Abstract
Classical late neuronal ceroid lipofuscinosis (LINCL) is a fatal neurodegenerative disease of children caused by mutations in TPP1, the gene encoding the lysosomal protease tripeptidyl peptidase 1. LINCL is characterized by lysosomal accumulation of storage material of which only a single protein component, subunit c of mitochondrial ATP synthase, has been well established to date. Identification of other protein constituents of the storage material could provide useful insights into the pathophysiology of disease and the natural substrates for TPP1. We have therefore initiated a proteomic analysis of storage material in brain from a LINCL mouse model. One protein, glial fibrillary acidic protein (GFAP), was found to be elevated in the LINCL mice compared to normal controls in both isolated storage bodies and a lysosome-enriched subcellular fraction that contains storage material. To determine whether GFAP accumulates within the lysosome in LINCL, we examined its intracellular distribution using subcellular fractionation and morphological methods. These experiments demonstrate that GFAP is not a component of the storage material in LINCL, suggesting that reports of GFAP storage in other NCLs may need to be reexamined. A number of other proteins were elevated in the storage material and/or lysosome-enriched fraction from the LINCL mice but it remains unclear whether these proteins are true constituents of the storage material or, like GFAP, if they associate with this material upon purification.
Keywords: glial fibrillary acidic protein (GFAP), classical late-infantile neuronal ceroid lipofuscinosis (LINCL), storage material, lysosome, mass spectrometry
INTRODUCTION
The neuronal ceroid lipofuscinoses (NCLs) are a group of hereditary neurodegenerative diseases that primarily affect children and adolescents [1]. Originally classified by age of onset and subcellular pathology, the identification of defects in multiple genes that can lead to NCL now allows for definitive diagnosis at the genetic level. To date, mutations in 8 different genes have been associated with the NCLs (reviewed in [2, 3]): congenital NCL caused by defects in cathepsin D; infantile NCL (INCL) caused by defects in palmitoyl protein thioesterase 1; classical late infantile NCL (LINCL) caused by defects in tripeptidyl peptidase 1 (TPP1); variant late infantile NCLs caused by defects in either of the CLN5, CLN6 or CLN8 proteins or the major facilitator superfamily domain-containing protein-8 (CLN7); and juvenile NCL (JNCL) caused by defects in the CLN3 protein. There are also individuals with adult onset NCLs that may represent attenuated forms of other NCLs or lysosomal storage diseases [4, 5], or which may have a genetically distinct but unknown basis.
The different forms of NCL share clinical characteristics with a progressive course that includes visual loss, seizures, mental retardation, movement disorders and shortened life span. Most of these symptoms reflect CNS involvement and there is a significant loss of neurons as well as astrocyte activation and gliosis accompanying neurodegeneration [6]. At the cellular level, NCLs are characterized by lysosomal accumulation of storage material, loosely termed ceroid lipofuscin, that is detectable using a wide range of excitation and emission wavelengths when viewed using a fluorescent microscope. When visualized by electron microscopy, the storage material exhibits characteristics that may be indicative of the particular disease: for example, granular osmiophilic deposits are found in INCL, curvilinear inclusions in LINCL and fingerprint profiles in JNCL (reviewed in [1]).
The precise composition of the NCL storage material is not clear but, based on biochemical analysis of “storage bodies” (insoluble material prepared by centrifugation), it does appear to consist mostly of protein [7]. The autofluorescence observed by microscopy is likely due to protein stacking rather than the presence of a stored fluorophore [8]. Analysis of a sheep NCL model linked to CLN6 [9, 10] revealed that a major component of the storage material is subunit c of mitochondrial ATP synthase (SCMAS) [11]. This small, highly hydrophobic protein was subsequently identified to be a major component of storage material in LINCL and most other NCLs except INCL and congenital ovine NCL [7, 11-14]. SCMAS also accumulates within lysosomes in a mouse model of mucopolysaccharidosis III B [15], a lysosomal storage disorder caused by mutations in the gene encoding alpha-N-acetylglucosaminidase that is not classified as an NCL, and has also been reported to accumulate in other lysosomal storage diseases [16, 17]. The widespread presence of SCMAS storage in multiple genetically-distinct diseases strongly suggests that in some cases its accumulation may be a secondary response to lysosomal dysfunction. However, the possibility that the missing gene products in these diseases, including TPP1 in LINCL, directly participate in the degradation of SCMAS cannot be discounted [18].
Lysosomal storage of several other proteins has also been reported in some NCLs. In INCL and congenital ovine NCL, the hydrophobic saposins A and D are a major protein component of storage material [19, 20]. The basis for their accumulation in these disorders is not clear, but saposins are endogenous lysosomal proteins that are upregulated in multiple lysosomal storage disorders [21]. In addition, in a recent study of an affected Tibetan terrier of unknown genetic etiology that represents an adult-onset NCL model, glial fibrillary acidic protein (GFAP) and histone H4 were reported to specifically accumulate within isolated storage bodies [22].
For lysosomal diseases in general, understanding the composition of the storage material can provide valuable insights into the physiopathology of disease and may reveal possible targets for therapeutic intervention. For LINCL, despite extensive characterization of the in vitro substrate specificity of TPP1 [23], little is known about the physiological substrates for the enzyme or, other than SCMAS, the composition of proteins and peptides that might be stored. In this study, we have used standard proteomics/mass-spectrometric methods to investigate storage material present in the brain of a mouse LINCL model. We find that a number of potential protein constituents including GFAP are elevated. However, further analyses clearly demonstrate that GFAP is not located within the lysosome but instead appears to adventitiously associate with lysosomes and storage bodies during isolation.
MATERIALS AND METHODS
Animals
All experiments and procedures involving live animals were conducted in compliance with approved Institutional Animal Care and Use Committee protocols. The Tpp1-targeted LINCL mouse, designated Tpp1−/−, contains ~0.2% residual TPP1 activity. The Npc2-targeted mouse, designated Npc2−/−, is also a hypomorph and contains <5% residual NPC2 protein. Both have been described previously [24, 25] and were used to create a Tpp1−/−, Npc2−/− double mutant. All mice were in a 129/SvEv strain background.
