Abstract
Most, if not all, animals coexist with a complement of prokaryotic symbionts that confer a variety of physiologic benefits. In humans, the interaction between animal and bacterial cells is especially important in the gastrointestinal tract. Technical and conceptual advances have enabled rapid progress in characterizing the taxonomic composition, metabolic capacity, and immunomodulatory activity of the human gut microbiota, allowing us to establish its role in human health and disease. The human host coevolved with a normal microbiota over millennia and developed, deployed, and optimized complex immune mechanisms that monitor and control this microbial ecosystem. These cellular mechanisms have homeostatic roles beyond the traditional concept of defense against potential pathogens, suggesting these pathways contribute directly to the well-being of the gut. During their coevolution, the bacterial microbiota has established multiple mechanisms to influence the eukaryotic host, generally in a beneficial fashion, and maintain their stable niche. The prokaryotic genomes of the human microbiota encode a spectrum of metabolic capabilities beyond that of the host genome, making the microbiota an integral component of human physiology. Gaining a fuller understanding of both partners in the normal gut-microbiota interaction may shed light on how the relationship can go awry and contribute to a spectrum of immune, inflammatory, and metabolic disorders and may reveal mechanisms by which this relationship could be manipulated toward therapeutic ends.
The human gut is in continuous contact with prokaryotes, whereas other tissues, such as the endothelium, must remain essentially sterile for local function and the health of the individual. The implications of this seemingly obvious statement reflect an emerging theme in medicine that has only recently come to the forefront of our general view of gastrointestinal function—that microbes affect our biology and our health in profound and perhaps previously unsuspected ways. Outside the field of medicine, biologists have long known that many, if not all, multicellular organisms thrive in commensal or symbiotic relationships with prokaryotes. It is important to consider the role of microbes in the gut through the lens of the evolutionary history of prokaryotic-eukaryote relations, depart from the usual paradigm of microbes as presumptive pathogens, and assume that prokaryotic-eukaryotic interactions in the gut are generally mutually beneficial. There are more than 2000 species of commensal bacterial organisms within our bodies, the vast majority in the gut, and only about 100 known species of pathogen.1 What are the known, suspected, and proposed contributions made by the gut microbiota to normal intestinal biology and thus health? This review considers consequences of deficiencies/dysbiosis of the microbiota and its role in inflammatory, immune, and infectious diseases of the gut (and potentially elsewhere) and addresses the issue of therapeutic manipulation of the gut microbiota, encompassing the prebiotic, probiotic, and postbiotic therapeutic rationales and potential clinical utility. To address these topics in light of basic biology, it is instructive to consider eukaryotic-prokaryotic interactions as reflected in our own evolutionary history.
Eukaryotic-Prokaryotic Interactions
Evolutionary History of the Gut and Its Associates
Eukaryotes existed in close alliance with microbes even before the appearance of multicellular life.2–4 Indeed, the very nature of the ancestral eukaryotic animal cell was radically changed by the capture and co-option of endosymbiotic prokaryotes that had the biochemical/metabolic capacity for oxidative phosphorylation, resulting in the formation of the mitochondria, intracellular organelles that are vital and defining features of eukaryotic animal cells.5 Symbiotic relationships, by definition, are mutually beneficial (in commensalism one party benefits while the other is unaffected); in this review, the human microbiota will be referred to as symbiotic. In most examples of eukaryotic-prokaryotic symbioses, the microbe benefits by acquisition of a stable nutrient supply and immediate environment, while the eukaryotic host gains extended metabolic/digestive ability and competitive exclusion of less-benign microbes, an arrangement that clearly applies to the intestine-microbiota system.
Planktonic bacteria originally associated and, to use a term that we will return to repeatedly, coevolved with simple marine invertebrates.2 Once the animal body plan reached sufficient size, exceeding the diffusion radius of molecules used as energy sources, a gut tube became necessary for capture, concentration, and extraction of soluble environmental nutrients. Not coincidentally, the gut structure developed into an attractive niche for otherwise free-living bacteria. Thus, as is widely exemplified across modern phyla, many animals acquired a gut-inhabiting prokaryotic microbiota. During long-standing interactions, mutual coevolution of host and microbe occurred, resulting in biochemical specialization of the ectosymbiotic microbes to most efficiently utilize energy sources provided (eaten) by host and adaptation of the host to assimilate and utilize the novel metabolic processes conferred by bacteria (providing the selective pressure for their continued presence).6 For example, ruminant (cud-chewing) mammals require a community of cellulolytic bacteria in the intestine to digest grassy food stuffs, without which they could not derive sufficient energy.7 Similarly, lignocellulose is a primary food source of termites (Nasutitermes) requiring the presence of a gut community of prokaryotes that hydrolyze cellulose and xylan.8 Thus, a wide variety of animals depend on novel biochemistries provided by gut symbionts for energy extraction.
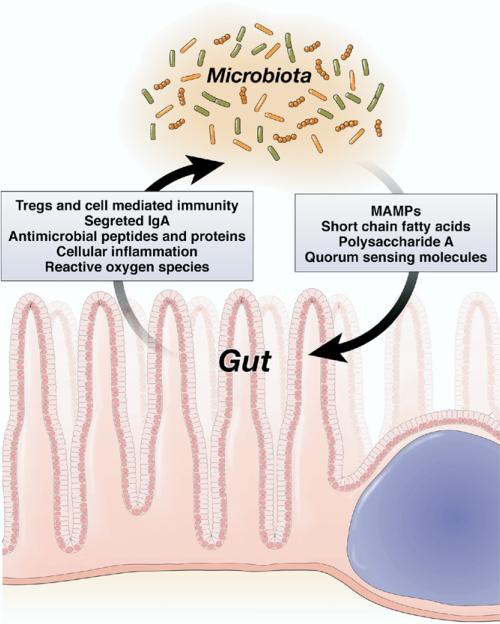
Both parties in the symbiotic dyad have established elaborate checks and balances to influence and regulate—but not harm—each other and thus ensure their continued “place at the table” (Figure 1). The immune system, broadly defined, is entrusted to modulate bacterial numbers and perhaps diversity, whereas the resident prokaryotes have their own means to deliberately modify host processes. This mutual crosstalk often involves reciprocal manipulation of growth, survival, and inflammatory controls and is another dimension of the gutmicrobe relationship, which when disturbed can result in a spectrum of intestinal disorders. The gut was designed for nutrient concentration and absorption in multicellular animals and became the dominant arena and proving ground of eukaryotic-prokaryotic interactions in humans. To consider the role of this relationship in human health and disease, it is necessary to ask, “What does a eukaryotic cell do when it encounters a prokaryotic cell?”
Figure 1.
Mechanisms of microbiota and gut crosstalk. Both parties in the symbiotic dyad possess means to alter and shape each other, resulting in a “negotiated settlement” at equilibrium. A breakdown on this crosstalk may result in a “dysbiotic” microbiota and clinical consequences.
Pattern Recognition and Microbial Perception
To tolerate a microbiota and realize its benefits, the eukaryotic host must maintain surveillance over the microbiota and control its number and composition. The human gut is equipped with both innate and adaptive immunity to perform these functions. The detailed mechanisms by which host nonadaptive immunity monitors and responds to the presence of microbes is a topic of intense interest and has been extensively reviewed.9–13 The general paradigm holds that the gut (among other cells) is equipped with pattern recognition receptors (PRRs), an operational term for transmembrane or intracytoplasmic receptors that are defined by the ability to specifically recognize and bind distinctive microbial macromolecular ligands designated as microbial-associated molecular patterns (MAMPs) such as lipopolysaccharide, flagellin, peptidoglycans, formylated peptides, and so on (Table 1). PRRs include the trans-membrane Toll-like receptors (TLRs), which scan the extracellular space, whereas the Nod-like receptors (NLRs) guard the intracellular cytoplasmic compartment. The association of mutant forms of the NLR Nod2 with Crohn's disease clearly underscores the importance of PRR monitoring in intestinal health. Functionally related RIG-like helicases and C-type lectin receptors are PRRs involved in detection of viral and fungal components, respectively. Formylated peptide receptors are a type of transmembrane PRR that is expressed in neutrophils, where they perceive bacterial cell wall products (formylated peptides) and stimulate neutrophil functions, such as reduced NADPH oxide (NOX)–dependent reactive oxygen generation and phagocytic motility.14 Recently, several paralogues of the neutrophil formylated peptide receptor have been characterized in intestinal epithelial cells, prompting interest that these epithelial receptors mediate microbial monitoring in the gut in a manner analogous to their traditional functions in phagocytes.15
Table 1.
