Abstract
Numerous proteins controlling cell cycle progression, apoptosis, and angiogenesis are degraded by the ubiquitin/proteasome system, which has become the subject for intense investigations for cancer therapeutics. Therefore, we used in silico and experimental approaches to screen compounds from the NCI chemical libraries for inhibitors against the chymotrypsin-like (CT-L) activity of the proteasome and discovered PI-083. Molecular docking indicates that PI-083 interacts with the Thr21, Gly47 and Ala49 residues of the β5 subunit and Asp114 of the β6 subunit of the proteasome. PI-083 inhibits CT-L activity and cell proliferation and induces apoptosis selectively in cancer cells (ovarian T80-Hras, pancreatic C7-Kras and breast MCF-7) as compared to their normal/immortalized counterparts (T80, C7 and MCF-10A, respectively). In contrast, Bortezomib, the only proteasome inhibitor approved by the Food and Drug Administration (FDA), did not exhibit this selectivity for cancer over non-transformed cells. In addition, in all cancer cells tested, including Multiple Myeloma (MM), breast, pancreatic, ovarian, lung, prostate cancer cell lines as well as fresh MM cells from patients, PI-083 required less time than Bortezomib to induce its antitumor effects. Furthermore, in nude mouse xenografts in vivo, PI-083, but not Bortezomib, suppressed the growth of human breast and lung tumors. Finally, following in vivo treatment of mice, PI-083 inhibited tumor, but not hepatic liver CT-L activity, whereas Bortezomib inhibited both tumor and liver CT-L activities. These results suggest that PI-083 is more selective for cancer cells and may have broader antitumor activity and therefore warrants further advanced preclinical studies.
Keywords: proteasome inhibitor, high throughput screening, cyclin-dependent kinase inhibitors, cell proliferation, apoptosis, nude mice
Introduction
Cancer is associated with deregulated cell cycle progression and/or decreased apoptosis. Both of these processes are regulated by a complex interplay of transcription, protein synthesis, protein-protein interactions, protein phosphorylation and protein degradation. More than 80 % of cellular proteins are degraded by the ubiquitin/proteasome system (UPS) 1. Deregulation of various components of the UPS resulting in increased degradation of cell cycle inhibitors or pro-apoptotic proteins (e.g. p21Cip1, p27Kip1, p53, Bax, IκBα) or decreased degradation of cell cycle stimulators or anti-apoptotic proteins (e.g. cyclins, Bcl-2) can contribute to the transformed phenotype 1–3. The UPS has two distinct steps: recognition/ubiquitination and degradation (reviewed in refs. 4, 5). Key components of this regulatory system were discovered in the early 1980s 6, 7. Ubiquitination of proteins consists of the transfer of multiple ubiquitin molecules, polypeptides of 76 amino acids, to the target protein and is accomplished by the concerted action of three enzymes termed ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2 and ubiquitin-protein ligase E3, where some degree of substrate specificity is provided by hundreds of different E3 enzymes. Polyubiquitin-flagged proteins are then recognized by the proteasome, a large multi-subunit complex found in the cytoplasm and the nucleus of all eukaryotic cells, which was first described in 1988 8. Degradation of proteins is mediated by the 20S catalytic complex of the 26S proteasome 9, 10, which is an enzyme consisting of three proteolytic activities, namely peptidyl glutamyl peptide hydrolase (PGPH), trypsin-like (T-L), and CT-L activities, residing in the β1, β2, and β5 catalytic subunits, respectively 2, 11.
In contrast to normal cells, which just require a low level of survival signals to stay alive 12, cancer cells typically have acquired a series of mutations that render them dependent on strong activation of one or more survival pathways 13. One of these is the UPS-dependent degradation of cellular proteins, which drives cell cycle progression and/or survival. Therefore, the UPS has become a promising target for anti-cancer strategies (reviewed in refs. 2, 3, 11, 14).
