Abstract
In eukaryotic cells, a specific set of proteins are modified by C-terminal attachment of 15-carbon farnesyl groups or 20-carbon geranylgeranyl groups that function both as anchors for fixing proteins to membranes and as molecular handles for facilitating binding of these lipidated proteins to other proteins. Additional modification of these prenylated proteins includes C-terminal proteolysis and methylation, and attachment of a 16-carbon palmitoyl group; these modifications augment membrane anchoring and alter the dynamics of movement of proteins between different cellular membrane compartments. The enzymes in the protein prenylation pathway have been isolated and characterized. Blocking protein prenylation is proving to be therapeutically useful for the treatment of certain cancers, infection by protozoan parasites and the rare genetic disease Hutchinson-Gilford progeria syndrome.
Isoprenoids are lipids made of five-carbon blocks, and they constitute one of the main classes of natural products. A subset of isoprenoids called prenyl groups are found on a variety of biological substances, including proteins. Prenylated proteins and peptides are found in most (if not all) eukaryotes. They arise from the post-translational attachment of 15-carbon farnesyl or 20-carbon geranylgeranyl groups to the C-terminal segment of proteins. Protein prenyl groups not only are hydrophobic elements that bind proteins to membranes, but, at least in some cases, they also function as molecular handles that bind to hydrophobic grooves on the surface of soluble protein factors; these factors then remove the prenylated protein from the membrane in a regulated manner.
Protein prenylation has been vigorously studied over the past ~15 years because it is found on several signaling proteins (including heterotrimeric G proteins) that connect cell surface receptors to intracellular effectors, and also on Ras proteins, one of the most common oncoproteins found in human tumors. Ras proteins serve as molecular switches at cellular membranes to control propagation of growth signals from cell surface receptors to nuclear transcription factors. Because Ras mutations are important in many human cancers, there has been extensive focus on Ras prenylation.
Discovery of protein prenyl groups and structural varieties
The first reports of prenylated proteins and peptides described the secreted pheromone peptides from jelly fungi1,2, whose structure resembles that of the well-known a-factor mating pheromone from baker's yeast (Saccharomyces cerevisiae), which contains a cysteine methylester farnesylated at the C terminus3. In the 1980s, studies on the timing of cholesterol biosynthesis with respect to the cell cycle in human cells led to the discovery that a compound derived from mevalonic acid other than cholesterol is incorporated into a specific set of proteins4. Rigorous structure-elucidation work showed that these proteins contain cysteine-linked farnesyl and geranylgeranyl groups5,6.
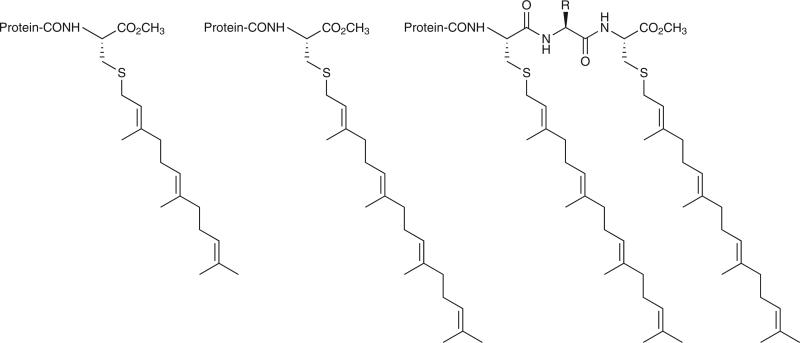
Figure 1 shows the structures of the C termini of prenylated proteins that have been reported so far. In the case of farnesylated proteins, the lipid is thioether linked to the cysteine sulfur, and the α-carboxyl group of the C-terminal farnesylated cysteine is methylated. Proteins containing a single geranylgeranyl group are similarly modified on cysteine. The third structural class of prenylated proteins contain two geranylgeranyl groups, with each lipid chain attached to separate cysteines that sometimes flank a single nonlipidated amino acid (Fig. 1)7. In some doubly geranylgeranylated proteins, the two lipidated cysteines are next to each other in the protein sequence and are followed by either zero or two additional amino acids, and in these cases there is no C-terminal carboxyl methylation (that is, C(geranylgeranyl)C(geranylgeranyl)-COOH or C(geranylgeranyl)C(geranylgeranyl)-XY-COOH)8.
Figure 1.
Structures of the C termini of prenylated proteins. Farnesylated protein (left); geranylgeranylated protein (middle); doubly geranylgeranylated protein (right).
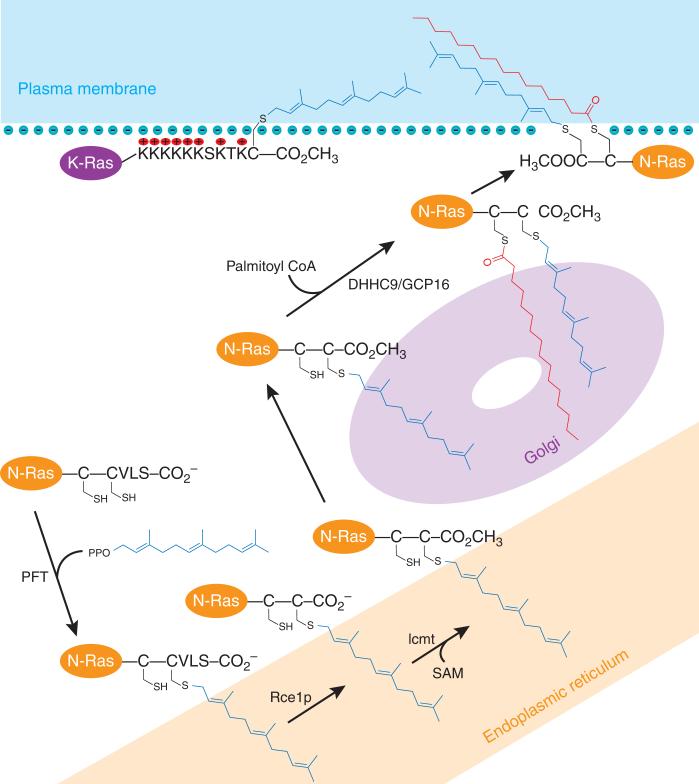
Known farnesylated proteins include nuclear lamins A and B, which form a filamentous structural layer on the inner side of the nuclear membrane; the γ subunit of the heterotrimeric G protein transducin, which functions in visual signal transduction in the retina; all three isoforms of Ras GTP-binding proteins, which function in cellular growth regulation; the large-antigen component of the hepatitis δ virus; and yeast a-factor. All farnesylated proteins contain a C-terminal sequence CaaX, where C is cysteine, a is usually but not necessarily an aliphatic amino acid and X is one of a variety of amino acids. After attachment of the farnesyl group via a thioether linkage to the cysteine residue, the last three amino acids, aaX, are removed by a prenyl protein–specific endoprotease, and the α-carboxyl group of the newly exposed farnesylated cysteine is methylated by a prenyl protein–specific methyltransferase (Fig. 2). Some prenylated proteins such as H-Ras and N-Ras contain a second lipid chain, a 16-carbon palmitoyl group, that is thioester linked to the SH group of a cysteine that is close in protein sequence to the farnesylated-cysteine C terminus (Fig. 2). Known geranylgeranylated proteins include the γ subunit of heterotrimeric G proteins that function at the plasma membrane, and many small GTP-binding proteins such as the Rho proteins. The Rab subfamily of GTP-binding proteins contain a pair of geranylgeranyl groups on adjacent or nearly adjacent cysteines at the C terminus of the protein (Fig. 1). Approximately 50 different prenylated proteins have been identified so far. Two-dimensional gel analysis of radiolabeled prenylated proteins suggests that there may be ~100–200 such proteins in mammalian cells.
Figure 2.
Enzymatic pathway for the modification of prenylated proteins. PFT attaches a farnesyl group to the C-terminal CaaX motif of specific cytosolic proteins. The farnesylated protein undergoes C-terminal aaX removal by the endoprotease Rce1p, and this is followed by S-adenosylmethionine (SAM)-dependent methylation of the COOH terminus by Imct; both modifications occur in the endoplasmic reticulum. Some proteins, such as N-Ras and H-Ras, are further modified after transfer to Golgi membranes by palmitoylation on one or two cysteines near the prenylated cysteine in a reaction catalyzed by DHHC9 and GCP16 in mammals (Erf2 and Erf4 in yeast endoplasmic reticulum membranes) and using palmitoyl coenzyme A (CoA) as the fatty-acid donor. After palmitoylation, fully lipidated N-Ras and H-Ras are transferred to the plasma membrane probably by vesicle transport. Other proteins, such as KRas, contain a polybasic region (KKKKKKSKTK) next to the prenylated C terminus that is thought to bind the protein to the cytosolic face of the plasma membrane through electrostatic interactions with acidic phospholipids. PPO, pyrophosphate. Farnesyl groups are shown in blue and palmitoyl groups are red.
