Abstract
More than 300 different types of protein post-translational modifications (PTMs) have been described, many of which are known to have pivotal roles in cellular physiology and disease. Nevertheless, only a handful of PTMs have been extensively investigated at the proteome level. Knowledge of protein substrates and their PTM sites is key to dissection of PTM-mediated cellular processes. The past several years have seen a tremendous progress in developing MS-based proteomics technologies for global PTM analysis, including numerous studies of yeast and other microbes. Modification-specific enrichment techniques combined with advanced MS/MS methods and computational data analysis have revealed a surprisingly large extent of PTMs in proteins, including multi-site, cooperative modifications in individual proteins. We review some of the current strategies employed for enrichment and detection of PTMs in modification-specific proteomics.
Keywords: Affinity chromatography, Microbiology, MS, Post-translational modification
1 Introduction
Post-translational modification (PTM) represents an important mechanism for diversifying and regulating the cellular proteome. In this review, PTM refers to a chemical event that converts a ribosomally coded amino acid residue into a non-standard amino acid residue by an enzymatic reaction. The identification of protein substrates and their PTM sites are fundamental to the biochemical dissection of PTM pathways (for example, the identification of enzymes that catalyse PTM), to studies of the role of PTM in the function of substrate proteins, to the establishment of substrate–enzyme(s) relationships, and to providing insights into the possible regulation of cellular physiology by PTM.
Protein lysine acetylation provides a good example of a critical role of substrate identification for functional characterization of a PTM-mediated pathway. Lysine acetylation was initially identified in histones in the 1960s [1]. Demonstration of its first non-histone substrate protein, p53, in 1997 [2] stimulated extensive studies of the roles of lysine acetylation in transcriptional regulation. Identification of diverse substrates in both cytosolic and mitochondrial fractions [3] opened new avenues for its functional studies in energy metabolism, signal transduction, and mitochondrial regulation.
Conventionally, PTM substrates have been identified by laborious biochemical approaches, including in vitro PTM reaction assays using radioactive isotope-labeled substrates, Western blot analysis, and more recently peptide and protein arrays [4, 5] (Table 1). While useful, these methods suffer from various shortcomings. For example, radio-isotopes of carbon and hydrogen are rather weak radio emitters (for example, 14C or 3H in the case of protein methylation and acetylation), which makes it difficult to efficiently detect their corresponding modified proteins. Antibody-based Western blot analysis has been successful for identifying candidate substrate proteins for certain types of PTM, such as tyrosine phosphorylation. However, the small size of the structural motifs of other common PTMs (for example, protein methylation and acetylation) makes it difficult to generate pan-specific antibodies, which recognize PTM peptides/proteins independent of its surrounding sequences, with good affinity for routine Western blotting.
Table 1.
Techniques for detection and identification of PTM substrates
| Method | In vitro or in vivo | Case studies | Advantages | Disadvantages |
|---|---|---|---|---|
| Radioactive isotope labeling | In vitro or in vivo | 32P (pSer, pThr, pTyr) 3H, or 14C for AcLys or MeK | Reagents accessible | Inconvenience/hazard low sensitivity |
| Western blotting | In vitro or in vivo | pTyr, AcLys or MeK | Good affinity | Moderate sensitivity |
| Peptide/protein array | In vitro | pSer/Thr/Tyr, AcLys or MeK | Rapid, global scale | Possibly non-specific, low sensitivity, requires verification |
| MS-proteomics | In vitro | pSer/Thr/Tyr, AcR or MeK | Specific, global scale | Need enrichment methods |
AcR, MeK, pSer, pThr, and pTyr, represent acetylated arginine, methyllysine, phosphorylated serine, threonine, and tyrosine residues, respectively.
Another valid approach for identifying protein substrates is based on the specificity of PTM-specific enzymes. For example, in vitro screens have been carried out using peptide or protein arrays to identify sequence motifs for a protein lysine methyltransferase [6] and for protein kinases [4]. Nevertheless, PTM substrate candidates identified by these approaches require further validation by MS analysis of the purified endogenous proteins. In summary, despite technical advances in the past few decades, more efficient and sensitive bioanalytical technologies are needed to address key bottlenecks in the identification of PTM substrate proteins, in mapping PTM sites, and to investigate in vivo PTM dynamics.
