Abstract
The transcription factor spalt4 is a key early-response gene in otic placode induction. Here, we characterize the cis-regulatory regions of spalt4 responsible for activation of its expression in the developing otic placode and report the isolation of a novel core enhancer. Identification and mutational analysis of putative transcription factor binding sites reveal that Pea3, a downstream effector of FGF signaling, and Pax2 directly activate spalt4 during ear development. Morpholino-mediated knock-down of each factor reduces or eliminates reporter expression. In contrast, combined over-expression of Pea3 and Pax2 drives ectopic reporter expression, suggesting that they function synergistically. These studies expand the gene regulatory network underlying early otic development by identifying direct inputs that mediate spalt4 expression.
Keywords: chick, Sall4, otic induction, inner ear, FGF8
Introduction
Ectodermal placodes are transient regions of thickened ectoderm of the head that contribute to the ear, nose, lens and cranial ganglia (Baker and Bronner-Fraser, 2001). Of these, the best studied is the otic placode, which is induced adjacent to the hindbrain by signals emanating from flanking tissues (Dominguez-Frutos et al., 2009; Kil et al., 2005; Kwon and Riley, 2009; Ladher et al., 2005; Park and Saint-Jeannet, 2008). Signals such as FGFs (Leger and Brand, 2002; Maroon et al., 2002; Vendrell et al., 2000; Wright and Mansour, 2003) Wnts (Freter et al., 2008; Ohyama et al., 2006) and BMPs (Kwon and Riley, 2009) are involved in otic placode induction (Martin and Groves, 2006) and activate specific patterns of gene expression (Litsiou et al., 2005). For example, over-expression of FGFs induces ectopic otic-like structures (Alvarez et al., 2003; Kil et al., 2005; Vendrell et al., 2000), while mutations in FGFs lead to defects in ear development (Alvarez et al., 2003; Ladher et al., 2005). Following induction, the columnar placode invaginates to form the otic vesicle, which subsequently differentiates into the complex inner ear, including the cochlea, vestibular system and endolymphatic sac.
The transcription factor spalt4, homolog of human SALL4 (Sweetman and Munsterberg, 2006), displays the correct localization pattern to be a key early-response gene in placode induction in the chick (Barembaum and Bronner-Fraser, 2007). It is initially expressed uniformly throughout the head ectoderm, overlapping with Six-Eya-Dach in the preplacodal domain. It then resolves to the presumptive otic and olfactory placode regions by stage 10, as non-placodal ectoderm loses competence to form otic placode (Baker and Bronner-Fraser, 2001; Groves and Bronner-Fraser, 2000). Expression of spalt4 in non-placodal ectoderm is sufficient to induce invagination or ingression and expression of a number of otic genes (Barembaum and Bronner-Fraser, 2007). Interestingly, the effects of its gain- and loss-of-function resemble those of FGF over-expression and mutations, respectively. This raises the intriguing possibility that spalt4 may be downstream of FGF and other inductive signals.
Numerous transcription factors in addition to spalt4, including Pax2 (Mackereth et al., 2005) Dlx3 (Esterberg and Fritz, 2009) and Dlx5 (Brown et al., 2005; Robledo and Lufkin, 2006), have been found to play important roles in otic development (Baker and Bronner-Fraser, 2001). Although the position and function of a few of these regulators are documented in otic development, their order, interrelationship and direct or indirect nature of their interactions are not yet known and currently under investigation (Esterberg and Fritz, 2009; Hans et al., 2007).
To better understand important gene regulatory interactions underlying ear development, we set out to characterize the cis-regulatory regions of spalt4 based on its key position in otic placode induction. We report the isolation and dissection of a novel spalt4 regulatory module, responsible for activation of spalt4 expression in the developing otic placode. We further interrogate this regulatory region to identify putative transcription factor binding sites and upstream regulators controlling its expression. The results reveal that Pea3, a downstream effector of FGF signaling, and Pax2 directly activate spalt4 during ear development. These studies expand our knowledge of known transcription factors and their direct interactions during development of the ear.
Materials and Methods
Cloning
Regions of non-coding genomic DNA in the vicinity of the spalt4 (Sall4) coding region were compared between chick and other vertebrates. Conserved regions were amplified from the BAC clone CH261-71D2 (BPRC, Oakland Research Institute, Oakland CA) and cloned into the pTK vector in front of a minimal TK promoter driving GFP expression (Uchikawa et al., 2004). Mutations were made by fusion PCR (Heckman and Pease, 2007). For over-expression experiments, the protein coding regions of chicken Pea3 and Pax2 were amplified by RT-PCR and cloned into the pCIG vector. Cerulean fluorescent protein was cloned into the pCAGS expression vector.
Electroporation
Stage 4–6 embryo were collected on Whatman filter rings and placed ventral side up in an electoporation chamber, with negative electrode at the bottom. Plasmid DNA, either at 1 mg/ml for enhancer constructs or 2 mg/ml for over expression constructs, was injected through the blastoderm into the space between the embryo and the vitelline membrane. The positive electrode was placed above the embryo and an electric current was applied of 4 pulses of 7 volts, 50 msec in duration with a 100 msec pause in between pulses. After electroporation, embryos were transferred ventral side up to a 35 mm dish with a thin layer of egg albumin at the bottom and incubated in a humidified incubator at 37° C for 24 h (Sauka-Spengler and Barembaum, 2008). Those embryos with high levels of fluorescence, indicating efficient eletroporation, were fixed in 4% formaldehyde overnight. Morpholino oligos were obtained from Gene-Tools (Philomath, OR) and dissolved in water at 1 mM or 3 mM concentrations. Plasmid DNA (pUC19) was added at a final concentration of 100 ng/µl prior to injecting into embryos. In order to electroporate the morpholino at 3 mM, the embryos were first electroporated with the enhancer construct then followed by the morpholino. The fluoresceinated morpholino oligos were made with the following sequence:
Pea3: 5’-CTG CTG GTC CAC GTA CCC CTT CAT C-3’
Pax2: 5’-GTC TGC CTT GCA GTG CAT ATC CAT G-3’
Control: 5’-CCT CTT ACC TCA GTT ACA ATT TAT A-3’
Implantation of beads
Stage 4 embryos were collected on Whatman filter paper rings and turned ventral side up in Ringer’s solution and electroporated as described above. A small slit was made in the area opaca next to the area pellucida (Litsiou et al., 2005). A bead soaked in 50 ug/ml Fgf8 (R&D Systems, Minneapolis MN) or BSA (bovine serum albumin) was inserted into the slit and incubated in modified New culture (Chapman et al., 2001) for 5–7 hours and then collected and fixed overnight in 4% paraformaldehyde.