Isolation of storage bodies
Storage bodies were isolated from mouse brains using a protocol based upon a previously described method [26]. Briefly, 16 week old mice were euthanized with a sodium pentobarbital/sodium phenytoin mixture (Euthasol, Delmarva Laboratories, Inc., Midlothian, VA) and saline-perfused by intracardiac puncture. Brains were removed and disrupted in 10 volumes of deionized water using a Polytron Homogenizer (Brinkman, Westburg, NY). This and subsequent procedures were conducted at 0-4°C. The homogenate was sonicated for 1 minute and filtered through glass wool. Solid CsCl was added to a final density of 1.18g/ml then the homogenate was centrifuged for 1hr at 48,000 × g. The pellet containing storage bodies was washed twice by resuspension in deionized water and centrifugation for 15min at 48,000 × g. The pellet was collected and stored at −80°C before further analysis.
Subcellular fractionation
Mice were fasted overnight, then killed and perfused as described above. Brains were removed, immersed in 2-3 volumes of ice-cold 0.25M sucrose, and homogenized by three passes of a motor-driven Potter device (1500rpm). All subsequent procedures were conducted at 4°C. Subcellular organelles were fractionated by differential centrifugation using minor modifications [27] of classical methods [28]. Sucrose density gradient centrifugation was conducted by bottom loading of a combined ML fraction (see Results) as described previously [27].
Mass spectrometry
Protein samples from storage bodies and ML fractions were prepared for mass spectrometry by in-gel reduction, alkylation and trypsin digestion [29]. Digests were analyzed by nanospray LC-MS/MS using an LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA) as described [30]. Peak lists were generated [30] and used to search the NCBI m37 ENSEMBL 49.37b build of the mouse proteome using the X!Tandem module of GPM-XE Manager version 2.2.1 [31] with published parameters [4]. Relative protein abundance was estimated by spectral counting [32] using duplicate LC-MS/MS runs for each storage body sample (individual preparations from two controls and two Tpp1−/− animals) and duplicate LC-MS/MS runs for each ML sample (one control and one Tpp1−/− animal).. Spectral counts from a given sample type were summed and statistical analysis performed using R version 2.8.0 (http://www.r-project.org/) as described previously [33].
Western blotting
Whole mouse brain homogenates were prepared from thawed frozen tissue at 4°C using a Polytron (see above) in 0.15M NaCl containing 0.1% Triton X-100. Homogenates and subcellular fraction samples were separated by SDS-PAGE and transferred to PVDF. SCMAS was detected using a rabbit polyclonal antibody (generously provided by Dr. E.F. Neufeld [15] with a HRP-goat anti-rabbit IgG (cat # A8275; Sigma, St. Louis, MO). GFAP was detected using a mouse monoclonal antibody (cat # G3893; Sigma) with a HRP-goat anti-mouse IgG (cat # A2304; Sigma). Signals were visualized using the Super-Signal West Pico chemiluminescent system (Pierce Biotechnology, Inc., Rockford, IL).
Enzyme assays
β-galactosidase and cytochrome c oxidase were measured as described previously [34]. Protein levels were measured using the Advanced Protein Assay reagent (Cytoskeleton Inc., Denver, CO).
Confocal microscopy
Mice were euthanized as above, perfused with saline followed by 4% paraformaldehyde, and brains fixed overnight in 4% paraformaldehyde. Brains were embedded in Shandon M-1 Embedding Matrix (Thermo Fisher Scientific Inc., Waltham, MA) and used to prepare 10 μm sagittal cryosections. SCMAS and GFAP were analyzed using the primary antibodies used for western blotting (see above). Mannose 6-phosphorylated glycoproteins (M6PGPs) were analyzed using a biotinylated cation-independent mannose 6-phosphate receptor derivative as described previously [35]. Primary reagents were visualized with Alexafluor conjugated goat anti-rabbit antibodies, goat anti-mouse antibodies or streptavidin (Invitrogen).
Fluorescence images were obtained using a confocal laser-scanning microscope (LSM510; Carl Zeiss, Jena, Germany). For images shown in Fig. 4, conditions were used to maximize and minimize autofluoresence signal in the green channel and red channels, respectively. The green channel represents data acquired using the 488nm laser line and a longpass emission filter (515 nm and above). The red channel represents data acquired using the 568nm laser line and a 590-610nm narrow bandpass emission filter to visualize Alexafluor 568 conjugated secondary antibodies or Alexafluor 555 conjugated strepavidin. For images shown in Fig. 5, conditions were chosen to maximize specific staining and minimize contribution of autofluorescence to both the red and green channels. Here, the green channel represents data acquired using the 488nm laser line and a 515-540nm bandpass emission filter to visualize AlexaFluor 488 conjugated reagents (secondary antibodies or streptavidin) while the red channel was as described above.
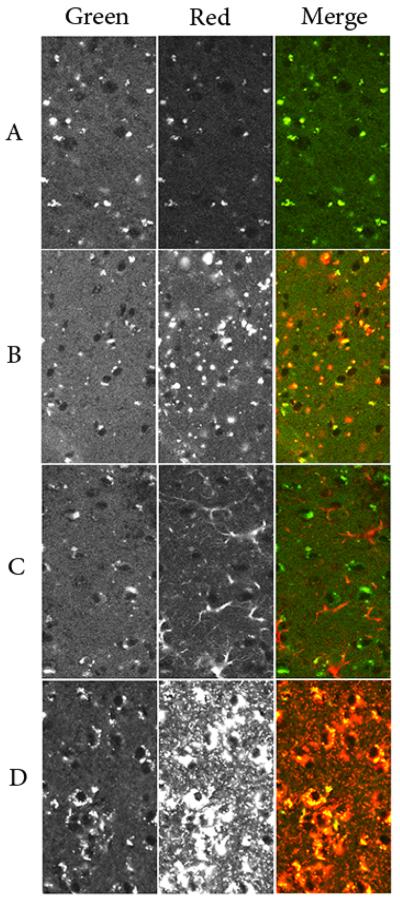
Figure 4. Autofluorescence and single-label confocal microscopy.
