Abstract
Mitochondrial diseases are clinically and genetically heterogeneous disorders, which make the exact diagnosis and classification difficult. The purpose of this study was to identify pathogenic mtDNA mutations in 61 Korean unrelated families (or isolated patients) with MELAS or MERRF. In particular, the mtDNA sequences were completely determined for 49 patients. From the mutational analysis of mtDNA obtained from blood, 5 confirmed pathogenic mutations were identified in 17 families, and 4 unreported pathogenically suspected mutations were identified in 4 families. The m.3243A>G in the tRNALeu(UUR) was predominantly observed in 10 MELAS families, and followed by m.8344A>G in the tRNALys of 4 MERRF families. Most pathogenic mutations showed heteroplasmy, and the rates were considerably different within the familial members. Patients with a higher rate of mutations showed a tendency of having more severe clinical phenotypes, but not in all cases. This study will be helpful for the molecular diagnosis of mitochondrial diseases, as well as establishment of mtDNA database in Koreans.
Keywords: DNA, mitochondrial; MELAS syndrome; MERRF syndrome; point mutation
Introduction
Mitochondrial diseases are clinically and genetically a very heterogeneous disorder group. Some mitochondrial disorders only affect a single organ, such as the eye in Leber hereditary optic neuropathy (LHON), but most diseases involve multiple organ systems and often typically manifest in tissues with high-energy demand, e.g., nerve and muscle. The multi-organ involvement and overlapping of symptoms among mitochondrial disorders make the exact diagnosis and classification difficult. Identifying mitochondrial DNA (mtDNA) mutations is now an important method in the diagnosis of patients with mitochondrial disorders. The mtDNA mutations are largely divided into two groups. The first group is comprised of point mutations in tRNA, rRNA, or protein coding genes, which are commonly maternal inheritance. The second group is composed of the rearrangements of mtDNA, such as duplication or large deletion, which are usually either maternally inherited or sporadic (Schmiedel et al., 2003). The frequent mitochondrial disorders caused by the point mutations of mtDNA are mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS, MIM# 540000), and myclonus epilepsy with ragged-red fibers (MERRF, MIM# 545000).
To date, several hundreds of different mtDNA mutations have been reported to be associated with various mitochondrial diseases in a Human Mitochondrial Genome Database (MITOMAP: http://www.mitomap.org) (Ruiz-Pesini et al., 2007). Of them, the m.3243A>G, m.3271T>C, m.3291T>C and m.10191T>C in MELAS, and m.8344A>G, m.8356T>C and m.8363G>A in MERRF are well-confirmed common point mutations (Goto et al., 1990, 1991, 1994; Shoffner et al., 1990; Silvestri et al., 1992; Santorelli et al., 1996; Taylor et al., 2001). Many MELAS and MERRF patients have shown one of these mtDNA mutations, however, no causative mutation has still been identified in a large part of patients.
The mtDNA mutational studies have not been actively performed in Korean patients (Kwon et al., 2004; Choi et al., 2008). Moreover, complete mtDNA sequencing data for Koreans are very limited. In the present study, the mtDNA sequence was analyzed to find the pathogenic mutations in 61 families that have MELAS or MERRF in Korea. In particular, the mtDNA sequences were completely determined for 49 patients. In our study, 9 mutations (5 confirmed and 4 suspected) were identified in 21 patients, and the correlation between the heteroplasmic rates and the severity of clinical phenotypes were conducted.
Results
Identification of causative mutations
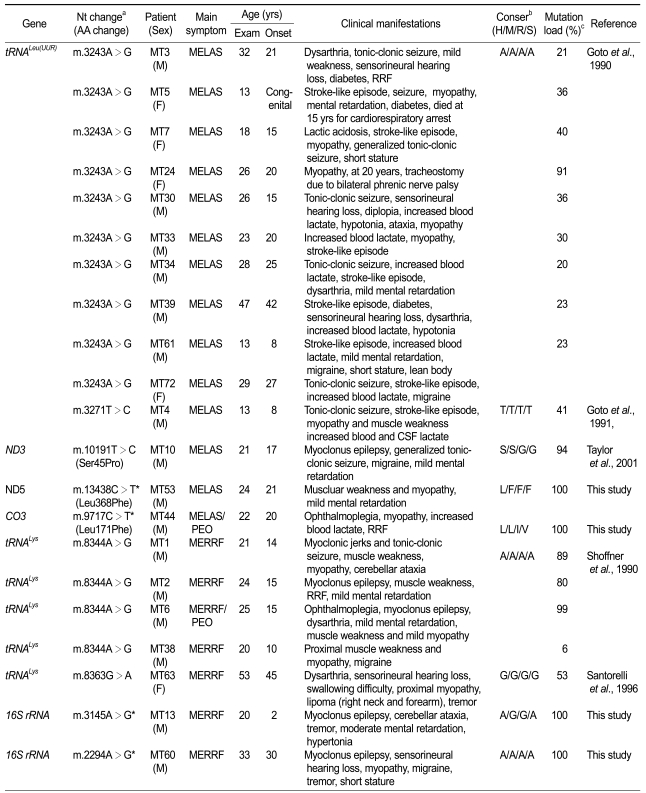
Five reported pathogenic point mutations were identified in 17 families from the screening of 61 families or isolated patients (27.4%). The m.3243A>G in the tRNALeu(UUR) was observed in 10 MELAS patients, followed by m.8344A>G in the tRNALys of 4 MERRF patients. Others were m.3271T>C in the tRNALeu(UUR), and m.10191T>C (Ser45Pro) in the ND3 of each MELAS patient, and m.8363G>A in the tRNALys of a MERRF patient. Details of the mutations and clinical phenotypes of the patients are listed in Table 1.