Enzymatic processing of prenylated proteins
So far, three distinct protein prenyltransferases that attach prenyl groups to proteins have been identified. Protein farnesyltransferase (PFT) transfers the farnesyl group from farnesyl diphosphate to the cysteine SH of the CaaX motif at the C terminus of proteins (Fig. 2). Protein geranylgeranyltransferase type I (PGGT-I) catalyzes a similar reaction using geranylgeranyl diphosphate as the prenyl donor. Protein geranylgeranyltransferase type II (also known as Rab geranylgeranyltransferase) catalyzes the transfer of both geranylgeranyl groups from geranylgeranyl diphosphate to two cysteine SH groups at the C terminus of Rab proteins. Much is known about the structure and catalytic properties of these enzymes9–14. All three are heterodimeric proteins that use active site–bound Zn2+ as a catalytic cofactor. Rab geranylgeranyltransferase recognizes Rab proteins bound to a carrier protein called Rab escort protein (REP).
The specificity rules that dictate which CaaX motifs are acted on by PFT, which are acted on by PGGT-I and which are not prenylated at all are not completely understood. The X residue of CaaX is probably the most important element for differential recognition by PFT versus PGGT-I (ref. 15), but the aa portion of CaaX is also important16. A poly-basic stretch of amino acids upstream of the CaaX motif can facilitate binding of proteins such as K-Ras to PFT (ref. 17). Several elements of Rab proteins are recognized by Rep, and only the Rab–Rep complex is acted on by PGGT-II (ref. 18).
The postprenylation CaaX processing machinery includes the endoprotease Rce1p (and in some cases Ste24p, see below), which releases an intact aaX tripeptide from the newly prenylated CaaX protein, and isoprenylcysteine carboxylmethyltransferase (Icmt), which transfers a methyl group from S-adenosylmethionine to the α-carboxyl group of the prenyl cysteine (Fig. 2). Two important breakthroughs based on the discovery of the genes encoding these enzymes in S. cerevisiae were the identification of the genes’ mammalian homologs and their recent targeted disruption in mice. Ras localization and transforming capacity was found to be markedly altered in mouse embryonic fibroblasts having null and conditional alleles of Rce1 and Icmt, which provided compelling evidence for the physiological importance of postprenylation CaaX processing steps in Ras oncogenesis19,20. These genetic studies, together with the recent appreciation that cross-prenylation by prenyltransferases can diminish the therapeutic value of PFT inhibitors (FTIs; discussed below), have spurred renewed interest in the postprenylation enzymes as attractive anticancer targets.
There are two gene products capable of mediating CaaX endoproteolysis in yeast: the Ras-converting enzyme Rce1p, and Ste24p (designated Zmpste24 in mammalian cells)21,22. Although functionally redundant for the endoproteolysis of the yeast a-factor CaaX motif, Rce1p and Ste24p have distinct, albeit overlapping, substrate specificities when tested in vivo against a panel of CaaX motifs23. Notably, the processing of Ras and most other CaaX proteins is solely dependent on Rce1p; a-factor is the only known physiological exception, though more may exist.
Rce1p and Ste24p are multispanning endoplasmic reticulum membrane proteins24. Because membrane spans are not commonly a feature of proteases, purification was important in providing proof that Ste24p is a bona fide CaaX protease25. Ste24p contains a canonical zinc metal-loprotease motif, and the purified enzyme is zinc dependent. It should be noted that yeast Ste24p, and its mammalian counterpart Zmpste24, have additional roles in an N-terminal proteolytic processing step for a-factor and prelamin A, respectively (see below); the unusual ability of Zmpste24 to cleave at two distinct sites remains puzzling.
Rce1p has been a focus of biochemical studies because of its key role in the CaaX processing of Ras proteins. Using defined synthetic substrates and membrane-associated or partially purified Rce1p, investigators have shown that Rce1p activity depends on the prenylation status of its substrate, and that Rce1p has a preference for particular CaaX motifs26–31. Unfortunately, Rce1p has eluded purification, and neither its amino acid sequence nor its biochemical properties reveal straightforward clues about its mechanism of action. Inhibitor and bioinformatic analyses suggest that Rce1p may be a serine protease or a metalloprotease, respectively29,32. However, mutagenesis of critical residues implicated by those studies does not affect enzyme activity33; further mutational analysis of residues conserved among Rce1p orthologs may reveal new clues about the reaction mechanism of Rce1p.
The embryonic lethality of the Rce1−/− mouse and the dilated cardiomyopathy resulting from specifically ablating Rce1 expression in the mouse heart indicate significant physiological roles for postprenylation processing34,35. Studies of mouse fibroblasts with either null or conditional Rce1 mutant alleles have provided compelling evidence that the membrane association, plasma membrane targeting and transformation capacity of Ras are all substantially lower when Rce1p function is lacking; in fact cells lacking Rce1 function are sensitized to FTI treatment34–36. These findings have spurred renewed interest in developing Rce1 inhibitors. So far, inhibitor studies have focused on substrate analogs, including prenylpeptide mimetics and related compounds that may act as competitive inhibitors27,37. Clearly, the identification of new Rce1p inhibitors presents a promising avenue for further research.
Like the endoproteases, the methyltransferase Icmt is a multispanning endoplasmic reticulum membrane protein, with its active site presumably facing the cytosol38. Topology studies have shown that the yeast Icmt (Ste14p) has six trans-membrane spans, and two additional spans have been predicted for the mammalian enzyme39. Because of its membrane spans and because it lacks a classical methyltransferase consensus motif, Icmt is an atypical member of the methyltransferase family of enzymes40. However, the recent purification of Ste14p from yeast, and its reconstitution in liposomes in an enzymatically active form, have provided conclusive evidence that it is the sole component comprising Icmt activity40,41. Using the purified enzyme, researchers showed that yeast Ste14p recognizes the farnesylated and geranylgeranyl substrates N-acetyl-S-farnesylcysteine and N-acetyl-S-geranylgeranylcysteine equivalently.
The Icmt−/− knockout mouse has an embryonic lethal phenotype, although it dies at an earlier stage than the Rce1−/− mouse, an observation underscoring the physiological importance of Icmt (ref. 42). As is the case for Rce1-deficient fibroblasts, null and conditional Icmt mutant fibroblasts have defects in the membrane association and plasma membrane targeting of Ras, and also in Ras-induced transformation efficiency43,44. Notably, the analysis of Ras-transformed Icmt–deficient fibroblasts highlights an additional and important twist: the unmethylated geranylgeranylated RhoA produced in the absence of Icmt is metabolically unstable, and the resulting low steady-state concentration of RhoA may provide an important contribution to inhibiting Ras-induced oncogenic transformation under these circumstances44. As this example with RhoA illustrates, carboxyl methylation has diverse and sometimes unanticipated roles, affecting different prenylated proteins in different ways. Proposed roles for the carboxyl methylation of prenylated proteins, for which there is evidence in particular cases, include influencing intracellular protein localization, membrane attachment, metabolic stability and interactions with other proteins19,20,45,46. Although carboxyl methylation of prenylated proteins has been suggested to be reversible, and therefore could represent a regulatory event, evidence for this reversibility in cells has not been forthcoming.
Given the importance of carboxyl methylation for Ras-induced onco-genesis shown by the Icmt mouse studies mentioned above, the development of Icmt inhibitors as anticancer drugs is of obvious interest19,41,47. The well-known chemotherapeutic compound methotrexate (an antifolate that blocks thymidine synthesis, which is necessary for tumor growth) causes accumulation of S-adenosylhomocysteine, a potent inhibitor of nearly all cellular methyltransferases; it has been suggested that at least some of methotrexate's antitumor activity may stem from its ability to inhibit Icmt, though specificity for Icmt is lacking48. It is worth noting that there were initial reservations that specific Icmt inhibitors might be toxic to cells because they would affect all prenylated proteins; these concerns have been significantly mitigated by recent work showing that although the localization of farnesylated proteins is strongly affected when methylation is blocked, the localization of geranylgeranylated proteins (the main class of prenylated proteins) is relatively unaffected49. Efforts to identify specific Icmt inhibitors are currently underway. Both the screening of compound libraries50 and the search for mechanism-based and substrate-analog inhibitors51 are beginning to yield candidate compounds.
Palmitoylation of prenylated proteins
S-Palmitoylation of proteins entails connection of a (C16) fatty-acid chain to the thiol functionality of a cysteine via a thioester bond. Palmitoylation of proteins has several possible functions, depending on the protein under scrutiny. Much like protein prenylation in general, protein palmitoylation enables or contributes to membrane binding and possibly localization52,53. Additionally, palmitoylation may modulate protein-protein or protein-lipid interactions and enzyme activity. In contrast to protein prenylation, palmitoylation is a reversible post-translational modification54. In the Ras superfamily, H-Ras and N-Ras are examples of proteins that are both prenylated and palmitoylated55. Palmitoylation of the C terminus of these Ras proteins occurs after the farnesylation of the C-terminal cysteine and at cysteine residues slightly upstream of the farnesylated cysteine (Fig. 2). Although inhibitors of Ras palmitoylation are known (2-bromopalmitate56 and cerulenin analogs57,58, for example), the mechanisms and selectivity steering this palmitoylation process are still unclear. Several palmitoylation motifs have been found in different classes of proteins and have been reviewed52. We focus on the recent progress in the elucidation of the enzymology and function of palmitoylation in Ras proteins.