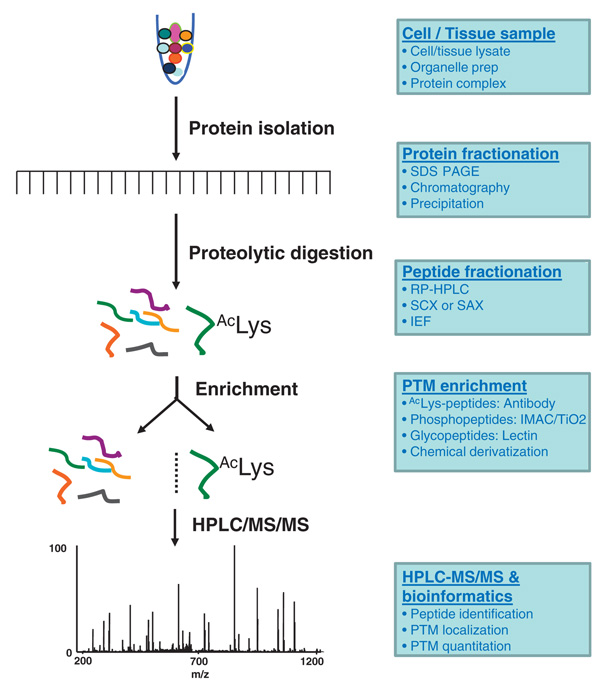
During the past decade, MS-based proteomics has been shown to be a powerful technique for proteome-wide identification of PTM substrates and mapping of PTM sites. Such studies typically involve four steps (Fig. 1). First, the protein lysate of interest is proteolytically digested, usually by a specific protease, such as trypsin. Second, the resulting proteolytic peptides are subjected to enrichment, using a suitable method, to separate the PTM peptides of interest from the rest of the proteolytic peptides. Third, The isolated PTM-peptides are then analyzed by nano-HPLC/MS/MS for peptide identification and precise localization of PTM sites. Finally, the peptide candidates are further evaluated by a manual or an automated verification method to ensure the accuracy and statistical significance of the identification [7]. In addition, a separation step can be included in the procedure to separate either proteins (before the proteolytic digestion) or peptides (after the proteolytic digestion) into multiple fractions to reduce sample complexity.
Figure 1.
Experimental procedure for PTM proteomics. Antibody-based affinity purification for lysine acetylated peptides is used as an example. The proteomics study of a PTM typically involves preparation of a protein sample, proteolytic digestion of proteins into peptides, enrichment of PTM peptides using an appropriate method, HPLC/MS/MS analysis of the enriched PTM peptides, and protein sequence database searching using the MS/MS data for identifying peptide sequences, mapping PTM sites, and quantification. When necessary, a protein/peptide fractionation step is included prior to PTM peptide enrichment in order to reduce the sample complexity and to enhance the yield of enrichment. Different methods can be selected for protein fractionation, peptide fractionation, and enrichment of PTM peptides. Abbreviation: SAX, strong anion exchange.
High sensitivity is desirable in PTM proteomics to detect substrate proteins that exist in low abundance in cells. The detection sensitivity of a PTM proteomics screening depends on four factors: (i) yield of affinity enrichment, (ii) level of contamination from irrelevant peptides, (iii) sensitivity of the HPLC/MS/MS system, and (iv) complexity of the peptide mixture. The PTM peptides are present in an ocean of non-PTM peptides and may be present in low stoichometry. Accordingly, without enrichment, mass spectrometric analysis has low efficiency to detect PTM peptides. Despite advances in the sensitivity of HPLC/MS systems and the development of more powerful algorithms for protein sequence database searching, the lack of efficient procedures for enrichment of PTM peptides has become a major bottleneck for PTM proteomic research.
Here, we review existing MS-based proteomics strategies for global PTM analysis, with a focus on enrichment methods for PTM peptides. We also discuss future challenges for comprehensive PTM analysis. Readers interested in general information about PTMs, mapping PTM sites in proteins and PTM quantification by MS are referred to several recent review articles [8–13].
2 Sample preparation
Before PTM peptides are enriched, the protein lysate of interest is typically prepared from cultured cells and/or tissues, and subsequently proteolytically digested. In some cases, cellular organelles and/or a protein complex are isolated and followed by proteolytic digestion and PTM analysis. A few key issues in this step include prevention of in vitro artificial PTM reaction, increase detection sensitivity by reducing complexity of a protein sample, and preparation of a protein sample that is biologically relevant.
2.1 Reducing sample complexity
To reduce the sample complexity, proteins can be resolved into fractions using various methods, including organelle separation, and fractionation of proteins and proteolytic peptides by electrophoretic techniques or multi-dimensional chromatography. A combination of these fractionation methods can further simplify peptide mixtures. Protein or peptide fractionation cannot only reduce the complexity, but can also enhance the yield of affinity enrichment by raising the peptide concentration in the peptide mixture and reducing competitive inhibition (for example, in the case of antibody affinity purification).