Table 1.
Phenotypic characteristics of patients with causative or unreported mtDNA mutations.
aAsterisks (*) have not been reported in the MITOMAP database (http://www.mitomap.org), bConservation: H, human; M, mouse; R, rabbit; S, sheep; cProband's heteroplasmic rates.
The m.3243A>G in the tRNALeu(UUR) (21.3%) and the m.8344A>G in the tRNALys (33.3%) were the most frequently identified in Korean MELAS and MERRF patients, respectively. However, the m.3243A>G has been identified in up to 80% of MELAS patients, since Goto et al. (1990) first reported it. Shoffner et al. (1995) also reported 90% of MERRF patients have the m.8344A>G mutation. The low detection frequency of causative mutations might be due that the mutational screen was done by using blood DNA instead of affected muscle DNA.
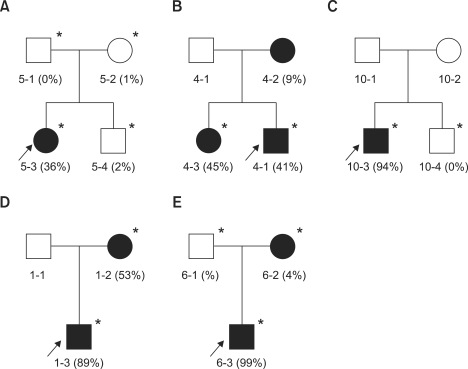
The m.3243A>G mutation found in 10 MELAS patients showed heteroplasmy in all the patients with a range of 20-91%. In the MT5 family, the proband revealed heteroplasmy of 36%, but her mother and younger brother showed barely 1-2% (Figure 1A). Her father showed no mutation. The proband showed typical MELAS phenotype, including stroke-like episodes, seizures, myopathy, mental retardation, and diabetes. When she was 15 years old, she died due to the cardiorespiratory arrest. However, her other family members have displayed no sign of MELAS symptom. The MT24 patient revealed the highest rate of heteroplasmy (91%) among the patients that had m.3243A>G in this study. When she was 20 years old, tracheostomy was done due to bilateral phrenic nerve palsy. She showed very severe myopathy, compared to other patients with m.3243A>G mutations. The m.3271T>C mutation was found in a MELAS family (MT4). The heteroplasmic rates were about 41%, 9%, and 45% for the proband, his mother, and elder sister, respectively (Figure 1B). The proband showed generalized tonic-clonic seizures, stroke-like episodes, moderate muscle weakness, and increased level of blood and CSF lactate. However, his elder sister and mother showed only mild muscle weakness. The m.10191T>C in the ND3 was found in a MELAS patient (MT10) with a 94% heteroplasmic rate. However, his brother showed neither mutation (0%) nor MELAS symptom (Figure 1C). This mutation has been previously reported in the epilepsy, stroke, optic atrophy, and cognitive decline (ESOC) and Leigh-like patients (Taylor et al., 2001; McFarland et al., 2004). However, the patient showed neither optic atrophy nor cognitive decline.
Figure 1.
Pedigree analysis and variable heteroplasmic rates among family members. Each family member showed very different heteroplasmic rates in most pedigrees. The available DNAs are indicated by asterisks (*). The filled (■, ●) and open symbols (□, ○) represent affected and unaffected members, respectively. The arrows indicate probands. (A) MT5 MELAS family with m.3243A>G, (B) MT4 MELAS family with m.3271T>C, (C) MT10 MELAS family with m.10191T>C, (D) MT1 MERRF family with m.8344A>G, and (E) MT6 MERRF/PEO overlapping family with m.8344A>G.
