Summary
Hsp70s mediate protein folding, translocation, and macromolecular complex remodeling reactions. Their activities are regulated by proteins that exchange ADP for ATP from the nucleotide-binding domain (NBD) of the Hsp70. These nucleotide exchange factors (NEFs) include the Hsp110s, which are themselves members of the Hsp70 family. We report the structure of an Hsp110:Hsc70 nucleotide exchange complex. The complex is characterized by extensive protein:protein interactions and symmetric bridging interactions between the nucleotides bound in each partner protein's NBD. An electropositive pore allows nucleotides to enter and exit the complex. The role of nucleotides in complex formation and dissociation, and the effects of the protein:protein interactions on nucleotide exchange, can be understood in terms of the coupled effects of the nucleotides and protein:protein interactions on the open-closed isomerization of the NBDs. The symmetrical interactions in the complex may model other Hsp70 family heterodimers in which two Hsp70s reciprocally act as NEFs.
Introduction
Hsp70s are a diverse family of chaperones involved in a variety of protein processing reactions, including protein import into the endoplasmic reticulum and mitochondrion, protein folding, and the remodeling or dissociation of defined protein complexes and heterogeneous protein aggregates (Mayer and Bukau, 2005; Sousa and Lafer, 2006; Young et al., 2003). All Hsp70s are composed of a nucleotide-binding domain (NBD) and a protein substrate-binding domain (SBD), with the latter subdivided into β sandwich (SBDβ) and α-helical (SBDα) subdomains. The helical subdomain is designated the “lid” because it caps the substrate-binding pocket of the SBDβ to block substrate release. Release of substrate is induced when ATP binds to the NBD (Bukau and Horwich, 1998; Schmid et al., 1994; Wei et al., 1995). The SBD can then bind a new substrate. Presentation of the substrate is carried out by a J protein cochaperone that also stimulates the Hsp70 to hydrolyze the bound ATP (Misselwitz et al., 1998; Szabo et al., 1994; Walsh et al., 2004). Upon ATP hydrolysis the SBD reverts to a conformation that releases substrate slowly, so the product of this step is a long-lived Hsp70*ADP*substrate complex (Brehmer et al., 2001; Szabo et al., 1994).
NEFs regulate the lifetime of the Hsp70*ADP*substrate complex by inducing ADP release from the NBD, allowing ATP to bind and induce release of substrate. The Hsp70 NEFs include the homologous GrpE (Packschies et al., 1997) and Mge1p (Deloche and Georgopoulos, 1996) found in prokaryotes and mitochondria, respectively, and the Bag (Sondermann et al., 2001) and HspBP1/Fes1/Sls1 (Shomura et al., 2005) families from eukaryotes. Structures of complexes of GrpE (Harrison et al., 1997) and Bag1 (Sondermann et al., 2001) with isolated NBDs from, respectively, E. coli Hsp70 (DnaK) or bovine Hsc70 reveal that, while GrpE and Bag are structurally unrelated, they use a similar mechanism to cause the Hsp70 NBD to release nucleotide. Binding of nucleotide to the NBD induces it to close around the nucleotide, and both GrpE and Bag bind to the NBD and induce it to open, so that nucleotide can dissociate. The HspBP1 proteins also bind and induce a conformational change in the Hsp70 NBD, but in this case, existing structures with HspBP1 encompass only a subdomain of the NBD (Shomura et al., 2005), so the nature of the changes induced to cause nucleotide release are less clear. The Hsp110s have recently been recognized as another class of eukaryotic NEFs. Hsp110s are themselves divergent members of the Hsp70 family: while they possess similar NBD and SBD architecture and can bind unfolded protein substrates to act as “holdases,” they do not display the same ATP-driven allosteric cycles as do the canonical members of the Hsp70 family (Dragovic et al., 2006; Raviol et al., 2006; Shaner et al., 2004). Instead their primary function in vivo seems to be to act as NEFs for the Hsp70s. In fact, inviability resulting from deletion of the yeast Hsp110s (Sse1 and Sse2) can be compensated for by expression of a structurally unrelated NEF such as Fes1 (Dragovic et al., 2006; Raviol et al., 2006; Shaner et al., 2006).
The mechanism by which Hsp110s induce nucleotide release is unclear but is distinct from how other NEFs work, in that an Hsp110 must first bind ATP to assume a conformation that allows it to bind Hsp70 (Shaner et al., 2006). The resulting Hsp110:Hsp70 complex is stable and dissociates only when ATP subsequently binds to Hsp70 (Andréassonetal., 2008). To better understand how Hsp110 regulates Hsp70, we have determined the crystal structure of yeast Hsp110 (Sse1) complexed with bovine Hsc70. This structure reveals extensive intermolecular contacts encompassing an entire face of each NBD, with numerous charge:charge pairs and direct H bonds, consistent with the remarkable stability of this complex. While the Hsp110 NBD contains tightly bound nucleotide and is in a closed conformation, the Hsc70 NBD is open with nucleotide more weakly bound in what probably represents a prerelease configuration. These findings provide a structural basis for understanding how a member of the Hsp70 chaperone family diverged to become a dedicated Hsp70 cochaperone.
Results
Yeast and Mammalian Hsp110 Are Functionally Interchangeable in a Clathrin Basket Dissociation Reaction Mediated by Mammalian Hsc70
Yeast Hsp110 forms stable complexes with either mammalian (bovine) Hsc70 oryeast Hsp70 (Ssa1) in an ATP-dependent fashion (Shaner et al., 2006). To determine if yeast Hsp110 could also functionally substitute for mammalian Hsp110 in a macromolecular complex dissociation reaction mediated by bovine Hsc70, we tested the effects of these NEFs on clathrin basket dissociation. Mixing of clathrin baskets with Hsc70, ATP, andaneuronal-specific J cochaperone (bovine auxilin) results in basket dissociation and can be followed by the decrease in light scattering as the large clathrin baskets are dissociated into clathrin triskelia (Figure 1A) (Jiang et al., 2005). When the Hsc70 concentration is substoichiometric relative to the clathrin, dissociation follows burst kinetics with an initial, rapid exponential phase and a subsequent, slower linear phase. Both the amplitude of the exponential phase and the slope of the linear phase are proportional to the amount of added Hsc70 (Figure 1 A; see legend for fitting details). When the Hsc70 concentration approaches or exceeds the clathrin concentration, basket dissociation is rapid with half the baskets dissociated in ∼29 s. These kinetics are due to formation of clathrin:Hsc70 complexes, which dissociate slowly and therefore limit the Hsc70's ability to participate in multiple rounds of basket dissociation (Greene and Eisenberg, 1990). Addition of increasing amounts of a NEF, such as Bag1, to a reaction with a low Hsc70 concentration increasingly accelerates the second, slow phase of the dissociation reaction (Figure 1B). Yeast and mammalian (human) Hsp110s also accelerate basket dissociation but have quantitatively similar effects as Bag1 at 5-fold lower concentrations (Figures 1C and 1D). These experiments used full-length Hsc70, so we ran similar reactions with a nonoligomerizing truncated variant (aa 1–554) of the Hsc70 that is more suitable for crystallization. We found that this truncated variant can also dissociate baskets and that its basket dissociation activity is stimulated by yeast Hsp110 (see Figure S1 available online).
Figure 1. Yeast and Human Hsp110 Are Functionally Interchangeable.