Analysis of embryos
Embryos were collected in Ringer’s solution and fixed in 4% paraformaldehyde overnight. Embryos were washed in PBT and embedded in gelatin for histochemical analysis or dehydrated in methanol for in situ hybridization. In situ hybridization was performed as described previously (Wilkinson, 1992). Antibodies to GFP (Abcam), Pax2 (Zymed), and fluorescein (Roche) were obtained commercially. Primary antibodies were visualized with Alexa Fluor 488-conjugated donkey anti-goat or Alexa Fluor 594-conjugated donkey anti-rabbit secondary antibodies (Molecular Probes). Cerulean fluorescent protein expression was distinguished from fluorescein signal using a Zeiss 510 META 2 inverted microscope.
Results
Dissection of putative spalt4 regulatory region
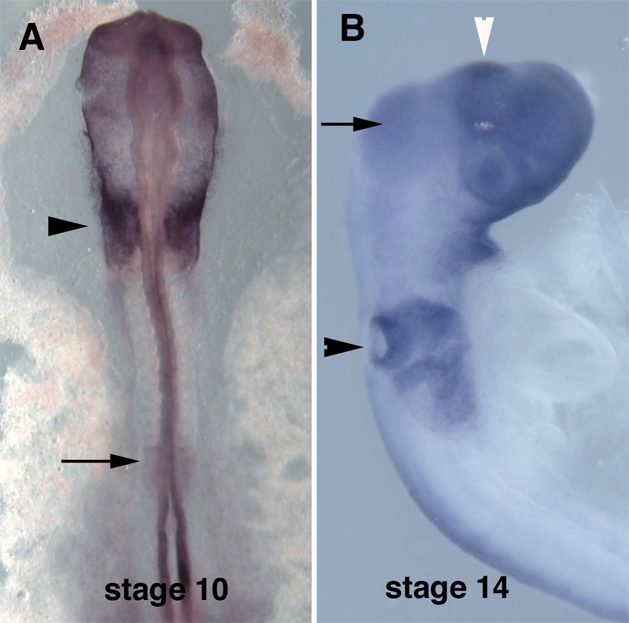
Spalt4 is initially expressed throughout the preplacodal domain but resolves to the otic placode by stage 9–10 (Fig. 1A) (Barembaum and Bronner-Fraser, 2007). By stage 14, it is robustly expressed in the otic pit and maintained during formation of the otic vesicle (Fig.1B). We set out to identify enhancer elements capable of driving reporter expression that recapitulated this spatiotemporal expression pattern.
Figure 1.
In situ hybridization of chicken embryos with spalt4 RNA probe. A. Stage 10 embryo showing signal in the otic placode (arrowhead) and presomitic mesodem (arrow). B. Stage 14 embryo showing signal in the otic pits (black arrowhead) and midbrain (arrow). The region that will give rise to the epiphysis also expresses spalt4 (white arrowhead). Other forebrain staining is background.
To identify the cis-regulatory elements responsible for spalt4 expression in developing ear, we probed the genomic region surrounding the spalt4 gene using comparative genomic analysis to isolate highly conserved genomic regions with putative regulatory activity. For our study, non-coding genomic regions in the upstream of the spalt4 coding region and the first intron were compared in silico using the UCSC gene browser (Fig. 2A) (Karolchik, et al., 2008). Fragments 1 kb to 5 kb in size containing the putative regulatory regions with the highest homology were amplified from a chicken BAC clone and cloned into an EGFP reporter vector containing a thymidine kinase basal promoter (Uchikawa et al., 2004).
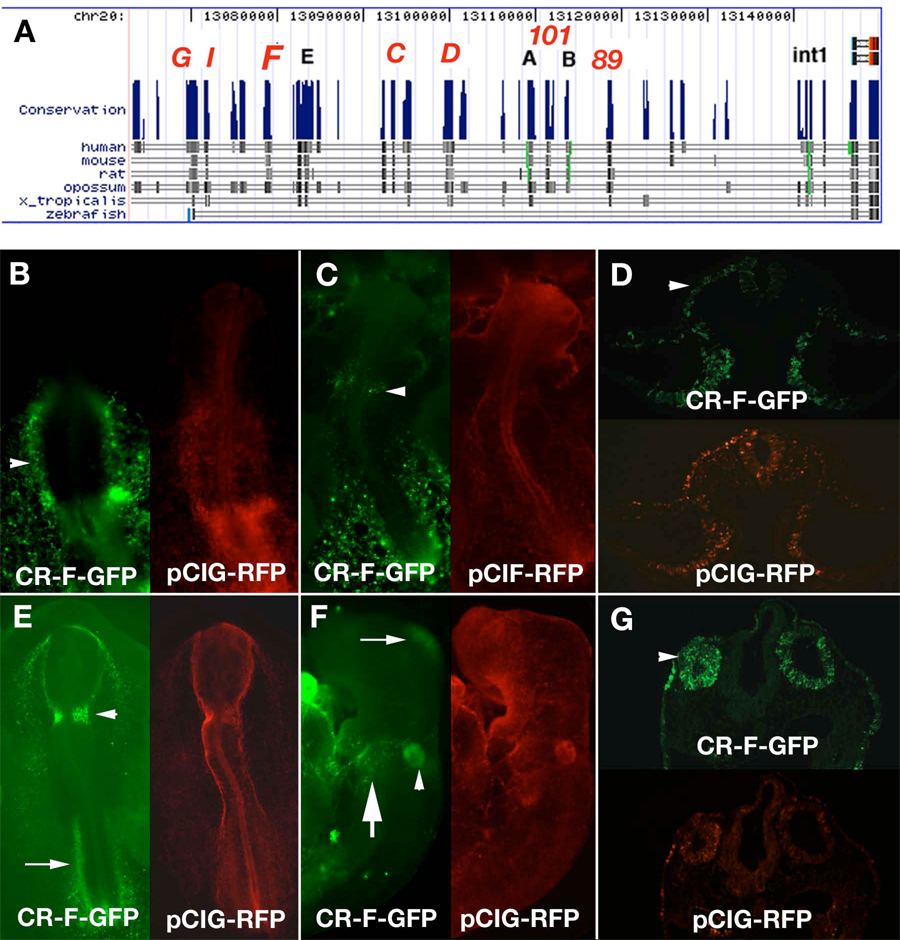
Figure 2.
Isolation of conserved regions adjacent to chicken spalt4. A. Map of the region adjacent to spalt4. The peaks show regions of conservation. The dark bands below each peak show the regions conserved between chicken and each of the other species examined. The red letters above each peak identify the regions that were cloned. Spalt4 coding region is at the extreme right of the map. B–G. Enhancer activity of conserved region F (CR-F). B. At stage 9, CR-F drives expression in the lateral plate mesoderm (arrowhead). C. At stage 10 CR-F drives GFP in the otic placode (arrowhead). D. A section through a stage 10 embryo electroporated with CR-F shows that the otic placode (arrowhead) and the lateral plate mesoderm express GFP. E. Stage 12 embryo with GFP in the otic pits (arrowhead). F. Stage 14 embryo shows GFP expression in the otic vesicle (arrowhead), epibranchial placodes (large arrow) and the midbrain small (arrow). G. Section through a stage 14 embryo showing GFP expression in the otic vesicles (arrowhead). Each embryo was co-electroporated with pCIG-RFP as an electroporation control.