The m.8344A>G in the tRNALys, which was found in 4 MERRF patients, showed 76% or higher heteroplasmic rate. In the MT1 MERRF family, the proband and his mother showed rates of about 89% and 53%, respectively (Figure 1D). The proband revealed the MERRF phenotypes with myoclonic jerks, generalized tonic-clonic seizures, and myopathy. His mother also revealed MERRF clinical symptoms, except for myopathy. In the MT6 MERRF/PEO overlapping family, the proband revealed about 99% of heteroplasmy. However, his mother showed only about 4%, and his father showed no mutation (Figure 1E). He showed ophthamoplegia, myopathy, epilepsy, and mental retardation. His mother and father were normal in appearance. The m.8363G>A in the tRNALys was found in a MERRF patient (MT63) who had symptoms of dysarthria, sensorineural hearing loss, swallowing difficulties, and proximal weakness. In addition, she also had lipomas on the right neck and forearm. The patient showed about 53% of heteroplasmy.
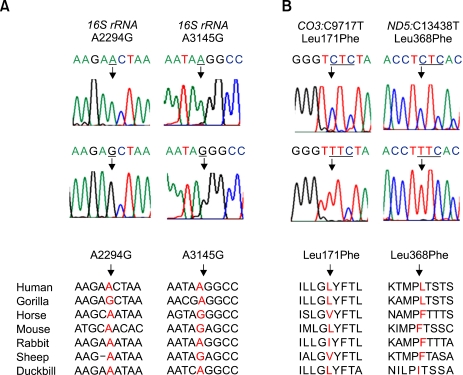
Four unreported mutations were also identified in each different patient: m.2294A>G and m.3145A>G in the 16S rRNA, m.9717C>T (Leu171Phe) in the CO3, and m.13438C>T (Leu368Phe) in the ND5 (Figure 2). These 4 mutations were not found in 200 controls. Therefore, they might be associated with the corresponding disease (Table 1). The homoplasmic m.9717C>T mutation was identified in a MELAS/PEO overlapping family (MT44). The family showed progressive ophthamoplegia, and also had typical MELAS features, including ragged-red fibers (RRF). The m.13438C>T was identified in a MELAS patient who had proximal myopathy and mental retardation (MT53). The m.3145A>G was identified in a MERRF patient with cerebellar ataxia, tremors, moderate mental retardation, and hypertonia. The m.2294A>G was identified in a MERRF patient (MT60) who also had an additional mutation of m.7119G>A (Asp406Asn) in the CO1 (Table 2). He showed myoclonus epilepsy, sensorineural hearing loss, and tremors.
Figure 2.
Sequencing chromatograms and their conservation for the unreported mutations. (A) Mutations in the rRNA gene. (B) Mutations in the coding genes.
Table 2.
Mutations reported as both pathogenic and polymorphic.
aH, human; M, mouse; R, rabbit; S, sheep; bPol, polymorphism; DEAF, maternally inherited DEAFness; NIDDM, non-insulin dependent diabetes mellitus; AD, Alzheimer's disease; PD, Parkinson's disease; DM, diabetes mellitus; MS, multiple sclerosis.
Mutations reported as both associative and polymorphic
Eleven mutations that have been previously reported to be both pathogenic (or as risk factor) and polymorphic were identified (Table 2). The m.827A>G, m.961T>C, and m.2835C>T were located on the non-coding genes, and m.3316G>A, m.3394T>C, m.3497C>T, m.5460G>A, m.7119G>A, m.9438G>A, m.12026A>G and m.13708G>A were located on the coding genes. All of these mutations showed homoplasmy. Of them, 9 mutations were also found in the controls with even lower frequencies, but m.7119G>A in the CO1, and m.9438G>A in the CO3 were not found in the controls.
The m.961T>C, m.3497C>T, m.5460G>A, and m.9438G>A were found from the patients who also had well confirmed pathogenic mutations. The m.961T>C in the 12S rRNA was identified from the MT34 MELAS family that also showed the m.3243A>G. The m.961T>C has been reported to be related to both maternally inherited DEAFness (Li et al., 2005). The m.3497C>T (Ala64Val) in the ND1 was identified in the MT6 MERRF/PEO family that had the m.8344A>G mutation. This mutation has been reported to be related to LHON (Matsumoto et al., 1999). The m.5460G>A (Ala331Thr) in the ND2 was identified in the MT7 MELAS patient who also had the m.3243A>G mutation. The m.5460G>A has been reported to be associated with Alzheimer's or Parkinson's patients (Lin et al., 1992; Schnopp et al., 1996). The m.9438G>A (Gly78Ser) in the CO3 was identified in the MT10 MELAS patient who also had the m.10191T>C mutation. This mutation has been reported to be associated with LHON (Oostra et al., 1995). The m.7119G>A (Asp406Asn) in the CO1 was found in a MERRF patient (MT60) who also had a suspected mutation m.2294A>G in the 16S rRNA. The m.7119G>A mutation was recently reported in multiple sclerosis patients associated with the haplogroup U (Ban et al., 2008).