Cerebral cortex from 16-week old mice was examined by confocal microscopy. Left panels represent the signal in the green channel derived from autofluorescence. Middle panels represent signal in the red channel derived from autofluoresence alone (Panel A) or autofluorescence plus labeling for SCMAS (Panel B), GFAP (Panel C) or M6PGPs (Panel D). Right Panels show the merged images. All images were acquired and displayed using identical conditions to facilitate comparison and filter sets were chosen to maximize and minimize autofluorescence signal in the green channel and red channels, respectively (see Materials and Methods).
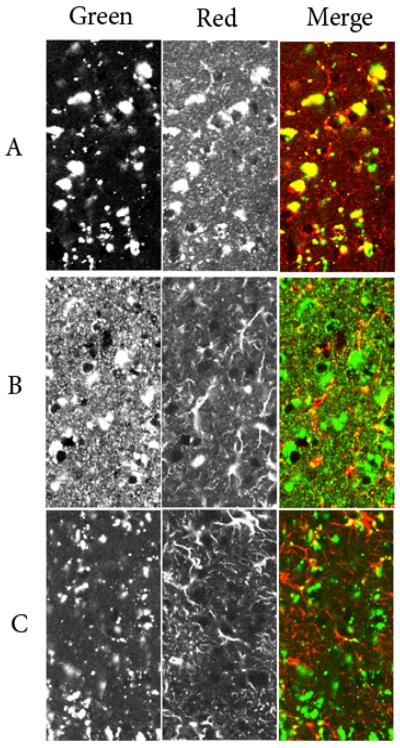
Figure 5. Double-label confocal microscopy.
Panel A: SCMAS - green channel, M6PGPs - red channel. Panel B, M6PGPs - green channel, GFAP - red channel. Panel C; SCMAS - green channel, GFAP - red channel. Conditions were as described in Materials and Methods.
RESULTS
Proteomic analysis of LINCL storage material
Our initial aim was to identify protein components of the storage material in LINCL. The approach was to prepare tryptic digests of fractions enriched in storage material from Tpp1−/−mouse brain and to identify peptides using mass spectrometric methods. Two different fractionation procedures were used. One procedure, the direct isolation of storage bodies from a sonicated whole brain homogenate [26], would be expected to yield protein aggregates that remain insoluble in aqueous solution in the absence of detergent. The other procedure, differential centrifugation to prepare an “ML” fraction (see below) enriched in lysosomes, would be expected to yield both insoluble and soluble storage material as well as a variety of resident proteins from lysosomes and other organelles. In addition, we conducted a parallel analysis on material derived from wild type mouse brain, with the expectation that peptides derived from storage material would be elevated in the Tpp1−/− samples compared to controls. A total of 630 proteins were confidently assigned when all data (i.e., storage body samples and ML fractions from control and Tpp1−/− mouse brains) were searched together in a “Mudpit” analysis (Supplemental Table 1).
A total of 596 proteins were confidently identified in the Tpp1−/− samples, 346 being found in both the ML fraction and storage samples, 135 being only detected in the ML fraction, and 115 only in the storage body samples (Supplemental Table 2). When using spectral counting to estimate relative protein abundance, a total of 15 proteins were found to be significantly elevated in the LINCL mouse model samples compared to controls (Table 1 and Supplemental Tables 2 and 3). Of these, only a single protein, glial fibrillary acidic protein (GFAP) was found to be elevated in both the ML fraction and isolated storage bodies.
Table 1. Proteins identified in storage body preparations or ML fractions that are elevated in the LINCL mouse compared to normal controls.
Only proteins that are elevated with statistical significance (q<=0.05, shaded) in one or both preparations are shown. Relative protein abundance is expressed in terms of spectral counts (see Materials and Methods and Supplemental Tables).
| ML fraction | Storage body | ||||
|---|---|---|---|---|---|
| Gene identifier | Protein name | TPP1−/− | TPP1+/+ | TPP1−/− | TPP1+/+ |
| ENSMUSG00000020460 | 40S ribosomal protein S27a | 1 | 0 | 70 | 11 |
| ENSMUSG00000022037 | Clusterin | 5 | 0 | 10 | 0 |
| ENSMUSG00000029131 | DnaJ homolog subfamily B member 6 | 0 | 0 | 12 | 1 |
| ENSMUSG00000020932 | Glial fibrillary acidic protein | 31 | 2 | 284 | 140 |
| ENSMUSG00000015656 | Heat shock cognate 71 kDa protein | 80 | 74 | 95 | 19 |
| ENSMUSG00000030120 | Myeloid leukemia factor 2 | 0 | 0 | 11 | 0 |
| ENSMUSG00000022055 | Neurofilament light polypeptide | 45 | 35 | 178 | 122 |
| ENSMUSG00000022565 | Plectin-1 | 2 | 0 | 22 | 5 |
| ENSMUSG00000069744 | Proteasome subunit beta type-3 | 0 | 0 | 9 | 0 |
| ENSMUSG00000022323 | Ribonuclease UK114 | 15 | 2 | 0 | 0 |
| ENSMUSG00000015837 | Sequestosome-1 | 0 | 0 | 15 | 0 |
| ENSMUSG00000026797 | Syntaxin-binding protein 1 | 54 | 69 | 70 | 38 |
| ENSMUSG00000036752 | Tubulin beta-2C chain | 227 | 236 | 351 | 273 |
| ENSMUSG00000050148 | Ubiquilin-2 | 0 | 0 | 14 | 0 |
| ENSMUSG00000026728 | Vimentin | 1 | 2 | 45 | 17 |
Elevation of GFAP in LINCL
A previous study reported that GFAP was abundant in storage body preparations from a dog model of adult-onset NCL and concluded that it represents a component of the storage material [22]. At first glance, our observations would be consistent with this finding. However, GFAP levels are frequently increased in neurodegenerative diseases, reflecting astrocyte activation and gliosis, thus another possibility is that this protein is simply a contaminant in the ML fraction and storage body samples from the LINCL mouse. To distinguish between these possibilities, we examined the cellular and subcellular distribution of GFAP in LINCL mouse brain.