The spalt4 gene is located on chicken chromosome 20 between genes ZFP64 and ATP9A. Because the May 2006 version of the chicken genome did not contain the sequence of the first exon, we cloned the DNA corresponding to a gap in the sequence from a BAC using GC rich PCR. This region contained the first exon of spalt4. At the 5’ end there is over 100 kb of sequence between spalt4 and ZFP64. We cloned several conserved regions upstream of the coding region, as well as in the first intron, which induced GFP expression in various regions of the embryo. These were denoted Conserved Region (CR) A–I (Fig. 2A).
To test if the conserved genomic regions were functional in vivo, they were individually electroporated into stage 4 chick embryos using an ex ovo technique (Uchikawa et al., 2004). Electroporation efficiency was assessed by co-electroporating a pCIG mRFP plasmid construct under control of a ubiquitous promoter. Embryos were collected 24–48 hrs (stages 10–20) after electroporation, fixed and analyzed for EGFP expression using fluorescence microscopy. Of the genomic regions analyzed, 13 had some enhancer activity, described below, though only CR-F had expression in the otic placode (Fig. 2B–G).
Identification of spalt4 genomic fragment with regulatory activity in the developing otic placode and other embryonic locations
The 1.3 kb fragment CR-F was conserved between chicken and mammals, but displayed no conservation with Xenopus or zebrafish. When electroporated into chicken embryos, CR-F drove GFP expression in the otic placode at stage 10 (Fig. 2C,D) with continued to be expression in the otic pit (Fig. 2E) and later in the otic vesicle (Fig. 2F,G) at least until stage 17. GFP was observed in the otic region as well as the contiguous lateral ectoderm that will give rise to the epibranchial placodes (Fig 2F,G). In addition it drove expression in the lateral plate mesoderm (Fig. 2B–E) and midbrain (Fig. 2F). The GFP in lateral plate mesoderm, which includes both the somatic and splanchnic mesoderm was expressed at least as early as stage 8 and included the heart field (Fig. 2A). The expression of GFP then became restricted caudally, so that it was only seen in the caudal trunk region at stage12 (Fig. 2E), but not in the developing heart. after stage 9 (Fig 2B).
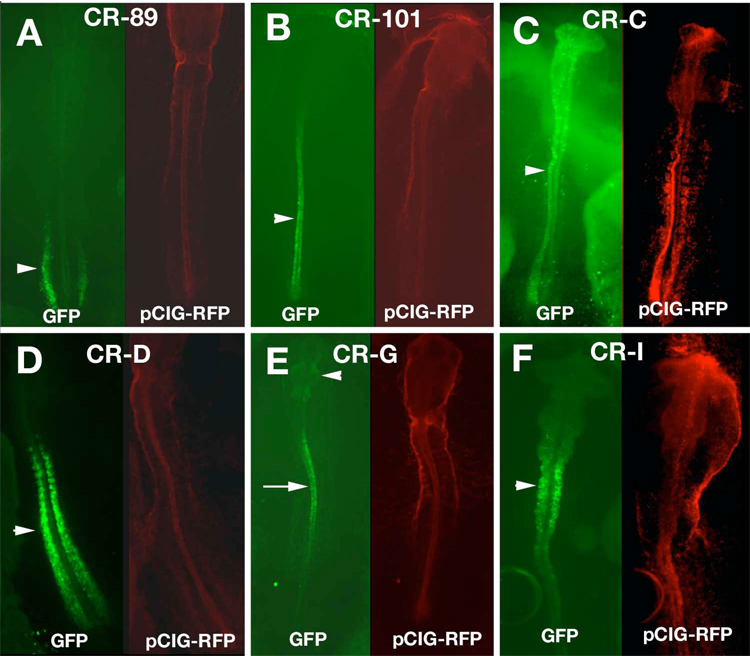
CR-A, CR-B, CR-E and CR-H induced GFP expression only in the blood islands, at the times and regions tested (data not shown). Two conserved regions besides CR-F drove expression of GFP in the trunk mesoderm. CR-89 was expressed in the intermediate mesoderm (Fig. 3A) and later in the developing pronephros. CR-C induced strong GFP expression in the presomitic mesoderm and the caudal somites (Fig. 3D). Several other conserved regions displayed complex expression patterns in the neural tube. CR-101 drove expression in the caudal neural tube, including the future hindbrain at stage 10, but only in the spinal cord at stage 12 and later (Fig. 3B). CR-C induced GFP throughout the neural tube (Fig. 3C) until stage 12 after which time GFP expression was reduced. CR-G expressed GFP in the midbrain, rostral hindbrain and rostral spinal cord, as well as in the presomitic mesoderm (Fig. 3E). In sections, only the ventral neural tube had GFP expression (data not shown). The most distal element, CR-I drove expression of GFP in the neural tube, though significantly higher expression was found in the hindbrain (Fig. 3F). Thus, much of the embryonic expression of spalt4 was recapitulated by the combined activity of the isolated enhancers. However, we were unable to detect a neural crest enhancer.
Figure 3.
Enhancer activity in other mesoderm and neural tube. A. Conserved region 89 (CR-89) drove GFP expression in the intermediate mesoderm (arrowhead). B. CR-101 in the caudal CNS (arrowhead). C. CR-C in the neural tube (arrowhead). D. CR-D in the presomitic mesoderm and the somites (arrowhead) E. CR-G in the midbrain (arrowhead) and the spinal cord (arrow). F. CR-I drove expression most strongly in the hindbrain (arrowhead). Each embryo was co-electroporated with pCIG-RFP as an electroporation control.