In yeast, the Erf2–Erf4 protein complex palmitoylates the yeast Ras homolog Ras2 (refs. 59,60). Until recently no Erf2 orthologs or other proteins featuring Ras S-palmitoylation activity had been found in the genomes of eukaryotes, including the human genome. However, two proteins or protein complexes (DHHC9 and GCP16) that may be involved in Ras palmitoylation have recently been discovered61,62.
The eukaryotic acyl protein thioesterase 1 (APT1) protein has been identified as participating in the removal of palmitate from proteins on the cytosolic face of membranes63. The relationship between Ras proteins and APT1 has therefore been further studied using a chemical biological approach64. In this investigation several semisynthetic Ras proteins, together with lipopeptides and newly developed APT1 inhibitors, were synthesized via combined chemical and biological methods and used as tools, without which the study would have been difficult62. These APT1 inhibitors prevented differentiation of PC12 cells by oncogenic N-Ras when N-Ras had to be finally processed within the cell (that is, when N-Ras lacked a palmitoyl anchor). The biological activity of an N-Ras lipoprotein having a complete but nonhydrolyzable membrane anchor was not reduced by the same inhibitors. Additional in vitro experiments with these inhibitors and semisynthetic N-Ras lipopeptides and proteins has shown that the inhibitors may inhibit Ras palmitoylation and depalmitoylation. This observation led to the suggestion that APT1 functions in vitro as a bidirectional enzyme, promoting either Ras palmitoylation or Ras depalmitoylation, depending on the local environment and on substrate availability. However, the possible involvement of APT1 in Ras palmitoylation in vivo still needs to be proven.
Yeast Erf2 is a protein that contains a DHHC cysteine-rich domain and functions in complex with Erf4 (ref. 60). An analogous human protein palmitoyltransferase complex has been discovered that consists of DHHC9, which contains a DHHC cysteine-rich domain, and GCP16, a palmitoylated membrane protein localized on the Golgi61. These two proteins colocalize in the Golgi apparatus and form a protein complex, and DHHC9 requires GCP16 for palmitoyltransferase activity. The complex also shows substrate selectivity for the C terminus of Ras proteins, which suggests that it is a human ortholog of the yeast palmitoyltransferase. The localization of the protein complex in the Golgi suggests that it is an important contributor to the control of Ras palmitoylation in vivo.
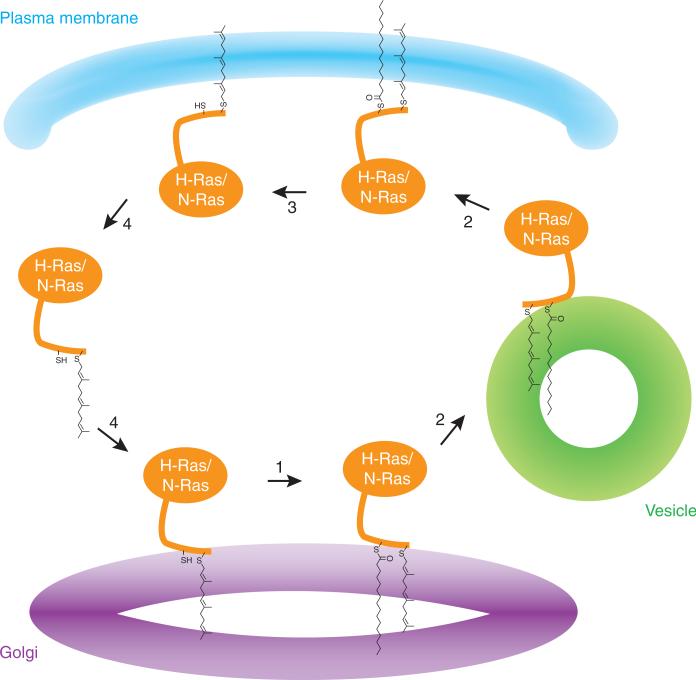
The importance of Ras palmitoylation has also been recently highlighted in studies on the regulation of its localization and activity65–68. Some of these studies used semisynthetic proteins, whereas others used fluorescently labeled proteins67. The specific subcellular distribution of palmitoylatable H-Ras and N-Ras isoforms is generated through a constitutive deacylation-reacylation cycle (Fig. 3). Palmitoylation status drives rapid exchange of Ras between the plasma membrane and the Golgi apparatus; depalmitoylated Ras protein exchanges rapidly between cytoplasm and membranes, and repalmitoylation occurs at the Golgi, where Ras signals or is redirected to the plasma membrane. In addition, the two individual palmitoyl residues of H-Ras have a distinct role in protein trafficking, localization and signaling69.
Figure 3.
Model for the trafficking of H-Ras and N-Ras from the Golgi complex to the plasma membrane and back67,68. S-Palmitoylation of the two Ras isoforms occurs at the Golgi (1), followed by directed vesicular transport to the plasma membrane (2), where the protein can be released after enzymatic hydrolysis of the thioester (3) and transported back via a nonvesicular pathway to the Golgi (4). APT1, DHHC9 and GCP16 may be involved in palmitoylation on the Golgi.
These results in regard to Ras palmitoylating enzymes and Ras processing in the cell are new starting points for the complete characterization of the Ras palmitoylation machinery and its functioning. Additional enzymes that are involved in protein palmitoylation are expected to be found, and the targeting mechanism of Ras via reversible post-translational modifications might be a paradigm for other types of proteins, palmitoylated or not. On top of that, use of the palmitoylation process may open up a new opportunity to target oncogenic H-Ras and N-Ras70.
Anchoring proteins to membranes: static and dynamic picture
The structure and lateral organization of lipids and proteins in biological membranes are under heavy scrutiny in the fields of membrane biochemistry and biophysics71,72. The existence of membrane subdomains with different lipid compositions and the relationship between lipid domain formation and the conformation and functional properties of membrane-anchored proteins are central topics in these fields. As is true for other lipidated proteins, the microlocalization and signaling of Ras depends on its lipidation pattern, although other factors, such as the amino acid sequence of the backbone and conformation of specific domains, also have prominent roles73. In addition, the position of the lipid functionalities seems to determine the membrane specificity74.
The different C-terminal amino acids and the concomitant lipidation motifs of Ras proteins are thought to target the different Ras iso-forms to different membrane microenvironments and thereby regulate their biological profiles. Experiments using green fluorescent proteins tagged with different types of lipid anchors and expressed in live cells have shown that myristoyl and palmitoyl groups promote clustering in cholesterol-rich liquid-ordered domains (‘rafts’), whereas isoprenyl groups do not and prefer the liquid-disordered phase75. Studies on fluorescently labeled N-Ras proteins and model membranes have shown that the complete protein has a preference for the liquid-disordered phase over the liquid-ordered and solid-ordered phases76, and NMR studies of this protein have shown a specific conformation of the lipidated C terminus when bound to the membrane77. Together with several studies on peptides, this indicates that the membrane localization of N-Ras is mainly governed by its lipidated C terminus, in contrast to H-Ras, for example, in which the GDP and GTP loading status also strongly regulates its localization69,78. Localization studies with atomic force microscopy (AFM) have shown that to a large extent the lipidated protein is actually located in the boundary region of the domains in mixed-phase liquid-ordered/liquid-disordered phases76. The specific localization and accumulation of N-Ras proteins in interfaces of lipid bilayer domains may have a special biological relevance, as it might serve, for example, as a vehicle for protein association. Recent computational modeling studies of immunogold spatial point patterns on intact plasma membrane sheets indicate that lipidated proteins on the inner plasma membrane are able to drive the formation of nanoclusters79. These findings are supported by independent studies on semisynthetic Ras proteins76. This indicates that Ras proteins are not passively targeted to microenvironments but may be actively involved in their generation80.
The K-Ras protein contains eight lysine residues just upstream of the farnesylated C terminus (Fig. 2). It is has been suggested that these cationic lysines form favorable electrostatic interactions with the cytosolic face of the plasma membrane; anionic phospholipids including phosphatidylserine and phosphatidylinositol are present at relatively high abundances on this membrane leaflet. An electrostatic basis for targeting of K-Ras to the plasma membrane (rather than a mechanism involving the binding of the C terminus to a putative receptor protein located in the plasma membrane) is consistent with the fact that mutation of all eight lysines to arginines81 or to D-lysines82 does not effect K-Ras targeting.