2.2 Context-dependent PTMs
A protein can be located in multiple cellular organelles and protein complexes; it is therefore highly likely that the protein will have a different spatio-temporal PTM profile and stoichiometry depending on the molecular and cellular context. A well-established example are the core histone proteins whose PTMs differ between heterochromatin and euchromatin. To gain biologically relevant PTM information, the protein of interest should be isolated from a biologically relevant cellular environment. For examples, EGF receptor can be located in either plasma membrane or nuclei [14]. The protein should therefore be independently isolated from both cellular organelles for PTM analysis.
3 PTM enrichment methods
3.1 Antibody-based affinity enrichment
Antibodies are widely used for the detection of PTMs in a protein by Western blot analysis. Immunoisolation of antigenic peptides using an antibody was combined with MS for identification of epitope peptides more than a decade ago [15]. An obvious extension is to use pan-PTM antibodies to isolate peptides bearing the PTM of interest. Isolation of PTM peptides from tryptic digests by immunoaffinity purification using a pan-PTM antibody should be simpler than isolating the corresponding proteins, because the PTM of interest is always exposed, and less non-specific binding is likely to occur for peptides than for proteins. This approach has been successfully used for global analysis of protein lysine acetylation [3, 16], arginine methylation [17], tyrosine nitration [18], and tyrosine phosphorylation [19, 20]. In addition to pan-specific antibodies, antibodies specific to PTM motifs in peptides have been used to identify kinase substrates and quantify their PTM changes [21]. The same approach should be applicable for identifying novel substrates where a consensus PTM motif exists for a PTM regulatory enzyme of interest.
However, high-quality antibodies are not always available for PTMs of interest. For example, due to their small sizes, it is not easy to generate high affinity antibodies against acetyllysine (AcLys) or methyllysine, and, not surprisingly, the available antibodies have low binding affinity. Nevertheless, the success of proteomic analysis of lysine acetylation [3] and arginine methylation [17] suggests that this approach is likely to be applicable to most, if not all, PTMs, provided that the PTM moiety of interest is antigenic and has reasonable size that is larger than a methyl or acetyl group.
A key issue for antibody-based affinity enrichment is quality control. In Western blot analysis, the signals detected by the antibody should be competitively abolished using a peptide library bearing a fixed PTM residue, or by a protein, such as BSA containing the PTM residues (generated through an in vitro chemical reaction). Those antibodies that can detect PTM-specific signals in a Western blot are likely to be suitable for affinity enrichment for proteomics screening.
It is not usually difficult to develop an antibody or to use a chemical/metabolic labeling method to isolate peptides bearing a PTM that has a large chemical moiety (see below). However, it can be a challenge to isolate PTM peptides in which the PTM induces a small change (for example, dehydration or monomethylation). Additionally, it can be difficult to generate antibodies against certain poorly antigenic PTMs. Nevertheless, new technologies are emerging for the generation of non-antibody affinity reagents that can be selected from randomized oligomer libraries, in which combinatorial binding sites are associated with a non-antibody scaffold (for example, methyllysine-binding domain) [22, 23]. Success of such technologies is likely to have a significant impact on PTM detection and proteomics studies.
3.2 Tagging PTMs by chemical derivatization
A range of chemical methods has been developed to tag PTMs, including in vitro chemical reactions and in vivo metabolic labeling. Azide, due to its small size and bioorthogonal nature, has been used for metabolic labeling of PTMs. The resulting chemically labeled PTM proteins can be subsequently conjugated to an affinity linker, such as biotin [24, 25]. This method was used successfully for the identification of protein farnesylation [26], O-GlcNAc modifications [24, 27], palmitoylation [28] and myristoylation.[29].
A PTM can also be converted, in vitro, into a tractable site for affinity labeling for the purpose of affinity enrichment. For example, β-elimination of O-phosphorylated residues (pSer and pThr) [30] or O-GlcNAcylated Ser/Thr residues [31] permits the introduction of a chemical with an affinity tag (e.g. biotin and fluorous affinity tag [32]). The introduced affinity tag enables the enrichment of PTM peptides of interest. Likewise, nitrotyrosine and S-nitrosylation modified residues have been derivatized with a chemical tag containing biotin for detection and affinity isolation [33, 34]. Similarly, chemical approaches to convert the S-palmitoyl group into a tractable tag for affinity-enrichment and subsequent HPLC/MS/MS analysis were demonstrated, and revealed numerous palmitoylated proteins in yeast [35] and in rat neuronal synapses [36].
Complex glycosylated proteins have been isolated by affinity enrichment methods using either lectin or chemical derivatization. Many proteomics studies of glycoproteins have been reported, which are based on lectin-based affinity enrichment [37]. In particular, combinations of different types of lectins seem promising for comprehensive analysis of the N-glycosylated proteome [38–41]. A major caveat of lectin-based enrichment is the low- and variable-binding affinity between lectins and glycoproteins. Recently, hydrophilic interaction liquid chromatography (HILIC) based methods for isolation of glycopeptides have emerged and proved their utility when combined with MS/MS [42].