Dissection of the CR-F fragment reveals a minimal enhancer region
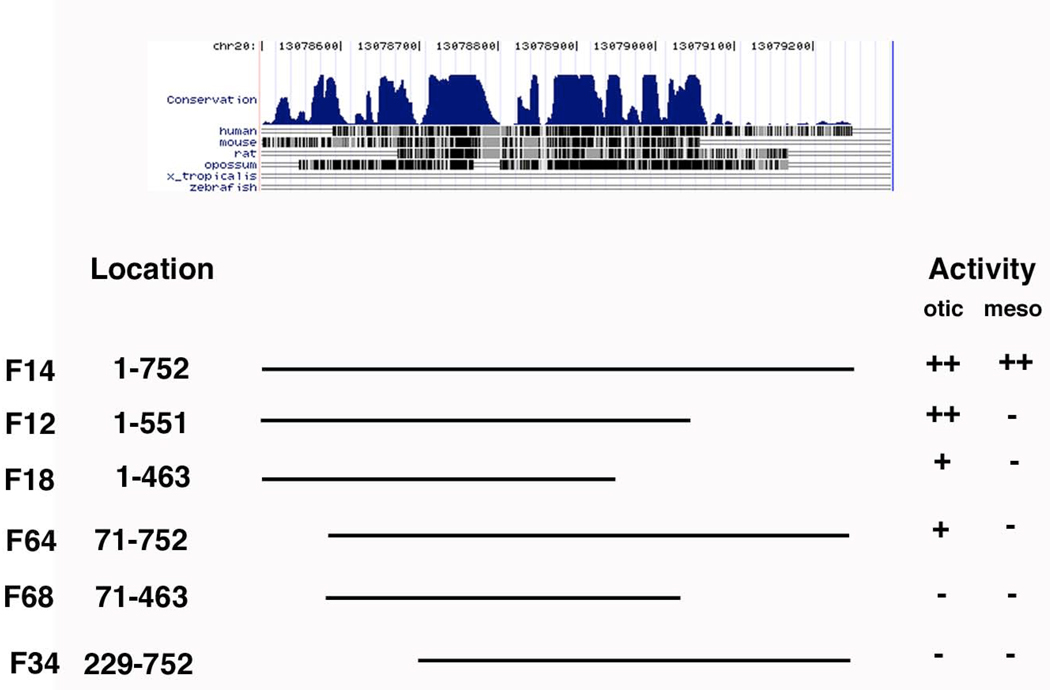
To better understand regulatory interactions underlying ear development, we further interrogated the CR-F fragment to determine a minimal otic enhancer element. For this purpose, we used PCR to generate a number of deletion constructs (Fig. 4). A highly conserved 752 bp subfragment (F14) retained both otic and mesodermal expression, similar to intact CR-F. Removing the 5’ 71 base pairs reduced GFP level in the otic placode and eliminated lateral mesoderm expression. Removing the 200 base pairs at the 3’ end reduced the mesoderm expression. Removing an additional 88 base pairs reduced the otic expression and eliminated the mesodermal expression. Loss of both the 5’ 71 base pairs and 3’ 200 base pairs eliminated all expression.
Figure 4.
Deletions made to CR-F. The level of conservation is seen at the top. Each deletion is identified by its location in the sequence in Figure 5. They were qualitatively assessed for their ability to drive expression in the otic placode (otic) or lateral plate mesoderm (meso) to determine whether they had the same level as the the full length (F14) construct, ++, had a reduced level, +, or an undetectable level (−).
The minimal fragment found to drive strong otic expression was 551 bp in length. Otic expression appears to depend on several regions spread along the length of this fragment. To determine putative transcription factor binding sites that could be tested by mutational analysis, we turned to a bioinformatics analysis.
Identification of transcription factor binding sites in CR-F
Examination of the 752 bp conserved sequence of CR-F using the JASPAR database (Wasserman and Sandelin, 2004) revealed a number of putative transcription factor recognition sites (Fig. 5). Of those most relevant to otic development, we found several potential Pax2 binding sites, as well as a number of putative Ets transcription factor binding sites, several of which are recognized by Pea3 type family members, known to act as downstream effectors of FGF signaling (Raible and Brand, 2001; Roehl and Nusslein-Volhard, 2001). Since Pax2 (Groves and Bronner-Fraser, 2000) and Pea3 (Lunn et al., 2007) both are expressed early in otic development, they represented good candidates for regulating expression of spalt4 in the otic placode.
Figure 5.
The sequence of CR-F14. Some of the consensus transcription factor recognition sites are shaded for identification.
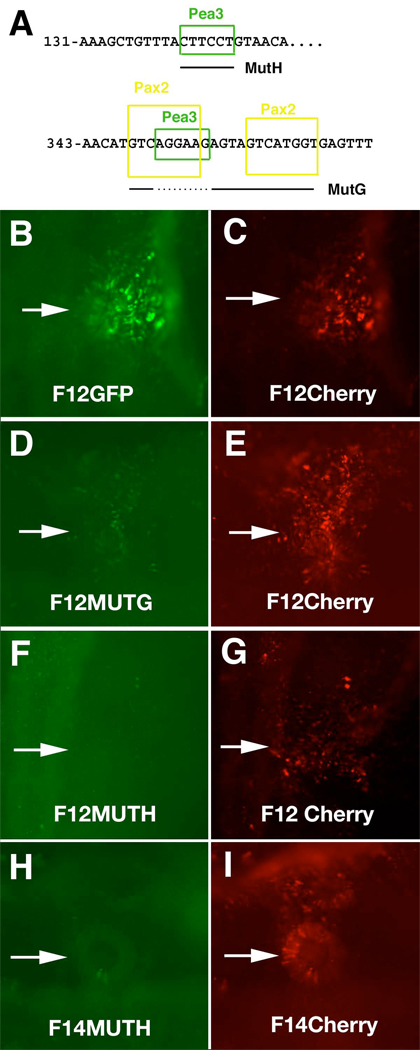
To test whether the putative Pax2 and the Pea3 binding sites are necessary for enhancer activity, a number of mutations were made in a minimal enhancer element by replacing the putative binding sequences by heterologous GFP sequences of the same size. The mutations were performed in the deletion construct CR-F12 since it maintains otic expression, though the mesodermal enhancer activity is reduced. We targeted both remaining Pea3 sites as well as the two Pax2 sites (fig. 6A). Mutation of the upstream putative Pea3 site (MutH, nucleotides 142–147 in Fig. 5) resulted in loss of otic enhancer activity (Fig 6F–I). We noted a reduction of otic enhancer activity by mutating the Pax2 sites (MutG, 348–358), but not the intervening Pea3 site (351–358) (Fig. 6D,E). The requirement for both the upstream Pea3 site and the Pax2 sites suggests a possible synergistic interaction between these two sites. If the F14 (1–752) construct has the mutation that removes both of the Pax2 sites and the intervening Pea3 site, otic enhancer activity is retained (data not shown). This may be due to the presence of additional transcription binding sites that also have activity. We determined that he Pea3 site is required in the larger fragment (Fig. 6H,I). Interestingly, the enhancer activity in the midbrain is unaffected by mutating the Pea3 site (data not shown). Mutations to other putative recognition sites, Otx and Tcf/Lef, have little or no effect on otic enhancer activity. However, these sites may be important for expression in other regions or at times other than those examined.
Figure 6.