Immunohistochemical analysis of LINCL mouse brain reveals robust GFAP staining throughout the brain of older mice [36] (also see below and data not shown). Here, increased levels of GFAP were demonstrated in whole brain homogenates by western blotting (Fig. 1, upper panel). In addition, a known component of the storage material, SCMAS, was also found to be increased in the LINCL mouse model (Fig. 1, lower panel).
Figure 1. GFAP levels are elevated in LINCL mouse brain.
GFAP (upper) and SCMAS (lower) were visualized by western blotting and chemiluminescence in brain homogenates (5 μg protein per lane) from 13 week-week old mice.
Subcellular localization of GFAP in LINCL
If the increased levels of GFAP in LINCL reflect lysosomal storage then it should be found within lysosomes in addition to its normal location within the cytoplasm. To investigate the cellular localization of GFAP, we used classical methods for subcellular fractionation, comparing the distribution of GFAP with that of markers for cellular organelles. As a first step, we used differential centrifugation to fractionate the homogenate into 4 fractions: N, composed primarily of the nucleus, unbroken cells, and large networks and aggregates; ML, the combined heavy and light mitochondrial fraction enriched in mitochondria, lysosomes, and peroxisomes; P, the microsomal fraction enriched in endoplasmic reticulum, plasma membrane and Golgi; and S, the high speed supernatant containing cytosol and the soluble contents of ruptured organelles.
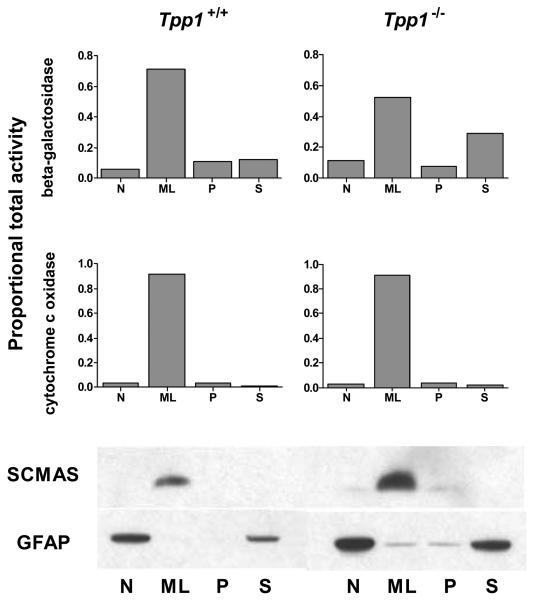
The only well-established storage marker in LINCL is SCMAS. This is normally a resident mitochondrial inner membrane protein but a substantial fraction of SCMAS accumulates within lysosomes in LINCL due to impaired lysosomal digestion following normal autophagic turnover of mitochondria [37]. As expected, SCMAS is found predominantly in the ML fractions in both Tpp1+/+ and Tpp1−/− mouse brain, consistent with its normal localization in mitochondria as well as its accumulation in lysosomes in LINCL. Although GFAP was found in significantly higher amounts in the TPP1-deficient sample, the overall distribution in the differential centrifugation fractions was similar to that of control (Fig 2, lower blot), with most of the GFAP being found in the N and S fractions. This is consistent with the existence of two forms of GFAP, an insoluble fibrillar form which is a component of the cytoskeleton and which sediments in the N fraction, and a soluble form that remains in the S fraction following high speed centrifugation [38]. While the ML fraction from Tpp1−/− mouse brain contains more GFAP than the wild-type control, the amount is still very low compared to levels in the N and S fractions and there is no evidence for specific accumulation.
Figure 2. Distribution of GFAP and SCMAS in wild-type and LINCL mouse brain after differential centrifugation.
Brains from 16 week-old wild type and Tpp1−/− mice were processed as described in Materials and Methods. Histograms show organellar markers were β-galactosidase (lysosomes) and cytochrome c oxidase (mitochondria). Western blotting was performed for SCMAS (upper blots) and GFAP (lower blots) using equivalent proportions of each fraction for the analysis.
While there was very little GFAP in the ML fraction, we could not exclude the possibility that a small amount accumulated in lysosomes. To further investigate the organellar association of GFAP within mouse brain ML fractions, we exploited the finding that deficiencies in NPC2, a resident lysosomal protein that binds cholesterol, result in an accumulation of lipids that alters the density of the lysosomes but not of other organelles [27]. This allows lysosomes to be readily distinguished from mitochondria and other cellular components after density gradient centrifugation. In preliminary experiments, the NPC2 deficiency was found to have no effect on the levels of SCMAS in wild-type and LINCL mice as determined by immunoblotting of differential centrifugation fractions (data not shown).
Sucrose density gradient centrifugation of ML fractions from Npc2−/−,Tpp1+/+ and Npc2−/−,Tpp1−/− mice are shown in Fig. 3 and profiles for mitochondria and lysosomes are determined by measurement of specific marker activities. Lysosomes are enriched in the fractions of lower density (~1.09-1.13g/ml) while mitochondria resolve to the higher density fractions (~1.15-1.20g/ml). Previous studies [27] indicate that NPC2-deficient lysosomes are well resolved from other cellular components (e.g., peroxisomes, endoplasmic reticulum, synaptic vesicles and plasma membrane) that are also located in the higher density fractions.
Figure 3. Distribution of GFAP and SCMAS in sucrose density gradients.