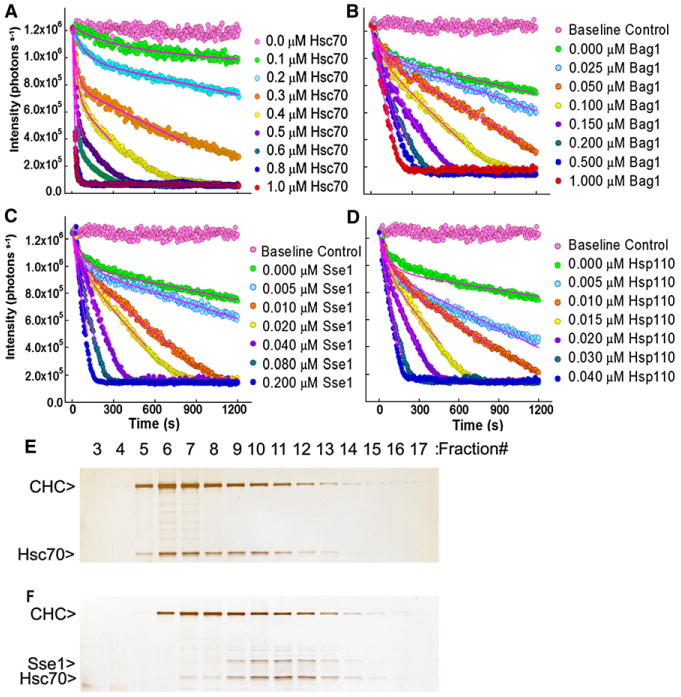
(A) Clathrin basket dissociation mediated by full-length Hsc70 and auxilin, and monitored by dynamic light scattering. The clathrin and auxilin concentrations were, respectively, 1.0 and 0.1 μM, and the Hsc70 concentration was varied as indicated. Basket dissociation was fit to y = yo + A1(exp(−t1*x) − 1) − k*x, where yo = total scatter at x = 0 s = ∼1,225,000 for 1 μM clathrin; A1 = amplitude of 1st (rapid) exponential phase; t1 = time constant of 1st exponential phase; k = rate of 2nd (slow) linear phase = decrease/s in number of scattered photons. Data were globally fit with yo held constant, since yo is determined by the constant amount of clathrin baskets added to each reaction.
(B–D) Basket dissociation reactions run with constant Hsc70 (0.2 μM), auxilin (0.1 μM), and clathrin (1 μM) concentrations but with varying concentrations of either Bag1 (B), yeast Hsp110 (Sse1; [C]), or human Hsp110 (D). Data in each panel were globally fit as in (A) with yo constant, but with A1 and t1 shared by all data sets since the model assumes that A1 and t1 are determined by the amount of Hsc70 added. Only data between 0%–80% completion of the dissociation reaction were used in fitting to reduce effects due to substrate depletion.
(E) Fractions from rapid gel exclusion chromatography of a basket dissociation reaction run with 0.6 μM clathrin (“CHC”), 0.1 μM auxilin, and 0.6 μM Hsc70.
(F) As in (E) but with 0.6 μM yeast Hsp110 (Sse1) added.
To determine if the effects of yeast Hsp110 on basket dissociation are, in fact, due to yeast Hsp110 causing release of clathrin from Hsc70, we analyzed dissociation reactions by rapid gel exclusion. When reactions were run in the absence of a NEF, the 70 kD Hsc70 coeluted with the much larger (650 kD) clathrin triskelia, indicating that these two proteins remained associated (Figure 1E). However, when the reaction was run in the presence of yeast Hsp110, the Hsc70 emerged in a separate peak with the Hsp110, consistent with its lower MW (Figure 1F), indicating that the NEF action of the yeast Hsp110 had released clathrin from its association with bovine Hsc70. We conclude that yeast Hsp110 can functionally substitute for a mammalian NEF in this reaction.
Crystallization and Structure Determination
Bovine Hsc70 (aa 1–554; “Hsc70ΔC”) missing its 10 kD C-terminal oligomerization domain (Jiang et al., 2005) was mixed with yeast Hsp110 (Sse1; aa 1–666) missing 27 residues from its flexible C terminus, and complex formation was induced by addition of 1 mM ADP*BeFx. SDS PAGE (data not shown) of crystals obtained in the presence of 30%–40% PEG 400 (Experimental Procedures) revealed both proteins at 1:1 stoichiometry. Molecular replacement using the isolated open NBD from the Hsc70ΔC structure (1YUW; Jiang et al., 2005) and the NBD + SBDβ of the yeast Hsp110 (2QXL; Liu and Hendrickson, 2007) was followed by manual placement of the Hsc70 SBD and Hsp110 SBDα, and refinement to an R/Rfree of 22.4/28.3 at a resolution of 3.1 Å (Table 1). With the exception of two and seven residues at, respectively, the N and C termini of the Hsc70, aa 385–389 of the Hsc70 interdomain linker, and 17 residues at the C terminus of the Hsp110, the entire complex was ordered and could be clearly traced.
Table 1. Data and Refinement Statistics.
| PDB Accession Code | 3C7N |
|---|---|
| Wavelength (Å) | 1.0720 |
| Beamline | ALS 4.2.2 |
| Space group | P2(1)2(1)2 |
| Unit cell dimensions (Å) | a = 123.5, b = 169.5, c = 87.7 |
| Diffraction resolution (Å) | 50–3.10 |
| No. of observations | 179, 719 |
| No. of unique reflections | 32, 644 |
| Completeness (%) | 97.2 (92.8)a |
| Mean I/σl | 8.8 (2.6)a |
| Rsym | 0.15 (0.55)a |
| No. of protein atoms | 9285 |
| No. of adenosine-5′-diphosphate (ADP) molecules | 2 |
| No. of berillium-trifluoride ions | 1 |
| No. of magnesium ions | 1 |
| No. of chloride ions | 3 |
| No. of sulfate ions | 4 |
| Rcryst | 0.224 (0.290)a |
| Rfree | 0.283 (0.324)a,b |
| RMSD bond lengths (Å) | 0.012 |
| RMSD bond angles (degrees) | 1.42 |
| Ramachandran plotc | |
| Favored (%) | 81.4 |
| Allowed (%) | 16.6 |
| Generous (%) | 1.5 |
| Disallowed (%) | 0.5 |
| Average B factors (Å2) | |
| Protein | 53 |
| ADP molecules | 53 |
| Berillium-trifluoride | 40 |
| Magnesium ion | 15 |
| Chloride ions | 61 |
| SO4 | 86 |
Values in parentheses are for the outer resolution shell of the data.
5% Rfree random test set used for crossvalidation.
The Ramachandran plot was generated with PROCHECK66.
Overall Structure and Interactions in the Heterodimer
A dramatic feature of the complex is the sheer extent of the intermolecular interactions (Figure 2), with a total of 15,900 Å2 of buried protein surface. Consistent with observations that the isolated NBD of Hsc70 can form a stable complex with Hsp110 (Shaner et al., 2006), the most extensive interactions are between the NBDs of the two proteins (aa 1–384 of each molecule): 9800 Å2 of protein surface are buried in an interaction that involves an entire face of each NBD (Figures 2–4). This interface exhibits extensive shape and chemical complementarity with a total of 533 intermolecular contacts (between 2.5 and 5 Å), 12 charge pairs, and 24 H bonds (Figures 2 and 3; Table 2).
Figure 2. Ribbon Models of the Hsp110:Hsc70 Nucleotide Exchange Complex.