One proteasome inhibitor that has been studied extensively is the dipeptide boronic acid analog PS-341 (Bortezomib) (for reviews, see refs. 1, 15). Preclinical studies have shown that Bortezomib induces apoptosis in different cancer cell lines including multiple myeloma 16, lung 17, 18 and prostate cancer 19, 20. Likewise, in xenografts implanted in nude mice, Bortezomib inhibits the growth of human prostate cancer 19, 21, squamous cell carcinoma 22, and ovarian cancer 23. However, in other tumors such as human A549 lung tumors 18 or MIA-PaCa2 pancreatic tumors 24, even when administered in combination with other agents, Bortezomib has only marginal effects. Currently, Bortezomib has been approved by the FDA for treatment of relapsed/refractory MM 25, 26, as a single agent or in combination with conventional therapies 27, 28, and is being investigated for solid tumors 29, including non-small cell lung cancer and prostate cancer (reviewed in refs. 30, 31). However, Bortezomib is associated with undesired side effects in MM patients 32 and does not display substantial clinical activity in other cancers 30, 31. We therefore sought to identify novel proteasome inhibitors. To this end, we have screened 3,229 compounds of the NCI Diversity, Natural Product, Challenge and Mechanistic Sets for inhibitors against the CT-L activity of the purified 20S proteasome and confirmed 8 lead compounds. One of these, PI-083, was synthesized in-house and used in this study. PI-083 could induce cell death and apoptosis efficiently in different cancer cells including MM cell lines and MM patients’ primary bone marrow cells. PI-083, but not Bortezomib, was more selective for cancer cells over normal cells. Furthermore, in vivo, PI-083, but not Bortezomib, inhibited the growth of human breast and lung tumors in the nude mouse xenograft model. Overall, our results suggest that PI-083 targets selectively cancer cells and that further investigation of PI-083 as a potential anticancer drug is warranted.
Results
High Throughput Screening (HTS) Identifies a Novel Proteasome Inhibitor, PI-083
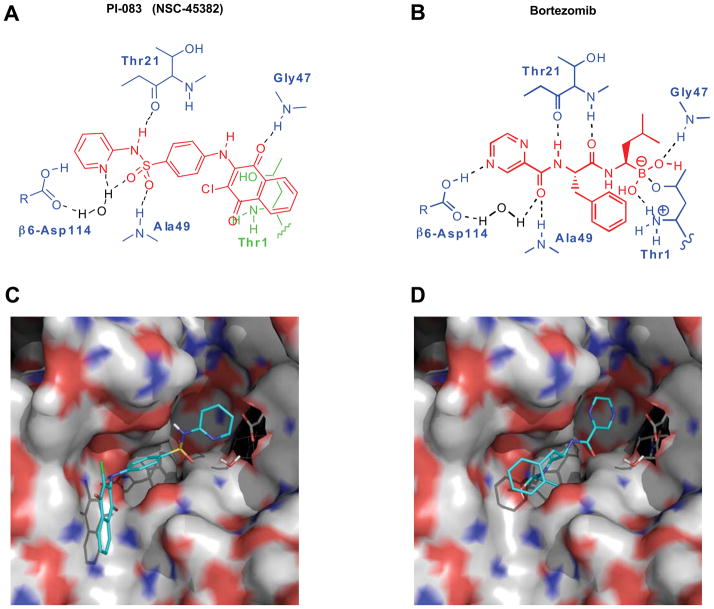
We have used both experimental and virtual (in silico) HTS to screen chemical libraries from the NCI. For the experimental portion, we screened the NCI Diversity Set (1,990 compounds), the Natural Product Set (235 compounds), and the Challenge and Mechanistic Sets (1,004 compounds) against the CT-L activity of the proteasome as described in Materials and Methods. Eight (5 of which were from the NCI diversity set) hits with IC50 values < 10 μM were identified. The most potent of these, PI-083 (IC50 = 1 μM), was confirmed with in-house synthesized material and is shown along with the structure of Bortezomib in Figs. 1A and B. For in silico screening, the GLIDE computer program, version 3.0 (Schrödinger, LLC, New York, NY), using default options and parameters for grid generation and docking, was employed to screen the NCI-3D Diversity Set database. Coordinates for the CT-L β5 subunit derived from the X-ray crystal structure of the yeast 20S proteasome determined at 3.0 Å resolution (PDB ID: 1JD2) were employed for the automated docking studies 33. Structurally, the yeast and mammalian 20S proteasome are similar and the