PRRs, Their MAMP Ligands, and Subcellular Expression Patterns
| PRR | Ligand MAMP | Cellular location |
|---|---|---|
| TLR1 | Lipopeptides | Surface membrane |
| TLR2 | Lipoprotein, lipoteichoic acid, others | Surface membrane |
| TLR3 | dsRNA | Endosome membrane |
| TLR4 | Lipopolysaccharide | Surface membrane |
| TLR5 | Flagellin | Surface membrane |
| TLR6 | Lipoprotein, lipoteichoic acid, others | Surface membrane |
| TLR7 | Viral RNA | Endosome membrane |
| TLR8 | Viral RNA | Endosome membrane |
| TLR9 | CpG DNA | Endosome membrane |
| TLR10 | Unknown | Surface membrane |
| TLR11 | Profilin | Surface membrane |
| Nod1 | Gram negative peptidoglycan | Cytoplasmic |
| Nod2 | Gram negative and positive peptidoglycan | Cytoplasmic |
| IPAF | Flagellin | Cytoplasmic |
| NALP3 | RNA | Cytoplasmic |
| FPR | Formylated peptides, ? | Surface membrane |
| FPRL 1–2 | Formylated peptides, ? | Surface membrane |
| RIG-like helicases | Viral RNA | Cytoplasmic |
| C-type lectins | Fungal carbohydrates | Surface membrane |
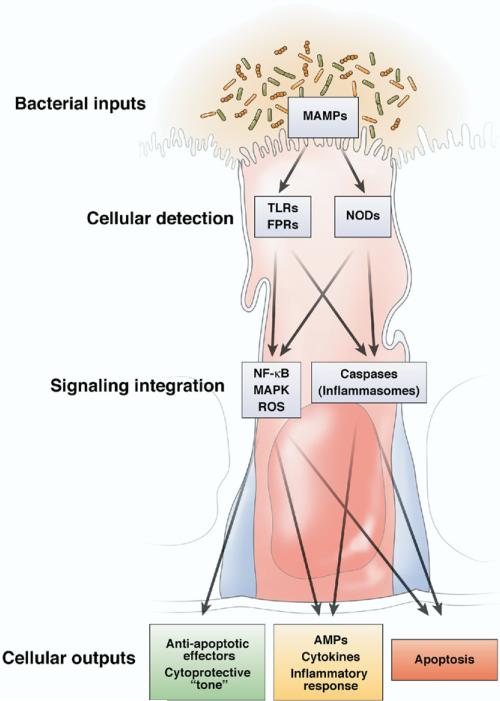
PRRs are in all multicellular organisms. Several components of the Toll pathway that are involved in microbe recognition have been characterized in nematodes.16 The TLRs themselves were first described in Drosophila and are necessary for defense against gram-positive bacteria.17 The NLRs serve as the primary microbial sensors in plant immunity. PRR-stimulated signaling pathways are also highly conserved. In the gut, activation of PRRs initiates regulatory pathways such as the mitogen-activated protein kinase (MAPK) and nuclear factor κB (NF-κB)/Rel pathways, as well as caspase-dependent signaling cascades such as the inflammasome.9,12,18,19 These systems represent intertwined cytoplasmic information relays, which when activated use rapid posttranslational events (covalent protein modifications and regulated protein degradation) to transduce PRR binding into transcriptional or posttranscriptional effector processes (Figure 2). Microbe-elicited signaling occurs through the Rel/NF-κB and mitogen-activated protein kinase pathways in Drosophila17,20 and in nematodes.21 Thus, evolution has conserved the mechanisms necessary to detect and respond to microbes, mechanisms that mediate both the benefits and the hazards of eukaryotic-prokaryotic interactions.
Figure 2.
Cellular consequences to bacterial stimuli. Bacterial MAMPs may stimulate pattern recognition receptors (including extracellular TLRs and formylated peptide receptors, or intracellular Nods). Intensity, duration, and spatial origin of the subsequent signaling responses are integrated by an intricate and interrelated network of transduction pathways that determine if MAMP perception warrants a “low gain” cytoprotective response, a “medium gain” inflammatory reaction, or “high gain” programmed cell death result.
Host Control of Microbes
Antimicrobial effectors (or effectors that influence microbes; not all responses are microbicidal) are also highly conserved. In invertebrates, the major effector arm is transcriptional up-regulation of small antimicrobial peptides.22 These molecules, observed in many multicellular organisms, are small cationic peptides that function by assembling and forming pores at the bacterial cell wall. In small intestinal Paneth cells, release of α-defensins or cryptdins is proposed to maintain the sterility of the proliferative stem cell compartment present in the small intestinal crypts23 and is necessary for defense against overt pathogens such as Salmonella.24 Secreted proteins such as RegIIIγ (a C-type lectin) and angiogenin 4 have also been shown to possess potent antimicrobial activity in vertebrates and are induced in the mucosa in response to contact with symbiotic bacteria.25,26 These highly conserved effectors generally exhibit selectivity toward either gram-negative or gram-positive bacteria, a property shared with secreted effectors in invertebrate immunity.
The rapid generation of reactive oxygen species (ROS) is a cardinal feature of the phagocyte reaction to both pathogenic and symbiotic bacteria; there is evidence that ROS are also physiologically elicited in other cell types, including intestinal epithelia, in response to microbial signals.27 ROS are short-lived reactive molecules that derive from the incomplete reduction of O2 and are considered microbicidal, although ROS also serve as critical second messengers in diverse intracellular signaling networks.28,29 Invertebrate phagocytes stimulated with formylated peptides generate ROS in the same manner as mammalian neutrophils.30 In Drosophila, commensal microbe-induced hydrogen peroxide (H2O2) maintains gut epithelial homeostasis31–33; plants induce ROS to modulate signal transduction pathways in response to bacterial pathogens and symbionts.34–36 Interestingly, experiments with several species of normal human gut bacteria have shown that the mammalian epithelium also responds to prokaryotic symbionts with rapid generation of epithelial ROS,37 again showing conservation of a response to microbes throughout evolution.
Enzymes that are homologous to phagocytic reduced NOX generate ROS in nonphagocytic eukaryotic cells and are widely distributed among multicellular organisms.27,38,39 Interestingly, 2 examples of this class of ROS-generating enzyme, NOX1 and DUOX2, are highly and specifically expressed in colonic epithelial cells,40 although a functional role for these enzymes has not been identified in the mammalian gut. It will be interesting to determine whether microbe-elicited ROS generation in the intestinal epithelia facilitates a microbicidal/microbistatic role, as in phagocytes, or mediates a signaling function (or both).29,41
Discrimination of Pathogen Versus Symbiont
When we consider the innate immune monitoring of the human gut, or for that matter any host-microbe symbiotic relationship, a major conundrum is how PRR monitoring and response distinguishes between the abundant normal microbiota and the rare pathogen. None of the conserved systems discussed that have evolved to monitor prokaryotes and are conserved across multicellular organisms discriminate between symbiotic and pathogenic organisms; in the case of many of the Enterobacteriaceae, the difference between symbiont and pathogen can be acquisition of a single plasmid. This realization has resulted in the current designation of PRR ligands as MAMPs, as opposed to the prior, more limited and “pathocentric” term “pathogen-associated molecular patterns.” Although some gut symbionts can limit or alter their surface MAMPs to evade PRR monitoring, so can pathogens.42 Similarly, PRR expression or signaling can be attenuated by various mechanisms, such as through down-regulation of TLR expression at transcriptional43 or posttranscriptional levels44,45 or tissue-specific expression of inhibitory signaling intermediates.46 Intestinal alkaline phosphatases may alter lipopolysaccharide structure and reduce the ability of this MAMP to stimulate eukaryotic cells.47 Although these mechanisms are likely important gut-specific specializations designed to allow the mucosa to avoid constitutive inflammation following contact with the microbiota and its products, they are not selective for specific microbes.
PRRs are strategically deployed; there is receptor-specific spatial distribution that contributes to discrimination between pathogen and symbiont.9 TLR5 seems to function as a primary epithelial sentry, monitoring the basolateral surface of the epithelium but not the apical luminal surface. TLRs 2 and 4 are found enriched on the surface of macrophages, less exposed to the MAMP-filled environment of the gut lumen, but poised to survey the less microbial-rich intercellular space. Expression of TLR9 is restricted to the cytoplasmic vacuolar compartments, sheltered from the extracellular environment. TLR9-induced signals may be different depending on whether the ligand (bacterial DNA) interacts with the apical or basolateral surface of the epithelial cells,41 suggesting that directional vesicular trafficking modulates MAMP-elicited responses. TLR expression patterns allow for bacteria detected in these areas to be perceived as pathogens and evoke overt inflammatory reactions. The fact that human gut microbes are symbionts in the intestinal lumen but can elicit fatal peritonitis or sepsis in other areas of the body illustrates this concept. Similarly, at the cellular level, MAMP detection by the intracellular NLRs evokes more severe cellular defensive reactions, up to and including programmed cell death, appropriate for the more dire situation caused by violation of cellular integrity by an invasive organism12,48 (Figure 2). Pathogens, impelled by their life cycles, utilize their virulence factors to enter areas where coevolution has not allowed the host to tolerate them and are caught. Yet, PRRs still signal the presence of nonpathogenic bacteria, albeit with a likely graded or dampened response. These tempered responses are not overtly inflammatory and may actually have salutary outcomes.
In summary, pattern recognition receptors and innate immune effector circuitry have evolved to allow the gut to perceive and respond to bacterial threats. However, these responses must not be so effective so as to eliminate the microbiota and the benefits it offers.