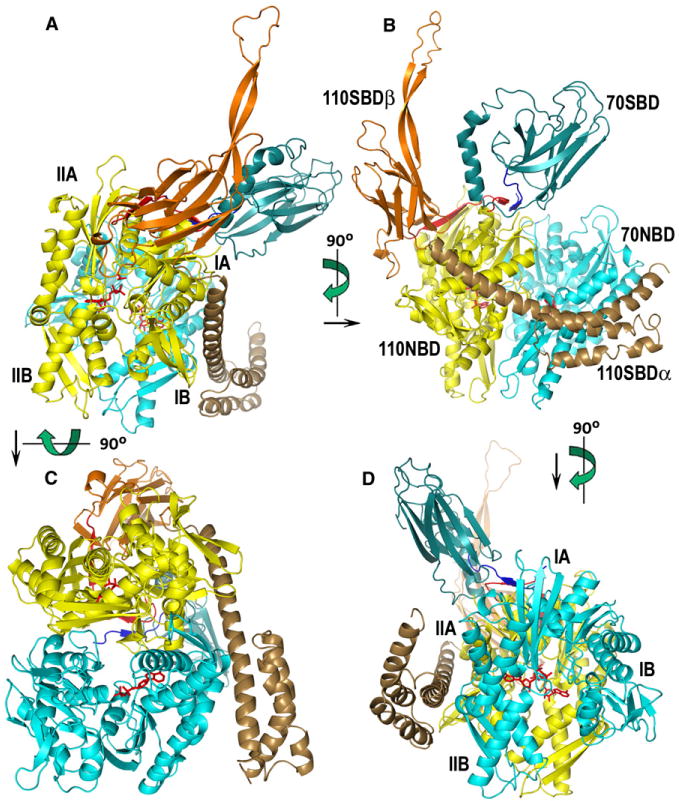
Hsc70 is colored in blue tones, and Hsp110 is colored in autumn tones (red, orange, yellow, brown). NBDs of Hsp110 and Hsc70 are colored yellow and light cyan, respectively. The interdomain linkers of Hsp110 and Hsc70 are red and blue, respectively. The Hsc70 SBD is dark cyan, and the Hsp110 SBDβ and SBDα are orange and brown, respectively. Subdomains IA–IIB of the NBDs are labeled in (A) and (D), and individual domains are labeled in (B); different views of the complex are related by the indicated rotations.
Figure 4. Conformation and Oligomeric State of the Hsp110:Hsc70 Complex in Solution.
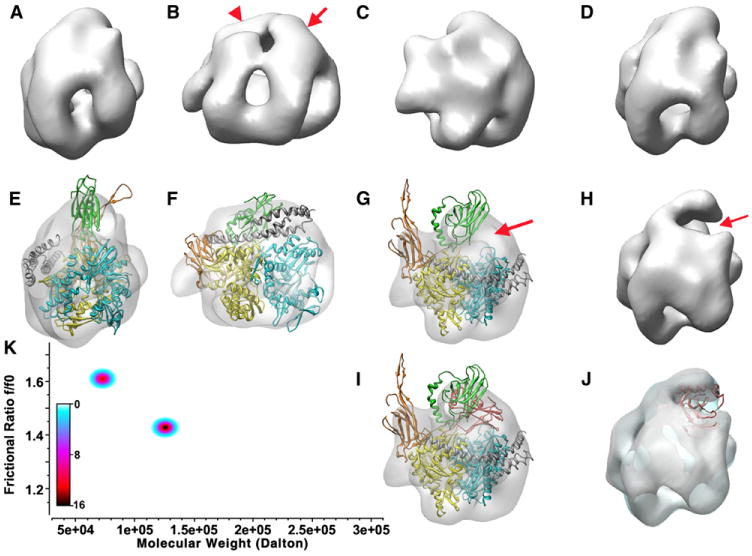
(A)–(G) and (I) show different views of the 3D reconstruction of the Hsp110:Hsc70 complex, while (H) and (J) show reconstructions of an Hsp110:Hsc70 NBD complex. (E)–(G) and (H) also show a ribbon model of the Hsp110:Hsc70 crystal complex (110 NBD: yellow; 110 SBDβ: orange; 110 SBDα: gray; 70 NBD: light cyan; 70 SBD: green) fit into the EM envelope. The model fits very well with the exception of the acidic insertion loop and the Hsc70 SBD. An alternative position for the Hsc70 SBD (red) suggested in (I) and (J) fits very well into the density that is missing in the complex in which the SBD is deleted (arrows in [G] and [H]). (K) Genetic algorithm Monte Carlo fit of the sedimentation data for the Hsp110:Hsc70 complex. Relative concentrations are shown as a color gradient (white = 0 μM, black = 16 μM). The predominant species (black center) corresponds to the dimer, while the minor species at ∼72 kDa corresponds to free Hsc70ΔC and Hsp110.
Figure 3. Crystal Packing Interactions and Induced Conformational Changes.
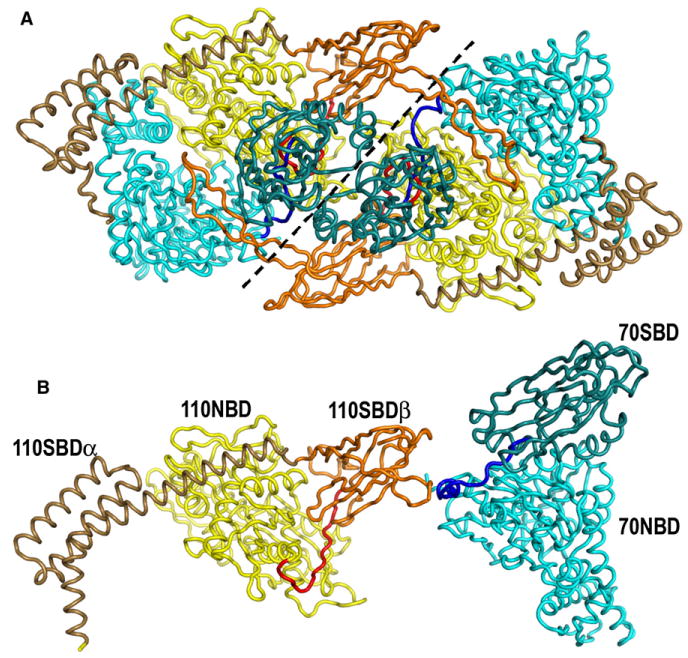
(A) Ribbon model of two crystallographically related Hsp110:Hsc70 heterodimers colored as in Figure 2. The dashed line demarcates the boundary between each heterodimer, but the acidic insertion loop of the Hsp110 SBDβ domain reaches across from one heterodimer to interact with the Hsc70 NBD of its neighbor.
(B) Ribbon models (colored and oriented as in [A]) of the Hsp110 (2QXL; Liu and Hendrickson, 2007) and Hsc70 (1YUW; Jiang et al., 2005) in the conformations seen when these proteins are not complexed with each other. The conformational differences between the proteins in isolation and in the nucleotide exchange complex include movement of the Hsp110 SBDα toward the Hsc70 NBD, ordering of the acidic insertion loop due to crystal packing interactions, and rotation of the Hsc70 SBD due to crystal packing and/or interactions between the interdomain linkers in the nucleotide exchange complex.
Table 2. Intermolecular Charge Pairs and H Bonds in the Hsp110:Hsc70 Complex.