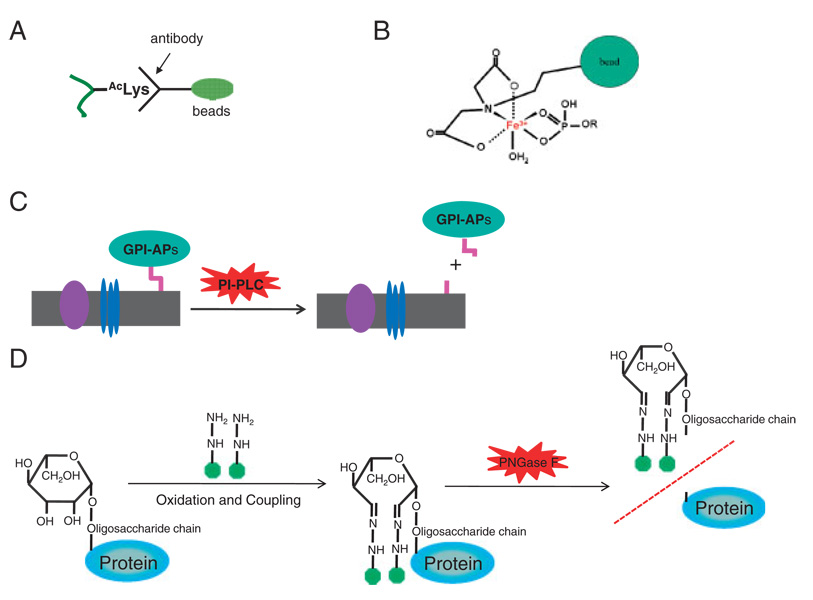
Chemical derivatization involving oxidation of the carbohydrate side chain and conjugation of glycopeptides to hydrazide resin provides an alternative approach to enrich glycosylated peptides (Fig. 2). The isolated glycopeptides can be subsequently released by treatment with the glycanspecific enzyme PNGase F and then identified by MS [43, 44]. This approach has been used to identify and quantify glycoproteins that are associated with plasma membrane, tissues and bodily fluids [44, 45].
Figure 2.
Example strategies for enriching PTM peptides. (A) Antibody-based affinity purification for isolation of lysine acetylated peptides. An antibody immobilized to a solid support can bind AcLys peptide for enrichment. (B) IMAC for enriching phosphopeptides. Ferric ion (Fe3+) immobilized in solid beads (indicated by a solid green circle) bind specifically phosphate group in a phosphopeptide. (C) Isolation of GPI-APs by enzyme-catalyzed specific release. The plasma membrane fraction is isolated. The GPI-APs are specifically released from the membrane when phosphatidylinositol phospholipase hydrolyzes the phosphatidylinositol. (D) Chemical derivatization and subsequent enzymatic cleavage for isolation of glycosylated proteins. To isolate glycoproteins, glycoproteins are first oxidized and conjugated to hydrazide beads, while non-glycoproteins can be removed. The glycoproteins are then released by PNGase F. Abbreviations: PI-PLC, phosphatidylinositol phospholipase C; PNGase F, peptide-N-glycosidase F.
While useful, chemical methods for introducing an affinity tag should be used with care (Table 2). Sample loss can be a problem as chemical derivatization reactions can be inefficient, and they often produce unwanted side products. Accordingly, a procedure involving multiple reactions is not desirable, unless all the chemical reaction steps are highly controlled and sample losses are minimized.
Table 2.
Strategies for enriching PTM peptides
| Method | Case studies | Advantages | Disadvantages |
|---|---|---|---|
| Antibody | AcR or MeK, pTyr, | High speed and specificity | Require good antibody |
| Metabolic tagging | O-GlcNAc, farnesylation | Artificial system, limited to bulk PTMs | |
| In vitro derivatization | pSer/Thr, O-GlcNAc, glycoproteins | Reagent accessible | Multiple reactions, reduced sensitivity, side reactions |
| Chemoenzymatic | O-GlcNAc | Reagent accessible | Multiple steps |
| Ionic interaction based | pSer/Thr/Tyr | Established method | |
| Other affinity reagents | Glycoproteins/lectin | Reagents accessible | Low-binding affinity |
AcR, MeR, pSer, pThr, pTyr represent acetylated arginine, methylated arginine, phosphorylated serine, threonine, and tyrosine residues, respectively.