catalytic site in the β5 subunit is highly conserved between the two species 34, 35. Of the 5 diversity set experimental hits, 4 (including PI-083) ranked within the top 125 compounds with the best docking scores. Subsequent to our initial docking studies, the X-ray structure of the yeast 20S proteasome complexed to Bortezomib became available 36. This structure revealed that the pyrazine ring in Bortezomib interacts via a direct hydrogen bond with Asp114 from the β6 subunit of the proteasome. Since PI-083 contains a pyridine ring we re-docked it to a new model that we derived from the Bortezomib-proteasome complex that included both the β5 and β6 subunits (PDB ID: 2F16). The structure of PI-083 docked to the CT-L subunit of the proteasome using this new model, is shown in Fig. 1C. Similar to the X-ray structure of Bortezomib (Fig. 1D), the computer model of PI-083 docked to the proteasome suggests key H-bonding interactions between the napthoquinone ring and Gly47 and between the sulfonamide and Thr21 and Ala49 (Fig. 1A). Moreover, the modeling suggests that the pyridine ring of PI-083 is H-bonded to Asp114 via an intervening water molecule located crystallographically in the Bortezomib structure and included in our model. We next examined the potency and selectivity of PI-083 and Bortezomib to inhibit CT-L, T-L and PGPH activities of the proteasome as described under Materials and Methods. Table 1 shows that PI-083 inhibited CT-L, T-L and PGPH with IC50 values of 1, 4.5 and 4.5 μM, respectively. Bortezomib inhibited these activities with IC50 values of 9 nM, 7.0 μM and 0.48 μM, respectively.
Figure 1. Molecular modeling of PI-083 (NSC-45382), a novel proteasome inhibitor.
Chemical structures of (A) PI-083 (NSC-45382), and (B) Bortezomib are shown in red. The hydrogen bonds formed between PI-083 or Bortezomib and the protein, via Thr1, Thr21, Ala49, Gly47, and Asp114 (from the β6 subunit) are shown schematically but not to scale. (C) Protein surface rendering of the CT-L subunits β5 (left) and β6 (right) of the 20S proteasome with PI-083 docked. The surface is colored according to atomic charge. Positively charged areas are colored in blue and negatively charged areas are colored in red. For PI-083, carbon atoms are colored in cyan, oxygen in red, nitrogen in blue, hydrogen in white, and sulfur in dark yellow. Asp114 from the β6 subunit is also shown (carbon atoms in gray) along with a crystallographically located water molecule that is H-bonded to Asp114. (D) Protein surface rendering of the CT-L subunits β5 (left) and β6 (right) of the 20S proteasome with Bortezomib bound (from the X-ray structure). Asp114 from the β6 subunit is also shown (carbon atoms colored in gray) along with the crystallographically located water molecule. Note that the pyrazine ring of Bortezomib is within H-bond distance from Asp114. Since the pKa of pyrazine is approximately 1.0, Asp114 is most likely protonated (not shown).
Table 1.
IC50 values (μM) of PI-083 and Bortezomib for CT-L, T-L and PGPH-like activities in vitro
| Compound | CT-L | T-L | PGPH |
|---|---|---|---|
| PI-083 | 1.0 ± 0.63 | 4.5 ± 1.4 | 4.5 ± 1.2 |
| Bortezomib | 0.009 ± 0.006 | 7.0 ± 0.24 | 0.48 ± 0.021 |
The values given are the means of 3 experiments ± standard error.