| Charge Pairs | Direct H Bonds |
|---|---|
| Hsp110NBD:Hsc70NBD | |
| R34:E367 | K56Nz:E283Oe2 |
| K56:E283 | T58Og1:D285Od2 |
| D140:K509 | I24O:K25Nz |
| K262:D285 | E30O:S276Og |
| R266:D285 | V31O:R272Ne |
| R266:D292 | N33Od1:R272NH2 |
| E289:R258 | N55Nd2:S281O |
| E289:R262 | T282Og1:E48Oe2 |
| K351:D32 | S287Og:R258Ne |
| E370:R36 | S59Og:E283Oe1 |
| K374:E27 | S287O:R258NH2 |
| D25:K25 | S290Og:S286Og |
| T386O:V396N | |
| F185O:H23Ne2 | |
| K276O:Q33Ne2 | |
| S279O:Q33Oe1 | |
| N281Od1:A54O | |
| N281Nd2:N57Od1 | |
| E289Oe1:R258Ne | |
| T364Og1:A133O | |
| K374Nz:E27Oe1 | |
| K374Nz:E27Oe2 | |
| Hsp110Linker:Hsc70Linker | |
| R388N:L394O | |
| R390:D390 | R388OL394N |
| R390N:L392O | |
| R390NH2:D390O | |
| R390NH2:D390N | |
| Hsp110NBD + Linker + SBDβ:Hsc70SBD | |
| R388NH2:T419Og1 | |
| R388NH1:N417O | |
| E494:K524 | S2OG:R509NE |
| D532:K524 | I382O:K512NZ3.16 |
| Hsp110Lid:Hsc70NBD | |
| D569:R299 | E568Oe2:Q279Og1 |
| E575:R301 | E568O:T278Og1 |
| E576:R299 | D569Od1:T278Og1 |
| E587:R311 | K571Nz:R301NH1 |
| N572Nd2:T298Og1 | |
| E575Oe1:R301Ne | |
| E575Oe1:R301NH1 | |
| E575Oe1:R301Ne |
In addition, the Hsp110 SBDα (aa 541–651) reaches out more than 40 Å from the rest of the Hsp110 to make a one-armed embrace around Hsc70 (Figures 2 and 3; Table 2). Residues 568–579 of the SBDα contact T278/Q279 and helix I residues 289–304/307 of subdomain IIB of the Hsc70 NBD and make four charge pairs and eight H bonds with these elements (Figure 2; Table 2). The flexible and functionally critical linkers (aa 385–398) that connect the NBDs and SBDs of each molecule interact with each other symmetrically, creating a short two-stranded antiparallel β sheet between Hsp110 residues 386–390 and Hsc70 residues 392–396 (Figures 2 and 3; Table 2).
The ability of yeast Hsp110 to form a functional complex with bovine Hsc70 likely reflects the high sequence identity between bHsc70 and yeast Hsp70 (Ssa1), which are 82% identical in the regions (aa 1–400, corresponding to the NBD and linker) most important for binding Hsp110. Residues at the intermolecular interface are even better conserved: 90% of the bHsc70 residues that form the interface are identical in Ssa1, and all substitutions are conservative and maintain the charge pairs listed in Table 2 (Figure S2). Mammalian (human) and yeast Hsp110 are more divergent, with an overall identity of 41%. Again, the regions forming the protein:protein interface in the nucleotide exchange complex are more highly conserved, exhibiting 61% identity and a high level of conservative substitutions (Figure S3).
Complex-Induced Conformational Changes
The largest changes seen in Hsp110 due to binding Hsc70 are in SBDα, which shifts by several Å (mean RMSD between free versus Hsc70-bound Hsp110 is 5.8 Å for the main chain of SBDα aa 568–651) to wrap around the Hsc70 NBD (Figures 2 and 3). Interactions with Hsc70 residues E48–M61 induce modest conformational changes in Hsp110 V277–F268 (mean RMSD of aa 277–268 in Hsc70-bound versus free Hsp110 is 3.1 Å), which are propagated into neighboring elements (aa 300–315, 347–356) of the Hsp110. In addition to these changes, which are induced by interactions in the heterodimer, crystal packing interactions with a neighboring complex induce ordering of the acidic insertion loop (Hsp110 aa 503–525), an element unique to the Hsp110s that is disordered in the structure of Hsp110 alone (Liu and Hendrickson, 2007) (Figure 3). However, apart from these changes, the conformation of the Hsp110 in the complex is similar that of the free molecule (Liu and Hendrickson, 2007) (mean RMSD = 1.2 Å).
The conformation of the Hsc70 is more drastically altered. The Hsc70 SBD (aa 399–547) undergoes a whole-body rotation and is shifted by 70 Å from its position in the structure of a two-domain Hsc70 (Jiang et al., 2005) (Figure 3). The new position of the Hsc70 SBD is stabilized by crystal packing interactions with the Hsc70 SBD and Hsp110 acidic insertion loop from a neighboring complex. However, apart from this whole-body movement, the conformation of the SBD is unaltered: SBD residues 400–547 of the free Hsc70 superimpose on the SBD from the Hsp110:Hsc70 complex with a mean RMSD of 1.1 Å. Significantly, aa 539–544 near the C terminus of the Hsc70 SBD remain bound in the Hsc70 substrate-binding pocket as seen in the structure of the isolated two-domain Hsc70. This interaction is believed to be a consequence of the C-terminal truncation in this Hsc70 construct, which results in destabilization of the terminal helix and subsequent binding of the unwound element in the substrate-binding pocket, mimicking the interaction of an extended polypeptide substrate with the Hsc70 (Wang et al., 1998). The observation that this interaction persists in the complex suggests that binding to Hsp110 does not affect protein substrate interactions with Hsc70.
Solution Conformation and Oligomeric State of the Complex
Given the potential for crystal packing interactions to generate spurious oligomeric states or alter interdomain interactions in these flexible proteins, we used analytical ultracentrifugation (AUC) to characterize the solution oligomeric state of the complex and 3D reconstruction of the Hsp110:Hsc70 complex using electron microscopy to corroborate the domain placement.
Sedimentation analysis of a sample used for crystallization revealed two major species with sedimentation coefficients of 3.81 ± 0.006S and 6.19 ± 0.005S. Genetic algorithm analysis (Figure 4K) revealed the MW and shape of the 3.81S species to be 72.4 kD (frictional ratio = 1.61), in good agreement with the molecular weight of either bHsc70ΔC (61.1 kD) or Hsp110 (74.4 kD). This may reflect dissociated or unassembled complex as well as uncomplexed bHsc70ΔC in the sample, since we use an excess of Hsc70 during crystallization to ensure saturation of the Hsp110. The calculated MW of the 6.19S species, which was the predominant component, was 125.6 kD (frictional ratio = 1.42), which matched the theoretical MW of the Hsp110:Hsc70 complex (135.4 kD). There was no evidence for higher-order oligomers.
For the EM reconstruction, complexes were negatively stained, and 16290 particles obtained at 0° and 25° tilt were selected. Consistent with the AUC, particles corresponding to heterotetramers were not seen, indicating that the heterodimer is the solution species. The 3D reconstruction reveals a compact structure (Figures 4A–4D), with the presence of a “V”-shaped mass that can be assigned to the NBD of the Hsc70, since a 3D reconstruction performed with 6557 particles of a complex between Hsp110 and an Hsc70 NBD points to the missing region (Hsc70 SBD; compare Figures 4D and 4H; see arrow in Figure 4H) as near this mass.
Opposite and parallel to the Hsc70 NBD, a second “V”-shaped density can be assigned to Hsp110 (arrow and arrowhead in Figure 4B, respectively), and this is supported by docking the crystal structure into the 3D reconstruction (Figures 4E–4G) which fits the two NBDs into the “V”-shaped masses of the EM-generated volume (Figure 4F). There are, however, differences between the EM and crystal structures. The first is the acidic insertion loop in the Hsp110 SBDβ, which protrudes from the 3D reconstruction (orange domain in Figures 4E, 4G, and 4I). The absence of density for this loop in the reconstruction is consistent with observations that it is disordered in the isolated Hsp110 (Liu and Hendrickson, 2007) and only becomes ordered in the crystal structure of the complex due to crystal packing interactions.
The largest difference resides in the Hsc70 SBD, which protrudes from the volume of the reconstruction (green domain in Figures 4E, 4G, and 4I), leaving empty a mass in which the Hsc70 SBD fits very well (see arrow in Figure 4G and red domain in Figure 4I), and which is absent in the reconstruction of the Hsc70 NBD:Hsp110 complex (Figures 4H and 4J). This indicates that the position of the Hsc70 SBD in the crystal is displaced by packing contacts from where it would otherwise sit. Significantly, the Hsc70 SBD is not simply delocalized, since in that case, the averaging of multiple particle images would have erased the density for this portion of the complex. Instead, the Hsc70 SBD seems positioned to interact with both the Hsp110 and Hsc70 NBDs near their vertices (NBD subdomains IA and IIA), though at the 21 Å resolution afforded by this reconstruction, we cannot be more specific about these contacts. Nevertheless, this suggests that the Hsc70 SBD may play a functional role in the complex, perhaps providing additional stabilizing interactions with the Hsp110 NBD.