In addition to the attachment of small chemical moieties, PTMs by conjugation of a polypeptide, such as ubiquitin or ubiquitin-like polypeptides, onto proteins have also been described. Global studies of these kinds of PTMs have been performed in various species, including yeast. Typically, an epitope tag, such as a histidine tag or HA tag, is introduced into the PTM moiety (for example, at the N-terminus of ubiquitin) to facilitate affinity purification using nickel beads (for histidine tag) or an anti-epitope antibody [46–50]. A single step purification generally yields a high proportion of non-specifically bound proteins (for example, proteins with multiple histidine residues in a short peptide sequence) along with conjugated proteins. This problem can be significantly alleviated by tandem purification using two affinity tags (for example, histidine tag and FLAG tag) for sequential enrichment [51].
3.3 Ionic interaction-based enrichment
The most successful analytical strategies for enriching phosphopeptides take advantage of the unique chemical characteristics of the phosphate group; that is, its negative charge and ability to interact with ion exchange beads and to participate in coordinate covalent bonding with immobilized metal ions. IMAC using Fe(III) was initially used to isolate phosphopeptides from a protein of interest for mapping phosphorylation sites. It was then extended to isolate phosphopeptides from a protein mixture. The method was used in the first few successful phosphoproteomics studies [52, 53]. The IMAC procedure has been continuously improved [54–56] and can be combined with other separation technologies. For example, peptide separation prior to IMAC has the advantage of improving enrichment efficiency, specificity and dynamic range. Accordingly, both strong anion exchange chromatography and strong cation exchange chromatography (SCX) have been used to prefractionate protein lysates prior to IMAC for phosphoproteomics studies [57–59]. Phosphoproteomic analysis of yeast (S. cerevisiae, S. pombe) under various conditions has been highly successful by using such methods [60–62] (Table 2).
In addition to IMAC beads, a titanium dioxide (TiO2)-based solid matrix has proven highly efficient and specific for enriching phosphopeptides [63–65]. As demonstrated by several recent phosphoproteomics studies [65, 66] TiO2 has been easier to implement and has proved to be more robust than IMAC for the analysis of complex protein samples [67].
Other methods have been used for enriching phosphopeptides. SCX, which takes advantages of charge-state differences between tryptic phosphopeptides and tryptic unphosphorylated peptides at low pH, was used to enrich phosphopeptides [68]. Other metal oxides, such as zirconium and aluminum, have also been explored for enriching phosphopeptides [69–71], but have not yet been widely used. Interestingly, the combined use of IMAC and TiO2 (called SIMAC) seems able to separate mono-phosphorylated peptides from multi-phosphorylated counterparts, thereby reducing sample complexity and leading to higher sensitivity [72]. These two enrichment techniques also demonstrated to be complementary in a study of S. pombe phosphoproteome [61]. Combinations of either IMAC or TiO2 with other complementary fractionation techniques, such as HILIC [73] and peptide IEF [74–76], may further improve our abilities to pursue comprehensive phosphoproteome analysis in the future.
3.4 Utility of PTM-specific enzymes
Modification-specific enzymes are attractive reagents for analysis of PTM substrates. This is nicely demonstrated by a modification-specific workflow for proteome-wide analysis of glycosylphosphatidylinositol-anchored proteins (GPIAPs) (Fig. 2). GPI-APs represent a special class of glycoproteins that are located at the extracellular surface of the plasma membrane. Modification-specific analysis of GPIAPs is facilitated by the availability of phospholipases that are highly specific for cleavage of the phosphatidylinositol linkage that tethers the protein to the cell surface. Using this method, GPI-APs from human and plant plasma membrane fractions were specifically released for subsequent proteomic analysis by LC-MS/MS [77, 78].
A chemo-enzymatic method was used to tag O-GlcNAc-modified proteins. This method uses an engineered galactosyltransferase enzyme to selectively label O-GlcNAc-modified proteins with a ketone-containing galactose analog followed by a conjugation reaction with an aminooxy biotin compound which attaches the biotin tag [79].
4 MS analysis and data evaluation
A MS/MS dataset generated from HPLC/MS/MS analysis of the enriched PTM peptides is subjected to protein sequence alignment for identifying PTM peptide sequences, PTM sites, and quantification. High sensitivity of a HPLC/MS system and a long HPLC elution gradient will facilitate identification of more PTM peptides. In addition, accurate identification and quantification of PTMs are of paramount importance.
4.1 Quality control of PTM data sets
Despite previous efforts to improve the accuracy of protein sequence database searches, the false identification of PTM peptides and their sites continues to be a problem, especially in the context of large-scale PTM analysis, or when multiple PTMs are included in the protein sequence database search. Given that datasets of PTM substrates and their PTM sites are likely to be widely used by the biomedical research community, it is highly important to ensure high MS data quality for accurate annotation of PTM sites.