The Microbiota in Health
Overview of Human Microbiota
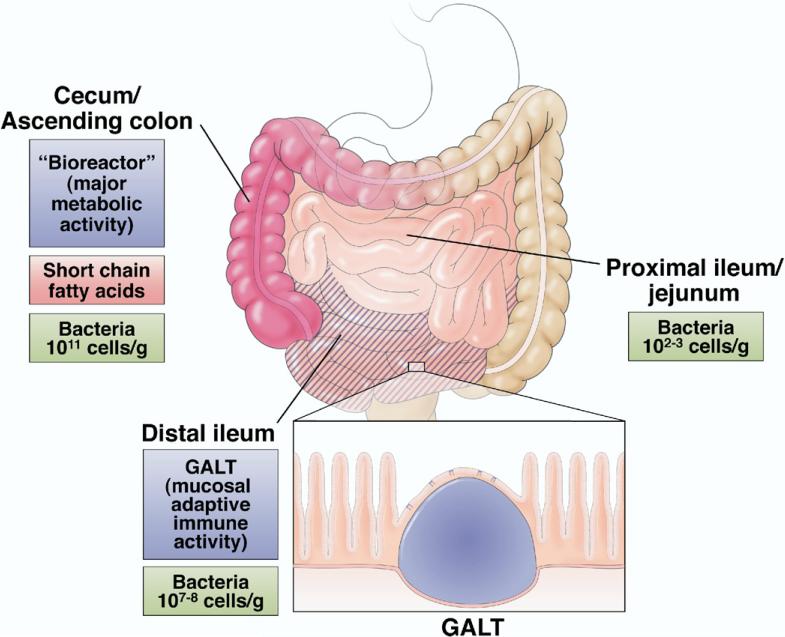
The human normal flora, or microbiota, is vast, both in its absolute quantitative mass and its qualitative diversity. A detailed inventory of the normal microbiota was long considered nearly unattainable by conventional microbiologic techniques; however, recent initiatives utilizing high-throughput sequencing and molecular taxonomic methodologies have greatly increased our understanding of the population composition, dynamics, and ecology of the gut microbiota (reviewed in several reports6,49–54). Humans have been proposed to be “meta-organisms” consisting of 10-fold greater numbers of bacterial than animal cells that are metabolically and immunologically integrated.52,53 The human meta-organism includes approximately 1014 prokaryotic organisms, with a biomass of >1 kg. The population composition is remarkably stable at different anatomic locations along the gut, but absolute numbers vary greatly, ranging from 1011 cells/g content in the ascending colon to 107–8 in the distal ileum and 102–3 in the proximal ileum and jejunum (Figure 3). Anaerobes are several orders of magnitude more abundant that aerobes in the bacterial community, and a majority of the population (60%–90%) are representatives of 2 divisions: the Bacteroidetes and Firmicutes. Interestingly, eukaryotic fungal species have also recently been identified as components of the microbiota.55 Most of the community is autochthonous, meaning indigenous and stable, although allochthonous, or transient, members are also to be found (certainly most enteric pathogens fall into this category). At birth, the gut is sterile and is colonized immediately, ultimately developing into a stable community, although there are marked variations in microbial composition between individuals.56 Host parameters that select for and ultimately shape the stable climax community are a topic of considerable interest.52,57
Figure 3.
Preferred sites of commensal/probiotic interaction with the gut. Cecum/ascending colon is a “bioreactor” with the greatest amounts of bacteria, metabolic activity, and SCFA fermentation. Concentration of SCFA diminishes along the colon. The distal ileum is enriched in GALT (Peyer's patches) and is the dominant site of luminal sampling and mucosal adaptive immune activity.
New molecular phylogenic approaches based on sequencing of bacterial 16S ribosomal RNA, or in higher-resolution, bulk genomic sequencing of entire communities/environments, allow census-based, non–culture-dependent inventories or “metagenomics” of the entire microbiota population composition and their component genes, which can be tabulated and correlated with host phenotypes.53,58 These approaches have been used to identify populations associated with obesity (discussed in the following text) and inflammatory bowel disease,59,60 although an altered configuration of the microbiota may be a consequence rather than a cause of these disorders. Lupp et al described changes in the composition of the microbiota in a mouse model of experimental colitis that were similar regardless of whether the proximate inciting event was infectious or chemical.61 Thus, an observed correlation between changes in microbiotal composition and a disease phenotype must be accompanied and supported by a plausible mechanism (eg, metabolic or immune alterations) if observed population changes are to be considered as causal.
Metabolic Roles of the Microbiota
Metagenomic approaches have highlighted impressive genetic diversity and the vast potential reservoir of metabolic capability of an established microbial community. The combined biochemical capacity of the microbiota has been called a “forgotten organ,”50 mediating diverse beneficial roles including vitamin synthesis, bile salt metabolism, and xenobiotic degradation,62 but likely the collective biochemical output, the “metabolome,” is involved in additional processes. “Metabolomics” is another system-based approach of high-throughput analysis of complex biological samples that has great promise in linking functional consequences to ecological changes in the microbiotal community. Researchers have used this approach to identify differences in the gut microbiotal populations and consequent metabolic processes in normal weight and obese mice. Backhed et al showed that mice raised under normal conditions have more body adipose tissue than germ-free animals, suggesting a role for the microbiota in enhancing energy homeostasis.63 Using metagenomic techniques to describe altered community composition (a reduction in Bacteroidetes and an increase in Firmicutes) in genetically obese (ob/ob) mice, Turnbaugh et al showed this microbiota had an augmented ability to liberate an additional energy source (short-chain fatty acids [SCFAs]) for host uptake.64 These population changes were correlated with observations in lean and obese humans65 and suggest that the microbiota of an obese person is more efficient at extracting energy from the diet than that of a lean individual. The recognition that the microbiota can facilitate biochemical processes in the meta-organism, coupled with the technical ability to study these events, has prompted studies tentatively linking microbiotal population changes to other presumptive metabolic conditions, such as non–insulin-dependent diabetes, nonalcoholic hepatosteatosis, and atherosclerosis.66,67 There is much more research to be done into the metabolic role of the microbiota in systemic health and disease.
Conclusions derived from “omic” methodologies should obtain empirical support from biochemical approaches. The normal microbiota thrives in a largely anaerobic luminal environment, generating its own energy through the fermentation of dietary complex carbohydrates. Complex carbohydrates (starches) are poorly digested by the human digestive system and require the microbiota for breakdown via fermentation. In the human gut, the end products of fermentation are a spectrum of organic acids, including SCFAs such as butyrate, succinate, and propionate, as well as other terminal products such as lactate.68,69 SCFAs are an important energy source for the colonic epithelium and the host, providing for an estimated 5%–15% of human energy requirements (far higher in ruminant mammals).70 The organisms most efficient at producing SCFAs (the microbes possessing the enzymatic capacity to ferment these substrates) are Firmicutes such as Clostridium species and Bifidobacterium species, which are enriched in the microbiota of obese mice and humans.64 Thus, SCFAs are bacterial products and well-known host energy sources that provide a link from community-level changes in microbiotal composition and their encoded metabolic machinery to a human phenotype.
SCFAs also show an intriguing ability to influence various aspects of gut physiology beyond functioning solely as a crude caloric source.71,72 For example, butyrate and other SCFAs have well-known differentiating and growth-promoting activities in vitro and in vivo, a biological effect ascribed to histone deacetylase activity. SCFAs have also been noted to have immunomodulatory effects, suppressing inflammatory cytokine secretion in cultured epithelial cells and ameliorating model colitis in mice, suggesting these molecules contribute to the ability of the mucosa to tolerate the presence of vast quantities of living microorganisms and associated MAMPs. Butyrate can induce epithelial production of ROS and subsequent redox-dependent signaling effects including NF-κB suppression.73 Furthermore, luminal instillation of butyrate has been shown to be a promising experimental therapy in human ulcerative colitis and related inflammatory disorders.71,74 SCFAs appear to be a class of effector molecule, produced preferentially by one subset of the microbiotal community. That a product of bacterial fermentation, an exogenous chemical from the perspective of the host, possesses such potent physiologic regulatory properties on eukaryotic cells beyond utilization as an energy source is an interesting illustration of how the host has coevolved dependency, or at least benefit, from its prokaryotic partners.
Microbial Effects on Epithelial Cell Function, Growth, and Survival
Gut epithelia have a surface area of 300 – 400 m2, comparable to that of a tennis court.75 Essentially all the physical and near-physical contact between host and microbe occurs at the epithelial (or epithelial-derived, eg, mucus layer) surfaces. Epithelial cells, by embryologic definition, are interfaces between the host and the environment (in a topological sense, the gut lumen is external to the body) and are equipped with a number of structural and biochemical modifications to mediate this function.76 They are “polarized” or adapted with apical surface specializations (microvilli, mucus production, vectorial ion secretion, intercellular junctions) to allow interface with the microbe-rich luminal environment, and the basolateral surfaces are optimized for interaction with the host itself (milieu intérieur). Impressively, the epithelia and complete mucosa must tolerate the normal microbiota, yet this tissue is obviously required to mediate digestive as well as fluid and nutrient absorptive functions.