PI-083 inhibits CT-L and cell proliferation selectively in cancer cells over their non-transformed counterparts
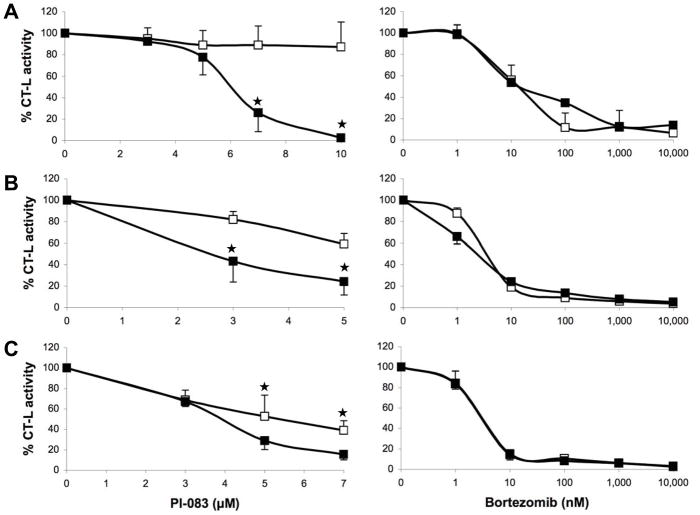
We next determined the ability of these two proteasome inhibitors to inhibit cell proliferation and induce apoptosis in tumor compared to non-transformed cells. To this end, we used 3 pairs of cell lines from ovarian, pancreatic, and breast origin. The first pair consists of T80 normal human ovarian cells immortalized with Large T-antigen (inactivates both p53 and pRb) and hTERT (human telomerase) based on the original Weinberg model 37, 38 and T80-Hras (T80 cells expressing oncogenic human V12-Hras) 39. The second pair of cell lines consists of C7 and C7-Kras (generated in a similar fashion to the T80/T80-Hras pair except that this pair originated from normal pancreatic duct epithelial cells and the oncogene is C7-Kras and not V12-Hras) 40. The third pair consists of MCF-7 (a human breast cancer cell line) and MCF-10A (an immortalized non-transformed breast cell line). First, we determined the ability of PI-083 and Bortezomib to inhibit CT-L in the 3 pairs of tumor/non-transformed cell lines as described under Materials and Methods. Fig. 2 shows that PI-083 inhibited more selectively the CT-L activity in tumor over non-transformed cells in the 3 pairs of cell lines. The selectivity was statistically significant (p < 0.05) and was more pronounced in the breast cell lines. In contrast, Bortezomib was non-selective and inhibited CT-L activity equally well in both cancer and non-transformed cells (Fig. 2).
Figure 2. Effects of PI-083 and Bortezomib on proteasomal CT-L activities in cancer and normal/immortalized cells.
Exponentially growing cancer cells (■) and normal cells (□) were treated with indicated concentrations of PI-083 (left panel) or Bortezomib (right panel) for 24 h, followed by measurement of CT-L activity in whole cell extracts. (A) MCF-7 breast cancer and MCF-10A breast epithelial cells. (B) T80-Hras ovarian cancer and T80 ovarian epithelial cells. (C) HPDE6-C7-Kras pancreatic cancer and HPDE6-C7 pancreatic epithelial cells. The graphs represent the means ± standard deviation of at least 3 independent experiments. Asterisks indicate statistical significance (p < 0.05).
Next, we determined the ability of PI-083 and Bortezomib to inhibit cell proliferation of the above 3 pairs of cell lines. To this end, cells were treated with various concentrations of either PI-083 or Bortezomib for 24 h and processed for Trypan Blue cell counting as described under Materials and Methods. Fig. 3 shows that PI-083 inhibited the proliferation of all three cancer cell lines (MCF-7, T80-Hras and C7-Kras) more potently than their non-transformed “normal” counterparts (MCF-10A, T80 and C7). This selectivity was statistically significant in all 3 pairs of cell lines (p < 0.05). In contrast, Bortezomib was either more potent towards normal over tumor cells (breast and ovarian, Fig. 3A and B) or equally potent (pancreatic, Fig. 3C). To confirm these results, we treated the 3 pairs of cell lines (breast, ovarian and pancreatic) as well as multiple myeloma, human prostate and lung cancer cell lines with either PI-083 or Bortezomib for 24 or 72 h and determined their effects on proliferation by MTT assays as described under Materials and Methods. Table 2 shows that at the 24 h time point, as with the Trypan Blue results of Fig. 3, PI-083 was more selective at inhibiting proliferation of cancer cells as compared to non-transformed cells. The difference in IC50 values between the cancer cells and their non-transformed counterparts was statistically significant for breast (p < 0.0001), pancreatic (p < 0.0013) and ovarian (p < 0.017) cells. In contrast, at 24 h, Bortezomib IC50 values were over 30 μM for all cancer and non-transformed cell lines except for the RPMI-8226 cell line (Table 2). Table 2 also shows that, after both 24 and 72 h, PI-083 inhibited the proliferation of human prostate cancer cells DU-145 (undetectable Bax levels) and LNCaP (Bax-over expressed) as well as that of lung cancer cells CaLu-1 (both p53 alleles deleted) and A549 (p53 present and wild type), suggesting that PI-083 does not require the two proteasome substrates Bax or p53 for inhibiting cancer cell proliferation. Similar results were obtained with Bortezomib except that, as seen with the other cell lines, PI-083 was more potent after 24 h whereas Bortezomib was more potent after 72 h of treatment. Finally, both PI-083 and Bortezomib inhibited proliferation in MM cells, with RPMI-8226 being much more sensitive especially to Bortezomib after 24 h of treatment.