Mice (8-9 week old) were NPC2-deficient in order to shift the buoyant density of lysosomes away from that of mitochondria [27]. ML fractions were bottom loaded on sucrose density gradients and each gradient separated into 12 fractions which were analyzed for a lysosomal marker (β-galactosidase), a mitochondrial maker (cytochrome c oxidase), SCMAS (upper blots) and GFAP (lower blots). Note that the amount of SCMAS accumulated in the Tpp1−/− compared to the Tpp1+/+ sample is less than that shown in Fig. 2. This reflects a difference in the age of analysis of these mice rather than an effect of the Npc2 genotype: the Npc2−/− mice have a shortened lifespan compared to Tpp1−/− mice, necessitating use of younger animals for this analysis.
Western blotting for SCMAS was performed on the sucrose density gradient fractions to determine its subcellular localization (Fig. 3. upper blots). In the control, SCMAS exclusively codistributed with the mitochondrial marker and was not detected in the lysosomal fractions, as expected. In the case of LINCL (Fig. 3), SCMAS was again found in the mitochondrial fractions but there was additional detectable protein that codistributed with the lysosomal marker. These data are consistent with the known presence of SCMAS as a component of the LINCL storage material.
Immunodetection of GFAP across the sucrose density gradient is shown in Fig. 3. While there was more GFAP present in the LINCL mouse brain gradient compared to the control, the relative distribution of GFAP was essentially the same in both gradients. Importantly, unlike SCMAS, GFAP was not detected in the low density lysosomal peak in the LINCL sample suggesting that GFAP is not stored within lysosomes.
Morphological localization of GFAP in LINCL mouse brain
As a complementary approach, we used immunostaining and confocal microscopy to investigate whether GFAP is a component of the lysosomal storage material in LINCL mouse brain. We used both autofluorescence and SCMAS as a marker for storage material. Note that endogenous mitochondrial SCMAS was not detectable using these experimental conditions as ascertained by staining of wild type sections (data not shown and [15]). Lysosomal glycoproteins containing mannose 6-phosphate (M6PGPs) were used as a marker for neuronal lysosomes [35]. Importantly, experimental conditions were chosen to minimize overlap of autofluorescence and staining of specific markers.
Autofluorescence of storage material without immunostaining is shown in Fig. 4A. While essentially no signal was detectable in the wild-type control (data not shown), the LINCL mouse sample showed perinuclear signal in both the red (Fig. 4A, middle panel) and green channels (Fig. 4A, left panel). While there is complete overlap, in the merged image (Fig. 4A, right panel), the signal is predominantly green, reflecting the low relative intensity in the red channel due to choice of parameters used for image acquisition and display.
Images obtained after probing for a single marker are shown in Fig. 4, Panels B-D. While there is some section-to-section variation, the signal in the green channel is comparable to that of Panel A, reflecting similar levels of autofluorescence. In contrast, the signals derived from the probes were far stronger than that arising from autofluorescence (compare the red channel in Fig 4, Panel A with Panels B-D).
Immunostaining for SCMAS clearly demonstrates its expected colocalization with the autofluorescent storage material (Fig. 4B). GFAP immunostaining reveals the typical morphology of astrocytes, but there is no evident colocalization with autofluorescent storage material (Fig. 4C). The morphological marker for neuronal lysosomes, M6PGPs, were detected in the cytoplasm around the nucleus, and extensively overlap with the autofluorescent storage material (Fig 4D).
Double-staining experiments provided further evidence that GFAP does not localize to the storage material in the LINCL mouse model. In these experiments, we used conditions to minimize autofluorescence signal in both green and red channels, and also used probes in the green channel that previously were shown to colocalize with autofluorescent storage material (see Fig. 4). Fig. 5A demonstrates that the extensive SCMAS staining (green) colocalized with the M6PGPs (red), indicating a lysosomal distribution for the known component of the storage material. Based on cell morphology, immunostaining for GFAP indicated association with astrocytes but not neurons and there is no apparent colocalization with either SCMAS or M6PGPs (Fig. 5B and C). Taken together, these data clearly demonstrate that GFAP is not localized within the neuronal lysosomes that represent the primary site of storage in the LINCL mouse brain.
DISCUSSION
This study was initiated to identify potential in vivo substrates for TPP1 that accumulate in a mouse model of LINCL. To this end, we prepared insoluble storage bodies or an ML differential centrifugation fraction enriched in storage material from Tpp1−/− brain and analyzed tryptic digests of both preparations by standard LC-MS/MS proteomic methods. Equivalent samples from control mouse brains were analyzed in parallel. GFAP was clearly elevated in both the preparations from the LINCL model as well as in total brain homogenates. However, additional experiments using both subcellular fractionation and confocal microscopy demonstrated that very little if any of the GFAP represented lysosomal storage material and it instead appeared to be an adventitious contaminant of the preparations used for mass spectrometric analysis.
GFAP was previously identified from mass spectrometric analysis of storage bodies prepared from frozen brain tissue derived from an adult-onset Tibetan terrier NCL dog model [22]. Additional experiments comparing signals from autofluorescence and anti-GFAP indirect immunofluorescence using conventional fluorescent microscopy indicated considerable GFAP staining in non-autofluorescent cells with morphology typical of astrocytes. In addition, there was apparent colocalization of the GFAP signal with autofluorescence in what appeared to be neurons, leading to the suggestion that GFAP is a component of the storage material in these cells [22]. The Tibetan terrier NCL model reportedly does not represent a TPP1-deficiency and thus our results cannot necessarily rule out lysosomal storage of GFAP in this case. However, given our results and the clear potential for confounding artifacts, it is important to stress the need for orthogonal approaches to establish whether a component actually is stored in the lysosome. In particular, there are inherent limitations when using morphological approaches alone. For instance, even when using appropriate filter sets and controls omitting primary antibodies, it is possible that a given primary antibody would non-specifically associate with storage material, or due to accessibility conditions, only detect a small subset of the protein of interest present in the cell. The latter situation is clearly illustrated here in the case of SCMAS, where the subset of protein in the storage material can be detected but the larger pool of protein within the mitochondria is not seen. For such reasons, as stressed by de Duve and others, following preparative fractionation, analytical fractionation employing quantitative balance sheet methods to determine the relative amount of a given protein in different fractions with reference to the total sample is critical for assigning proteins to a given intracellular location [39]. With these considerations in mind, it would be of interest to revisit storage in the Tibetan terrier using additional biochemical and morphological approaches.