Mutational analysis of CR-F12. A. Two different mutation constructs were made. MutH had a mutation in the Pea3 site located at 142–147. MutG mutated both Pax2 sites between 348 and 368 and the Pea3 site at 351–356 was not mutated (dotted line under the sequence). Embryos were co-electroporated with the mutant constructs driving GFP (D,F,H) and non-mutant constructs driving Cherry fluorescent protein (C, E, G,I) as controls. Arrows point to the otic placode. B. Embryo electroporated with the non-mutated CR-F12 driving GFP had otic expression. C. Embryo in (B) showing comparable levels of Cherry. D. MutG construct showing much reduced levels of GFP in the otic compared to E. F. F12MutH construct had undetectable levels of GFP in the otic while the control had normal levels G. H. F14MutH had undetectable levels of GFP in the otic, while the F14 control had normal levels I.
Additionally, using the CR-F14 construct that contains the full conserved region, we have identified a putative Tbx binding site (512–520) that is required for enhancer activity in the lateral plate mesoderm. Mutating this region resulted in the loss of enhancer activity in the lateral plate mesoderm (data not shown).
Ectopic FGF, Pea3 or Pax2 induce CR-F expression
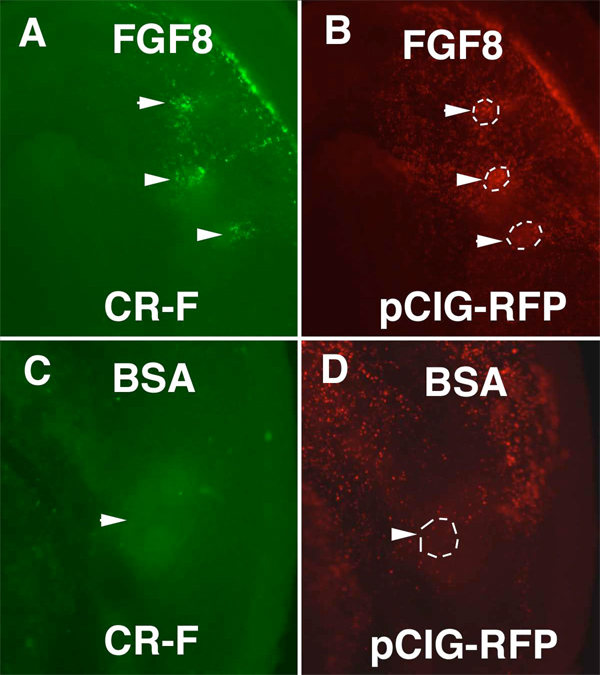
The Ets transcription factors Pea3 and Erm are both expressed in the otic placode and are part of the FGF signaling pathway (Lunn et al., 2007). To test if CR-F could be activated by FGF, we inserted beads soaked in FGF8 in the area opaca adjacent to the area pellucida in stage 4 embryos that had been previously electroporated with a CR-F pTK construct. In these embryos, the cells surrounding the beads expressed GFP, indicating that FGF can induce the activity of the CR-F enhancer (Fig. 7). Previously, in situ hybridization revealed spalt4 transcripts were induced in the cells surrounding FGF coated beads (Barembaum and Bronner-Fraser, 2007). Electroporation of Pea3 over-expression construct together with the CR-F Cherry reporter caused ectopic Cherry expression co-localized in the extraembryonic ectoderm with ectopic Pea3 expression (Fig 8A,B). However, Pea3 overexpressing cells in the embryonic ectoderm did not induce enhancer activity over the embryo. Thus the ability of Pea3 to induce enhancer activity is limited.
Figure 7.
FGF8 induced CR-F activity. Embryos were electroporated with CR-F-pTK and pCIG-RFP. Beads soaked in FGF8 (A,B) or BSA (C,D) were implanted in the area opaca. A. Ectoderm above the FGF8 soaked beads (arrowheads) contained GFP. B. Same embryo as in (A) showing broad RFP as an electroporation control. The positions of the beads are outlined. C. Ectoderm above a BSA soaked bead (arrowhead) had no visible GFP positive cells. D. Same embryo as in (C) showing the cells above the bead were electroporated (arrowhead). The position of the bead is outlined.
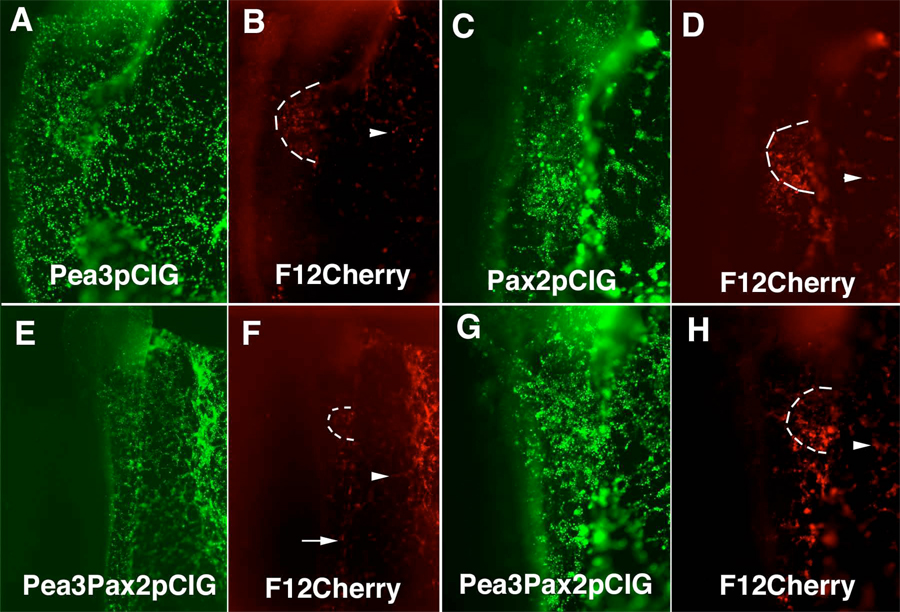
Figure 8.
Pea3 and Pax2 can induce CR-F activity in the extraembryonic ectoderm and together can induce CR-F activity throughout the embryo. Embryos co-electroporated with Pea3pCIG and CR-F12 cherry (A,B), Pax2pCIG and CR-F12 Cherry (C,D) and Pea3pCIG, Pax2pCIG and CR-F12 cherry (E–H). A. Pea3 expressed throughout the embryo. B. Cherry driven by CR-F was expressed in the otic placode and in the extraembryonic ectoderm (arrohead). C. Pax2 was expressed throughout the embryo. D. Cherry driven by CR-F was expressed in the otic placode and the extraembryonic ectoderm (arrowhead). E. G. combined expression of Pea3 and Pax2 throughout the ectoderm. F. H. Cherry driven by CR-F was detected in the trunk ectoderm (arrow) as well as the extraembryonic ectoderm (arrowhead). (E) and (F) are lower magnification pictures of the embryo in (G) and (H). The otic placode is outlined in (B), (D), (F), and (H).