Figure 3. Effects of PI-083 and Bortezomib on proliferation in cancer and normal/immortalized cells.
Exponentially growing cancer cells (■) and normal cells (□) were treated with indicated concentrations of PI-083 (left panel) or Bortezomib (right panel) for 24 h, followed by determination of viable cells. (A) MCF-7 breast cancer and MCF-10A breast epithelial cells. (B) T80-Hras ovarian cancer and T80 ovarian epithelial cells. (C) HPDE6-C7-Kras pancreatic cancer and HPDE6-C7 pancreatic epithelial cells. The graphs represent the means ± standard deviation of at least 3 independent experiments. Asterisks indicate statistical significance (p < 0.05).
Table 2.
IC50 values (μM) of PI-083 and Bortezomib for cell viability measured by MTT assay in different cancer and normal cell lines
| Cancer | Cell line | 24 h | 72 h | ||
|---|---|---|---|---|---|
| PI-083 | Bortezomib | PI-083 | Bortezomib | ||
| Breast | MCF-7 | 4.5 ± 0.82 * (0.0001) | >30 | 2.2 ± 1.1 * (0.04) | 8.6 ± 1.41 |
| MCF-10A | 17± 0.86 | >30 | 4.2 ± 2.1 | 0.15 ± 0.074 * (0.0015) | |
| Pancreas | C7-Kras | 2.4 ± 0.23 * (0.0013) | >30 | 1.9 ± 0.2 * (0.007) | 0.028 ± 0.006 (0.51) |
| C7 | 5.1 ± 0.53 | >30 | 4.2 ± 1.6 | 0.032 ± 0.001 | |
| Ovary | T80-Hras | 1.5 ± 0.57 * (0.017) | >30 | 2.1 ± 0.4 | 0.024 ± 0.01 |
| T80 | 2.7 ± 0.53 | >30 | 1.7 ± 0.39 (0.15) | 0.014 ± 0.001 (0.29) | |
| Prostate | DU-145 | 8.6 ± 0.63 | >30 | 5.0 ± 0.72 | 0.025 ± 0.002 |
| Prostate | LNCaP | 14 ± 2.8 | >30 | 5.2 ± 1.1 | 0.057 ± 0.004 |
| Lung | CaLu-1 | 7.3 ± 0.43 | >30 | 4.9 ± 1.2 | 0.022 ± 0.002 |
| Lung | A549 | 41 ± 17 | >30 | 11 ± 6.7 | 0.41 ± 0.07 |
| Multiple Myeloma | U266 | 23 ± 1.7 | >30 | 7.0 ± 1.5 | 0.012 ± 0.003 |
| Multiple Myeloma | RPMI-8226 | 7.4 ± 0.38 | 0.049 ± 0.025 | 10 ± 4.0 | 0.026 ± 0.003 |
The values given are the means of 3 to 5 experiments ± standard deviation. Statistically significant differences between cancer and non-transformed cells were established by performing a t-test
p < 0.05.