The Nucleotide Exchange Mechanism
From the perspective of nucleotide exchange the most important conformational effects are on the open versus closed state of the NBDs. Consistent with the observation that Hsp110 must bind ATP or ADP*BeF3− (Shaner et al., 2006) before it can bind Hsp70, the Hsp110 NBD is closed with ADP*BeF3− tightly bound (Figures 2 and 6). The Hsc70 NBD, however, is in an open conformation similar to those seen in nucleotide-free NBD structures in a two-domain Hsc70 (Jiang et al., 2005) or in a complex with Bag (Sondermann et al., 2001), but the NBD in the Hsp110 complex is even more open: the angle formed by the Cα atoms of K250/G190/E110 is 37.9° in a closed ATP*NBD (Flaherty et al., 1990), 43.2° in the nucleotide-free 2 domain Hsc70 (Jiang et al., 2005), 44.9° in the complex with Bag (Sondermann et al., 2001), and 45.9° in the Hsp110 complex (Figure 5A).
Figure 6. Nucleotide Interactions and Nucleotide Entry/Exit Pore.
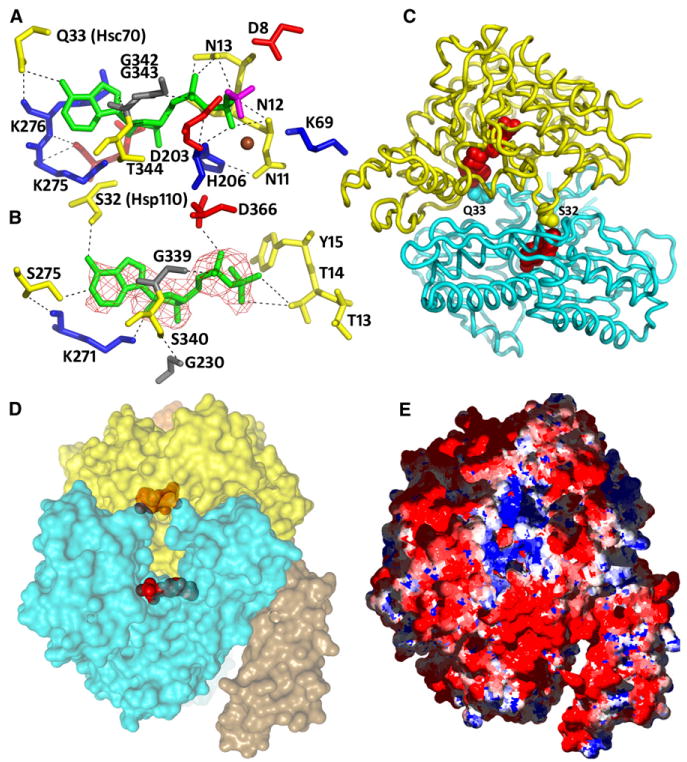
(A) The nucleotide-binding site in the Hsp110 NBD. The ADP is green, BeF3− is magenta, Mg2+ is brown, and residues interacting with these ligands are blue, red, yellow, and gray for positive, negative, polar, and hydrophobic. The BeF3− is bound by D8, D203, K69, and the Mg2+. The Mg2+ is also liganded by the N of N11, the NZ of K69, and an Oβ from the ADP. Protein:ADP H bonds include the following: four between the β-phosphate and the amides of N11, N12, N13, and H206 and one to the N13 carbonyl O; two between the α-phosphate and H206 and G343 amides and one to the N13 side-chain O; three ribose O4 and O2 bonds to, respectively, the T344 amide and Oε2 and NZ of E272 and K275; five between the base N3, N9, and N6 atoms and the NZ and carbonyl oxygens of K276 and G343 and to the Oε of Hsc70 Q33.
(B) The nucleotide-binding site in the Hsc70 NBD. Colors as in (A). Electron density from an Fo-Fc map phased with a model missing the nucleotide is shown contoured at 3.0σ. Protein:ADP H bonds include the following: one between the β-phosphate and Oγ1 of T14; two between the α-phosphate and OΔ1 of D366 and the G202 amide; five between the ribose O2, O3, and O4 atoms and the NZ of K271, the Oε1 of E268, and the amide and Oγ of S340; and three between the base N6 and N7 and R272 carbonyl and the Oγ of Hsp110 S32.
(C) Ribbon models of Hsp110 (yellow) and Hsc70 (cyan) NBDs with nucleotides (red) and side chains (Hsc70 Q33 and Hsp110 S32) that bridge one NBD to the adenine N6 of the nucleotide in the partner in space-filling representation.
(D) Transparent surface model of the complex with nucleotides in space-filling representation (coloring and orientation as in [C]) reveals a pore that connects the nucleotide-binding sites to the bulk solvent.
(E) Coloring of the surface according to charge (blue = positive; red = negative) reveals that the pore forms an electropositive well embedded in an electronegative surround.
Figure 5. Effects of Hsc70:Hsp110 Interactions and Nucleotides on the Open-Closed Isomerization of the NBDs.
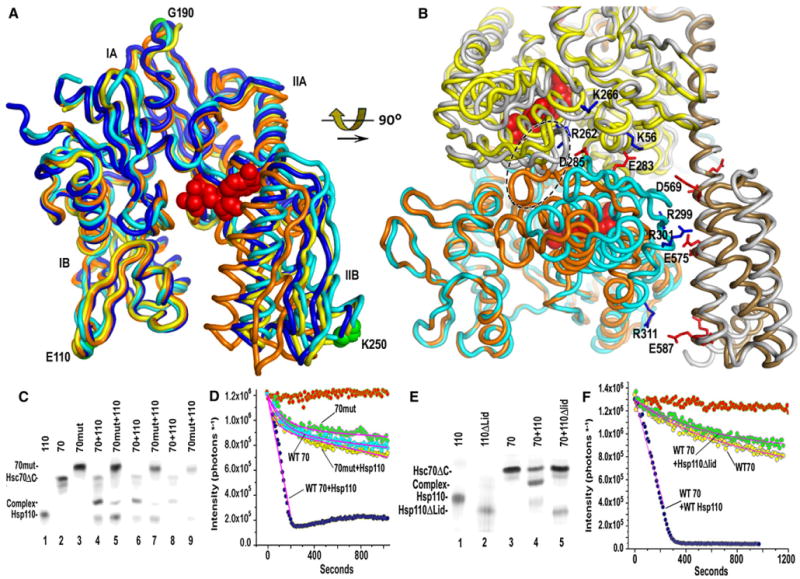
(A) Superimposed ribbon models of closed ATP bound Hsc70 NBD (1KAX; Flaherty et al., 1990; orange), the NBD from a nucleotide free two-domain Hsc70 (1YUW; Jiang et al., 2005; blue), from an Hsc70 NBD:Bag1 complex (1HIX; Sondermann et al., 2001), yellow), and from the Hsp110:Hsc70 complex (light cyan). Increased opening of the NBD occurs as subdomain II swings progressively farther from IB and is greatest in the Hsp110:Hsc70 complex (the Hsc70-bound ADP from the Hsp110:Hsc70 complex is shown in red space-filling representation).
(B) Ribbon models of a closed Hsc70 NBD (orange) and free Hsp110 (light gray) superimposed on the Hsp110:Hsc70 complex (colored as in Figure 2). Selected residues forming ionic interactions (blue for positively charged; red for negative) between the proteins are in stick representation. The loops formed by residues 285–292 and 289–296 of, respectively, the Hsc70 and Hsp110 NBDs are enclosed with a dashed ellipse. Closing of the Hsc70 NBD would create steric clashes between these loops and rupture the illustrated ionic interactions.