Studies with gnotobiotic mice have shown that the enteric microbiota is not functionally insulated from the mucosa; in contrast, these bacteria influence epithelial metabolism, proliferation and survival, and barrier function.51,77–80 The small intestinal villi of the germ-free gut are relatively longer, whereas crypts are atrophic, show a slower turnover of the epithelial cells,33 and exhibit defective angiogenesis.81 Hooper et al reported robust transcriptional responses of gnotobiotic mice monocolonized with a single gut symbiont species (Bacteroides thetaiotaomicron).80 In further experiments in gnotobiotic mice, Sonnenburg et al showed that colonization with this strain and supplemented with a second species (Bifidobacterium longum) elicited a different and distinct host cell transcriptional response, illustrating the complexity of interpreting host responses to whole microbial communities (and the difficulty of predicting potential beneficial effects of a given strain).82 Interestingly, Rawls et al performed expression profiling of epithelial responses after reciprocal transplantation of natural fish and mouse microbiota into germ-free mice and germ-free zebrafish and showed shared and conserved host transcriptional responses, suggesting a common program of vertebrate genomic regulatory responses to symbiotic bacteria.57 How a taxonomically complex and ecologically dynamic symbiotic microbial community can influence epithelial signaling is a fascinating question for future study.
Bacterial products such as SCFAs can act as effectors to influence mucosal and perhaps systemic homeostasis. Additionally, microbial perception via the primordial PRR pathways can have effects beyond the activation of innate immunity (meaning acute inflammation in the histopathologic sense). Gene expression elicited by PRR signaling can positively affect homeostasis in the gut. In a seminal report, Rakoff–Nahoum et al showed that mice with intestines cleared of the normal microbiota and thus deficient in luminal bacteria and MAMPs were markedly more sensitive to dextran sodium sulfate–induced colitis; the mucosal injury mediated by this compound could be ameliorated by oral administration of isolated MAMPs such as lipopolysaccharide and LTA.83 Additionally, this study showed that these cytoprotective effects were lost in TLR2- and TLR4-null mice, implicating TLR signaling in the protective mechanism. Pull et al showed that regenerative responses to colonic injury were markedly attenuated in germ-free animals, indicating a discernable role of the microbiota in induction of epithelial proliferation and response to injury.33 Additionally, restitution was similarly reduced in MyD88-null (a signaling intermediate required by multiple TLRs) mice, reinforcing the concept that PRR-mediated signaling is necessary for trophic/restitutive effects. TLR2/MyD88-dependent signaling has also been shown to increase physiologic epithelial barrier integrity.84 Other investigators have found that gut anti-inflammatory/cytoprotective effects could be mediated by isolated MAMPs; purified unmethylated bacterial DNA has been shown to ameliorate dextran sodium sulfate–induced colitis.85 These and related observations in mice with defects in epithelial NF-κB pathway components86–89 have suggested that some degree of PRR signaling is necessary for gut homeostasis, presumably because of the tonic up-regulation of cytoprotective genes (gene products with antiapoptotic, chaperone/stress response, and antioxidant effects).86 In vivo, symbiotic bacteria induce expression of cytoprotective genes such as heat shock proteins83 and RELMβ,90 as well as antimicrobial peptides and lectins.25 There might be a “graded response” that distinguishes and responds to symbiont and pathogen and that when activated at “high gain” elicits acute inflammation with neutrophil influx, yet at “low gain” elicits a more limited (and possibly distinct) transcriptional response (Figure 2). Thus, while in keeping with the overall mission of immunity, protection from potential injurious stimuli, PRR signaling stimulated by the microbiota does not activate tissue responses that we would interpret as a disease state. This notion of “basal inflammatory tone” thus explains at a molecular level why a bacterial presence, within the proper boundaries and of the proper constitution and intensity, may make a positive contribution to intestinal homeostasis and health.
An MAMP-PRR interaction with special relevance to the gut is found in the flagellin/TLR5 ligand receptor pair. As an MAMP, flagellin is recognized by plants, invertebrates, and mammals and appears to be a dominant epitope in sera from patients with inflammatory bowel disease (IBD).91,92 TLR5 knockout mice are the only TLR-null mice that develop spontaneous colitis.93 Flagellin elicits a cytoprotective response in the gut94,95; TLR5 is not widely expressed outside the gastrointestinal tract, and systemic shock does not occur if flagellin enters the vascular space, as occurs with lipopolysaccharide. Several studies have illustrated the potential of exploiting flagellin as a systemic agent for preventing effects of detrimental stimuli to the gut and perhaps other tissues.96–98
Small peptides can also function as MAMPs. Formylated peptides have been shown to stimulate mitogen-activated protein kinase signaling pathways and up-regulate cytoprotective genes in vitro.99 The discovery of multiple paralogues of the classic neutrophil formylated peptide receptor in the intestine indicates that these bacterially derived molecules may contribute to epithelial homeostasis.14,15 Finally, bacterial quorum-sensing molecules, small molecules that mediate interbacterial chemical communication, might also function as MAMPs and elicit cytoprotective effects.100 Thus, PRR-mediated signaling, first recognized as an antimicrobial, innate immune function, seems to have additional physiologic and beneficial activities that could ultimately be developed as therapeutics for patients with intestinal disorders.
Microbial Effects on Inflammatory Signaling
The epithelia has specific mechanisms, including dampening TLR signaling or limiting/sequestering TLR expression, to suppress or moderate the potent antimicrobial arsenal that has necessarily evolved to manage explicit pathogenic threats (as well as prevent induction of overt inflammation by MAMPs). Individual members of the microbiota can also influence signaling intensity.101–103 Nonpathogenic prokaryotes, including natural commensals and those with proposed probiotic function, are able to suppress eukaryotic inflammatory signaling pathways and inflammatory effector functions; these suppressive effects are mediated either by intact viable organisms or by secreted products.104–107 The mammalian intestinal symbiont B thetaiotaomicron inhibits NF-κB pathways by regulating cytoplasmic to nuclear translocation of the p65 subunit.108 Symbiotic bacteria are able to influence inflammatory pathways, and probably other cell regulatory processes, by manipulating the ubiquitin system.102,109–111 Ubiquitination is a covalent modification that regulates a wide spectrum of biochemical processes, generally by targeting modified proteins for controlled degradation via the proteasome. A key example of a signaling component regulated by ubiquitination is the inhibitory component of the NF-κB pathway, IκB,112 and there are numerous examples of pathogens that utilize preformed effector proteins to influence IκB ubiquitination and thus innate immunity.113–115 Symbiotic bacteria that interact with epithelial cells in vitro are capable of blocking IκB ubiquitination and thus NF-κB activation by interference with the function of the IκB ubiquitination ligase, SCFβTrCP.109,116,117 Furthermore, these events are mediated by transient oxidant-induced inactivation of the ubiquitination enzymatic machinery as a consequence of prokaryotic elicited ROS production.37,117 Transient oxidative inactivation of a wide spectrum of regulatory enzymes is an increasingly recognized mechanism for influencing cellular homeostasis.28,39 Many multicellular organisms generate ROS in response to microbial interactions, and specifically in response to MAMPs,29 making this a general and nonspecies selective mechanism by which a complex microbiotal community could influence a wide range of host signaling and homeostatic processes.117
Regulation of Adaptive Immunity
An additional facet of the “negotiated settlement” that occurs between host and microbe is adaptive immunity, specifically the gut/mucosal arm of the adaptive immune system, which provides humoral and cell-mediated immunity against ingested antigens and luminal organisms. Adaptive immunity features the selective ability to respond to or ignore individual, specific antigens based on past encounters. Thus, the mucosal immune system can develop tolerance to ingested (or gut resident) antigens; repeat or continual exposure to the same stimulus does not elicit the immune response that it does in a naive animal. Adaptive immunity is only present in vertebrates.118 As we have discussed, innate (PRR)-based immune pathways recognize and respond to microbial patterns reproducibly and predictably, regardless of past exposure (with some short-term exceptions such as receptor down-regulation). This is sufficient for the host-microbial interactions with the invertebrate gut, which are of low complexity, usually composed of a single or small number of highly specialized symbionts. McFall–Ngai suggested that the evolutionary emergence of adaptive immunity endowed the host with an additional and more selective means of managing microbial symbionts.1 This evolutionary advance allowed vertebrates to tolerate multigenera gut microbial “consortia,” ultimately increasing the microbial diversity in the gut, expanding its immunologic and metabolic roles (and consequences). The occasional dysregulation of this arrangement, such as during the pathogenesis of IBD and perhaps other immune and metabolic disorders, may be the price paid for an extended metabolic ability (and other still uncharacterized benefits) provided by a normal microbiota.