To further study the effect of Pax2 on CR-F induction in the ear, we elecroporated a Pax2 expression construct and a pTK construct using CR-F12 driving the expression of Cherry fluorescent protein in the ectoderm. In these experiments, the reporter expression reflected the extent of Pax2 misexpression in the extraembryonic ectoderm (Fig. 8C,D). We also noted ectopic Cherry expression in the caudal hindbrain ectoderm, but did not detect cherry fluorescent protein in the rostral head or in the trunk ectoderm. Ectopic Pax2 can thus drive ectopic CR-F induction of GFP only in limited regions of the embryo. In situ hybridization with a spalt4 mRNA probe showed that these regions with ectopic Pax2 and CR-F12 driven cherry expression also expressed spalt4 transcript (Supplementary Figure 1A).
Since the mutation experiments show that both Pax2 and Pea3 binding sites are required for CR-F12 activity, we co-electroporated Pax2 and Pea3 expression constructs along with the reporter. This resulted in Cherry expression in cells expressing Pax2 and Pea3 at all rostrocaudal levels, even in the trunk ectoderm that does not normally form sensory placode cells (Fig. 8E–H). The combination of Pax2 and Pea3 misexpression yielded much broader enhancer activity than the sum of the effects of Pax2 and Pea3 overexpression individually, demonstrating synergism between Pax2 and Pea3 in CR-F enhancer activity.
Effects of Pea3 and Pax2 antisense morpholinos on CR-F expression and endogenous ear development
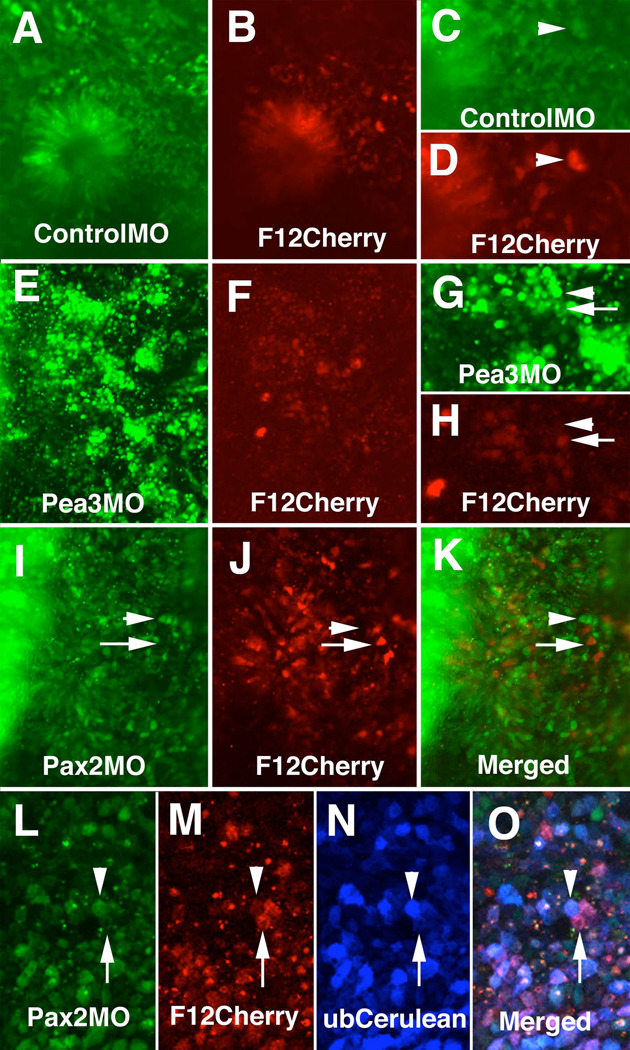
To study the loss-of function effect of Pea3, we used a morpholino oligomer to knockdown expression of Pea3 in the otic placode. Electroporation of 1 mM Pea3 morpholino, directly tagged with fluorescein, along with the CR-F12 enhancer construct resulted in the profound loss of enhancer activity (Fig. 9E–H) as well as defects in ear development. In contrast, embryos electroporated with control morpholino plus CR-F12-Cherry retained enhancer activity (Fig A–D) and normal appearing otic placodes.
Figure 9.
Morpholino oligos to Pax2 and Pea3 reduce CR-F activity in the otic placode. A. Fluorescein in embryo treated with 3 mM control morpholino. B. Embryo in (A) showing CR-F enhancer activity driving Cherry fluorescent protein expression. C. Higher magnification of (A) showing cells with high levels of control morpholino (arrowhead). D. Same region as in (C) showing the cells with high levels of control morpholino had high levels of CR-F activity (arrowhead). E. Fluorescein in embryo treated with 1 mM Pea3 morpholino. F. Embryo in (E) showing CR-F12 enhancer activity driving Cherry fluorescent protein expression. G. Higher magnification of (A) showing cells with either high levels of Pea3 morpholino (arrowhead) or low levels (arrow). H. Same region as in (G) showing the cells with high levels of Pea3 morpholino had low levels of CR-F activity (arrowhead) and cells with low levels of morpholino had high levels of CR-F activity (arrow). I. Fluorescein in embryo treated with 3 mM Pax2 morpholino. J. Embryo in (I) showing CR-F enhancer activity driving Cherry fluorescent protein expression. K. Merged image of (I) and (J). Arrows in (I–K) show cells with high enhancer activity and low levels of fluorescein tagged Pax2 morpholino. Arrowheads point to cells with high morpholino levels and low enhancer activity. L-O. Embryo electroporated with F12Cherry enhancer reporter construct and Cerulean ubiquitous expression vector followed by a second electroporation with fluorescein tagged Pax2 morpholino and imaged with a Zeiss 510 META inverted microscope. L. Fluorescein signal showing cells that contained the Pax2 morpholino. M. Cherry signal showing cells that have F12 enhancer activity. N. Cerulean signal showing the cells that were electroporated. O. Merged image of (L–M). Arrowheads in (L–O) point to a cell that had high levels of fluorescein tagged morpholino had little enhancer activity though still expressing Cerulean. Arrow points to a cell with high level of enhancer activity and Cerulean expression, but had low levels of fluorescein tagged morpholino.