Toward this goal, PTM sites should be precisely mapped and all the major peaks in MS/MS spectra of PTM-peptides should be assigned. For some types of PTM, exact location of a PTM site is critical. For example, protein methylation (identified with a mass shift of + 14 Da) can be present on several amino acid residues (for example, Arg, Lys, Asp, Glu, Asn, Cys, His, and Ser). Likewise, similar challenges exist for protein acetylation (at Lys, Ser, Thr, and Tyr) and for protein phosphorylation (at Ser, Thr, Tyr, and His). The existing protein database search algorithms often have difficulty in distinguishing those PTM isomers that have adjacent PTM sites. Although laborious and not often practical, manual verification is always useful to precisely locate the PTM sites with high confidence. Accordingly, for biology researchers who use the dataset, it is critical to carefully examine the MS/MS data of interest, by using objective scoring and evaluation criteria and by working closely with a mass spectrometrist to ensure the accuracy of PTM identification before investing significant efforts in functional studies. The fact that some PTMs, such as pSer, pThr, and O-GlcNAc, are labile and may (partially) decompose during LC-MS/MS analysis further complicates the situation and challenges data interpretation.
4.2 Quantification of global PTM changes
Dynamic studies of a PTM can provide insights into the role of the PTM in a biological process, and can identify substrates of a PTM regulatory enzyme. Integration of enrichment methods for PTM peptides with quantitative MS provides the means for determining changes of a PTM on a global scale. Quantitative PTM proteomics can be achieved using methods with or without stable isotope labels [13, 80]. (reviewed in [9]). Label-free quantification is primarily based on the intensities of the MS signals generated from HPLC/MS analysis. Labeling of peptides with stable isotopes can be achieved either in vitro or in vivo. Isotopic labeling methods offer the advantage of higher accuracy, while label-free quantification techniques enjoy more flexibility in sample source (for example, multiple samples from cultured cells or animal tissues).
4.3 PTM cross-talk
Studies in histones and p53 demonstrate that multiple PTMs can be present in proteins and that PTMs operate in a cooperative fashion. However, the vast majority of cellular proteins have not been carefully examined for the possible existence of multiple PTMs. Previous studies in PTM proteomics have focused on a particular type of PTM. Accordingly, those proteins bearing more than a few different PTMs have not been examined at a global scale. Recent advances in non-restrictive sequence alignment algorithms provide a possible means for identifying all types of PTMs in a protein [81–84]. Identification of PTM peptides with more than one type of PTM in a short proteolytic peptide is likely to reveal important insights into the principles that govern PTM cross-talk, such as synergy or inhibition [85, 86]. Recent advances in MS fragmentation techniques – electron capture dissociation [87] and electron transfer dissociation [88] – facilitate efficient fragmentation of large peptides and small protein domains. Thus, it is becoming feasible to study multi-site and cooperative PTMs within small protein domains.
5 Perspectives and concluding remarks
In summary, tremendous progress has been made in the past several years in developing PTM-specific enrichment methods and MS-based proteomics technologies for PTM analysis in prokaryotes and eukaryotes, including yeasts. Yeast, i.e. Saccharomyces cerevisiae, remains one of the favorite model organisms for developing and implementing quantitative proteomics technologies for comprehensive mapping of PTMs. Given the high number of PTMs and high abundance of some of the common ones (for example, phosphorylation, ubiquitination, and lysine acetylation), PTMs likely constitute the most complex and delicate regulatory networks in eukaryotic cells.
The future challenges for PTM proteomics include achieving higher sensitivity and a wider dynamic range of protein abundance for detection of low-abundance PTM events, improving the accuracy of PTM identification and localization, developing robust quantitative methods for studying PTM dynamics in tissues, and developing methods for analysis of all types of PTMs in proteins. In addition, the emerging importance of multisite, cooperative PTM events in proteins necessitates the development of novel strategies for full-spectrum PTM identification not only in proteolytic peptides, but also in regulatory protein domains and intact proteins.
Acknowledgments
We want to thank Yoshida Oda, Hui Zhang, and members of Zhao and Jensen laboratories for critically reading this paper and for helpful suggestions. This work was supported by NIH grants to Y.Z. and by grants from the Lundbeck Foundation and the Danish Research Agency to O.N.J. We apologize to the colleagues whose works cannot be cited in this review paper due to space limitations.
Abbreviations
- AcLys
acetyllysine
- GPI-APs
glycosylphosphati-dylinositol-anchored protein
- SCX
strong cation exchange
Footnotes
The authors have declared no conflict of interest.