So far we have focused on the membrane anchoring ability of protein prenyl groups. Methylation of the prenylated cysteine is expected to increase the hydrophobicity of the protein's C terminus, thereby facilitating membrane anchoring. Studies also show that in some cases the prenyl group serves as a molecular handle to allow extraction of prenylated proteins from membranes by other proteins. For example, the Rab3a GTP-binding protein cycles between the membrane of synaptic vesicles and the cytosol during regulated release of neurotransmitters83. The physiological significance of this cycling is not yet understood. Membrane extraction and delivery of prenylated proteins of the Rab and Rho families is mediated by their respective GDP dissociation inhibitors (GDIs). Although Rab GDIs and Rho GDIs differ in structure, they have similar biological regulatory activities84. X-ray crystallography and NMR studies have provided insight into the binding of Rho GTPases to Rho GDI85. Together with kinetic studies, this has resulted in a proposed two-step mechanism for the release of, for example, the Rho GTPase Cdc42 or of Rab proteins from membranes86. In the first fast step, Rho GDI binds to the switch regions of Cdc42, enabling the hydrophobic pocket to bind the hypervariable region of the GTPase. In the second step, the geranylgeranyl moiety is isomerized and inserted into the hydrophobic pocket, resulting in release from the membrane. Back delivery of the GTPase into the membrane is thought to involve several factors such as phosphorylation status, phospholipid composition and protein displacement factors.
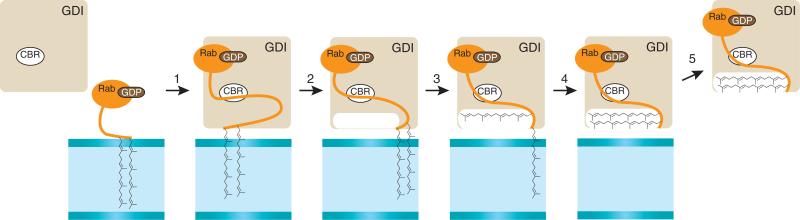
Data obtained on the structural and kinetic properties of the GDI-Rab interaction have allowed investigators to formulate a comparable GDI-Rab membrane extraction model (Fig. 4)87. In short, GDI recognizes the GDP-bound form of Rab through an initial interaction that involves only the globular part of the GTPase. Subsequent docking of the Rab C terminus onto GDI leads to stepwise extraction of the prenyl moieties from the membrane, a process that is driven by binding of the lipid groups in the highly hydrophobic GDI binding site. Logically, the mechanism for delivering Rab back into the membrane is a reversal of the extraction process, but it probably involves an additional membrane-bound factor (termed GDI displacement factor) having specificity for specific Rab molecules. An important number of these displacement factors belong to the group of Yip/PRL proteins (one of the proteins in this group, Yip1, is an essential protein in yeast that interacts with Rab proteins)88. Rab escort proteins (REPs) that shuttle the Rab protein to and from the Rab geranylgeranyltransferase share structural features with GDIs, including a hydrophobic groove for binding geranylgeranyl groups.
Figure 4.
Model for extraction of prenylated Rab proteins from membranes via GDI, as previously formulated87. (1) The initial recognition occurs via the low-affinity interaction of the Rab protein with the C-terminal–binding region (CBR) of GDI. Then, the lipid-binding site of GDI is positioned over the prenyl functionalities of the Rab protein (2), the first geranylgeranyl moiety is transferred (3) and the second geranylgeranyl moiety is transferred (4). (5) This finally results in the dissociation of the complex from the membrane.
These models for extraction and delivery of prenylated proteins provide guidance for additional studies that will hopefully lead to an understanding of specific membrane targeting of these proteins.
Semisynthetic tools for studying lipidated proteins
Progress in the area of protein ligation methods has given researchers access to a broad spectrum of methods for the semisynthetic production of post-translationally modified proteins, including lipidated proteins. These methods yield either native bonds (for example, prior thiol capture, native chemical ligation and expressed protein ligation) or non-native bonds (for example, imine capture ligation, oxime ligation and maleimidocaproic acid ligation). This combination of organic chemistry and molecular biology gives scientists access to modified proteins (with both natural and non-natural modifications) that are generally not accessible through other (for example, enzymatic) processes89.
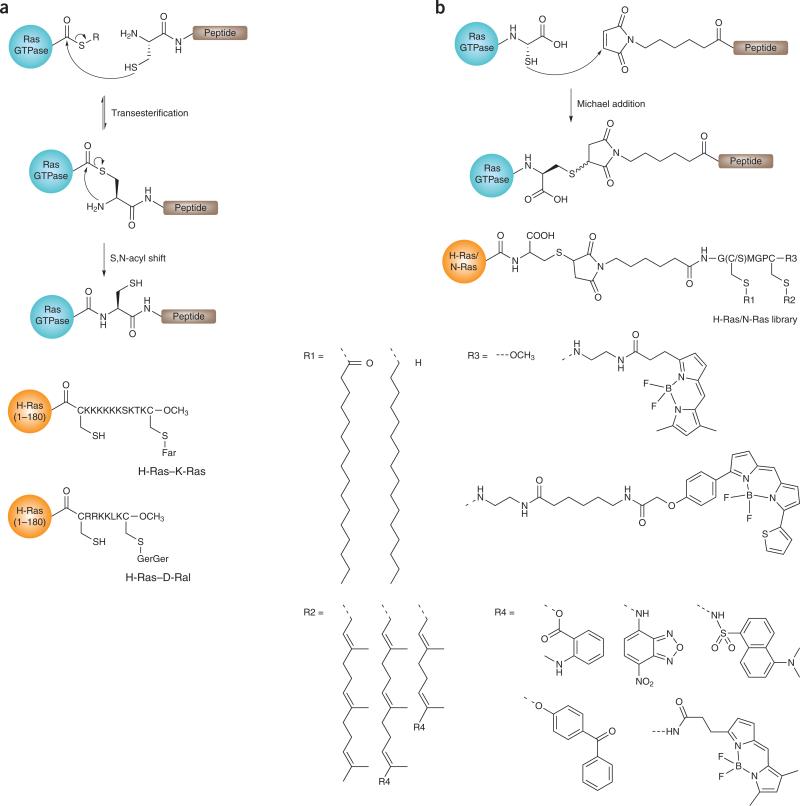
Two chemical ligation approaches have been used to obtain fully functional Ras GTPases (Fig. 5). The first approach, expressed protein ligation (EPL), connects a lipidated peptide having an N-terminal cysteine to the C terminus of a thioester-tagged Ras GTPase via a native peptide bond. The second approach incorporates synthetic lipidated peptides into the GTPases using maleimidocaproyl (MIC)-controlled ligation. This ligation requires an accessible (that is, C-terminal-free) thiol group on the protein, usually on a cysteine, to connect the N-terminally MIC-modified peptide.
Figure 5.
Schematic representation of methods used to introduce natural and modified lipidated C termini on Ras GTPases, and a collection of semisynthetic neo-Ras proteins synthesized via these methods58,62,64,67,76. (a) EPL. H-Ras (1–180) denotes the first 180 amino acids of this protein. (b) MIC ligation. Far, farnesyl; Ger, geranyl.
Development of FTIs as cancer therapeutics
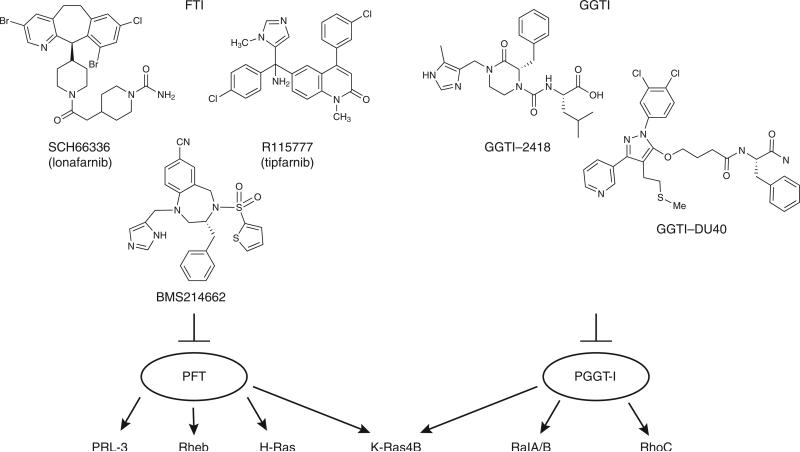
Over the past ~15 years, a variety of small-molecule inhibitors of protein farnesyltransferase have been developed90–92. These include compounds that compete with CaaX peptide substrates or with farnesyl diphosphate, as well as compounds that coordinate to zinc, an essential metal involved in catalysis. Computer modeling has also aided in the improvement and design of FTIs. Three compounds, R115777 (tipifarnib), SCH66336 (lonafarnib) and BMS214662 (Fig. 6), are currently being evaluated in anticancer clinical trials. These compounds show high potency and selectivity against PFT (>1,000-fold preference over PGGT-I). This high specificity seems to be a result of selective stacking interactions between the FTI compound and aromatic residues in the binding site92. In addition to these highly specific compounds, there are FTI compounds that have dual specificity. One example is L-778,123, which inhibits both PFT and PGGT-I (ref. 93). Notably, this compound assumes two different modes of interaction with the two enzymes94. Other examples of dual-specificity inhibitors include the Bristol-Myers Squibb compounds BMS1, BMS2, BMS3 and BMS4, which inhibit both PFT and PGGT-II (ref. 95). These compounds are strongly proapoptotic, which distinguishes them from other FTI compounds. It is speculated that the apoptotic activity of these BMS compounds is a result of PGGT-II inhibition affecting Rab protein function.