The topic of mucosal immunity is vast and well reviewed,119–121 but it is important to emphasize that the anatomical and functional components of this system are found predominantly in the small bowel and, as might be expected, are strongly influenced by the microbiota. The collective gut-associated lymphoid tissue (GALT) is the largest immune organ in the body, and much of the GALT is organized into discrete structures termed Peyer's patches, which are essentially mucosal lymph nodes overlaid with a specialized epithelial cell type, the M cell, which possesses the endocytotic machinery that takes up particulate antigens from the gut lumen (Figure 3). Members of the normal microbiota, along with nonviable particulate antigens and all-too-viable pathogens, are continually sampled by the M cells that lie over the Peyer's patches and perhaps other portals122 for processing by local dendritic cells and subsequent education of regulatory CD4+ T-cell populations (Tregs).120 Tolerance results from induction of Tregs that prevent immune responses toward that tolerizing antigen via elaboration of cytokines (interleukin [IL]-10, transforming growth factor β, and so on) that are suppressive of effector lymphocytes. Importantly, these processes are restricted to the GALT and vicinal mesenteric lymph nodes, allowing the systemic adaptive immune system to remain ignorant of the ongoing interactions with the normal microbiota and preventing autoimmune responses.
The GALT is grossly hypoplastic in germ-free animals. Furthermore, germ-free animals have reduced total CD4 T-cell populations and an inappropriate balance of TH cell subsets, which can be rectified within weeks upon conventionalization with a representative member of the normal microbiota (Bacteroides fragilis).123 Commensal bacteria are necessary for the up-regulation of the IL-17 family member IL-25, which serves to repress expression of IL-23 and subsequent development of proinflammatory IL-17–producing Th17 CD4+ T cells.124 These results indicate the role of the microbiota in development of the mucosal adaptive immune system. Stimulation of immune development can be mediated via recognition of bacterial capsular polysaccharides. A specific polysaccharide (polysaccharide A) product of B fragilis has been identified that is recognized by dendritic cells and serves to stimulate development of Tregs with the ability to attenuate pathogen- or chemical-induced colitis.125 These data indicate that specific carbohydrate moieties on symbiotic bacteria can initiate suppressive regulatory effects on effector lymphocytes. This cytoprotective effect of a bacterial product acts via the adaptive immune system, rather than the more ancient PRR-based innate/inflammatory signaling and cellular cytoprotective pathways.
Another mechanism by which the mucosal adaptive immune system can mediate inflammatory and immune tolerance toward the microbiota is humoral immunity via secretory immunoglobulin (Ig) A.126 Strain-specific IgA is also stimulated by sampling of symbionts in the lumen, again with antigen presentation and B-cell selection restricted to the confines of the mucosal GALT.127,128 In experiments with gnotobiotic immunodeficient (Rag-null) mice, animals responded to colonization with a defined, limited microbiota with increased innate responses, confirming a role for adaptive immunity in modulation of mucosal inflammation. This inflammatory hyperresponsiveness was reversed by supplementation with the appropriate strain-specific IgA, indicating the immunomodulatory role of humoral immunity.129 Thus, the adaptive immune system monitors the microbiota and contributes to homeostasis by supervising T-cell responses and provision of local mucosal IgA secretion. Microbial sampling and the subsequent processing through the regulatory circuits of the adaptive immune system could be relevant to health in tissues far removed from the gut.
The Microbiota in Deficiency: Symbiosis Out of Balance
The Hygiene Hypothesis
Although it is important to consider the evolution of prokaryotic-eukaryotic interactions in understanding the mechanisms of gut-microbe interactions, clinical and epidemiologic studies have revealed the importance of this relationship to human health. Although great improvements in human health and longevity stem largely from 19th-century advances in sanitation and public health that reduced mortality from epidemics of infectious disease, recent observations have indicated that not all exposure to microbes is deleterious to human health. First proposed by Strachan in 1986, the “hygiene hypothesis” suggests that inadequate exposure to microbes and their products may be detrimental and is based on the observation that increasing rates of allergic or atopic disorders are positively associated with increased household cleanliness and socioeconomic status.130 In 2002, Bach reported that declines in rates of endemic infectious diseases correlated within the same population with increases in autoimmune disease or conditions of immune dysregulation, including asthma, multiple sclerosis, and IBD.131 Less-industrialized nations have consistently lower incidences of such immune/inflammatory conditions. The pathophysiologic mechanisms that account for these population-based observations are unclear, although they are unlikely to result solely from reduced exposure to overt pathogens; the same phenomenon has been observed in rural residents of developed countries with low rates of infectious morbidity/mortality.132 Quantitative or qualitative deficiencies in the normal microbiota, especially in early life during development of the immune system, may underlie these measurable changes in disease patterns. The term “old friends” has been used to describe the gut-inhabiting microbes originating from soil, plants, and especially domesticated animals that humans have coevolved with before our transition to a relatively sterile urban environment.133 It is possible that an insufficient/inadequate microbiota (lacking “old friends”) leads to dysregulation of immune effector cells, accounting for changes in systemic, nonintestinal allergic conditions.134,135 Symbionts modulate innate immune responses; similarly, nonpathogens have been shown to modulate adaptive immunity in murine models by induction of Treg differentiation and anti-inflammatory cytokine secretion.136,137 Additionally, at the mucosal level, a reduced MAMP-driven inflammatory “tone,” or relative underrepresentation of bacterial species with significant intrinsic immunoinflammatory modulatory activities, could permit or promote epithelial dysfunction and result in intestinal inflammation. Symbiotic bacteria do have variable degrees of epithelial immunosuppressive properties37 as well as differential abilities to stimulate immunosuppressive T-cell development,125 hinting at potential and plausible features of a health-promoting microbiota. Thus, an optimal composition of microbiota is necessary for health but in specific cases can be considered ill adaptive for the meta-organism, or “dysbiotic.”
Dysbiosis of the Microbiota
A dysbiotic microbiota is an ecological disorder of the bacterial community; the concept is often associated with the pathogenesis of IBD (for review, see Sartor138 and Strober et al139). One study has shown that dysbiotic microbiota, or “colitogenic” microbiota, can cause IBD; Garrett et al showed that immunocompromised mice with homozygous disruption of the gene encoding the lymphocyte specific transcription factor T-bet, a regulator of immune cell differentiation, developed an inflammatory colitis resembling human ulcerative colitis.140 This colitis could be eliminated with antibiotic treatment, implicating the prokaryotic microbiota in its pathogenesis. The investigators made the fascinating observation that the colitic phenotype could be transmitted vertically to the progeny of the affected parents and horizontally to unrelated animals (by foster rearing of pups). The colitic phenotype was shown to be transmissible to wild-type mice, indicating that an abnormal microbiota is sufficient for the development of colitis. However, transmission of the disease was more severe and penetrant in immunodeficient animals, consistent with the current model presuming underlying intrinsic (genetic) host defects in forms of human IBD.139 Thus, the concept of “colitogenic” microbiota is gaining firmer ground, although it has no clear microbiological definition. A recent report that the PSA capsular antigen is a bacterial moiety with adaptive immunoregulatory properties indicates that this molecule might be a participant.125 Metagenomic analyses of human IBD might identify additional proinflammatory and anti-inflammatory determinants of the microbiota and yield potential biomarkers and perhaps provide a rationale for the formulation of corrective probiotic supplements or replacements.141
Even in the limited gut microbiota in invertebrates, a deleterious dysbiotic microbiota has been described. In Drosophila, the transcription factor caudal represses antimicrobial peptide section, presumably as a mechanism to dampen excessive antimicrobial responses and to promote an optimal microbiota. Genetic disruption of this factor increased AMP expression and altered the microbiota in a quantitative and qualitative manner, resulting in overgrowth of an autochthonous microbe that elicited epithelial apoptosis and death of the flies.142 Notably, these experiments illustrate that dysbiosis can be a direct result of innate immune dysfunction. It is hoped the unique genetic tractability of the Drosophila system will contribute to the analysis of gut-symbiont interactions and facilitate identification of potential harmful determinants that are present in members of a normal microbiota. Taken together, these experiments illustrate that in widely diverse gut-microbiota systems, host immune regulatory processes are vital in shaping an optimal microbiota; disturbance of host regulation creates a dysbiotic microbiota and consequent disease.
Microbiota and Enteric Pathogens
Enteric infectious diseases, caused by common pathogens such as Salmonella, Shigella, various strains of enteropathogenic Escherichia coli, and Yersinia, are of great public health consequence in the developed and developing worlds. Infection is generally conceptualized as invasion by foreign agents; this notion is not incorrect, but the microbiota is also an important component of overall resistance to infection. Germ-free mice have increased susceptibility to a variety of enteric pathogens, an observation that led to the concept of “colonization resistance.”143,144 This presumptive role of the microbiota in suppressing encounters with overt pathogens is likely multifactorial. The normal microbiota may compete for access to adhesive sites on the epithelial surface or stimulate increased mucin production. SCFAs may be bacteriostatic for a subset of bacterial species, either directly or by reducing pH. Some members of the microbiota also generate bacteriocins, small peptide molecules with microbicidal or microbistatic properties.145 In addition, there is increasing interest in the effects of microbial-microbial signaling on the overall equilibrium of optimal microbiotal ecosystems.146 Within a prokaryotic species, secretion of soluble chemicals is used by individual bacterial cells to communicate with one another, inducing coordinated gene regulation when environmental conditions are appropriate (quorum sensing). A subset of these small diffusible molecules that mediate intraspecies signaling can be perceived by other strains of bacteria, allowing interspecies communication. Thus, the normal microbiota produces a carefully balanced combination of interspecific and intraspecific chemical signals that could suppress pathogenic invaders, as well as optimize the composition and numbers of appropriate members of the microbiota. A dysbiotic microbiota might also involve some degree of internecine dysfunction, a deleterious combination of chemical signals that disorder the microbiotal community structure.