PI-083, but not Bortezomib, induces apoptosis more selectively in cancer cells over non-transformed cells
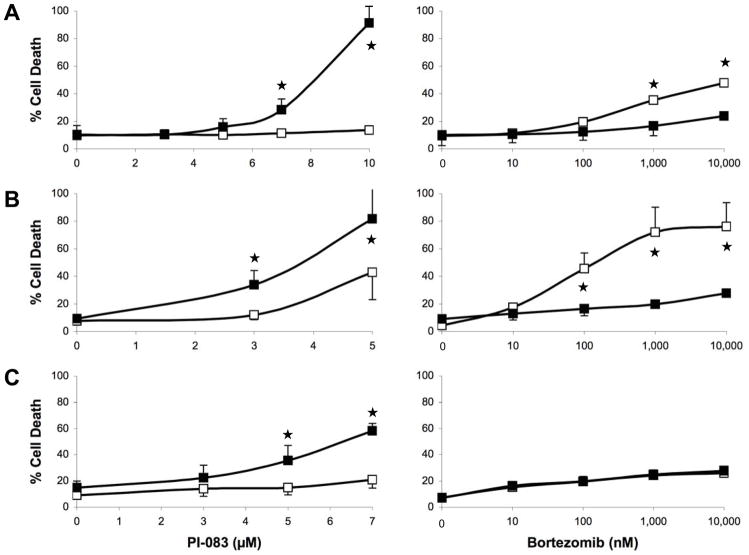
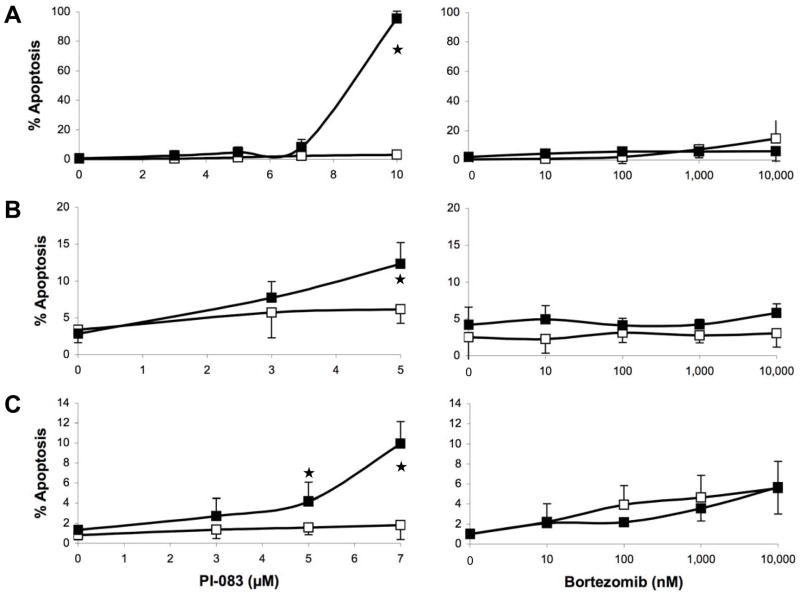
Fig. 3 and Table 2 show major differences between PI-083 and Bortezomib, with PI-083, but not Bortezomib, being more selective towards inhibiting the proliferation of cancer over non-transformed cells. We next determined the ability of PI-083 and Bortezomib to induce cell death (Trypan Blue) and apoptosis (TUNEL) in the above 3 pairs of cancer/non-transformed cell lines as described under Materials and Methods. As seen with proliferation assays, PI-083 was much more effective in increasing cell death in all 3 cancer cell lines over their non-transformed counterparts, whereas Bortezomib was not (Fig. 4A–C). These results were confirmed by TUNEL assay: Fig. 5A–C shows that PI-083 induced apoptosis in all 3 cancer cell lines MCF-7, T80-Hras and C7-Kras, with MCF-7 being the most sensitive. In contrast, PI-083 did not induce apoptosis in non-transformed MCF-10A and C7 and induced very little apoptosis in T80 cells. In contrast to PI-083, Bortezomib induced little apoptosis in any of these cell lines (Fig. 5A–C). Taken together, Tables 1 and 2 and Figs. 2–5 show that after 24 h of treatment, PI-083 inhibited proliferation and induced cell death and apoptosis with IC50 values similar to those that inhibited the CT-L activity of the proteasome. In contrast, Bortezomib after 24 h of treatment inhibited potently the CT-L activity with very little effect on proliferation, cell death and apoptosis. However, after 72 h of treatment both PI-083 and Bortezomib inhibited proliferation at concentrations that inhibited the CT-L activity.
Figure 4. Effects of PI-083 and Bortezomib on cell death in cancer and normal/immortalized cells.
Exponentially growing cancer cells (■) and normal cells (□) were treated with indicated concentrations of PI-083 (left panel) or Bortezomib (right panel) for 24 h, followed by determination of dying/dead cells by the Trypan Blue exclusion assay. (A) MCF-7 breast cancer and MCF-10A breast epithelial cells. (B) T80-Hras ovarian cancer and T80 ovarian epithelial cells. (C) HPDE6-C7-Kras pancreatic cancer and HPDE6-C7 pancreatic epithelial cells. The graphs represent the means ± standard deviation of at least 3 independent experiments. Asterisks indicate statistical significance (p < 0.05).