The morpholino to Pax2 was effective at 3 mM concentrations, resulting in a reduction of Pax2 protein as assayed of using an anti-Pax2 antibody. Cells containing high levels of morpholino, as determined by the amount of fluorescein signal, did not contain Pax2 protein (data not shown). As expected, Pax2 morpholino resulted in some defects in ear development. The placodes on the morpholino-electroporated side were much thinner than the untreated side, and failed to invaginate. We noted that electroporations of 3 mM morpholino resulted in a decrease of enhancer activity in control and experimental embryos (data not shown), likely due to the high morpholino concentration reducing the efficiency of plasmid transfection. To circumvent this problem, we electroporated the enhancer construct first, followed by the morpholino electroporation. Cells that received high levels of Pax2 morpholino lacked strong F12 enhancer activity (Fig. 9I–K). Since not every cell that is transfected with the morpholino is also transfected by the enhancer construct we co-electroporated a ubiquitous Cerulean expression construct with the F12 enhancer reporter followed by a second electroporation with the fluorescein tagged Pax2 morpholino. We were able to detect that cells that had Cerulean expression and fluorescein labeling had no expression from the F12 enhancer reporter. We also found that most cells expressing Cerulean and Cherry did not have high levels of fluorescein tagged Pax2 Morpholino (Fig. 9L–O). With control morpholino, however, many cells with high levels of control fluorescein tag had high F12 enhancer activity (Fig. 9A–D). Thus, cells that contain high levels of Pax2 morpholinos, lacking Pax2 protein, also lack enhancer activity, whereas cells with lower levels of Pax2 morpholino, as seen by lower fluorescein levels, had some reporter expression. Spalt4 in situ hybridization also show reduced expression in the otic placode of the morphant embryos (Supplementary Fig. 1B). These results are consistent with our experiments with the mutated Pax2 sites that show that Pax2 is necessary for CR-F enhancer activity in the ear.
DISCUSSION
In depth analysis of the conserved regulatory regions of a key early-response gene in placode induction has provided important information regarding the gene regulatory inputs involved in chick otic development. By dissecting the chick spalt4 enhancer, we show that the synergistic input of two transcription factors, Pea3 and Pax2, drives spalt4 expression in the developing otic placode. Spalt4, which itself plays a role in otic formation, is expressed early in the placode and later in the otic vesicle. Like FGFs, ectopic spalt4 is sufficient to cause non-placodal ectoderm to invaginate and form vesicles that express many of the genes characteristic of the ear (Barembaum and Bronner-Fraser, 2007).
Precursors for all the paired sensory placodes, including the otic placode, are derived from the preplacodal domain, a horseshoe shaped region surrounding the anterior neural plate and marked by Six-Eya gene expression (Schlosser, 2006; Streit, 2007). Later, FGF signaling induces ectodermal cells within the preplacodal region to form otic placode (Martin and Groves, 2006). The source of inductive signals appears to emanate from both the hindbrain (Kil et al., 2005; Park and Saint-Jeannet, 2008) and underlying mesoderm (Kwon and Riley, 2009; Ladher et al., 2005). Studies in several species show that FGFs have the proper spatiotemporal distribution to be involved in otic placode induction (Schimmang, 2007). In the chick, FGF8 (Ladher et al., 2005) and FGF19 (Ladher et al., 2000) are expressed in the mesoderm and FGF3 in the hindbrain (Mahmood et al., 1996). Furthermore, adding ectopic FGF3, 19 and 8 can induce ectopic vesicles and expand the otic vesicles (Kil et al., 2005; Vendrell et al., 2000). Inhibition of FGF signaling blocks Pax2 expression (Martin and Groves, 2006) and knockdown of FGF8 or FGF3 reduces or eliminates the otic placode (Ladher et al., 2005; Zelarayan et al., 2007). FGF signaling activates the Ets genes, Pea3 and Erm, through the action of the MAP kinase ERK (Raible and Brand, 2001; Roehl and Nusslein-Volhard, 2001; Firnberg and Neubuser, 2002). Both Pea3 and Erm transcription factors are expressed in the otic placode (Lunn et al., 2007), and thus may be involved in transcriptional control of downstream genes.
Similarly, genes of the Pax2/5/8 family are expressed in the developing otic placode (Groves and Bronner-Fraser, 2000; Lawoko-Kerali et al., 2002)(Pfeffer et al., 1998). In mice, Pax8 is expressed earliest followed by Pax2. In zebrafish, loss of Pax8 leads to defects in otic induction that are enhanced by also reducing FGF signaling; furthermore, loss of Pax8 combined with loss of Pax2a and Pax2b results in loss of the otic vesicle (Mackereth et al., 2005). In zebrafish, Pax 2 and Pax8 may be downstream of Foxi1 and Dlx3b (Hans et al., 2004). However, knockout mice lacking Pax8 have no apparent otic phenotype (Mansouri et al., 1998), whereas Pax2 knockouts present agenesis of the cochlea and vestibuloacoustic ganglia (Torres et al., 1996).
The present results show that a non-coding region of spalt4 conserved between mammals and birds (CR-F) is sufficient to drive reporter expression that recapitulates endogenous spalt4 in the otic placode at stage 10 and later in the otic vesicle. Similarly, expression of spalt4 in the CNS can be recapitulated by the sum of at least four separate conserved regions, each of which is responsible for a distinct subset of expression patterns in the brain and spinal cord. There are also three different regions that have enhancer activity in different mesodermal regions.
The CR-F region contains a number of consensus transcription factor binding sites for potential regulators of otic placode genes: Pea3/Erm which are effectors of FGF signaling, Pax2 which is expressed in the ectoderm prior to its thickening into the otic placode, and TCF-Lef which is a component of the Wnt signaling pathway. While mutation of the TCF-Lef site had no apparent effect on reporter expression, mutations of either the Pax2 sites or the upstream Pea3/Erm site reduced or abolished enhancer activity in the otic placode and vesicle. Thus, each site is necessary for activity and they appear to act synergistically.
While we have shown that the Pax2 and Pea3/Erm binding sites are required, we cannot exclude the possibility that other binding sites and other transcription factors are also involved in activation of CR-F otic enhancer activity. To address whether the endogenous transcription factors are required for the enhancer activity, we performed loss-of-function analysis. Accordingly, a Pea3 morpholino completely eliminated enhancer activity. Similarly, cells electroporated with high levels of the Pax2 morpholino also lacked CR-F activity. In addition, both Pea3 and Pax2 morpholinos reduced the levels of the endogenous spalt4 mRNA. These data are consistent with the mutational experiments in suggesting that Pea3 and Pax2 are direct inputs to the CR-F enhancer.
Furthermore, Pea3 and Pax2 can induce CR-F activity when expressed in ectopic locations such as the extraembryonic ectoderm. We can also detect CR-F activity in the hindbrain level ectoderm caudal to the otic placode when Pax2 is over-expressed in this location. However, many regions of the embryo, such as the trunk, do not have CR-F activity when Pax2 or Pea3 are expressed by themselves. Simultaneous electroporation of both Pax2 and Pea3 expression constructs results in ectopic CR-F enhancer activity in the trunk ectoderm and throughout the hindbrain level ectoderm. Thus, Pax2 and Pea3 together are sufficient to induce ectopic CR-F throughout the embryo, whereas neither alone is sufficient. This supports the idea that these two transcription factors act synergistically.
In summary, we have interrogated the regulatory region of a key gene involved in specification of the otic placode. Our results place the synergistic interaction of Pax2 and Pea3 directly upstream of spalt4, which in turn feeds back to activate Pax2 (Barembaum and Bronner-Fraser, 2007). Establishing direct inputs into the regulatory region of spalt4 that drives its otic expression provides important insights into the gene regulatory network underlying induction of the chick inner ear.