Polymorphisms identified by complete mtDNA analysis
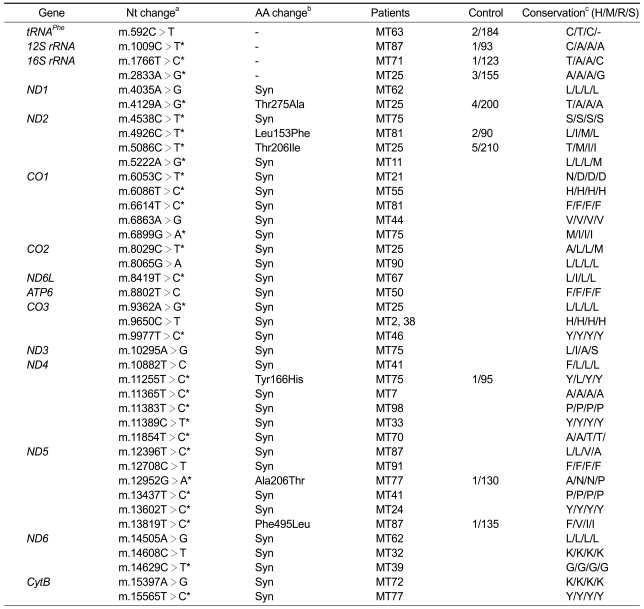
Whole mtDNA sequencing analysis (49 patients) identified 40 unreported variations in the MITMAP website (Ruiz-Pesini et al., 2007). The complete sequencing data and corresponding haplobroup of each sample were provided as Supplemental data Table S1. As shown in Table 3, 13 variations were even not reported in the mtDB (http://www.genpat.uu.se/mtDB) (Ingman and Gyllensten, 2006). Most variations in the coding genes were synonymous, but six were missense mutations (m.4129A>G (Thr275Ala) in the ND1, m.4926C>T (Leu153Phe) and m.5086C>T (Thr206Ile) in the ND2, m.11255T>C (Tyr166His) in the ND4, m.12952G>A (Ala206Thr) and m.13819T>C (Phe495Leu) in the ND5). All the 6 missense mutations and 4 mutations in the non-coding genes exhibited homoplasmy and were also found in controls. Therefore, they were regarded as not the genetic causes of the mitochondrial diseases, but rare polymorphisms.
Table 3.
Unreported mtDNA polymorphisms found in MELAS or MERRF patients.
aAsterisks (*) have not been reported in the MITOMAP (http://www.mitomap.org), but referred in the Human Mitochondrial Genome Database (http://www.genpat.uu.se/mtDB), bSyn, synonymous mutation; cH, human; M, mouse; R, rabbit; S, sheep. MtDNA mutations in MELAS and MERRF patients.
Pathological findings
Muscle biopsies were performed in 23 patients (MELAS 15, MERRF 6, MELAS/PEO 1, MERRF/PEO 1) of 61 studied patients from the biceps brachii (n = 21) or the quadriceps (n = 2). Among them, ragged-red fibers were found in 21 patients (91%). However, electron microscopic examination showed mitochondrial proliferation or enlarged abnormal mitochondria in all patients. In addition, some patients (52%) displayed mitochondria with over abundant or branched cristae.
Discussion
Mitochondrial DNA mutations in 61 patients clinically diagnosed as MELAS or MERRF were examined. The 12 well-confirmed common pathogenic mutations were first screened for all the samples, and then the whole mtDNA sequence was determined in 49 patients. From the analysis, 5 confirmed pathogenic mutations were identified in 17 patients (27.9%), and 4 pathogenically suspected unreported mutations were identified in 4 patients (Table 1). The detection rate of two major pathogenic mutations (A3243G and A8344G) was 22.9%, which is well consistent with the previous Korean study (23.1%) by Kwon et al. (2004). Ten mutations that have been reported as both pathogenic and polymorphic were found in 18 patients (Table 2). Thirteen novel polymorphisms which have not been reported in either of the MITOMAP (http://www.mitomap.org) or the mtDB (http:www.genpat.uu.se/mtDB) were also identified (Ingman and Gyllensten, 2006; Ruiz-Pesini et al., 2007).