Additionally, a microbiota could also be quantitatively deficient, as in the iatrogenic suppression that occurs following antibiotic use. Suppression of microbial numbers would disturb the normal mechanisms of community regulation and pathogen colonization resistance (not to mention the effects of reducing PRR-mediated cytoprotective tone). A microbiota in ecological collapse could permit emergence of autochthonous bacteria that blur the distinction between symbiont and pathogen, exemplified by Clostridia difficile proliferation following vancomycin treatment, which can result in pseudomembranous colitis.147–149 In murine models, antibiotic administration suppressed expression of the commensal-induced antimicrobial protein RegIIIγ and allowed pathogen overgrowth. Additionally, supplementation of mice with TLR ligands reinduced RegIIIγ and corrected innate immune defects.150 Together, these data illustrate the important role of the microbiota in protection from enteric pathogens.
The Microbiota as Therapeutic Target
Probiotics
There is currently much interest in deliberately manipulating the normal microbiota to accrue health benefits through an approach known as “probiotics.” Probiotics are defined as “live microorganisms which when administered in adequate amounts confer a health benefit on the host.”151 Clearly, the conceptual basis of probiotics is well grounded.121,137,152 Therapeutic bacteria could presumably provide the same beneficial functions and activities that have evolved for the normal microbiota. For example, symbionts can expand nutrient utilization and thus metabolic range of the host. Likewise, at the cellular and tissue level, symbiotic bacteria can suppress innate immune signaling. Symbionts can stimulate protective T-cell responses. MAMP-elicited signaling has clear effects on epithelial cytoprotection, survival/proliferation pathways, and barrier function. Normal gut inhabitants can competitively exclude or chemically suppress pathogens.
However, at the level of the individual, where medicine is ultimately grounded, data from probiotics remain preliminary. Probiotic approaches are and will always be confounded by the diversity of the human microbiota and its plasticity in the face of varied human diets and genetic backgrounds. Nevertheless, based on data from animal models, probiotics offer great potential benefits and might be used to treat intestinal functional and inflammatory disorders, systemic immune/allergic conditions, and metabolic syndromes, as well as to even modulate intestinal nociception, psychological stress responses, and longevity (an original claim made for probiotics by Elie Metchnikoff a century ago).152 Clinical evidence indicates that probiotics are effective in the treatment or prevention of acute viral gastroenteritis, postantibiotic-associated diarrhea, certain pediatric allergic disorders, necrotizing enterocolitis, and IBD such as Crohn's and postsurgical pouchitis.153
Although most of these conditions are gastrointestinal in nature, some, such as allergic disorders, are systemic. When contemplating how probiotics could alter host physiology and function as therapies, it must be remembered that the great difference in the normal anatomical distributions of bacterial numbers and their consequent metabolic and immunologic relationships with the host may have implications for probiotic dosing and delivery schemes (Figure 3). Could/should exogenous bacteria be incorporated as stable members of the microbiota or mediate a transient pharmacologic effect? Vast numbers of bacteria thrive in a colon, especially the right (ascending) colon and cecum, a “bioreactor” where indigestible complex carbohydrates are fermented within the large-bore, slow-transit colonic lumen to SCFAs. Indeed, the concentration of butyrate, with all its known bioactive properties, in these regions reaches 100 mmol/L.72 Systemic metabolic changes from altered microbiota, such as changes in energy homeostasis, could be mediated by stably manipulating bacteria at this site. Additionally, the effects of bacteria and their products on suppressing innate immune pathways may occur by direct anti-inflammatory action on the colon epithelia; the beneficial effects in pouchitis or perhaps ulcerative colitis and functional bowel disorders support this model. Of note, idiopathic ulcerative colitis is a disease of the distal colon, with inflammation generally diminishing in more proximal regions, a distribution inverse to the numbers of bacteria and the luminal concentration of SCFAs found in the colon. Thus, colonic disorders may be more effectively addressed by long-term changes in the composition of the microbiota by supplementation of live microbes. In contrast, the bulk of organized lymphoid tissue and microbial sampling by immune cells occurs in the ileum, which is permanently colonized with far fewer organisms (by a factor of several orders of magnitude). Thus, although the host interface with metabolic outputs of the microbiotal community is likely to occur predominantly in the colon, the role of the microbiota in mediating systemic immune and allergic phenomena might be primarily mediated by the GALT of the small bowel.154 Supplementing qualitatively and quantitatively optimized bacteria to these sites, over the long term, may provide a beneficial stimulus to Treg development and consequent immunoregulation. Finally, although it might be challenging to select probiotics that would improve the ecological health of the microbiota and convert dysbiosis to eubiosis, it might be more immediately practical to provide supplementary bacteria to restore a quantitative deficiency state following treatment with antibiotics. This would prevent the expansion of opportunistic pathogens and stimulate cytoprotective responses.
Prebiotics and “Postbiotics”
Another approach to therapeutic exploitation of the microbiota involves manipulating its energy sources.152 Alterations in diet affect the composition and, more importantly, the collective metabolic output of the microbiota. Thus, deliberate dietary supplementation with bacterial fermentative substrates, usually complex carbohydrates (eg, inulin, oligofructose), can increase luminal concentrations of SCFAs, including butyrate, and indirectly modify immune function.153 Such dietary supplementation is termed “prebiotics,” defined as nondigestible (by the host) food ingredients that have a beneficial effect through their selective metabolism in the intestinal tract.156 Prebiotic substrates could be administered with live bacteria most able to exploit that energy source, an approach called “synbiotics.” Obviously, the emerging ability to couple metagenomics of bacterial populations with resultant metabolomics will facilitate rational attempts to manipulate endogenous gut microbiota by dietary changes.
Finally, isolated bacterial components could also be administered as therapeutics. The gut lumen could be supplemented with bacterial products, “postbiotics,” such as butyrate and other SCFAs. Although this approach extends beyond the official definition of probiotics (defined as living microbes), it does not differ much in concept from vaccination, the administration of microbial components to stimulate the adaptive immune system. The sheer quantity of MAMPs within the lumen might have a tonic effect on the gut via PRR signaling, and in cases of reduced or dysbiotic microbiota, supplementation of individual components might provide at least some of the cytoprotective effects of symbionts on the gut. The use of nonviable bacteria or bacterial components would also avoid the risk of sepsis or other potential complication of probiotics, particularly in a critically ill patient with compromised intestinal barrier function.157,158 Toward this end, luminal or even systemic administration of MAMPs has shown striking cytoprotective effects in mammalian experimental gut injury models.83,85,96,98
Summary
An intricate symbiotic relationship has evolved between humans and microbes. We more fully understand the degree to which gastrointestinal biology is intertwined with microbiology, that the relationship between host and microbe is required for health, and that this relationship might be manipulated therapeutically (Table 2). Perhaps one day, an optimal microbiota will be considered one aspect of nutrition that is as amenable to intervention. Finally, with a fuller understanding of the host-microbe interaction in the gut, perhaps the pathogenic side of this relationship can be suppressed, whereas the beneficial effects of the gastrointestinal microbiota can be maintained and restored.
Table 2.
Influences of the Microbiota
| 1. Metabolic/nutritional/energy utilization |
| • Vitamin synthesis |
| • SCFA as energy source—role in obesity |
| 2. Innate Immune Regulation |
| • Dampening of inflammatory responses |
| 3. Adaptive immune Regulation, |
| • Induction of Immunosuppressive T cells (Tregs) |
| 4. Epithelial development and survival |
| • Stimulation of proliferation, angiogenesis, epithelial restitution |
| • Cytoprotective effects of PRR signaling |
| 5. Competitive exclusion of pathogens |
Acknowledgments
The author thanks Lora Hooper and Andrew Gewirtz for a critical reading of the manuscript, as well as members of the Neish Laboratory and the Epithelial Pathobiology Division in the Department of Pathology at the Emory University School of Medicine.
The authors disclose the following: Supported by National Institutes of Health grants DK071604 and AI064462.