Our data indicate that GFAP is not stored in neuronal lysosomes. However, we do find elevated amounts of GFAP in the storage body samples and ML fractions from Tpp1−/− mouse brain and the basis for this is unknown. GFAP is the principle component of intermediate filaments in astrocytes and is synthesized in increased amounts during reactive gliosis accompanying neurodegeneration in multiple neurodegenerative diseases including NCLs. It is likely that a small amount of intermediate filaments containing GFAP are entrapped in the storage body and ML fractions during preparation. This well may have also occurred in the analysis of the Tibetian terrier model [22]. Indeed, it has been previously reported that GFAP constitutes a major fraction of the insoluble material in INCL brain autopsy specimens that initially cosediments with autofluorescent storage material but can be separated from this by further manipulations [40]. It is worth noting that vimentin, another intermediate filament protein, was also elevated in the storage body sample from the LINCL mouse, and this may represent the same phenomenon.
In addition to GFAP, a number of other proteins were also significantly elevated in the Tpp1−/− storage body samples compared to controls (Table 1). These proteins can be grouped according to their functions, and most fall into several categories. Many of these proteins are cytoskeletal/structural (e.g., plectin, GFAP, vimentin, neurofilament light polypeptide and tubulin beta-2C chain) whereas others are associated with the ubiquitin-proteosome degradation system (e.g., sequestosome-1, ubiquilin-2, proteasome subunit beta type-3) or heat stress response (e.g., heat shock cognate 71 kDa protein, DnaJ homolog subfamily B member 6). It is worth noting that some of these proteins have been reported to be elevated in neurodegenerative disease: for example, sequestosome-1 and ubiquilin-2 accumulate in neurofibrillary tangles in Alzheimer disease and in Lewy bodies of neurons in Parkinson disease [41, 42]. For most of these proteins, it is unclear whether any of these represent bona fide constituents of the storage material or if they are simply abundant or “sticky” proteins that are adventitiously associated with the storage body preparation. For heat shock proteins, however, there is evidence to indicate that some may be specifically associated with the lysosome [43]. Thus, further studies are warranted to investigate whether the elevated proteins identified here represent artifacts or true lysosomal constituents.
Finally, it is important to stress that the failure to detect a given protein using the mass spectrometric methods employed in our study does not mean that it is not present. For instance, SCMAS, a known major component of storage material in LINCL, was not identified in this study despite immunoblotting data to show that it was elevated in the LINCL mouse brain ML samples (Fig. 2) and storage body preparations (data not shown) used for our LC-MS/MS analysis. This can be rationalized in that detection of SCMAS and other highly hydrophobic proteins and peptides are often not seen using the typical trypsin digestion and LC separation methods used in many proteomics workflows, and that specialized conditions may be required for their identification [44]. In addition, extremely hydrophilic peptides or polypeptides lacking tryptic cleavage sites might also escape identification using standard conditions. Future studies using alternate sample preparation and analytical methods may therefore provide valuable insights into potential TPP1 substrates accumulating in LINCL.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grants NS37918 (P.L.) and FRFC grant 2.4543.08. (MJ). We would like to thank Dr. Elizabeth Neufeld (UCLA) for generously providing the SCMAS antibody, Drs. Loren W. Runnels, Donald Winkelmann and Mengqing Xiang for help with microscopy, and Dr. Haiyan Zheng for expert assistance with mass spectrometry.
Abbreviations
- GFAP
glial fibrillary acidic protein
- NCL
neuronal ceroid lipofuscinosis
- INCL
infantile NCL
- JNCL
juvenile NCL
- LINCL
classical late-infantile NCL
- TPP1
tripeptidyl peptidase 1
- SCMAS
subunit c of mitochondrial ATP synthase
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- M6PGPs
mannose 6-phosphorylated glycoproteins
REFERENCES
- 1.Hofmann SL, Peltonen L. The neuronal ceroid lipofuscinoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic & molecular bases of inherited disease. McGraw-Hill; New York: 2001. pp. 3877–3894. [Google Scholar]
- 2.Kohlschutter A, Schulz A. Towards understanding the neuronal ceroid lipofuscinoses. Brain Dev. 2009;31:499–502. doi: 10.1016/j.braindev.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Jalanko A, Braulke T. Neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta. 2009;1793:697–709. doi: 10.1016/j.bbamcr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Sleat DE, Ding L, Wang S, Zhao C, Wang Y, Xin W, Zheng H, Moore DF, Sims KB, Lobel P. Mass spectrometry-based protein profiling to determine the cause of lysosomal storage diseases of unknown etiology. Mol. Cell. Proteomics. 2009;8:1708–1718. doi: 10.1074/mcp.M900122-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Diggelen OP, Thobois S, Tilikete C, Zabot MT, Keulemans JL, van Bunderen PA, Taschner PE, Losekoot M, Voznyi YV. Adult neuronal ceroid lipofuscinosis with palmitoyl-protein thioesterase deficiency: first adult-onset patients of a childhood disease. Ann. Neurol. 2001;50:269–272. doi: 10.1002/ana.1103. [DOI] [PubMed] [Google Scholar]