Most pathogenic mutations showed heteroplasmy, and the rates were considerably different within the family members (Figure 1). For instance, proband and his sibling of the MELAS family (MT10) showed 94% and 0%, respectively, and proband and his mother of the MERRF/PEO overlapping family (MT6) showed 99% and 4%, respectively. Patients with a higher rate of mutations exhibited a tendency of more severe clinical phenotypes, but not in all cases. The m.3243A>G mutation showed a 20-40% ratio of mutants in most patients, but the MT24 patients showed a very high heteroplasmic rate (91%). This patient showed MELAS symptoms with an additional phenotype of severe respiratory difficulty. Individuals with 1-2% heteroplasmy showed no sign of MELAS symptoms in the MT5 family. The m.8344A>G mutation showed 50-90% ratios in the MERRF patients, but MT6-1 revealed almost homoplasmic mutation (99%). The MT6-1 patient showed the MERRF with an additional phenotype of ophthamoplegia, which is a frequent symptom of PEO. In the MT1 MERRF family, the proband (89%) and his mother (53%) revealed similar MERRF phenotypes. However, the proband showed epilepsy, but his mother had no symptoms of epilepsy. However, the MT4 MELAS family with m.3271T>C mutation showed a slightly different aspect. The proband (40%) and his mother (9%) showed stroke-like episodes and generalized tonic clonic seizure, but his elder sister (45%) revealed a less severe phenotype with only lactic acidosis. Several studies have suggested the heteroplasmic rates are associated with the severity of clinical phenotypes (Koga et al., 2000; Salpietro et al., 2003), but others have suggested no or weak relation (Harrison et al., 1997; Dobrowolski et al., 2009). It is known that the heteroplasmic rates of pathogenic mutations are depending on the tissue kinds, even if they originated from an individual.
The 4 unreported mutations suspected for genetic defects of mitochondrial diseases were identified as homoplasmy. The m.2294A>G in the 16S rRNA was identified in a MERRF patient (MT60) who also had the m.7119G>A (Asp406Asn) in the CO1, which has been recently suggested as an association with multiple sclerosis as a rare polymorphism (Ban et al., 2008). Both mutations were well conserved in different species and were not found in the 200 Korean controls (Tables 1 and 2), although they have been reported as rare polymorphism (http:www.genpat.uu.se/mtDB). Thus, it is likely that the two combined mutations may function as a genetic risk factor for MERRF. Although the m.3145A>G and m.13438C>T (Leu368Phe) in the MELAS patients (MT53), and m.9717C>T (Leu171Phe) in the MELAS/PEO overlapping patient were not found in 200 controls, the mutation sites were less conserved in different species. Thus, it appears that they might be associated with the disease as indirect risk factors rather than as primary genetic defects. Several secondary mutations (m.4435A>G, m.12192G>A, and m.15951A>G) have been reported to elevate clinical symptoms in the LHON patients (Mimaki et al., 2003; Qu et al., 2006).
This study identified several mutations that have been reported in both pathogenic and polymorphism (Table 3). Since most of them were also found in controls, they might be not pathogenic, but polymorphic in the Korean population. However, the m.9438G>A, which was not found in controls, might function as genetic risk factors or secondary mutations for the mitochondrial diseases. Carelli et al. (2003) suggested a two-locus genetic model that involves a primary mitochondrial mutation and a nuclear modifier. Several decades of polymorphic variants that have been not reported in the MITMAP (Table 3) were also identified. Of them, 13 mutations were not reported, even in the haplogroup studies.
Since reports that LHON patients are preferentially associated with the haplogroup J (Hofmann et al., 1997; Torroni et al., 1997), association studies have become an important approach to uncover the function of mtDNA variations. Although the association study was not performed due to the small size of patient groups, the whole mtDNA sequencing data would be used for further haplogroup studies of Korean mitochondrial patients. Because this study was performed using blood DNA, the suggested rates of causative mutation identification and heteroplasmic loads would be different from the affected muscles. If further mutational study is performed using DNAs from affected tissues, the mutational distribution will be characterized more exactly in Korean MELAS and MERRF patients. This study would be helpful for the molecular diagnosis of mitochondrial diseases, as well as for the basic establishment of the mtDNA database in Korea.
Methods
Patients
This study included 61 independent mitochondrial patients of Korean origin where 43 patients had MELAS, and 18 patients had MERRF (47 isolated cases and 14 families). Of them, 2 patients showed MELAS/PEO (progressive external ophthalmoplegia) overlapping syndrome, and 1 patient demonstrated MERRF/PEO overlapping phenotype. The study also included about 200 healthy controls who had no clinical features and family history of mitochondrial disorders. All participants included in this study provided written informed consent according to the protocol approved by the Ethics committee of Ewha Woman's University, School of Medicine.