References
- 1.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. Natl. Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 3.Kim SC, Sprung R, Chen Y, Xu Y, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Klemic JF, Chang S, Bertone P, et al. Analysis of yeast protein kinases using protein chips. Nat. Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- 5.Reimer U, Reineke U, Schneider-Mergener J. Peptide arrays: from macro to micro. Curr. Opin. Biotechnol. 2002;13:315–320. doi: 10.1016/s0958-1669(02)00339-7. [DOI] [PubMed] [Google Scholar]
- 6.Rathert P, Dhayalan A, Murakami M, Zhang X, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat. Chem. Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- 8.Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Jensen ON. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell. Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 10.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 11.Cantin GT, Yates JR., III Strategies for shotgun identification of post-translational modifications by mass spectrometry. J. Chromatogr. A. 2004;1053:7–14. doi: 10.1016/j.chroma.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 12.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 14.Lin SY, Makino K, Xia W, Matin A, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Muir TW, Kent SB, Tischer E, et al. Mapping protein-protein interactions by affinity-directed mass spectrometry. Proc. Natl. Acad. Sci. USA. 1996;93:4020–4024. doi: 10.1073/pnas.93.9.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Sprung R, Pei J, Tan X, et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 18.Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal. Biochem. 2006;354:279–289. doi: 10.1016/j.ab.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 19.Rikova K, Guo A, Zeng Q, Possemato A, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 20.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. USA. 2000;97:179–184. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 22.Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr. Opin. Struct. Biol. 2007;17:481–487. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat. Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 24.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, et al. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, et al. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kho Y, Kim SC, Jiang C, Barma D, et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprung R, Nandi A, Chen Y, Kim SC. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res. 2005;4:950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 28.Kostiuk MA, Corvi MM, Keller BO, Plummer G, et al. Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J. 2008;22:721–732. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin DD, Vilas GL, Prescher JA, Rajaiah G, et al. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 2008;22:797–806. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oda Y, Nagasu T, Chait BT. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 31.Wells L, Gao Y, Mahoney JA, Vosseller K, et al. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic beta-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002;277:1755–1761. doi: 10.1074/jbc.m109656200. [DOI] [PubMed] [Google Scholar]
- 32.Go EP, Uritboonthai W, Apon JV, Trauger SA, et al. Selective metabolite and peptide capture/mass detection using fluorous affinity tags. J. Proteome Res. 2007;6:1492–1499. doi: 10.1021/pr060608s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Qian WJ, Knyushko TV, Clauss TR, et al. A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. J. Proteome Res. 2007;6:2257–2268. doi: 10.1021/pr0606934. [DOI] [PubMed] [Google Scholar]
- 34.Derakhshan B, Wille PC, Gross SS. Unbiased identification of cysteine S-nitrosylation sites on proteins. Nat. Protoc. 2007;2:1685–1691. doi: 10.1038/nprot.2007.210. [DOI] [PubMed] [Google Scholar]
- 35.Roth AF, Wan J, Bailey AO, Sun B, et al. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang R, Wan J, Arstikaitis P, Takahashi H, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaji H, Saito H, Yamauchi Y, Shinkawa T, et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 38.Xiong L, Andrews D, Regnier F. Comparative proteomics of glycoproteins based on lectin selection and isotope coding. J. Proteome Res. 2003;2:618–625. doi: 10.1021/pr0340274. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin. Chem. 2006;52:1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 40.Madera M, Mann B, Mechref Y, Novotny MV. Efficacy of glycoprotein enrichment by microscale lectin affinity chromatography. J. Sep. Sci. 2008;31:2722–2732. doi: 10.1002/jssc.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sparbier K, Asperger A, Resemann A, Kessler I, et al. Analysis of glycoproteins in human serum by means of glycospecific magnetic bead separation and LC-MALDITOF/TOF analysis with automated glycopeptide detection. J. Biomol. Tech. 2007;18:252–258. [PMC free article] [PubMed] [Google Scholar]
- 42.Hagglund P, Bunkenborg J, Elortza F, Jensen ON, et al. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- 43.Tian Y, Zhou Y, Elliott S, Aebersold R, et al. Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 45.Ramachandran P, Boontheung P, Xie Y, Sondej M, et al. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 46.Peng J, Schwartz D, Elias JE, Thoreen CC. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, Kwon SW, Anselmo A, Kaur K, et al. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J. Biol. Chem. 2004;279:20999–21002. doi: 10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 48.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., III Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 49.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, et al. Large-scale analysis of the human ubiquitin-related proteome. Proteomics. 2005;5:4145–4151. doi: 10.1002/pmic.200401280. [DOI] [PubMed] [Google Scholar]