(C) Native PAGE of complex formation between Hsp110 (aa 1–666; “110”) and either WT Hsc70DC (aa 1–554; “70”) or Hsc70ΔC E283K/D285K (“70mut”). Lanes 1–3: individual indicated proteins at 10 μM; lanes 4 and 5: indicated proteins mixed at 10 μM; lanes 6 and 7: indicated proteins mixed at 5 μM; lanes 8 and 9: indicated proteins at 2.5 μM. All reactions contained 1 mM ATP.
(D) Effects of Hsp110 on basket dissociation with either full-length WT or E283K/D285K Hsc70. Red data points: no Hsc70 added; cyan: WT Hsc70 alone; green: E283K/D285K Hsc70 alone; dark blue:WT Hsc70+0.1μM Hsp110; yellow: E283K/D285K Hsc70+0.1 μM Hsp110. Reactions were run with1 μM clathrin, 0.2 μM Hsc70, and 0.1 μM auxilin and fit as in Figure 1.
(E) Native PAGE of complex formation between Hsc70ΔC and either Hsp110 (aa 1–666) or Hsp110ΔLid (aa 1–567; “110ΔLid”). Lanes 1–3: individual indicated proteins; lane 4: Hsp70ΔC + Hsp110; lane 5: Hsc70ΔC + Hsp110ΔLid (all proteins at 10 μM).
(F) Basket dissociation reactions as in (D), but run with either full-length Hsc70 alone (yellow), Hsc70 + Hsp110 (blue), or Hsc70 + Hsp110ΔLid (green).
The extensive interactions between the two proteins are only possible with the Hsp110 NBD closed and the Hsc70 NBD open. If a closed Hsc70 NBD is superimposed on the complex, interactions are lost and steric clashes are created. For example, in the open conformation residues D285 and E283 of Hsc70 subdomain IIB make ionic interactions with a cluster of positively charged Hsp110 NBD residues (K56/K110/R262), and Hsc70 NBD residues R299/R301/R311 interact with the negatively charged face of the Hsp110 SBDα (Figure 5B). Closure of the Hsc70 NBD involves swinging of subdomain IIB toward subdomain IB and would rupture these ionic interactions and create steric clashes between the loop (aa 285–292) connecting Hsc70 β strands 14 and 15 and the corresponding loop (aa 289–296) of Hsp110 (Figure 5B). The effects of the open-closed isomerization of the Hsc70 NBD on the interactions between the two proteins can therefore account for the observed effects of nucleotides on Hsp110:Hsp70 complex formation and dissociation as well as nucleotide exchange. Binding of ATP to Hsp110 induces the closed conformation, which can then bind Hsc70 and induce the latter to open, facilitating nucleotide release. Subsequently, ATP can bind to Hsc70 and induce it to close, rupturing NBD:NBD interactions and creating steric clashes that cause complex dissociation (see Movie S1).
To test the importance of these interactions, we generated an E283K/D285K mutant in the Hsc70 and a deletion of residues C-terminal to aa 567 in the Hsp110 (Hsp110ΔLid). As measured by native PAGE, complex formation between the E283K/D285K Hsc70 and Hsp110 was markedly reduced relative to that seen with WT Hsc70 (Figure 5C). The clathrin basket dissociation activity of the E283K/D285K Hsc70 was similar to that of the WT enzyme (Figure 5D); the ATPase rate of the mutant was also similar to WT (Figure S4), but while the basket dissociation activity of WT Hsc70 was strongly stimulated by Hsp110, the E283K/D285K mutant was only weakly stimulated (Figure 5D). The effects of deletion of Hsp110 residues 567–666 were even more severe. Complex formation was completely abrogated by this deletion (Figure 5E) and the Hsp110ΔLid mutant did not stimulate basket dissociation (Figure 5F). These results indicate, consistent with the X-ray structure, that Hsp110 SBDα and Hsc70 residues E283 and D285 make interactions critical for complex formation and function.
Nucleotide Interactions in, and between, the NBDs
A large number of interactions anchor the ADP*BeF3− in the Hsp110 NBD (detailed in the legend to Figure 6A). The BeF3− is H bonded to D8 and D203, forms an ionic interaction with K69, and is coordinated by a Mg2+ ion that is also bound by an oxygen from the β-phosphate. Two other ligands for the Mg2+ are the Nz of K69 and the N of N11. However, at this resolution we did not attempt to place water molecules in the structure, and it is likely that missing waters may also participate in Mg2+ coordination. Many of the interactions made with the ADP are identical to interactions seen in structures of Hsc70 NBDs complexed with ATP or ADP (Flaherty et al., 1990, 1994), and even where the sequences have diverged, analogous interactions are made. For example, a stretch of asparagines (N11–N13) makes an almost identical set of H-bonding interactions with the phosphate oxygens as is made by T13-T14-Y15 of Hsc70.
Inspection of the maps revealed electron density for an ADP in the Hsc70 NBD (Figure 6B). This is surprising because the Hsc70 NBD is open, and an open NBD with bound nucleotide has never been observed. This reveals that the strength of the Hsp110:Hsc70 interaction can hold the Hsc70 open against the binding energy of an ADP, which would usually induce the NBD to close. However, this ADP is more weakly bound than the ADP*BeF3− in the Hsp110 or the ADPs seen in previous NBD:ADP structures. There is no BeF3− or Mg2+ present to provide coordinating interactions with the phosphates, and there are few protein interactions with the phosphates because the nucleotide has, together with subdomain IIB, swung away from subdomains IB and IA with which the phosphates interact in the closed NBD (there are eight H bonds to the ADP phosphates in the closed Hsp110 NBD, but only three in the open Hsc70 NBD).
The adenine N6 groups of the ADPs in each NBD also make symmetric H bonds to the partner NBD in the complex, with the ADP in the Hsp110 H bonding to Hsc70 Q33 (Figure 6A) and the ADP in the Hsc70 interacting with Hsp110 S32 (Figure 6B). The nucleotides therefore drive complex formation not merely by inducing an Hsp110 conformation that can bind the Hsc70, but also by providing direct bridging interactions between the two NBDs.
A Nucleotide Entry/Exit Pore
The proposed mechanism for completion of the nucleotide exchange cycle requires that ATP bind to the Hsc70 to induce complex dissociation (Andréasson et al., 2008). Nucleotides must therefore have a mechanism for entry and exit from the complex. Inspection of the structure reveals a pore formed by subdomains IB and IIB of each NBD that is large enough to accommodate an ATP and that connects the nucleotide-binding sites to the bulk solvent (Figure 6D). There is no other visible mechanism of access to the nucleotide-binding sites, so this pore probably allows nucleotides to enter and exit the complex. Coloring of the surface according to electrostatic charge reveals that this pore forms an electropositive well within an electronegative surround (Figure 6E). Such a charge distribution may serve to both attract and orient the incoming ATP to accelerate its productive binding to the complex.
Discussion
While Hsp110 must bind ATP to assume a conformation that allows it to bind Hsc70 (Shaner et al., 2006), once the complex is formed it is kinetically trapped and ATP can be removed from the Hsp110 without causing complex dissociation (Andréasson et al., 2008). The extensive protein:protein interface seen in the complex appears to be consistent with this observation, as once formed, its spontaneous dissociation may be unlikely. This extensive interface primarily involves the NBDs of both proteins, but Hsp110 SBDα:Hsc70 NBD interactions also contribute to complex stability, consistent with observations that the Hsp110 SBD is also functionally important for the interaction with Hsc70 (Dragovic et al., 2006; Shaner et al., 2004).