DNA isolation
Total DNA was extracted from the whole peripheral blood samples by using the QIAamp Blood DNA mini kit (Qiagen, Hilden, Germany). The paternity and maternity were confirmed in familial samples by the genotyping of 15 microsatellite markers that were provided in the PowerPlex 16 kit (Promega, Madison, WI).
PCR amplification and sequence analysis
Six primer pairs were designed to detect 12 common mutations (m.3243A>G, m.3291T>C, m.3460G>A, m.3271T>C, m.8344A>G, m.8356T>C, m.8363G>A, m.8993T>G/C, m.10158T>C, m.10191T>C, m.11778G>A and m.14484T>C). The primer sequences and PCR conditions used are available on request to corresponding author. The entire mitochondrial genome was sequenced by the PCR by using 46 primer sets of the MitoSEQr resequencing system (Applied Biosystems, Foster city, CA). The PCR amplification condition consisted of initial denaturation at 96℃ for 5 min, followed by 35 cycles at 94℃ for 30 s, 60℃ for 45 s, 72℃ for 45 s, and a final extension at 72℃ for 10 min. PCR products were purified by the treatment of exonuclease I-shrimp alkaline phosphatase (Fermentas, Canada) and sequenced by an automatic genetic analyzer ABI3100 using the big dye terminator cycle sequencing ready reaction kit (Applied Biosystems). Sequence variations were confirmed by analyzing both strands of DNA. The sequences were compared with the revised Cambridge reference sequence (NC_012920) by using the SeqScape software (Ver. 2.1, Applied Biosystems) (Andrews et al., 1999).
Determination of heteroplasmy
The heteroplasmic rates of the mtDNA mutations were determined by screening bacterial clones that had the corresponding mtDNA fragment (Uusimaa et al., 2004). The DNA fragments, obtained by PCR amplification, were subcloned into the pGEM-T Easy vector (Promega), which was then used to transform E. coli (DH5α). Plasmid DNA was isolated from 100 colonies/mutation by using a plasmid DNA isolation kit (Intron Biotechnol., Korea), and the mutation was determined by the sequencing of the insert DNA. The rate of heteroplasmy was measured by the counting of clones that had a mutant sequence.
Clinical and pathological assessments
Careful clinical information and neurological examinations were obtained, and the MELAS and MERRF were diagnosed to meet the clinical criteria (Bernier et al., 2002). All patients underwent brain MRIs and electrophysiological studies. Whole brains were scanned by using a slice thickness of 7 mm and a 2-mm interslice gap to produce 16 axial images (Siemens Vision: Siemens, Germany). Muscle biopsies were done under local anesthesia, and cross-sections of the biopsy tissue were stained with hematoxylin-eosin, modified Gomori-trichrome, cytochrome c oxidase, and succinate dehydrogenase. For the electron microscopic observation, the specimen was fixed in 2% glutaraldehyde in 25 mM cacodylate buffer at pH 7.4, and processed for semithin and ultrathin studies.
Supplemental data
Supplemental Data include a table and can be found with this article online at http://e-emm.or.kr/article/article_files/SP-42-6-05.pdf.
Acknowledgments
This study was supported by the Mid-career Researcher Program through NRF funded by the MEST (R01-2008-000-20604-0), and by Korea Healthcare Technology R&D Project through KHIDI funded by the Ministry of health, welfare and family Affairs (A090500).
Abbreviations
- LHON
Leber hereditary optic neuropathy
- MELAS
mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes
- MERRF
myoclonus epilepsy with ragged-red fibers
- mtDNA
mitochondrial DNA
- rCRS
revised Cambridge reference sequence
- PEO
progressive external ophthalmoplegia
- RRF
ragged-red fibers
Supplementary Material
Supplemental Data
References
- 1.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 2.Ban M, Elson J, Walton A, Turnbull D, Compston A, Chinnery P, Sawcer S. Investigation of the role of mitochondrial DNA in multiple sclerosis susceptibility. PLoS ONE. 2008;3:e2891. doi: 10.1371/journal.pone.0002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology. 2002;59:1406–1411. doi: 10.1212/01.wnl.0000033795.17156.00. [DOI] [PubMed] [Google Scholar]