Figure 6.
FTI and GGTI compounds. Three FTI compounds used in clinical trials are shown. These compounds are highly selective inhibitors of PFT and may be effective in inhibiting cancer and metastasis arising from the overexpression or overactivation of H-Ras, Rheb and PRL-3 proteins. Two GGTI compounds are shown. They may be effective in inhibiting the function of RhoC, RalA/B and K-Ras4B.
FTIs have Ras-independent clinical activities
FTIs have been extensively examined as potential anticancer agents, and surprisingly they are well tolerated in clinical settings90,91. Clinical activity has been detected with hematologic malignancies including acute myeloid leukemia (AML), chronic myeloid leukemia (CML), multiple myeloma and myelodysplastic syndrome. In a phase 1 study of AML, about 30% of subjects responded to tipifarnib96, and this led to the current, multicentered, large-scale trial of tipifarnib for treating AML. In a trial involving individuals with multiple myeloma, disease stabilization was observed in about 60% of individuals taking tipifarnib97. Some clinical activity toward advanced breast cancer has been detected. However, effects of FTIs on solid tumors have been limited. Combining FTIs with other chemotherapeutic agents has produced some promising results. In combination with paclitaxel, lonafarnib showed clinical activity in people with non–small cell lung cancer98. Although FTIs have clinical activities, the effects seem to be largely independent of the inhibition of Ras protein; no correlations with Ras mutation status or with Ras inhibition have been observed. This is likely because FTIs are incapable of inhibiting the function of K-Ras4B, a predominant iso-form of K-Ras; K-Ras4B can be geranylgeranylated when farnesylation is inhibited99,100. Because there are many farnesylated proteins in the cell, FTIs have the potential to inhibit many proteins. Thus, the anticancer action of FTIs may reflect combinatorial effects on several farnesylated target proteins.
Toward rational uses of FTIs
Recently, human disorders in which a particular farnesylated protein is overactivated or overexpressed have been reported. In these situations, the effects of FTIs may be a result of their inhibiting the function of these particular proteins. H-Ras activation is observed in a small but substantial percentage of human cancer. Approximately 20% of samples from salivary gland tumors and bladder cancer contain H-Ras mutations101,102. Germline mutations of H-Ras have been identified in approximately 90% of people with Costello syndrome, a genetic disorder associated with characteristic craniofacial features, developmental delay and cardiac and skeletal anomalies103–105. Importantly, people with Costello syndrome have a predisposition to neoplasia, including transitional cell carcinoma and neuroblastoma. In these cases, FTIs may inhibit the function of H-Ras. Tuberous sclerosis syndrome (TSC) is a genetic disorder that is associated with the appearance of benign tumors called hamartomas in a variety of organs, such as kidney, skin and brain106. TSC1 gene products Tsc1 and Tsc2 form a complex that acts as a negative regulator of the farnesylated GTPase Rheb. Thus, overactivation of Rheb underlies the molecular basis of this disease. FTIs inhibit Rheb (refs. 107,108) and reverse transformed phenotypes of cells lacking Tsc. The effects of FTIs, in this case, parallel the effects of rapamycin, as Rheb is an activator of mammalian target of rapamycin (mTOR)109,110. TSC1 mutations also lead to lymphangioleiomyomatosis (LAM) disease, which is characterized by diffuse infiltration of the pulmonary parenchyma with benign smooth muscle–like cells111. A different farnesylated protein, PRL-3, is implicated in cancer metastasis. This protein is overexpressed in metastatic colon cancer as a result of chromosomal amplification112. Expression of PRL-3 in colon cancer cell lines results in greater invasive property than in healthy colon cells, and this is associated with the decrease of Rac and increase of RhoC and RhoA proteins113. Thus, it is worth exploring the possibility that FTIs may influence the metastatic potential of human cancer cells.
GGTIs: the next frontier in prenyltransferase inhibition
PGGT-I has recently emerged as an important target for cancer therapy development (Fig. 6). This is mainly a result of three lines of investigation. First, the geranylgeranylated protein RalA acts downstream of Ras to transform cells in several cancers114. In the case of pancreatic cancer, K-Ras activation leads to activation of RalGDS and then RalA. Inhibition of RalA by small interfering RNA (siRNA) inhibits transformed phenotypes of pancreatic cancer cells. Second, GGTIs can inhibit the geranylgeranylation of K-Ras4B that occurs when PFT is inhibited (as noted above). Thus, GGTIs may be used in combination with an FTI to block K-Ras function. Third, another geranylgeranylated protein, RhoC, has emerged as a critical protein in cancer metastasis. Studies of RhoC knockout mice show that loss of RhoC results in inhibition of metastasis115. Overexpression of RhoC in metastatic cancer116 suggests that GGTI may be explored as an antimetastasis agent.
Small-molecule inhibitors of PGGT-I have been developed based on the C-terminal CaaL motif found in many geranylgeranylated proteins117. Replacing the central dipeptide portion with different scaffolds led to the development of a series of inhibitors. Examples are GGTI-2154, which contains a 2-aryl-4-aminobenzoic acid scaffold, and GGTI-2418, which contains a piperazine scaffold. Recently, further improvement has been made by using a benzoyleneurea scaffold117. GGTIs have shown cellular effects including inhibition of Rho signaling, cell cycle arrest at G0 and G1, and apoptosis induction. The first nonpeptidomimetic GGTI, called GGTI-DU40, has recently been identified from a high-throughput screen of a chemical compound library118. This compound is highly potent, with a half-maximal inhibitory concentration (IC50) of 8 nM, and requires more than 250-fold higher concentration to inhibit PFT. Additional GGTIs are expected to be identified, and these will provide valuable reagents for studying and therapeutically inhibiting geranylgeranylation.
Progeria therapeutics based on protein prenylation
A new and unanticipated potential use for FTIs has emerged from recent studies of the rare premature-aging disease Hutchinson-Gilford progeria syndrome (HGPS). Children with HGPS have many characteristics of accelerated aging, including growth retardation, bone problems, hair loss and a receding mandible, and they die of heart disease, generally by about age 13. HGPS maps to LMNA, the gene encoding lamin A, which is a key component of the scaffold that underlies the nuclear membrane119.
The biogenesis pathway leading to mature lamin A involves several steps and provides a framework for understanding HGPS120. The prelamin A precursor undergoes CaaX prenylation, proteolysis and methylation. Then, an endoproteolytic cleavage step (unusual among mammalian CaaX proteins) removes the C-terminal 15 amino acids, including the newly generated carboxylmethylated farnesylcysteine. The reason that cells go to the trouble of C-terminally modifying prelamin A and then removing these modifications is not understood. The endoproteolytic cleavage of prelamin A is mediated by the zinc metal-loprotease Zmpste24 (refs. 121,122). In S. cerevisiae, the Ste24p protease mediates N-terminal endoproteolytic cleavage (and also CaaX processing, functioning redundantly with Rce1p at this step, as discussed above) of the yeast a-factor precursor123,124.
HGPS is commonly caused by a dominant de novo point mutation in LMNA that activates a cryptic splice site and results in the production of a mutant form of lamin A (called progerin) having a 50-amino-acid deletion that includes the Zmpste24 site119. Thus, progerin is not a substrate for further endoproteolytic cleavage and instead remains persistently farnesylated and carboxylmethylated125,126. Several lines of evidence indicate that accumulation of aberrantly CaaX-modified prelamin A accounts for the progeroid phenotypes characteristic of the HGPS mutation120. At the cellular level, the hallmark of fibroblasts from people with HGPS is their highly misshapen (folded, lobed and severely blebbed) nuclei119,127. Mouse and human fibroblasts expressing wild-type lamin A but lacking Zmpste24 activity owing to mutational inactivation have a similar aberrant nuclear phenotype. Persistently farnesylated and carboxymethylated prelamin A is thought to stick ‘too tightly’ to the nuclear envelope, thereby limiting remodeling of the nuclear scaffold and weakening support of the nuclear envelope, which in turn results in blebbed nuclei.
Evidence has recently been presented by several investigators showing that FTIs can block, and possibly even reverse, the aberrant nuclear morphology resulting from the expression of progerin in fibroblasts from people with HGPS, in mouse fibroblasts and in HeLa cells125,126,128–130. These findings have generated considerable excitement because they provide proof of principle, at the cellular level, that FTIs may be a useful therapy for HGPS.
