. Using species-specific primers and probe, P. vivax was detected by real-time PCR in all of the DNA samples. The fractional cycle number reflecting a positive PCR result is called the cycle threshold (Ct). Cycle threshold values were found to be between 19.8 and 29.2 with median threshold of 25.1 cycles. According to the results of the real-time PCR result, 60.9% (56/92) of samples were identified as positive for P. vivax (Fig. 3). Furthermore, the sensitivity-specificity of nested PCR and real-time PCR according to microscopic examination of thick blood films was calculated in this study. It was found that sensitivities were 97.7%, and 100%, respectively. The specificity of nested PCR and real-time PCR were found 81.2% and 75%, respectively (Table 2).
Fig. 1.
Results of genus-specific nested-PCR. Lane 1, 100-3,000 bp DNA lader; Lanes 2-4, Plasmodium spp. isolates.
Fig. 2.
Results of species-specific nested-PCR. Lane 1, 100-3,000 bp DNA lader; Lanes 2-4, P. vivax isolates.
Fig. 3.
Results of species-specific real-time PCR.
Table 2.
Results of nested PCR and real-time PCR according to microscopic examination of thick blood films
ME, Microscopic examination.
DISCUSSION
Routine laboratory diagnosis of malaria is usually based on microscopic examination of Giemsa-stained thick and thin blood films, immunochromatographic tests, and serological methods [6]. The advantages offered by these tests is, the fact that a result can be obtained within half an hour [11,16,17]. On the other hand, sensitivity decreases with these tests as parasitemia falls below 100 parasites/µl and false negatives are observed [11,18,19].
The real-time PCR and nested PCR with specific primers and a labeled probe of the 18S rRNA gene had been developed for the detection of malaria parasites [3,6,18,19]. The standardized 18S rRNA gene real-time PCR sensitivity was 5 times more sensitive than nested PCR (5-10 parasite/µl), 20 times more sensitive than microscopic examination (10-50 parasites/µl) [9].
The real-time PCR has more advantages than the other diagnosis molecular methods, such as nested PCR [11,16]. The risk of contamination at the real-time PCR is far lower than the risk in nested PCR because it is a closed-tube reaction [15,16,20]. The real-time PCR assay is also rapid and can reliably distinguish between 2 alleles following a single 3-hr experiment [16,21,22]. It has a higher reported sensitivity from 98% to 100% [11,22]. Boonma et al. [5] have studied multiplex PCR, nested PCR and real-time PCR assay for detected P. vivax, they have found that the real-time PCR is more sensitivity than the other tests. Farcas et al. [15] and Keen et al. [23] have reported that the sensitivity of real-time PCR (100%) over than the sensitivity of nested PCR. Similar results were found for the sensitivity of real-time PCR (100%) regarding the diagnosis of malaria in this study. The sensitivities of real-time PCR and nested PCR according to microscopic examination of thick blood films, were also compared and it was found that real-time PCR is more sensitive (100%) than the other tests.
The real-time PCR has been reported to be more useful than conventional laboratory methods, such as microscopic examination, immunochromatographic tests, and PCR, for the diagnosis of low-level parasitemia, and the correct diagnosis and treatment of malaria parasite species [5,22]. This study confirms these findings; displaying that real-time PCR was useful method when compared with microscopy of thick blood films and nested PCR.
References
- 1.Aslan G, Seyrek A, Kocagoz T, Ulukanligil M, Erguven S, Gunalp A. The diagnosis of malaria and identification of Plasmodium species by polymerase chain reaction in Turkey. Parasitology Int. 2007;56:217–220. doi: 10.1016/j.parint.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Zeyrek YF, Kurcer MA, Zeyrek D, Simsek Z. Parasite density and serum cytokine levels in Plasmodium vivax malaria in Turkey. Parasite Immunol. 2006;28:201–207. doi: 10.1111/j.1365-3024.2006.00822.x. [DOI] [PubMed] [Google Scholar]
- 3.Sundar S, Rai M. Laboratory diagnosis of visceral leishmaniasis. Clin Diagn Lab Immunol. 2002;9:951–958. doi: 10.1128/CDLI.9.5.951-958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farcas GA, Soeller R, Zhong K, Zahirieh A, Kain KC. Real-time polymerase chain reaction assay for the rapid detection and characterization of chloroquine-resistant Plasmodium falciparum malaria in returned travelers. Clin Infect Dis. 2006;42:622–627. doi: 10.1086/500134. [DOI] [PubMed] [Google Scholar]