- 51.Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 53.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Kokubu M, Ishihama Y, Sato T, Nagasu T, et al. Specificity of immobilized metal affinity-based IMAC/C18 tip enrichment of phosphopeptides for protein phosphorylation analysis. Anal. Chem. 2005;77:5144–5154. doi: 10.1021/ac050404f. [DOI] [PubMed] [Google Scholar]
- 55.Lee J, Xu Y, Chen Y, Sprung R, et al. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol. Cell. Proteomics. 2007;6:669–676. doi: 10.1074/mcp.M600218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishihama Y, Wei FY, Aoshima K, Sato T, et al. Enhancement of the efficiency of phosphoproteomic identification by removing phosphates after phosphopeptide enrichment. J. Proteome Res. 2007;6:1139–1144. doi: 10.1021/pr060452w. [DOI] [PubMed] [Google Scholar]
- 57.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell. 2004;16:2394–2405. doi: 10.1105/tpc.104.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruhler A, Olsen JV, Mohammed S, Mortensen P, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J. Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 62.Li X, Gerber SA, Rudner AD, Beausoleil SA, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 63.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, et al. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2-D-nanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 64.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, et al. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Thingholm TE, Larsen MR, Ingrell CR, Kassem M, et al. TiO(2)-based phosphoproteomic analysis of the plasma membrane and the effects of phosphatase inhibitor treatment. J. Proteome Res. 2008;7:3304–3313. doi: 10.1021/pr800099y. [DOI] [PubMed] [Google Scholar]
- 66.Olsen JV, Blagoev B, Gnad F, Macek B, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 67.Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun. Mass Spectrom. 2007;21:3635–3645. doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- 68.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kweon HK, Hakansson K. Selective zirconium dioxidebased enrichment of phosphorylated peptides for mass spectrometric analysis. Anal. Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 70.Zhou H, Xu S, Ye M, Feng S, et al. Zirconium phosphonate- modified porous silicon for highly specific capture of phosphopeptides and MALDI-TOF MS analysis. J. Proteome Res. 2006;5:2431–2437. doi: 10.1021/pr060162f. [DOI] [PubMed] [Google Scholar]
- 71.Wolschin F, Wienkoop S, Weckwerth W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC) Proteomics. 2005;5:4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- 72.Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics. 2008;7:661–671. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- 73.McNulty DE, Annan RS. Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol. Cell. Proteomics. 2008;7:971–980. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 74.Xu Y, Sprung R, Kwon SW, Kim SC, et al. Isolation of phosphopeptides by pI-difference-based electrophoresis. J. Proteome Res. 2007;6:1153–1157. doi: 10.1021/pr060498p. [DOI] [PubMed] [Google Scholar]
- 75.Cantin GT, Venable JD, Cociorva D, Yates JR., III Quantitative phosphoproteomic analysis of the tumor necrosis factor pathway. J. Proteome Res. 2006;5:127–134. doi: 10.1021/pr050270m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hung CW, Kubler D, Lehmann WD. p I-based phosphopeptide enrichment combined with nanoESI-MS. Electrophoresis. 2007;28:2044–2052. doi: 10.1002/elps.200600678. [DOI] [PubMed] [Google Scholar]
- 77.Elortza F, Nuhse TS, Foster LJ, Stensballe A, et al. Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics. 2003;2:1261–1270. doi: 10.1074/mcp.M300079-MCP200. [DOI] [PubMed] [Google Scholar]
- 78.Elortza F, Mohammed S, Bunkenborg J, Foster LJ. Modification-specific proteomics of plasma membrane proteins: identification and characterization of glycosylphosphatidylinositol-anchored proteins released upon phospholipase D treatment. J. Proteome Res. 2006;5:935–943. doi: 10.1021/pr050419u. [DOI] [PubMed] [Google Scholar]
- 79.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. USA. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oda Y, Huang K, Cross FR, Cowburn D. Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Y, Chen W, Cobb MH, Zhao Y. PTMap – a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc. Natl. Acad. Sci. USA. 2009;106:761–766. doi: 10.1073/pnas.0811739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gupta N, Tanner S, Jaitly N, Adkins JN, et al. Whole proteome analysis of post-translational modifications: Applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Savitski MM, Nielsen ML, Zubarev RA. ModifiComb, a new proteomic tool for mapping substoichiometric posttranslational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 84.Hansen BT, Davey SW, Ham AJ, Liebler DC. P-Mod: an algorithm and software to map modifications to peptide sequences using tandem MS data. J. Proteome Res. 2005;4:358–368. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- 85.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 86.Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N-termini. Proc. Natl. Acad. Sci. USA. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zubarev RA, Zubarev AR, Savitski MM. Electron capture/transfer versus collisionally activated/induced dissociations: solo or duet? J. Am. Soc. Mass Spectrom. 2008;19:753–761. doi: 10.1016/j.jasms.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, et al. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]