But what about at the organismal level? New results from Yang et al.131 and Fong et al.132 suggest that FTIs may indeed reverse disease pheno-types resulting from the accumulation of persistently CaaX-modified prelamin A in the whole animal as well. The investigators found that FTI treatment of Zmpste24−/− and HGPS mice substantially improved progeroid phenotypes with respect to longevity, body weight and bone integrity. These studies have laid the groundwork for serious consideration of clinical trials of FTIs for children with HGPS120,131,132. Though caution is warranted, the minimal toxicity associated with FTIs, together with the encouraging results from the cellular and mouse studies discussed above, suggest the compelling possibility that FTIs may be beneficial for children with HGPS.
FTIs as tropical parasitic disease therapeutics
A new use of FTIs is in the treatment of malaria and African sleeping sickness caused by the parasites Plasmodium falciparum and Trypanosoma brucei, respectively. FTIs are cytotoxic to these protozoa, perhaps because they contain PFT but seem to lack PGGT-I (refs. 133–136). In the case of malaria, the target of FTI toxicity is almost certainly PFT, given that parasites that have become resistant to FTIs contain mutant PFTs that bind FTIs less tightly137. The tetrahydroquinoline series of FTIs, originally developed by Bristol-Myers Squibb as anticancer agents, are the most promising antimalarial FTIs reported so far. They kill parasites in culture in the low nanomolar range and can cure rodents infected with malaria when dosed orally138. This illustrates the ‘piggyback medicinal chemistry’ approach of taking advantage of work done by large pharmaceutical companies and extending it for use in treating neglected diseases.
Concluding remarks
The attachment of prenyl groups to proteins in eukaryotic cells was discovered about two decades ago either through traditional natural-product isolation and structure elucidation (of fungal pheromones) or through careful observations centered on the timing of cholesterol biosynthesis during the mammalian cell cycle. Interest in understanding protein farnesylation for the purpose of cancer therapy grew out of the finding that one of the most common cancer-promoting elements in cells, the Ras proteins, require farnesylation to transform cells into tumor progenitors. Early studies showing that PFT inhibitors are very effective at shrinking human tumor masses implanted into experimental animals have led to preclinical drug development of such inhibitors at many pharmaceutical companies. After the first round of clinical trials, it is likely that one or two of these inhibitors will emerge as a well-tolerated agent for treating a subset of leukemias.
Further anticancer clinical trials of PFT inhibitors, especially in combination with existing anticancer drugs, are expected in the near future. The possibility that PGGT-I inhibitors may have beneficial anticancer properties is currently under investigation. Also, future studies may reveal the true anticancer basis of these compounds, as the original view that they act against cancer simply by blocking Ras farnesylation cannot be the whole story. The anticancer potential of inhibitors of other enzymes that are often required for processing prenylated proteins (such as endoproteases, methyltransferases and palmitoyltransferases) is a ripe area for new efforts in medicinal chemistry. Protein prenylation has revealed new features about the ways in which small GTP-binding proteins control many vital intracellular processes, including trafficking of membrane compartments within eukaryotic cells and cytoskeleton function. The analysis of these prenyl groups has set the stage for the discovery of soluble protein factors that move GTP-binding proteins from membranes into the soluble cellular compartment. There is much to be learned about the ways in which this translocation is coupled to processes such as cell morphology changes and cell movement and to transport processes such as protein secretion and neurotransmitter release.
Finally, it is estimated that a mere ~30% of prenylated proteins in eukaryotic cells have been identified. Thus, there may be other unexplored roles of protein prenyl groups beyond the well-established membrane-anchoring and molecular-handle functions.
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health grants AI054384 to M.H.G., GM41223 to S.M. and CA32737 and CA41996 to F.T.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Sakagami Y, et al. Isolation of a novel sex hormone tremerogen A-10, controlling conjugation tube formation in Tremella mesenterica fries. Agric. Biol. Chem. 1978;42:1093–1094. [Google Scholar]
- 2.Tsuchiya E, Fukui S, Kamiya Y, Sakagami Y, Fujino M. Requirements of chemical structure for hormonal activity of lipo-peptidyl factors inducing sexual differentiation in vegetative cells of heterobasidiomycetous yeasts. Biochem. Biophys. Res. Commun. 1978;85:459–463. doi: 10.1016/s0006-291x(78)80064-3. [DOI] [PubMed] [Google Scholar]
- 3.Anderegg RJ, Betz R, Carr SA, Crabb JW, Duntze W. Structure of Saccharomyces cerevisiae mating hormone a-factor. Identification of S-farnesyl cysteine as a structural component. J. Biol. Chem. 1988;263:18236–18240. [PubMed] [Google Scholar]
- 4.Glomset JA, Gelb MH, Farnsworth CC. Prenyl proteins in eukaryotic cells: a new type of membrane anchor. Trends Biochem. Sci. 1990;15:139–142. doi: 10.1016/0968-0004(90)90213-u. [DOI] [PubMed] [Google Scholar]
- 5.Farnsworth CC, Wolda SL, Gelb MH, Glomset JA. Human lamin B contains a farnesylated cysteine residue. J. Biol. Chem. 1989;264:20422–20429. [PMC free article] [PubMed] [Google Scholar]
- 6.Farnsworth CC, Gelb MH, Glomset JA. Identification of geranylgeranyl-modified proteins in HeLa cells. Science. 1990;247:320–322. doi: 10.1126/science.2296721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farnsworth CC, Seabra MC, Ericsson LH, Gelb MH, Glomset JA. Rab geranylgeranyl transferase catalyzes the geranylgeranylation of adjacent cysteines in the small GTPases, Rab1A, Rab3A, and Rab5A. Proc. Natl. Acad. Sci. USA. 1994;91:11963–11967. doi: 10.1073/pnas.91.25.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelb MH, Reiss Y, Ghomashchi F, Farnsworth CC. Exploring the specificity of prenyl protein-specific methyltranferase with synthetic prenylated rab peptides. Bioorg. Med. Chem. Lett. 1995;8:881–886. [Google Scholar]
- 9.Yokoyama K, Goodwin GW, Ghomashchi F, Glomset J, Gelb MH. Protein prenyltransferases. Biochem. Soc. Trans. 1992;20:489–494. doi: 10.1042/bst0200489. [DOI] [PubMed] [Google Scholar]
- 10.Casey PJ, Seabra MC. Protein prenyltransferases. J. Biol. Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 11.Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. 2003;22:5963–5974. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long SB, Casey PJ, Beese LS. Reaction path of protein farnesyltransferase at atomic resolution. Nature. 2002;419:645–650. doi: 10.1038/nature00986. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Seabra MC, Deisenhofer J. Crystal structure of Rab geranylgeranyltransferase at 2 angstrom resolution. Structure. 2000;8:241–251. doi: 10.1016/s0969-2126(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 14.Hicks KA, Hartman HL, Fierke CA. Upstream polybasic region in peptides enhances dual specificity for prenylation by both farnesyltransferase and geranylgeranyltransferase type I. Biochemistry. 2005;44:15325–15333. doi: 10.1021/bi050951v. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama K, Goodwin GW, Ghomashchi F, Glomset JA, Gelb MH. A protein geranylgeranyltransferase from bovine brain: implications for protein prenylation specificity. Proc. Natl. Acad. Sci. USA. 1991;88:5302–5306. doi: 10.1073/pnas.88.12.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J. Mol. Biol. 2004;343:417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 17.Long SB, Casey PJ, Beese LS. The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures. Structure. 2000;8:209–222. doi: 10.1016/s0969-2126(00)00096-4. [DOI] [PubMed] [Google Scholar]
- 18.Leung KF, Baron R, Seabra MC. Thematic review series: lipid posttranslational modifications. Geranylgeranylation of Rab GTPases. J. Lipid Res. 2006;47:467–475. doi: 10.1194/jlr.R500017-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 20.Young SG, Clarke S, Bergo MO, Philips M, Fong LG. In: The Enzymes. Clarke SG, Tamanoi F, editors. Vol. 24. Academic/Elsevier; San Diego: 2006. pp. 273–301. [DOI] [PubMed] [Google Scholar]
- 21.Tam A, et al. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J. Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 23.Trueblood CE, et al. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol. Cell. Biol. 2000;20:4381–4392. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam A, Schmidt WK, Michaelis S. The multispanning membrane protein Ste24p catalyzes CAAX proteolysis and NH2-terminal processing of the yeast a-factor precursor. J. Biol. Chem. 2001;276:46798–46806. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- 26.Ma YT, Chaudhuri A, Rando RR. Substrate specificity of the isoprenylated protein endoprotease. Biochemistry. 1992;31:11772–11777. doi: 10.1021/bi00162a014. [DOI] [PubMed] [Google Scholar]