Supplementary Material
Acknowledgements
We thank Dr. Tatjana Sauka-Spengler for her expert help and advice through all aspects of this project. We would like to thank Dr. Hisato Kondoh for providing the pTK vectors. We also want to thank Tatiana Hochgreb for helping with the confocal microscope. This work was supported by USPHS grant DE16459.
REFERENCES
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D, Schimmang T. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–6338. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Barembaum M, Bronner-Fraser M. Spalt4 mediates invagination and otic placode gene expression in cranial ectoderm. Development. 2007;134:3805–3814. doi: 10.1242/dev.02885. [DOI] [PubMed] [Google Scholar]
- Brown ST, Wang J, Groves AK. Dlx gene expression during chick inner ear development. J Comp Neurol. 2005;483:48–65. doi: 10.1002/cne.20418. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Collignon J, Schoenwolf GC, Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev Dyn. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Dominguez-Frutos E, Vendrell V, Alvarez Y, Zelarayan LC, Lopez-Hernandez I, Ros M, Schimmang T. Tissue-specific requirements for FGF8 during early inner ear development. Mech Dev. 2009 doi: 10.1016/j.mod.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Esterberg R, Fritz A. dlx3b/4b are required for the formation of the preplacodal region and otic placode through local modulation of BMP activity. Dev Biol. 2009;325:189–199. doi: 10.1016/j.ydbio.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnberg N, Neubuser A. FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev Biol. 2002;247:237–250. doi: 10.1006/dbio.2002.0696. [DOI] [PubMed] [Google Scholar]
- Freter S, Muta Y, Mak SS, Rinkwitz S, Ladher RK. Progressive restriction of otic fate: the role of FGF and Wnt in resolving inner ear potential. Development. 2008;135:3415–3424. doi: 10.1242/dev.026674. [DOI] [PubMed] [Google Scholar]
- Groves AK, Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–3499. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
- Hans S, Christison J, Liu D, Westerfield M. Fgf-dependent otic induction requires competence provided by Foxi1 and Dlx3b. BMC Dev Biol. 2007;7:5. doi: 10.1186/1471-213X-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Liu D, Westerfield M. Pax8 and Pax2a function synergistically in otic specification, downstream of the Foxi1 and Dlx3b transcription factors. Development. 2004;131:5091–5102. doi: 10.1242/dev.01346. [DOI] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Kober KM, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A, Trumbower H, Wang T, Zweig AS, Haussler D, Kent WJ. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil SH, Streit A, Brown ST, Agrawal N, Collazo A, Zile MH, Groves AK. Distinct roles for hindbrain and paraxial mesoderm in the induction and patterning of the inner ear revealed by a study of vitamin-A-deficient quail. Dev Biol. 2005;285:252–271. doi: 10.1016/j.ydbio.2005.05.044. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Riley BB. Mesendodermal signals required for otic induction: Bmp-antagonists cooperate with Fgf and can facilitate formation of ectopic otic tissue. Dev Dyn. 2009;238:1582–1594. doi: 10.1002/dvdy.21955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher RK, Anakwe KU, Gurney AL, Schoenwolf GC, Francis-West PH. Identification of synergistic signals initiating inner ear development. Science. 2000;290:1965–1967. doi: 10.1126/science.290.5498.1965. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–613. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawoko-Kerali G, Rivolta MN, Holley M. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J Comp Neurol. 2002;442:378–391. doi: 10.1002/cne.10088. [DOI] [PubMed] [Google Scholar]
- Leger S, Brand M. Fgf8 and Fgf3 are required for zebrafish ear placode induction, maintenance and inner ear patterning. Mech Dev. 2002;119:91–108. doi: 10.1016/s0925-4773(02)00343-x. [DOI] [PubMed] [Google Scholar]
- Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–4062. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- Lunn JS, Fishwick KJ, Halley PA, Storey KG. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev Biol. 2007;302:536–552. doi: 10.1016/j.ydbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Mackereth MD, Kwak SJ, Fritz A, Riley BB. Zebrafish pax8 is required for otic placode induction and plays a redundant role with Pax2 genes in the maintenance of the otic placode. Development. 2005;132:371–382. doi: 10.1242/dev.01587. [DOI] [PubMed] [Google Scholar]
- Mahmood R, Mason IJ, Morriss-Kay GM. Expression of Fgf-3 in relation to hindbrain segmentation, otic pit position and pharyngeal arch morphology in normal and retinoic acid-exposed mouse embryos. Anat Embryol (Berl) 1996;194:13–22. doi: 10.1007/BF00196311. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Chowdhury K, Gruss P. Follicular cells of the thyroid gland require Pax8 gene function. Nat Genet. 1998;19:87–90. doi: 10.1038/ng0598-87. [DOI] [PubMed] [Google Scholar]
- Maroon H, Walshe J, Mahmood R, Kiefer P, Dickson C, Mason I. Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development. 2002;129:2099–2108. doi: 10.1242/dev.129.9.2099. [DOI] [PubMed] [Google Scholar]
- Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2006;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Mohamed OA, Taketo MM, Dufort D, Groves AK. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 2006;133:865–875. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- Park BY, Saint-Jeannet JP. Hindbrain-derived Wnt and Fgf signals cooperate to specify the otic placode in Xenopus. Dev Biol. 2008;324:108–121. doi: 10.1016/j.ydbio.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063–3074. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev. 2001;107:105–117. doi: 10.1016/s0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Lufkin T. Dlx5 and Dlx6 homeobox genes are required for specification of the mammalian vestibular apparatus. Genesis. 2006;44:425–437. doi: 10.1002/dvg.20233. [DOI] [PubMed] [Google Scholar]
- Roehl H, Nusslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol. 2001;11:503–507. doi: 10.1016/s0960-9822(01)00143-9. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- Schimmang T. Expression and functions of FGF ligands during early otic development. Int J Dev Biol. 2007;51:473–481. doi: 10.1387/ijdb.072334ts. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Munsterberg A. The vertebrate spalt genes in development and disease. Dev Biol. 2006;293:285–293. doi: 10.1016/j.ydbio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Gruss P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development. 1996;122:3381–3391. doi: 10.1242/dev.122.11.3381. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Takemoto T, Kamachi Y, Kondoh H. Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech Dev. 2004;121:1145–1158. doi: 10.1016/j.mod.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Vendrell V, Carnicero E, Giraldez F, Alonso MT, Schimmang T. Induction of inner ear fate by FGF3. Development. 2000;127:2011–2019. doi: 10.1242/dev.127.10.2011. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Wholemount in situ hybridizationof vertebrate embryos. In: Wilkinson DG, editor. In Situ Hybridization: A Practical Approach. Oxford: IRL Press; 1992. pp. 75–83. [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol. 2007;308:379–391. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.