- 5.Boonma P, Christensen PR, Suwanarusk R, Price RN, Russell B, Lek-Uthai U. Comparison of three molecular methods for the detection and speciation of Plasmodium vivax and Plasmodium falciparum. Malar J. 2007;6:124. doi: 10.1186/1475-2875-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider P, Wolters L, Schoone G, Schallig H, Sillekens P, Hermsen R, Sauerwein R. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J Clin Microbiol. 2005;43:402–405. doi: 10.1128/JCM.43.1.402-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veron V, Simon S, Carme B. Multiplex real-time PCR detection of Plasmodium falciparum, P. vivax and P. malariae in human blood samples. Exp Parasitol. 2009;121:346–351. doi: 10.1016/j.exppara.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 9.Gama BE, Silva-Pires Fdo E, Lopes MN, Cardoso MA, Britto C, Torres KL, de Mendonça Lima L, de Souza JM, Daniel-Ribeiro CT, Ferreira-da-Cruz Mde F. Real-time PCR versus conventional PCR for malaria parasite detection in low-grade parasitemia. Exp Parasitol. 2007;116:427–432. doi: 10.1016/j.exppara.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 10.McKenzie FE, Sirichaisinthop J, Miller RS, Gasser RA, Jr, Wongsrichanalai C. Dependence of malaria detection and species diagnosis by microscopy on parasite density. Am J Trop Med Hyg. 2003;69:372–376. [PMC free article] [PubMed] [Google Scholar]
- 11.Safeukui I, Millet P, Boucher S, Melinard L, Fregeville F, Receveur MC, Pistone T, Fialon P, Vincendeau P, Fleury H, Malvy D. Evaluation of FRET real-time PCR assay for rapid detection and differentiation of Plasmodium species in returning travelers and migrants. Malar J. 2008;7:70. doi: 10.1186/1475-2875-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews L, Andersen RF, Webster D, Dunachie S, Walther RM, Bejon P, Hunt-Cooke A, Bergson G, Sanderson F, Hill AV, Gilbert SC. Quantitative real-time polymerase chain reaction for malaria diagnosis and its use in malaria vaccine clinical trials. Am J Trop Med Hyg. 2005;73:191–198. [PubMed] [Google Scholar]
- 13.Elsayed S, Plewes K, Church D, Chow B, Zhang K. Use of molecular beacon probes for real-time PCR detection of Plasmodium falciparum and other Plasmodium species in peripheral blood specimens. J Clin Microbiol. 2006;44:622–624. doi: 10.1128/JCM.44.2.622-624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vo TK, Bigot P, Gazin P, Sinou V, De Pina JJ, Huynh DC, Fumoux F, Parzy D. Evaluation of a real-time PCR assay for malaria diagnosis in patients from Vietnam and in returned travelers. Trans R Soc Trop Med Hyg. 2007;101:422–428. doi: 10.1016/j.trstmh.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Farcas GA, Zhong KJ, Mazzulli T, Kain KC. Evaluation of the Real-Art Malaria LC real-time PCR assay for malaria diagnosis. J Clin Microbiol. 2004;42:636–638. doi: 10.1128/JCM.42.2.636-638.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perandin F, Manca N, Calderaro A, Piccolo G, Galati L, Ricci L, Medici MC, Arcangeletti MC, Snounou G, Dettori G, Chezzi C. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J Clin Microbiol. 2004;42:1214–1219. doi: 10.1128/JCM.42.3.1214-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurukumar KR, Priyadarshini D, Patil JA, Bhagat A, Singh A, Shah PS, Cecilia D. Development of real-time PCR for detection and quantitation of dengue viruses. Virol J. 2009;6:10. doi: 10.1186/1743-422X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cropley IM, Lockwood DN, Mack D, Pasvol G, Davidson RN. Rapid diagnosis of falciparum malaria by using the ParaSight F test in travellers returning to the United Kingdom: prospective study. Br Med J. 2000;321:484–485. doi: 10.1136/bmj.321.7259.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills CD, Burgess DC, Taylor HJ, Kain KC. Evaluation of a rapid and inexpensive dipstick assay for the diagnosis of Plasmodium falciparum malaria. Bull World Health Organ. 1999;77:553–559. [PMC free article] [PubMed] [Google Scholar]
- 20.Alker AP, Mwapasa V, Meshnick SR. Rapid real-time PCR genotyping of mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2004;48:2924–2929. doi: 10.1128/AAC.48.8.2924-2929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee MA, Tan CH, Aw LT, Tang CS, Singh M, Lee SH, Chia HP, Yap EP. Real time fluorescence-based PCR for detection of malaria parasites. J Clin Microbiol. 2002;40:4343–4345. doi: 10.1128/JCM.40.11.4343-4345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, Thomson RB, Jr, Peterson LR, Kaul KL. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol. 2005;43:2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keen J, Farcas GA, Zhong K, Yohanna S, Dunne MW, Kain KC. Real-time PCR assay for rapid detection and analysis of PfCRT haplotypes of chloroquine-resistant Plasmodium falciparum isolates from India. J Clin Microbiol. 2007;45:2889–2893. doi: 10.1128/JCM.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]