- 6.Haltia M. The neuronal ceroid-lipofuscinoses: from past to present. Biochim. Biophys. Acta. 2006;1762:850–856. doi: 10.1016/j.bbadis.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Jolly RD, Palmer DN, Dalefield RR. The analytical approach to the nature of lipofuscin (age pigment) Arch. Gerontol. Geriatr. 2002;34:205–217. doi: 10.1016/s0167-4943(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 8.Palmer DN, Oswald MJ, Westlake VJ, Kay GW. The origin of fluorescence in the neuronal ceroid lipofuscinoses (Batten disease) and neuron cultures from affected sheep for studies of neurodegeneration. Arch. Gerontol. Geriatr. 2002;34:343–357. doi: 10.1016/s0167-4943(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 9.Broom MF, Zhou C, Broom JE, Barwell KJ, Jolly RD, Hill DF. Ovine neuronal ceroid lipofuscinosis: a large animal model syntenic with the human neuronal ceroid lipofuscinosis variant CLN6. J. Med. Genet. 1998;35:717–721. doi: 10.1136/jmg.35.9.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tammen I, Houweling PJ, Frugier T, Mitchell NL, Kay GW, Cavanagh JA, Cook RW, Raadsma HW, Palmer DN. A missense mutation (c.184C>T) in ovine CLN6 causes neuronal ceroid lipofuscinosis in Merino sheep whereas affected South Hampshire sheep have reduced levels of CLN6 mRNA. Biochim. Biophys. Acta. 2006;1762:898–905. doi: 10.1016/j.bbadis.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Palmer DN, Martinus RD, Cooper SM, Midwinter GG, Reid JC, Jolly RD. Ovine ceroid lipofuscinosis. The major lipopigment protein and the lipid-binding subunit of mitochondrial ATP synthase have the same NH2-terminal sequence. J. Biol. Chem. 1989;264:5736–5740. [PubMed] [Google Scholar]
- 12.Palmer DN, Fearnley IM, Medd SM, Walker JE, Martinus RD, Bayliss SL, Hall NA, Lake BD, Wolfe LS, Jolly RD. Lysosomal storage of the DCCD reactive proteolipid subunit of mitochondrial ATP synthase in human and ovine ceroid lipofuscinoses. Adv. Exp. Med. Biol. 1989;266:211–222. doi: 10.1007/978-1-4899-5339-1_15. discussion 223. [DOI] [PubMed] [Google Scholar]
- 13.Fearnley IM, Walker JE, Martinus RD, Jolly RD, Kirkland KB, Shaw GJ, Palmer DN. The sequence of the major protein stored in ovine ceroid lipofuscinosis is identical with that of the dicyclohexylcarbodiimide-reactive proteolipid of mitochondrial ATP synthase. Biochem. J. 1990;268:751–758. doi: 10.1042/bj2680751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyynela J, Suopanki J, Santavuori P, Baumann M, Haltia M. Variant late infantile neuronal ceroid-lipofuscinosis: pathology and biochemistry. J. Neuropathol. Exp. Neurol. 1997;56:369–375. doi: 10.1097/00005072-199704000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Ryazantsev S, Yu WH, Zhao HZ, Neufeld EF, Ohmi K. Lysosomal accumulation of SCMAS (subunit c of mitochondrial ATP synthase) in neurons of the mouse model of mucopolysaccharidosis III B. Mol. Genet. Metab. 2007;90:393–401. doi: 10.1016/j.ymgme.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elleder M, Sokolova J, Hrebicek M. Follow-up study of subunit c of mitochondrial ATP synthase (SCMAS) in Batten disease and in unrelated lysosomal disorders. Acta Neuropathol. 1997;93:379–390. doi: 10.1007/s004010050629. [DOI] [PubMed] [Google Scholar]
- 17.Kida E, Wisniewski KE, Golabek AA. Increased expression of subunit c of mitochondrial ATP synthase in brain tissue from neuronal ceroid lipofuscinoses and mucopolysaccharidosis cases but not in long-term fibroblast cultures. Neurosci. Lett. 1993;164:121–124. doi: 10.1016/0304-3940(93)90872-i. [DOI] [PubMed] [Google Scholar]
- 18.Ezaki J, Takeda-Ezaki M, Kominami E. Tripeptidyl peptidase I, the late infantile neuronal ceroid lipofuscinosis gene product, initiates the lysosomal degradation of subunit c of ATP synthase. J. Biochem. 2000;128:509–516. doi: 10.1093/oxfordjournals.jbchem.a022781. [DOI] [PubMed] [Google Scholar]
- 19.Tyynela J, Palmer DN, Baumann M, Haltia M. Storage of saposins A and D in infantile neuronal ceroid-lipofuscinosis. FEBS Lett. 1993;330:8–12. doi: 10.1016/0014-5793(93)80908-d. [DOI] [PubMed] [Google Scholar]
- 20.Tyynela J, Sohar I, Sleat DE, Gin RM, Donnelly RJ, Baumann M, Haltia M, Lobel P. A mutation in the ovine cathepsin D gene causes a congenital lysosomal storage disease with profound neurodegeneration. EMBO J. 2000;19:2786–2792. doi: 10.1093/emboj/19.12.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang MH, Bindloss CA, Grabowski GA, Qi X, Winchester B, Hopwood JJ, Meikle PJ. Saposins A, B, C, and D in plasma of patients with lysosomal storage disorders. Clin. Chem. 2000;46:167–174. [PubMed] [Google Scholar]
- 22.Katz ML, Sanders DN, Mooney BP, Johnson GS. Accumulation of glial fibrillary acidic protein and histone H4 in brain storage bodies of Tibetan terriers with hereditary neuronal ceroid lipofuscinosis. J. Inherit. Metab. Dis. 2007;30:952–963. doi: 10.1007/s10545-007-0683-y. [DOI] [PubMed] [Google Scholar]
- 23.Tian Y, Sohar I, Taylor JW, Lobel P. Determination of the substrate specificity of tripeptidyl-peptidase I using combinatorial peptide libraries and development of improved fluorogenic substrates. J. Biol. Chem. 2006;281:6559–6572. doi: 10.1074/jbc.M507336200. [DOI] [PubMed] [Google Scholar]