Abbreviations used in this paper
- GALT
gut-associated lymphoid tissue
- IL
interleukin
- MAMP
microbial-associated molecular pattern
- NF-κB
nuclear factor κB
- NLR
Nod-like receptor
- PRR
pattern recognition receptor
- ROS
reactive oxygen species
- SCFA
short-chain fatty acid
- TLR
Toll-like receptor
References
- 1.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 2.McFall-Ngai MJ. Identifying “prime suspects”: symbioses and the evolution of multicellularity. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:711–723. doi: 10.1016/s1096-4959(01)00406-7. [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- 4.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Hickman C. How have bacteria contributed to the evolution of multicellular animals? In: McFall-Ngai MJ, editor. The influence of cooperative bacteria on animal host biology. Cambridge University Press; New York: 2005. pp. 3–32. [Google Scholar]
- 6.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flint H. The rumen microbial ecosystem—some recent developments. Trends Microbiol. 1997;2:483–488. doi: 10.1016/S0966-842X(97)01159-1. [DOI] [PubMed] [Google Scholar]
- 8.Warnecke F, Luginbuhl P, Ivanova N, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 9.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 11.Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- 12.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 13.Ishii KJ, Koyama S, Nakagawa A, et al. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006;17:501–519. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Babbin BA, Jesaitis AJ, Ivanov AI, et al. Formyl peptide receptor-1 activation enhances intestinal epithelial cell restitution through phosphatidylinositol 3-kinase-dependent activation of Rac1 and Cdc42. J Immunol. 2007;179:8112–8121. doi: 10.4049/jimmunol.179.12.8112. [DOI] [PubMed] [Google Scholar]
- 16.Pujol N, Link EM, Liu LX, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 17.Ferrandon D, Imler JL, Hetru C, et al. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 18.Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol. 2005;174:4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 19.Karrasch T, Kim JS, Muhlbauer M, et al. Gnotobiotic IL-10–/–; NF-kappa B(EGFP) mice reveal the critical role of TLR/NF-kappa B signaling in commensal bacteria-induced colitis. J Immunol. 2007;178:6522–6532. doi: 10.4049/jimmunol.178.10.6522. [DOI] [PubMed] [Google Scholar]
- 20.Jones RM, Wu H, Wentworth C, et al. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Tzou P, Ohresser S, Ferrandon D, et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 23.Ayabe T, Satchell DP, Wilson CL, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 24.Salzman NH, Ghosh D, Huttner KM, et al. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 25.Cash HL, Whitham CV, Behrendt CL, et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper LV, Stappenbeck TS, Hong CV, et al. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 27.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 28.Terada LS. Specificity in reactive oxidant signaling: think globally, act locally. J Cell Biol. 2006;174:615–623. doi: 10.1083/jcb.200605036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogier-Denis E, Mkaddem SB, Vandewalle A. NOX enzymes and Toll-like receptor signaling. Semin Immunopathol. 2008;30:291–300. doi: 10.1007/s00281-008-0120-9. [DOI] [PubMed] [Google Scholar]
- 30.Schneewei[ss] H, Renwrantz L. Analysis of the attraction of haemocytes from Mytilus edulis by molecules of bacterial origin. Dev Comp Immunol. 1993;17:377–387. doi: 10.1016/0145-305x(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 31.Ha E-M, Oh C-T, Ryu J-H, et al. An antioxidant system required for host protection against gut infection in drosophila. Dev Cell. 2005;8:125–132. doi: 10.1016/j.devcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Ha E-M, Oh C-T, Bae YS, et al. A direct role for dual oxidase in drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 33.Pull SL, Doherty JM, Mills JC, et al. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotchoni SO, Gachomo EW. The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J Biosci. 2006;31:389–404. doi: 10.1007/BF02704112. [DOI] [PubMed] [Google Scholar]
- 35.Pauly N, Pucciariello C, Mandon K, et al. Reactive oxygen and nitrogen species and glutathione: key players in the legume-Rhizobium symbiosis. J Exp Bot. 2006;57:1769–1776. doi: 10.1093/jxb/erj184. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka A, Christensen MJ, Takemoto D, et al. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell. 2006;18:1052–1066. doi: 10.1105/tpc.105.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar A, Wu H, Collier-Hyams LS, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 39.Chiarugi P, Buricchi F. Protein tyrosine phosphorylation and reversible oxidation: two cross-talking posttranslation modifications. Antioxid Redox Signal. 2007;9:1–24. doi: 10.1089/ars.2007.9.1. [DOI] [PubMed] [Google Scholar]
- 40.Rokutan K, Kawahara T, Kuwano Y, et al. NADPH oxidases in the gastrointestinal tract: a potential role of Nox1 in innate immune response and carcinogenesis. Antioxid Redox Signal. 2006;8:1573–1582. doi: 10.1089/ars.2006.8.1573. [DOI] [PubMed] [Google Scholar]
- 41.Lee J, Mo JH, Katakura K, Alkalay I, et al. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. 2006;8:1327–1336. doi: 10.1038/ncb1500. [DOI] [PubMed] [Google Scholar]
- 42.Guo L, Lim KB, Gunn JS, Bainbridge B, et al. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 43.Melmed G, Thomas LS, Lee N, et al. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implications for host-microbial interactions in the gut. J Immunol. 2003;170:1406–1415. doi: 10.4049/jimmunol.170.3.1406. [DOI] [PubMed] [Google Scholar]
- 44.Lotz M, Gutle D, Walther S, et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otte JM, Cario E, Podolsky DK. Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. 2004;126:1054–1070. doi: 10.1053/j.gastro.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Xiao H, Gulen MF, Qin J, et al. The Toll-interleukin-1 receptor member SIGIRR regulates colonic epithelial homeostasis, inflammation, and tumorigenesis. Immunity. 2007;26:461–475. doi: 10.1016/j.immuni.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Bates JM, Akerlund J, Mittge E, et al. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao EA, Andersen-Nissen E, Warren SE, et al. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin Immunopathol. 2007;29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- 49.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 50.Marchesi J, Shanahan F. The normal intestinal microbiota. Curr Opin Infect Dis. 2007;20:508–513. doi: 10.1097/QCO.0b013e3282a56a99. [DOI] [PubMed] [Google Scholar]
- 51.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Mahowald MA, Ley RE, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Backhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 55.Scupham AJ, Presley LL, Wei B, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peterson DA, Frank DN, Pace NR, et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 62.Hooper LV, Bry L, Falk PG, et al. Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays. 1998;20:336–343. doi: 10.1002/(SICI)1521-1878(199804)20:4<336::AID-BIES10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 63.Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 65.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 66.Dumas M-E, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cani PD, Delzenne NM. Gut microflora as a target for energy and metabolic homeostasis. Curr Opin Clin Nutr Metab Care. 2007;10:729–734. doi: 10.1097/MCO.0b013e3282efdebb. [DOI] [PubMed] [Google Scholar]
- 68.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 69.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 70.Bergman E. Energy contribution of volitile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 71.Wachtershauser A, Stein J. Rationale for the luminal provision of butyrate in intestinal diseases. Eur J Nutr. 2000;39:164–171. doi: 10.1007/s003940070020. [DOI] [PubMed] [Google Scholar]
- 72.Hamer HM, Jonkers D, Venema K, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 73.Kumar A, Collier-Hyams LS, Wu H, et al. The bacterial fermentation product butyrate influences epithelial signaling via ROS mediated changes in Cullin-1 neddylation. J Immunol. doi: 10.4049/jimmunol.182.1.538. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vernia P, Annese V, Bresci G, et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur J Clin Invest. 2003;33:244–248. doi: 10.1046/j.1365-2362.2003.01130.x. [DOI] [PubMed] [Google Scholar]
- 75.Madara JL, Nash S, Moore R, et al. Structure and function of the intestinal epithelial barrier in health and disease. Monogr Pathol. 1990;31:306–324. [PubMed] [Google Scholar]
- 76.Laukoetter MG, Nava P, Nusrat A. Role of the intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2008;14:401–407. doi: 10.3748/wjg.14.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ismail AS, Hooper LV. Epithelial cells and their neighbors. IV. Bacterial contributions to intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2005;289:G779–784. doi: 10.1152/ajpgi.00203.2005. [DOI] [PubMed] [Google Scholar]
- 78.Madsen K, Cornish A, Soper P, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 79.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 80.Hooper LV, Wong MH, Thelin A, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 81.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 84.Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology. 2007;132:1359–1374. doi: 10.1053/j.gastro.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 85.Katakura K, Lee J, Rachmilewitz D, et al. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J Clin Invest. 2005;115:695–702. doi: 10.1172/JCI22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zaph C, Troy AE, Taylor BC, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 87.Nenci A, Becker C, Wullaert A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 88.Ben-Neriah Y, Schmidt-Supprian M. Epithelial NF-kappaB maintains host gut microflora homeostasis. Nat Immunol. 2007;8:479–481. doi: 10.1038/ni0507-479. [DOI] [PubMed] [Google Scholar]