- 27.Ma YT, Gilbert BA, Rando RR. Inhibitors of the isoprenylated protein endoprotease. Biochemistry. 1993;32:2386–2393. doi: 10.1021/bi00060a033. [DOI] [PubMed] [Google Scholar]
- 28.Jang GF, Gelb MH. Substrate specificity of mammalian prenyl protein-specific endoprotease activity. Biochemistry. 1998;37:4473–4481. doi: 10.1021/bi972289b. [DOI] [PubMed] [Google Scholar]
- 29.Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry. 2000;39:4096–4104. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Ma YT, Rando RR. Solubilization, partial purification, and affinity labeling of the membrane-bound isoprenylated protein endoprotease. Biochemistry. 1996;35:3227–3237. doi: 10.1021/bi952529s. [DOI] [PubMed] [Google Scholar]
- 31.Otto JC, et al. Cloning and characterization of a mammalian prenyl protein-specific endoprtease. J. Biol. Chem. 1999;274:8379–8382. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 32.Pei J, Grishin NV. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 2001;26:275–277. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- 33.Plummer LJ, et al. Mutational analysis of the ras converting enzyme reveals a requirement for glutamate and histidine residues. J. Biol. Chem. 2006;281:4596–4605. doi: 10.1074/jbc.M506284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergo MO, et al. On the physiological importance of endoproteolysis of CAAX proteins: heart-specific RCE1 knockout mice develop a lethal cardiomyopathy. J. Biol. Chem. 2004;279:4729–4736. doi: 10.1074/jbc.M310081200. [DOI] [PubMed] [Google Scholar]
- 35.Kim E, et al. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J. Biol. Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- 36.Bergo MO, et al. Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol. Cell. Biol. 2002;22:171–181. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlitzer M, Winter-Vann A, Casey PJ. Non-peptidic, non-prenylic inhibitors of the prenyl protein-specific protease Rce1. Bioorg. Med. Chem. Lett. 2001;11:425–427. doi: 10.1016/s0960-894x(00)00685-5. [DOI] [PubMed] [Google Scholar]
- 38.Romano JD, Schmidt WK, Michaelis S. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol. Biol. Cell. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romano JD, Michaelis S. Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Mol. Biol. Cell. 2001;12:1957–1971. doi: 10.1091/mbc.12.7.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson JL, Hrycyna CA. In: The Enzymes. Clarke SG, Tamanoi F, editors. Vol. 24. Academic/Elsevier; San Diego: 2006. pp. 245–272. [DOI] [PubMed] [Google Scholar]
- 41.Anderson JL, Frase H, Michaelis S, Hrycyna CA. Purification, functional reconstitution, and characterization of the Saccharomyces cerevisiae isoprenylcysteine carboxylmethyltransferase Ste14p. J. Biol. Chem. 2005;280:7336–7345. doi: 10.1074/jbc.M410292200. [DOI] [PubMed] [Google Scholar]
- 42.Bergo MO, et al. Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J. Biol. Chem. 2001;276:5841–5845. doi: 10.1074/jbc.C000831200. [DOI] [PubMed] [Google Scholar]
- 43.Bergo MO, et al. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J. Biol. Chem. 2000;275:17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- 44.Bergo MO, et al. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J. Clin. Invest. 2004;113:539–550. doi: 10.1172/JCI18829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sapperstein S, Berkower C, Michaelis S. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol. Cell. Biol. 1994;14:1438–1449. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright LP, Philips MR. CAAX modification and membrane targeting of Ras. J. Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Philips MR. Methotrexate and Ras methylation: a new trick for an old drug? Sci. STKE. 2004;2004:pe13. doi: 10.1126/stke.2252004pe13. [DOI] [PubMed] [Google Scholar]
- 48.Winter-Vann AM, et al. Targeting Ras signaling through inhibition of carboxyl methylation: an unexpected property of methotrexate. Proc. Natl. Acad. Sci. USA. 2003;100:6529–6534. doi: 10.1073/pnas.1135239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michaelson D, et al. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol. Biol. Cell. 2005;16:1606–1616. doi: 10.1091/mbc.E04-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter-Vann AM, et al. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc. Natl. Acad. Sci. USA. 2005;102:4336–4341. doi: 10.1073/pnas.0408107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson JL, Henriksen BS, Gibbs RA, Hrycyna CA. The isoprenoid substrate specificity of isoprenylcysteine carboxylmethyltransferase: development of novel inhibitors. J. Biol. Chem. 2005;280:29454–29461. doi: 10.1074/jbc.M504982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 53.Dietrich LEP, Ungermann C. On the mechanism of protein palmitoylation. EMBO Rep. 2004;5:1053–1057. doi: 10.1038/sj.embor.7400277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Linder ME, Deschenes RJ. New insights into the mechanisms of protein palmitoylation. Biochemistry. 2003;42:4311–4320. doi: 10.1021/bi034159a. [DOI] [PubMed] [Google Scholar]
- 55.Hancock JF. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 56.Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 57.Lawrence DS, Zilfou JT, Smith CD. Structure-activity studies of cerulenin analogues as protein palmitoylation inhibitors. J. Med. Chem. 1999;42:4932–4941. doi: 10.1021/jm980591s. [DOI] [PubMed] [Google Scholar]
- 58.Reents R, Wagner M, Kuhlmann J, Waldmann H. Synthesis and application of fluorescence-labeled Ras-proteins for live-cell imaging. Angew. Chem. Int. Edn Engl. 2004;43:2711–2714. doi: 10.1002/anie.200353265. [DOI] [PubMed] [Google Scholar]
- 59.Bartels DJ, Mitchell DA, Dong XW, Deschenes RJ. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6775–6787. doi: 10.1128/mcb.19.10.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 61.Swarthout JT, et al. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 62.Deck P, et al. Development and biological evaluation of acyl protein thioesterase 1 (APT1) inhibitors. Angew. Chem. Int. Edn Engl. 2005;44:4975–4980. doi: 10.1002/anie.200462625. [DOI] [PubMed] [Google Scholar]
- 63.Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 64.Bader B, et al. Bioorganic synthesis of lipid-modified proteins for the study of signal transduction. Nature. 2000;403:223–226. doi: 10.1038/35003249. [DOI] [PubMed] [Google Scholar]
- 65.Dudler T, Gelb MH. Palmitoylation of Ha-Ras facilitates membrane binding, activation of downstream effectors, and meiotic maturation in Xenopus oocytes. J. Biol. Chem. 1996;271:11541–11547. doi: 10.1074/jbc.271.19.11541. [DOI] [PubMed] [Google Scholar]
- 66.Silvius JR. Mechanisms of Ras protein targeting in mammalian cells. J. Membr. Biol. 2002;190:83–92. doi: 10.1007/s00232-002-1026-4. [DOI] [PubMed] [Google Scholar]
- 67.Rocks O, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 68.Goodwin JS, et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roy S, et al. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol. Cell. Biol. 2005;25:6722–6733. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wittinghofer A, Waldmann H. Ras - a molecular switch involved in tumor formation. Angew. Chem. Int. Edn Engl. 2000;39:4193–4214. doi: 10.1002/1521-3773(20001201)39:23<4192::AID-ANIE4192>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 71.Edidin M. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 72.Silvius JR. Lipidated peptides as tools for understanding the membrane interactions of lipid-modified proteins. Peptide-Lipid Interactions. 2002;52:371–395. [Google Scholar]
- 73.Hancock JF, Parton RG. Ras plasma membrane signalling platforms. Biochem. J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang TY, Leventis R, Silvius JR. Partitioning of lipidated peptide sequences into liquid-ordered lipid domains in model and biological membranes. Biochemistry. 2001;40:13031–13040. doi: 10.1021/bi0112311. [DOI] [PubMed] [Google Scholar]
- 75.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 76.Nicolini C, et al. Visualizing association of N-Ras in lipid microdomains: influence of domain structure and interfacial adsorption. J. Am. Chem. Soc. 2006;128:192–201. doi: 10.1021/ja055779x. [DOI] [PubMed] [Google Scholar]
- 77.Reuther G, et al. Structural model of the membrane-bound C terminus of lipid-modified human N-Ras protein. Angew. Chem. Int. Edn Engl. 2006;45:5387–5390. doi: 10.1002/anie.200504266. [DOI] [PubMed] [Google Scholar]
- 78.Rotblat B, et al. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol. Cell. Biol. 2004;24:6799–6810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Plowman SJ, Hancock JF. Ras signaling from plasma membrane and endo-membrane microdomains. Biochimica et Biophysica Acta-Molecular Cell Research. 2005;1746:274–283. doi: 10.1016/j.bbamcr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Hancock JF, Cadwallader K, Paterson H, Marshall CJA. CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okeley NM, Gelb MH. A designed probe for acidic phospholipids reveals the unique enriched anionic character of the cytosolic face of the mammalian plasma membrane. J. Biol. Chem. 2004;279:21833–21840. doi: 10.1074/jbc.M313469200. [DOI] [PubMed] [Google Scholar]