- 4.Cardaioli E, Dotti MT, Hayek G, Zappella M, Federico A. Studies on mitochondrial pathogenesis of Rett syndrome: ultrastructural data from skin and muscle biopsies and mutational analysis at mtDNA nucleotides 10463 and 2835. J Submicrosc Cytol Pathol. 1999;31:301–304. [PubMed] [Google Scholar]
- 5.Carelli V, Giordano C, d'Amati G. Pathogenic expression of homoplasmic mtDNA mutations needs a complex nuclear-mitochondrial interaction. Trends Genet. 2003;19:257–262. doi: 10.1016/S0168-9525(03)00072-6. [DOI] [PubMed] [Google Scholar]
- 6.Choi BO, Hwang JH, Kim J, Cho EM, Cho SY, Hwang SJ, Lee HW, Kim SJ, Chung KW. A MELAS syndrome family harboring two mutations in mitochondrial genome. Exp Mol Med. 2008;40:354–360. doi: 10.3858/emm.2008.40.3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobrowolski SF, Gray J, Miller T, Sears M. Identifying sequence variants in the human mitochondrial genome using high-resolution melt (HRM) profiling. Hum Mutat. 2009;30:891–898. doi: 10.1002/humu.21003. [DOI] [PubMed] [Google Scholar]
- 8.Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 9.Goto Y, Nonaka I, Horai S. A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) Biochim Biophys Acta. 1991;1097:238–240. doi: 10.1016/0925-4439(91)90042-8. [DOI] [PubMed] [Google Scholar]
- 10.Goto Y, Tsugane K, Tanabe Y, Nonaka I, Horai S. A new point mutation at nucelotide pair 3291 of the tRNALeu(UUR) gene in a patient with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) Biochem Biophys Res Commun. 1994;202:1624–1630. doi: 10.1006/bbrc.1994.2119. [DOI] [PubMed] [Google Scholar]
- 11.Harrison TJ, Boles RG, Johnson DR, LeBlond C, Wong LJ. Macular pattern retinal dystrophy, adult-onset diabetes, and deafness: a family study of A3243G mitochondrial heteroplasmy. Am J Ophthalmol. 1997;124:217–221. doi: 10.1016/s0002-9394(14)70787-1. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann S, Jaksch M, Bezold R, Mertens S, Aholt S, Paprotta A, Gerbitz KD. Population genetics and disease susceptibility: characterization of central European haplogroups by mtDNA gene mutations, correlation with D loop variants and association with disease. Hum Mol Genet. 1997;6:1835–1846. doi: 10.1093/hmg/6.11.1835. [DOI] [PubMed] [Google Scholar]
- 13.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–D751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koga Y, Akita Y, Takane N, Sato Y, Kato H. Heterogeneous presentation in A3243G mutation in the mitochondrial tRNA(Leu(UUR)) gene. Arch Dis Child. 2000;82:407–411. doi: 10.1136/adc.82.5.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon SJ, Park SS, Kim JM, Ahn TB, Kim SH, Kim J, Lee SH, Ha CK, Ahn MY, Jeon BS. Investigation of common mitochondrial point mutations in Korea. Ann N Y Acad Sci. 2004;1011:339–344. doi: 10.1007/978-3-662-41088-2_34. [DOI] [PubMed] [Google Scholar]
- 16.Lam CW, Yang T, Tsang MW, Pang CP. Homoplasmic 3316G-->A in the ND1 gene of the mitochondrial genome: a pathogenic mutation or a neutral polymorphism? J Med Genet. 2001;38:E10. doi: 10.1136/jmg.38.3.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Li R, Chen J, Liao Z, Zhu Y, Qian Y, Xiong S, Heman-Ackah S, Wu J, Choo DI, Guan MX. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Hum Genet. 2005;117:9–15. doi: 10.1007/s00439-005-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin FH, Lin R, Wisniewski HM, Hwang YW, Grundke-Iqbal I, Healy-Louie G, Iqbal K. Detection of point mutations in codon 331 of mitochondrial NADH dehydrogenase subunit 2 in Alzheimer's brains. Biochem Biophys Res Commun. 1992;182:238–246. doi: 10.1016/s0006-291x(05)80136-6. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto M, Hayasaka S, Kadoi C, Hotta Y, Fujiki K, Fujimaki T, Takeda M, Ishida N, Endo S, Kanai A. Secondary mutations of mitochondrial DNA in Japanese patients with Lebers hereditary optic neuropathy. Ophthalmic Genet. 1999;20:153–160. doi: 10.1076/opge.20.3.153.2281. [DOI] [PubMed] [Google Scholar]
- 20.McFarland R, Kirby DM, Fowler KJ, Ohtake A, Ryan MT, Amor DJ, Fletcher JM, Dixon JW, Collins FA, Turnbull DM, Taylor RW, Thorburn DR. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann Neurol. 2004;55:58–64. doi: 10.1002/ana.10787. [DOI] [PubMed] [Google Scholar]