- 89.Chen LW, Egan L, Li ZW, et al. The two faces of IKK and NF-kappaB inhibition: prevention of systemic inflammation but increased local injury following intestinal ischemia-reperfusion. Nat Med. 2003;9:575–581. doi: 10.1038/nm849. [DOI] [PubMed] [Google Scholar]
- 90.McVay LD, Keilbaugh SA, Wong TM, et al. Absence of bacterially induced RELMbeta reduces injury in the dextran sodium sulfate model of colitis. J Clin Invest. 2006;116:2914–2923. doi: 10.1172/JCI28121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gewirtz AT. Flag in the crossroads: flagellin modulates innate and adaptive immunity. Curr Opin Gastroenterol. 2006;22:8–12. doi: 10.1097/01.mog.0000194791.59337.28. [DOI] [PubMed] [Google Scholar]
- 92.Zeng H, Carlson AQ, Guo Y, et al. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J Immunol. 2003;171:3668–3674. doi: 10.4049/jimmunol.171.7.3668. [DOI] [PubMed] [Google Scholar]
- 93.Vijay-Kumar M, Sanders CJ, Taylor RT, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vijay-Kumar M, Wu H, Jones R, et al. Flagellin suppresses epithelial apoptosis and limits disease during enteric infection. Am J Pathol. 2006;169:1686–1700. doi: 10.2353/ajpath.2006.060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeng H, Wu H, Sloane V, et al. Flagellin/TLR5 responses in epithelia reveal intertwined activation of inflammatory and apoptotic pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G96–G108. doi: 10.1152/ajpgi.00273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burdelya LG, Krivokrysenko VI, Tallant TC, et al. An agonist of Toll-like receptor 5 has radioprotective activity in mouse and primate models. Science. 2008;320:226–230. doi: 10.1126/science.1154986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vijay-Kumar M, Aitken JD, Kumar A, et al. Toll-like receptor 5-deficient mice have dysregulated intestinal gene expression and nonspecific resistance to Salmonella-induced typhoid-like disease. Infect Immun. 2008;76:1276–1281. doi: 10.1128/IAI.01491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vijay-Kumar M, Aitken JD, Sanders CJ, et al. Flagellin treatment protects against chemicals, bacteria, viruses, and radiation. J Immunol. 2008;180:8280–8285. doi: 10.4049/jimmunol.180.12.8280. [DOI] [PubMed] [Google Scholar]
- 99.Carlson RM, Vavricka SR, Eloranta JJ, et al. fMLP induces Hsp27 expression, attenuates NF-kappaB activation, and confers intestinal epithelial cell protection. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1070–G1078. doi: 10.1152/ajpgi.00417.2006. [DOI] [PubMed] [Google Scholar]
- 100.Fujiya M, Musch MW, Nakagawa Y, et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1:299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 101.Kelly D, Conway S, Aminov R. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 2005;26:326–333. doi: 10.1016/j.it.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 102.Iyer C, Kosters A, Sethi G, et al. Probiotic Lactobacillus reuteri promotes TNF-induced apoptosis in human myeloid leukemia-derived cells by modulation of NF-kB and MAPK signalling. Cell Microbiol. 2008;10:1441–1452. doi: 10.1111/j.1462-5822.2008.01137.x. [DOI] [PubMed] [Google Scholar]
- 103.Neish AS. Microbial interference with host inflammatory responses. In: Hecht G, editor. Microbial pathogenesis and the intestinal epithelial cell. ASM Press; Washington, DC: 2003. pp. 175–190. [Google Scholar]
- 104.Yan F, Cao H, Cover TL, et al. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Menard S, Candalh C, Bambou JC, et al. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53:821–828. doi: 10.1136/gut.2003.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pena JA, Versalovic J. Lactobacillus rhamnosus GG decreases TNF-alpha production in lipopolysaccharide-activated murine macrophages by a contact-independent mechanism. Cell Microbiol. 2003;5:277–285. doi: 10.1046/j.1462-5822.2003.t01-1-00275.x. [DOI] [PubMed] [Google Scholar]
- 107.Madsen KL, Doyle JS, Jewell LD, et al. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116:1107–1114. doi: 10.1016/s0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- 108.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 109.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 110.Tien M-T, Girardin SE, Regnault B, et al. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. 2006;176:1228–1237. doi: 10.4049/jimmunol.176.2.1228. [DOI] [PubMed] [Google Scholar]
- 111.Petrof EO, Kojima K, Ropeleski MJ, et al. Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127:1474–1487. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 112.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 113.Kim DW, Lenzen G, Page AL, et al. The Shigella flexneri effector OspfG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Angot A, Vergunst A, Genin S, et al. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rytkonen A, Holden DW. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe. 2007;1:13–22. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Collier-Hyams LS, Sloane V, Batten BC, et al. Cutting edge: bacterial modulation of epithelial signaling via changes in neddylation of Cullin-1. J Immunol. 2005;175:4194–4198. doi: 10.4049/jimmunol.175.7.4194. [DOI] [PubMed] [Google Scholar]
- 117.Lee WJ. Bacterial-modulated signaling pathways in gut homeostasis. Sci Signal. 2008;1:pe24. doi: 10.1126/stke.121pe24. [DOI] [PubMed] [Google Scholar]
- 118.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 119.Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 120.Neutra MR, Mantis NJ, Kraehenbuhl JP. Collaboration of epithelial cells with organized mucosal lymphoid tissues. Nat Immunol. 2001;2:1004–1009. doi: 10.1038/ni1101-1004. [DOI] [PubMed] [Google Scholar]
- 121.Kelly D, King T, Aminov R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res. 2007;622:58–69. doi: 10.1016/j.mrfmmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 122.Jang MH, Kweon M-N, Iwatani K, et al. Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proc Natl Acad Sci U S A. 2004;101:6110–6115. doi: 10.1073/pnas.0400969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mazmanian SK, Liu CH, Tzianabos AO, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 124.Zaph C, Du Y, Saenz SA, et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 126.Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115:153–162. doi: 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Macpherson AJ, Gatto D, Sainsbury E, et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 128.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 129.Peterson DA, McNulty NP, Guruge JL, et al. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 130.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bach J-F. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 132.von Mutius E. Allergies, infections and the hygiene hypothesis—the epidemiological evidence. Immunobiology. 2007;212:433–439. doi: 10.1016/j.imbio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 133.Guarner F, Bourdet-Sicard R, Brandtzaeg P, et al. Mechanisms of disease: the hygiene hypothesis revisited. Nat Clin Pract Gastroenterol Hepatol. 2006;3:275–284. doi: 10.1038/ncpgasthep0471. [DOI] [PubMed] [Google Scholar]
- 134.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 135.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562–568. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 136.Di Giacinto C, Marinaro M, Sanchez M, et al. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J Immunol. 2005;174:3237–3246. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- 137.Boirivant M, Strober W. The mechanism of action of probiotics. Curr Opin Gastroenterol. 2007;23:679–692. doi: 10.1097/MOG.0b013e3282f0cffc. [DOI] [PubMed] [Google Scholar]
- 138.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 139.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hecht GA. Inflammatory bowel disease—live transmission. N Engl J Med. 2008;358:528–530. doi: 10.1056/NEJMcibr0707718. [DOI] [PubMed] [Google Scholar]
- 142.Ryu J-H, Kim S-H, Lee H-Y, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 143.Stecher B, Hardt W-D. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 144.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Corr SC, Li Y, Riedel CU, et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kaper JB, Sperandio V. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun. 2005;73:3197–3209. doi: 10.1128/IAI.73.6.3197-3209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994;330:257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 148.Top J, Willems R, Bonten M. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol. 2008;52:297–308. doi: 10.1111/j.1574-695X.2008.00383.x. [DOI] [PubMed] [Google Scholar]
- 149.Pamer EG. Immune responses to commensal and environmental microbes. Nat Immunol. 2007;8:1173–1178. doi: 10.1038/ni1526. [DOI] [PubMed] [Google Scholar]
- 150.Brandl K, Plitas G, Mihu CN, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.World Health Organization Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. 2001 Available at: http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf.
- 152.Hord NG. Eukaryotic-microbiota crosstalk: potential mechanisms for health benefits of prebiotics and probiotics. Annu Rev Nutr. 2008;28:1–17. doi: 10.1146/annurev.nutr.28.061807.155402. [DOI] [PubMed] [Google Scholar]
- 153.Park J, Floch MH. Prebiotics, probiotics, and dietary fiber in gastrointestinal disease. Gastroenterol Clin North Am. 2007;36:47–63. doi: 10.1016/j.gtc.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 154.Ouwehand AC. Antiallergic effects of probiotics. J Nutr. 2007;137:794S–797S. doi: 10.1093/jn/137.3.794S. [DOI] [PubMed] [Google Scholar]
- 155.Seifert S, Watzl B. Inulin and oligofructose: review of experimental data on immune modulation. J Nutr. 2007;137:2563S–2567S. doi: 10.1093/jn/137.11.2563S. [DOI] [PubMed] [Google Scholar]
- 156.Gibson GR, Probert HM, van Loo JAE, et al. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nut Res Rev. 2004;17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 157.Besselink MG, van Santvoort HC, Buskens E, et al. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet. 2008;371:651–659. doi: 10.1016/S0140-6736(08)60207-X. [DOI] [PubMed] [Google Scholar]
- 158.Land MH, Rouster-Stevens K, Woods CR, et al. Lactobacillus sepsis associated with probiotic therapy. Pediatrics. 2005;115:178–181. doi: 10.1542/peds.2004-2137. [DOI] [PubMed] [Google Scholar]