- 83.Ishizaki H, et al. Role of rab GDP dissociation inhibitor alpha in regulating plasticity of hippocampal neurotransmission. Proc. Natl. Acad. Sci. USA. 2000;97:11587–11592. doi: 10.1073/pnas.97.21.11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 85.Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 87.Pylypenko O, et al. Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI mediated Rab:membrane interaction. EMBO J. 2006;25:13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfeffer S, Aivazian D. Targeting RAB GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 89.Nilsson BL, Soellner MB, Raines RT. Chemical synthesis of proteins. Annu. Rev. Biophys. Biomol. Struct. 2005;34:91–118. doi: 10.1146/annurev.biophys.34.040204.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tamanoi F, Gau CL, Jiang C, Edamatsu H, Kato-Stankiewicz K. Protein farnesylation in mammalian cells: effects of farnesyltransferase inhibitors on cancer cells. Cell. Mol. Life Sci. 2001;58:1636–1649. doi: 10.1007/PL00000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Basso A, Kirshmeier P, Bishop WR. Farnesyltransferase inhibitors. J. Lipid Res. 2006;47:15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- 92.Reid TS, Beese LS. Crystal structures of the anticancer clinical candidates R115777 (Tipifarnib) and BMS-214662 complexed with protein farnesyltransferase suggest a mechanism of FTI selectivity. Biochemistry. 2004;43:6877–6884. doi: 10.1021/bi049723b. [DOI] [PubMed] [Google Scholar]
- 93.Lobell RB, et al. Preclinial and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol. Cancer Ther. 2002;1:747–758. [PubMed] [Google Scholar]
- 94.Reid TS, Long SB, Beese LS. Crystallographic analysis reveals that anticancer clinical candidate L-778,123 inhibits protein farnesyltransferase and geranylgeranyltransferase-I by different binding modes. Biochemistry. 2004;43:9000–9008. doi: 10.1021/bi049280b. [DOI] [PubMed] [Google Scholar]
- 95.Lackner MR, et al. Chemical genetics identifies Rab geranylgeranyl transferase as an apoptotic target of farnesyl transferase inhibitors. Cancer Cell. 2005;7:325–336. doi: 10.1016/j.ccr.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 96.Karp JE, et al. Clinical and biologic activity of the farnesyltransferase inhibitor R115777 in adults with refractory and relapsed acute leukemia: a phase I clinical-laboratory correlative trial. Blood. 2001;97:3361–3369. doi: 10.1182/blood.v97.11.3361. [DOI] [PubMed] [Google Scholar]
- 97.Alsina M, et al. Farnesyltransferase inhibitor tipifarnib is well tolerated, induces stabilization of disease and inhibits farnesylation and oncogenic/tumor survival pathways in patients with advanced multiple myeloma. Blood. 2004;103:3271–3277. doi: 10.1182/blood-2003-08-2764. [DOI] [PubMed] [Google Scholar]
- 98.Khuri FR, et al. Phase I study of the farnesyltransferase inhibitor lonafarnib with paclitaxel in solid tumors. Clin. Cancer Res. 2004;10:2968–2976. doi: 10.1158/1078-0432.ccr-03-0412. [DOI] [PubMed] [Google Scholar]
- 99.James GL, Goldstein JL, Brown MS. Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J. Biol. Chem. 1995;270:6221–6226. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 100.Whyte DB, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 101.Yoo J, Robinson RA. H-ras gene mutations in salivary gland mucoepidermoid carcinomas. Cancer. 2000;88:518–523. doi: 10.1002/(sici)1097-0142(20000201)88:3<518::aid-cncr4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 102.Burchill SA, Neal DE, Lunec J. Frequency of H-ras mutations in human bladder cancer detected by direct sequencing. Br. J. Urol. 1994;73:516–521. doi: 10.1111/j.1464-410x.1994.tb07636.x. [DOI] [PubMed] [Google Scholar]
- 103.Aoki Y, et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 104.Estep AL, Tidyman WE, Teitell MA, Cotter PD, Rauen KA. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am. J. Med. Genet. A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- 105.Gripp KW, et al. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am. J. Med. Genet. A. 2006;140:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- 106.Gomez M, Sampson J, Whittemore V. The Tuberous Sclerosis Complex. Oxford University Press; Oxford: 1999. [Google Scholar]
- 107.Gau C-L, et al. Farnesyltransferase inhibitors reverse altered growth and distribution of actin filaments in Tsc-deficient cells via inhibition of both rapamycin-sensitive and –insensitive pathways. Mol. Cancer Ther. 2005;4:918–926. doi: 10.1158/1535-7163.MCT-04-0347. [DOI] [PubMed] [Google Scholar]
- 108.Basso AD, et al. The farnesyltransferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J. Biol. Chem. 2005;280:31101–31108. doi: 10.1074/jbc.M503763200. [DOI] [PubMed] [Google Scholar]
- 109.Urano J, et al. Identification of novel single amino acid changes that result in hyperactivation of the unique GTPase, Rheb, in fission yeast. Mol. Microbiol. 2005;58:1074–1086. doi: 10.1111/j.1365-2958.2005.04877.x. [DOI] [PubMed] [Google Scholar]
- 110.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 111.Carsillo T, Astrindis A, Henske EP. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc. Natl. Acad. Sci. USA. 2000;97:6085–6090. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bardelli A, et al. PRL-3 expression in metastatic cancers. Clin. Cancer Res. 2003;9:5607–5615. [PubMed] [Google Scholar]
- 113.Fiordalisi JJ, Keller PJ, Cox AD. PRL tyrosine phosphatases regulate rho family GTPases to promote invasion and motility. Cancer Res. 2006;66:3153–3161. doi: 10.1158/0008-5472.CAN-05-3116. [DOI] [PubMed] [Google Scholar]
- 114.Lim K-H, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 115.Hakem A, et al. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Clark EA, Golub T, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 117.Carrico D, Blaskovich MA, Bucher C, Sebti S, Hamilton AD. Design, synthesis, and evaluation of potent and selective benzoyleneurea-based inhibitors of protein geranylgeranyltransferase-I. Bioorg. Med. Chem. 2005;13:677–688. doi: 10.1016/j.bmc.2004.10.053. [DOI] [PubMed] [Google Scholar]
- 118.Peterson YK, Kelly P, Weinbaum CA, Casey PJ. A novel protein geranylgeranyltransferase-I inhibitor with high potency, selectivity and cellular activity. J. Biol. Chem. 2006;281:12445–12450. doi: 10.1074/jbc.M600168200. [DOI] [PubMed] [Google Scholar]
- 119.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Young SG, Fong LG, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria–new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 121.Bergo MO, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pendas AM, et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 123.Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J. Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tam A, et al. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J. Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Capell BC, et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Glynn MW, Glover TW. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum. Mol. Genet. 2005;14:2959–2969. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- 127.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mallampalli MP, Huyer G, Bendale P, Gelb MH, Michaelis S. Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2005;102:14416–14421. doi: 10.1073/pnas.0503712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Toth JI, et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. USA. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang SH, et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. USA. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang SH, et al. Farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J. Clin. Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fong LG, et al. A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science. 2006;311:1621–1623. doi: 10.1126/science.1124875. [DOI] [PubMed] [Google Scholar]
- 133.Buckner FS, et al. Cloning, heterologous expression, and distinct substrate specificity of protein farnesyltransferase from Trypanosoma brucei. J. Biol. Chem. 2000;275:21870–21876. doi: 10.1074/jbc.M000975200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eastman RT, Buckner FS, Yokoyama K, Gelb MH, Van Voorhis WC. Thematic review series: lipid posttranslational modifications. Fighting parasitic disease by blocking protein farnesylation. J. Lipid Res. 2006;47:233–240. doi: 10.1194/jlr.R500016-JLR200. [DOI] [PubMed] [Google Scholar]
- 135.Buckner FS, Eastman RT, Yokoyama K, Gelb MH, Van Voorhis WC. Protein farnesyl transferase inhibitors for the treatment of malaria and African trypanosomiasis. Curr. Opin. Investig. Drugs. 2005;6:791–797. [PubMed] [Google Scholar]
- 136.Gelb MH, et al. Protein farnesyl and N-myristoyl transferases: piggy-back medicinal chemistry targets for the development of antitrypanosomatid and antimalarial therapeutics. Mol. Biochem. Parasitol. 2003;126:155–163. doi: 10.1016/s0166-6851(02)00282-7. [DOI] [PubMed] [Google Scholar]
- 137.Eastman RT, et al. Resistance to a protein farnesyltransferase inhibitor in Plasmodium falciparum. J. Biol. Chem. 2005;280:13554–13559. doi: 10.1074/jbc.M413556200. [DOI] [PubMed] [Google Scholar]
- 138.Nallan L, et al. Protein farnesyltransferase inhibitors exhibit potent antimalarial activity. J. Med. Chem. 2005;48:3704–3713. doi: 10.1021/jm0491039. [DOI] [PubMed] [Google Scholar]