- 21.Mimaki M, Ikota A, Sato A, Komaki H, Akanuma J, Nonaka I, Goto Y. A double mutation (G11778A and G12192A) in mitochondrial DNA associated with Leber's hereditary optic neuropathy and cardiomyopathy. J Hum Genet. 2003;48:47–50. doi: 10.1007/s100380300005. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell AL, Elson JL, Howell N, Taylor RW, Turnbull DM. Sequence variation in mitochondrial complex I genes: mutation or polymorphism? J Med Genet. 2006;43:175–179. doi: 10.1136/jmg.2005.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oostra RJ, Van den Bogert C, Nijtmans LG, van Galen MJ, Zwart R, Bolhuis PA, Bleeker-Wagemakers EM. Simultaneous occurrence of the 11778 (ND4) and the 9438 (COX III) mtDNA mutations in Leber hereditary optic neuropathy: molecular, biochemical, and clinical findings. Am J Hum Genet. 1995;57:954–957. [PMC free article] [PubMed] [Google Scholar]
- 24.Puomila A, Hamalainen P, Kivioja S, Savontaus ML, Koivumaki S, Huoponen K, Nikoskelainen E. Epidemiology and penetrance of Leber hereditary optic neuropathy in Finland. Eur J Hum Genet. 2007;15:1079–1089. doi: 10.1038/sj.ejhg.5201828. [DOI] [PubMed] [Google Scholar]
- 25.Qu J, Li R, Zhou X, Tong Y, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX. The novel A4435G mutation in the mitochondrial tRNA-Met may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest Ophthalmol Vis Sci. 2006;47:475–483. doi: 10.1167/iovs.05-0665. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz-Pesini E, Lott MT, Procaccio V, Poole J, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salpietro CD, Briuglia S, Merlino MV, Di Bella C, Rigoli L. A mitochondrial DNA mutation (A3243G mtDNA) in a family with cyclic vomiting. Eur J Pediatr. 2003;162:727–728. doi: 10.1007/s00431-003-1280-1. [DOI] [PubMed] [Google Scholar]
- 28.Santorelli FM, Mak SC, El-Schahawi M, Casali C, Shanske S, Baram TZ, Madrid RE, DiMauro S. Maternally inherited cardiomyopathy and hearing loss associated with a novel mutation in the mitochondrial tRNA(Lys) gene (G8363A) Am J Hum Genet. 1996;58:933–939. [PMC free article] [PubMed] [Google Scholar]
- 29.Schmiedel J, Jackson S, Schäfer J, Reichmann H. Mitochondrial cytopathies. J Neurol. 2003;250:267–277. doi: 10.1007/s00415-003-0978-3. [DOI] [PubMed] [Google Scholar]
- 30.Schnopp NM, Käsel S, Egensperger R, Graeber MB. Regional heterogeneity of mtDNA heteroplasmy in parkinsonian brain. Clin Neuropathol. 1996;15:348–352. [PubMed] [Google Scholar]
- 31.Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 32.Shoffner JM, Bialer MG, Pavlakis SG, Lott M, Kaufman A, Dixon J, Teichberg S, Wallace DC. Mitochondrial encephalomyopathy associated with a single nucleotide pair deletion in the mitochondrial tRNALeu(UUR) gene. Neurology. 1995;45:286–292. doi: 10.1212/wnl.45.2.286. [DOI] [PubMed] [Google Scholar]
- 33.Silvestri G, Moraes CT, Shanske S, Oh SJ, DiMauro S. A new mtDNA mutation in the tRNA(Lys) gene associated with myoclonic epilepsy and ragged-red fibers (MERRF) Am J Hum Genet. 1992;51:1213–1217. [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor RW, Singh-Kler R, Hayes CM, Smith PE, Turnbull DM. Progressive mitochondrial disease resulting from a novel missense mutation in the mitochondrial DNA ND3 gene. Ann Neurol. 2001;50:104–107. doi: 10.1002/ana.1084. [DOI] [PubMed] [Google Scholar]
- 35.Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 36.Uusimaa J, Finnilä S, Remes AM, Rantala H, Vainionpää L, Hassinen IE, Majamaa K. Molecular epidemiology of childhood mitochondrial encephalomyopathies in a Finnish population: sequence analysis of entire mtDNA of 17 children reveals heteroplasmic mutations in tRNAArg, tRNAGlu, and tRNALeu(UUR) genes. Pediatrics. 2004;114:443–450. doi: 10.1542/peds.114.2.443. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, Koczan D, Sulonen AM, Akkad DA, Kroner A, Comabella M, Costa G, Corongiu D, Goertsches R, Camina-Tato M, Thiesen HJ, Nyland HI, Mork SJ, Montalban X, Rieckmann P, Marrosu MG, Myhr KM, Epplen JT, Saarela J, Ibrahim SM. mtDNA nt13708A variant increases the risk of multiple sclerosis. PLoS ONE. 2008;3:e1530. doi: 10.1371/journal.pone.0001530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data