Interestingly, while the individual domains of Hsp110 are generally very similar to the corresponding elements in Hsc70, the SBDα constitutes one area where these differ. In the Hsp70s there is a bend between the first two helices of the SBD, which positions the second helix to act as a lid over the substrate-binding pocket (Zhu et al., 1996). In Hsp110 these helices are fused into one long helix (Liu and Hendrickson, 2007). The structure of the complex explains this: the long helix allows SBDα to extend more than 40 Å out from the Hsp110 to establish interactions with the NBD of the Hsc70. These interactions may be important not only for complex stability, but also for opening the Hsc70, since the interactions between the Hsp110 SBDα and Hsc70 NBD subdomain IIB require that subdomain IIB hinge outward toward the SBDα (Figure 5B). In concert with this outward movement, the Hsp110 SBDα moves toward subdomain IIB, away from its lower energy conformation in the isolated Hsp110. The SBDα is therefore probably under some tension in the complex, effectively pulling on subdomain IIB to help open the Hsc70 NBD.
However, the shift in the SBDα is modest, and the conformation of the rest of the Hsp110 is almost unaltered relative to its conformation in isolation. This is consistent with observations that interdomain interfaces in Hsp110 are robust and stable (Liu and Hendrickson, 2007) so that allostery (changes in interdomain interactions due to induced conformational changes in one domain) in this molecule is limited. For example, ATP binding to Hsp110 leads to changes in the trypsin sensitivity of its NBD but does not alter the trypsin sensitivity of the interdomain linker or SBD (Liu and Hendrickson, 2007). In contrast, ATP binding to the Hsc70 NBD primarily induces changes in the trypsin sensitivity of the linker and SBD (Wei et al., 1995). Binding of the J cochaperone to Hsc70 also requires displacement of NBD:SBD interactions (Jiang et al., 2007). Such observations are consistent with relatively weak NBD:SBD interactions in the Hsc70 (Liu and Hendrickson, 2007) that can be modulated by binding of nucleotides or other ligands. The lability of these interactions allows Hsc70 to act as an allosteric machine, while the stability of the Hsp110 interdomain interactions may limit its allosteric flexibility. In this study we observe two locations for the Hsc70 SBD. One of these is affected by crystal packing contacts and is unlikely to be physiological. However, the position of the Hsc70 SBD in the EM reconstruction suggests that it could interact with the Hsp110 (Figure 4). The observed interactions between the linkers of the two proteins could also be functionally important. In particular, since the ATP-driven conformational changes in the Hsc70 involve allosteric changes in the linker and SBD, they could contribute to ATP-induced complex dissociation by disrupting interactions between the Hsc70 linker or SBD and Hsp110.
The structure of the Hsp110:Hsc70 complex also explains how ATP initially stimulates Hsp110 binding to Hsc70 and, subsequently, causes dissociation of this complex by binding to the Hsc70 NBD to cause the latter to close. The ability of Hsp110 to induce ADP release from Hsc70 is also explained by the effect of the Hsp110 interaction on stabilizing the open state of the Hsc70 NBD, which it is able to do even when the latter has ADP bound. Interestingly, while the Hsc70 NBD is open, we observe ADP bound to it in a configuration not previously seen in any Hsp70 NBD nucleotide complex. The interaction of the ADP appears specific, for while it retains only a subset of the H bonds to the NBD that have been observed in previously described Hsp70 NBD*ADP complexes, those H bonds that persist are identical to those seen in those previously described complexes (Flaherty et al., 1994), and even the H bond made by the Hsp70 ADP to Hsp110 S32 recapitulates the bond made to Hsp70 Q33 by the ADP in the Hsp110 NBD (Figures 6A–6C).
It is therefore likely that what is captured here is a prerelease complex: the Hsp110 has bound Hsc70*ADP and induced opening of the Hsc70 NBD, but the ADP has not yet dissociated. The Hsp110:Hsp70 complex does not dissociate when ADP is released (Andreasson et al., 2008), implying that release of the ADP proceeds through the proposed nucleotide entry/exit pore (Figure 6D). Subsequently, ATP binding to the Hsp70 to induce complex dissociation would require that ATP enter through the pore, whose electrostatic character (Figure 6E) may serve to recruit and orient the incoming nucleotide to increase the frequency of productive binding events.
The Hsp110:Hsp70 complex is not the only functional Hsp70 family heterodimer that has been reported. The Hsp70s in the yeast ER, Lhs1 (Grp170) and Kar2 (BiP), form a complex in which each acts as a NEF for the other (Steel et al., 2004). The coordination provided through this coupling is essential for the function of these chaperones, as mutations that eliminate this reciprocal activation have the same effect as deletion of the Grp170. It has been proposed that such reciprocal activation by pairs of Hsp70s may be a common feature of this group of chaperones (Meunier et al., 2002; Steel et al., 2004). The Hsp110:Hsc70 structure may provide a model for these widespread Hsp70 heterodimer interactions.
Experimental Procedures
Protein Preparation and Crystallization
Bovine Hsc70ΔC (aa 1–554) was expressed and purified as described (Jiang et al., 2005). Proteolysis of complexes with yeast Hsp110 (Sse1) revealed that the ∼ 30 C-terminal amino acids of the Hsp110 were protease sensitive, so we prepared a construct expressing aa 1–666 of the Hsp110 and linked at its N terminus by a tobacco etch virus (TEV) protease site to an 8-histidine tag. The tagged Hsp110 was expressed as described (Shaner et al., 2006) and purified on nickel resin. To remove adhering proteins the Hsp110 on the resin was extensively washed with 6 M urea, 50 mM Tris (pH 8.0), 1 M NaCl, and 0.1% Triton X-100, followed by washing and on-column cleavage with TEV protease in 50 mM Tris (pH 8.0). Following elution with the same buffer, complexes were formed by mixing the Hsp110 and Hsc70 with Hsc70 in excess to ensure saturation of the Hsp110 (1:1.5) and then dialyzing the proteins into 10 mM Tris (pH 8.0), 1 mM DTT, 1 mM EDTA, 50 mM KCl, and 5 mM MgCl2. Following dialysis, the sample was supplemented with ADP, BeCl2, and NaF to concentrations of 1 mM, 1.2 mM, and 6 mM, respectively, and concentrated to 20–40 mg/ml by ultrafiltration through membranes with a 100 kD MW cutoff. Crystals grew over 2–3 months by vapor diffusion against 0.2 M LiSO4, 40% PEG 400 at 4°C in sitting drops prepared by mixing complexes 1:1 with reservoir solution. Before data collection, crystals were soaked in reservoir solution + 15% glycerol and flash cooled in liquid nitrogen.
Analysis of Hsc70:Hsp110 Complex Formation and Clathrin Basket Dissociation Assays
Native PAGE of Hsp110:Hsc70 complex formation was as described (Shaner et al., 2006). Clathrin baskets (1 μM CHC) made from bovine clathrin were mixed with 0.1 μM bovine auxilin in 20 μM imidazole, 25 mM KCl, 10 mM (NH4)2SO4, 2 mM Mg acetate, and 3 mM DTT (pH 7.0) at 25°C. Dissociation was initiated by adding bovine Hsc70 (0.2–1 μM) preincubated with 1 mM ATP and monitored by DLS as described (Jiang et al., 2005). Rapid gel filtration analyses were performed in spin columns with superose 6, immediately after mixing the reactions at the indicated concentrations.
X-Ray Data Collection and Structure Determination
Data were collected at ALS beamline 4.4.2, on a NOIR-1 MBC detector. The crystal-to-detector distance was 160 mm; the oscillation range was 0.5° for a total of 139° of data. Data were processed to 3.1 Å using HKL2000 (Otwinowski et al., 2003), and the P21212 SG was determined by examination of systematic absences. Molecular replacement (MR) was performed with Molrep (Vagin and Teplyakov, 2000). Hsp110 NBD + SBDβ (residues 3–540 from 2QXL; Liu and Hendrickson, 2007) was used as the first search model. Once this domain was placed, it was fixed while the second model (bHsc70 NBD) from 1YUW (Jiang et al., 2005) (residues 1–384) was used. The R factor from MR was 0.516 with a CC of 0.475. Rigid-body refinement in Refmac (Murshudov et al., 1996) using each subdomain as a rigid element reduced the R/Rfree to 0.44/0.45. Electron density maps showed clear density for the missing bHsc70 SBD and Hsp110 SBDα domains, which were placed by hand using Coot (Emsley and Cowtan, 2004), which was also used for all model building. Simulated annealing, positional, and residue grouped B factor refinement in PHENIX (Adams et al., 2002) gave a final refined structure with an R/Rfree of 0.224/0.283.