- 24.Sleat DE, Wiseman JA, El-Banna M, Kim KH, Mao Q, Price S, Macauley SL, Sidman RL, Shen MM, Zhao Q, Passini MA, Davidson BL, Stewart GR, Lobel P. A mouse model of classical late-infantile neuronal ceroid lipofuscinosis based on targeted disruption of the CLN2 gene results in a loss of tripeptidyl-peptidase I activity and progressive neurodegeneration. J. Neurosci. 2004;24:9117–9126. doi: 10.1523/JNEUROSCI.2729-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sleat DE, Wiseman JA, El-Banna M, Price SM, Verot L, Shen MM, Tint GS, Vanier MT, Walkley SU, Lobel P. Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5886–5891. doi: 10.1073/pnas.0308456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer DN, Martinus RD, Barns G, Reeves RD, Jolly RD. Ovine ceroid-lipofuscinosis. I: Lipopigment composition is indicative of a lysosomal proteinosis. Am. J. Med. Genet. Suppl. 1988;5:141–158. doi: 10.1002/ajmg.1320310618. [DOI] [PubMed] [Google Scholar]
- 27.Della Valle MC, Sleat DE, Sohar I, Wen T, Pintar JE, Jadot M, Lobel P. Demonstration of lysosomal localization for the mammalian ependymin-related protein using classical approaches combined with a novel density shift method. J. Biol. Chem. 2006;281:35436–35445. doi: 10.1074/jbc.M606208200. [DOI] [PubMed] [Google Scholar]
- 28.de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 1955;60:604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sleat DE, Della Valle MC, Zheng H, Moore DF, Lobel P. The mannose 6-phosphate glycoprotein proteome. J. Proteome Res. 2008;7:3010–3021. doi: 10.1021/pr800135v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sleat DE, Wang Y, Sohar I, Lackland H, Li Y, Li H, Zheng H, Lobel P. Identification and validation of mannose 6-phosphate glycoproteins in human plasma reveal a wide range of lysosomal and non-lysosomal proteins. Mol. Cell. Proteomics. 2006;5:1942–1956. doi: 10.1074/mcp.M600030-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Beavis RC. Using the global proteome machine for protein identification. Methods Mol. Biol. 2006;328:217–228. doi: 10.1385/1-59745-026-X:217. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 33.Qian M, Sleat DE, Zheng H, Moore D, Lobel P. Proteomics analysis of serum from mutant mice reveals lysosomal proteins selectively transported by each of the two mannose 6-phosphate receptors. Mol. Cell. Proteomics. 2008;7:58–70. doi: 10.1074/mcp.M700217-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Sleat DE, Sohar I, Lackland H, Majercak J, Lobel P. Rat brain contains high levels of mannose-6-phosphorylated glycoproteins including lysosomal enzymes and palmitoyl-protein thioesterase, an enzyme implicated in infantile neuronal lipofuscinosis. J. Biol. Chem. 1996;271:19191–19198. doi: 10.1074/jbc.271.32.19191. [DOI] [PubMed] [Google Scholar]
- 35.Jadot M, Lin L, Sleat DE, Sohar I, Hsu MS, Pintar J, Dubois F, Wattiaux-De Coninck S, Wattiaux R, Lobel P. Subcellular localization of mannose 6-phosphate glycoproteins in rat brain. J. Biol. Chem. 1999;274:21104–21113. doi: 10.1074/jbc.274.30.21104. [DOI] [PubMed] [Google Scholar]
- 36.Chang M, Cooper JD, Sleat DE, Cheng SH, Dodge JC, Passini MA, Lobel P, Davidson BL. Intraventricular enzyme replacement improves disease phenotypes in a mouse model of late infantile neuronal ceroid lipofuscinosis. Mol. Ther. 2008;16:649–656. doi: 10.1038/mt.2008.9. [DOI] [PubMed] [Google Scholar]
- 37.Ezaki J, Wolfe LS, Higuti T, Ishidoh K, Kominami E. Specific delay of degradation of mitochondrial ATP synthase subunit c in late infantile neuronal ceroid lipofuscinosis (Batten disease) J. Neurochem. 1995;64:733–741. doi: 10.1046/j.1471-4159.1995.64020733.x. [DOI] [PubMed] [Google Scholar]
- 38.Inagaki M, Nakamura Y, Takeda M, Nishimura T, Inagaki N. Glial fibrillary acidic protein: dynamic property and regulation by phosphorylation. Brain Pathol. 1994;4:239–243. doi: 10.1111/j.1750-3639.1994.tb00839.x. [DOI] [PubMed] [Google Scholar]
- 39.de Duve C. Tissue Fraction-Past and Present. J. Cell Biol. 1971;50:20. [PMC free article] [PubMed] [Google Scholar]
- 40.Paetau A, Elovaara I, Paasivuo R, Virtanen I, Palo J, Haltia M. Glial filaments are a major brain fraction in infantile neuronal ceroid-lipofuscinosis. Acta Neuropathol. 1985;65:190–194. doi: 10.1007/BF00686997. [DOI] [PubMed] [Google Scholar]
- 41.Massey LK, Mah AL, Ford DL, Miller J, Liang J, Doong H, Monteiro MJ. Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J. Alzheimers Dis. 2004;6:79–92. doi: 10.3233/jad-2004-6109. [DOI] [PubMed] [Google Scholar]
- 42.Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, Wooten MC. Signaling, Polyubiquitination, Trafficking, and Inclusions: Sequestosome 1/p62's Role in Neurodegenerative Disease. J. Biomed. Biotechnol. 2006;2006:62079. doi: 10.1155/JBB/2006/62079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bandyopadhyay U, Cuervo AM. Entering the lysosome through a transient gate by chaperone-mediated autophagy. Autophagy. 2008;4:1101–1103. doi: 10.4161/auto.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carroll J, Altman MC, Fearnley IM, Walker JE. Identification of membrane proteins by tandem mass spectrometry of protein ions. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14330–14335. doi: 10.1073/pnas.0706817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.