Electron Microscopy and Image Processing
Complexes between Hsc70ΔC (Hsc70) and Hsp110 were prepared by incubating the proteins in a 1:1 molar ratio in 10 mM Tris (pH 8.0), 1 mM DTT, 5 mM MgCl2, 25 mM KCl, and 1 mM ADPBeFx. The complex was isolatedby size exclusion chromatography on Superose 6. Fractions were analyzed by SDS PAGE, and a few microliters of fractions containing the complex were adsorbed to carbon grids and negatively stained with 1% uranyl acetate. Micrographs at 50,000× were taken with a JEOL 1230 operating at 100 kV under low-dose conditions. To increase the angular sampling, images were obtained at 0° and 25° tilt. Micrographs were digitized using a ZEISS scanner, and particles were extracted using XMIPP software (Sorzano et al., 2004). Data from tilted and untilted samples were averaged to 2.8 Å per pixel and processed without any a priori assumption. Several volumes were prepared as starting templates for angular refinement in EMAN (Ludtke et al., 1999). First, images were classified into homogeneous groups using reference-free methods (refine2d.py command in EMAN and maximum-likelihood approaches implemented in XMIPP), and selected averages were used to build reference volumes using common lines. We also used artificial noisy ellipses and Gaussian blobs with the rough dimensions of the protein as starting models (Rivera-Calzadaet al., 2007). Projections from template volumes were confronted to the data set using angular refinement in EMAN (Sorzano et al., 2004). The different strategies converged to similar solutions, and one of the models was selected to complete the refinement. Similar methods were used to analyze the data from a complex formed between Hsp110 and the Hsc70 NBD. In this case, the 3D reconstruction of the complex between Hsp110 and bHsc70ΔC was also tested as starting reference for refinement. The resolution of the final structures was estimated by FSC (eotest command in EMAN) to be 22 and 21 Å (using the 0.5 cross-correlation coefficient criteria) for, respectively, reconstructions of Hsp110:Hsc70ΔC and Hsp110:Hsc70NBD. A difference map between these two reconstructions was calculated using proc3d after alignment of the two structures using the align3d command in EMAN and normalization to a comparable grayscale. The density maps and atomic structures were visualized with UCSF Chimera (Pettersen et al., 2004). Thresholds were chosen to account for approximately 100% of the protein mass. The atomic structure of the Hsc70ΔC:Hsp110 complex was manually fit within the EM density, and the handedness providing the best fit was chosen to render the EM reconstruction.
Analytical Ultracentrifugation and Bead Modeling
Sedimentation experiments were performed in a Beckman Optima XLI analytical ultracentrifuge. A 30 μM sample of Hsp110:Hsc70 in 1 mM ADP*BeF3, 5 mM MgCl2, 50 mM KCl, 20 mM sodium phosphate (pH 7.5) was scanned at 35,000 rpm and 20°C using interference optics. Data were analyzed using UltraScan 9.5.1 (Demeler, 2005) Time-invariant noise was removed with the 2D spectrum analysis (2DSA) (Brookes and Demeler, 2006), and diffusion-corrected sedimentation coefficient distributions were calculated with the enhanced van Holde-Weischet method (Demeler and van Holde, 2004). Parsimonious regularization of the 2DSA results and MW determinations were performed by genetic algorithm and Monte Carlo analysis (Brookes and Demeler, 2007). Partial specific volumes calculated using UltraScan were 0.7347 ccm/g for bovine Hsc70, 0.7355 ccm/g for Hsp110, and 0.7351 ccm/g for the complex. Bead-modeling was performed with HydroPro (Garcia De La Torre et al., 2000).
Supplementary Material
Acknowledgments
Supported by GM-52522, AQ-1486, and 0755057Y from the NIH, Welch Foundation, and AHA, respectively (to R.S.); NS029051 from NINDS (to E.M.L.); NIH T32AG021890-03 (to J.P.S.); AQ-1399 from the Welch Foundation (to P.J.H.); BFU2007-62382/BMC from MEC and the EU-grant “3D repertoire” (LSHG-CT-2005-512028) (to J.M.V.); and SAF2005-00775 from MEC (O.L.). Support for the X-ray Crystallography, Mass Spectroscopy, and UTHSCSA Center for Macromolecular Interaction cores from the UTHSCSA ERC and SACI (5P30CA5174) is acknowledged. We thank Jay C. Nix and Virgil Schirf for, respectively, X-ray and AUC data collection. To mark his becoming Emeritus, this paper is dedicated to Merle Olson, who had the vision of developing a structural biology program in San Antonio.
Footnotes
Accession Numbers: Coordinates and structure factors have been deposited in the RCSB Protein Data Bank with accession code 3C7N.
Supplemental Data: The Supplemental Data include four figures and one movie and can be found with this article online at http://www.molecule.org/cgi/content/full/31/2/232/DC1/.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Andréasson C, Fiaux J, Rampelt H, Mayer MP, Bukau B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J Biol Chem. 2008;283:8877–8884. doi: 10.1074/jbc.M710063200. [DOI] [PubMed] [Google Scholar]
- Brehmer D, Rudiger S, Gassler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 2001;8:427–432. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- Brookes E, Demeler B. Genetic Algorithm Optimization for obtaining accurate Molecular Weight Distributions from Sedimentation Velocity Experiments. Vol. 137. New York: Springer; 2006. [Google Scholar]
- Brookes E, Demeler B. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments. Paper presented at GECCO Proceedings ACM 978-1-59593-697-4/07/0007.2007. [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Deloche O, Georgopoulos C. Purification and biochemical properties of Saccharomyces cerevisiae's Mge1p, the mitochondrial cochaperone of Ssc1p. J Biol Chem. 1996;271:23960–23966. doi: 10.1074/jbc.271.39.23960. [DOI] [PubMed] [Google Scholar]
- Demeler B. UK: Royal Society of Chemistry; 2005. A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments. UltraScan (ver. 9.5.1; http://www.ultrascan.uthscsa.edu) [Google Scholar]
- Demeler B, van Holde KE. Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. J Biol Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LE, Eisenberg E. Dissociation of clathrin from coated vesicles by the uncoating ATPase. J Biol Chem. 1990;265:6682–6687. [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hendrickson WA. Insights into hsp70 chaperone activity from a crystal structure of the yeast hsp110 sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Staeck O, Rapoport TA. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- Murshudov G, Vagin A, Dodson E. Refinement of protein structures. Paper presented at Proceeding of Daresbury Study Weekend.1996. [Google Scholar]
- Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- Packschies L, Theyssen H, Buchberger A, Bukau B, Goody RS, Reinstein J. GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry. 1997;36:3417–3422. doi: 10.1021/bi962835l. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56–62. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J Biol Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Sousa R, Lafer EM. Keep the traffic moving: mechanism of the Hsp70 motor. Traffic. 2006;7:1596–1603. doi: 10.1111/j.1600-0854.2006.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kurochkin AV, Pang Y, Hu W, Flynn GC, Zuiderweg ER. NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction. Biochemistry. 1998;37:7929–7940. doi: 10.1021/bi9800855. [DOI] [PubMed] [Google Scholar]
- Wei J, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- Young JC, Barral JM, Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28:541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