Figure 5. Effects of PI-083 and Bortezomib on apoptosis in cancer and normal/immortalized cells.
Exponentially growing cancer cells (■) and normal cells (□) were treated with indicated concentrations of PI-083 (left panel) or Bortezomib (right panel) for 24 h, followed by determination of the degree of apoptosis. (A) MCF-7 breast cancer and MCF-10A breast epithelial cells. (B) T80-Hras ovarian cancer and T80 ovarian epithelial cells. (C) HPDE6-C7-Kras pancreatic cancer and HPDE6-C7 pancreatic epithelial cells. The graphs represent the means ± standard deviation of at least 3 independent experiments. Asterisks indicate statistical significance (p < 0.05).
Effects of PI-083 and Bortezomib on bone marrow cells isolated from multiple myeloma patients
We next determined the ability of PI-083 and Bortezomib to inhibit proliferation and induce cell death in bone marrow cells from MM patients as described under Materials and Methods. Table 3 shows that treatment of the cells from 10 patients with either PI-083 or Bortezomib for 24 h resulted in inhibition of the CT-L activity of the proteasome with IC50 values of 1.3 ± 0.19 or 0.012 ± 0.006 μM, respectively. With PI-083, this 24 h treatment resulted in inhibition of proliferation and induction of cell death with IC50 values of 3.7 ± 0.86 and 4.0 ± 0.82 μM, respectively. In contrast, although Bortezomib was more potent than PI-083 at inhibiting CT-L activity, it was less potent at inhibiting proliferation and at inducing cell death after 24 h of treatment (IC50 values over 10 μM for all 10 patients). However, the MM cells became more sensitive to Bortezomib with increasing length of time of treatment with IC50 values to inhibit cell viability (MTT assay) of >10, 0.14 ± 0.08 and 0.046 ± 0.008 μM after 24, 48, and 72 h, respectively. The ability of PI-083 to inhibit cell viabilty was rapid and improved only slightly over time with IC50 values of 2.2 ± 0.27, 1.8 ± 0.43, and 1.6 ± 0.32 μM, after 24, 48, and 72 h of PI-083 treatment.
Table 3.
Effects of PI-083 and Bortezomib on primary multiple myeloma cells isolated from patients’ bone marrow (IC50 values, μM)
| Patient No. | CT-L Activity | Proliferation | Cell Death | Viability | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 24 h | 24 h | 24 h | 48 h | 72 h | |||||||
| PI-083 | Bortezomib | PI-083 | Bortezomib | PI-083 | Bortezomib | PI-083 | Bortezomib | PI-083 | Bortezomib | PI-083 | Bortezomib | |
| 1 | 2.1 | 0.0054 | 5 | > 10 | 7.3 | > 10 | ND | ND | ND | ND | ND | ND |
| 2 | 1.5 | 0.062 | 2.9 | > 10 | 3.2 | > 10 | > 10 | > 10 | ND | ND | ND | ND |
| 3 | 1.4 | 0.0015 | 6.6 | > 10 | 6.2 | > 10 | > 10 | > 10 | ND | ND | ND | ND |
| 4 | 0.68 | 0.0031 | 1.3 | > 10 | 2. 7 | > 10 | 2.2 | > 10 | 2.6 | 0.45 | 2.2 | 0.052 |
| 5 | 0.57 | 0.0046 | 2.2 | > 10 | 1.1 | > 10 | 3.4 | > 10 | 3.6 | 0.079 | 2.6 | 0.074 |
| 6 | 1.2 | 0.015 | 2.2 | > 10 | 2.7 | > 10 | 2.1 | > 10 | 0.9 | 0.055 | 1.4 | 0.054 |
| 7 | 0.57 | 0.0034 | 2.6 | > 10 | 2.4 | > 10 | 1.8 | > 10 | 1.4 | > 10 | 1.3 | 0.023 |
| 8 | 2.1 | 0.004 | 1.7 | > 10 | 1.7 | > 10 | 2.0 | 0.19 | 1.5 | 0.064 | 1.8 | 0.052 |
| 9 | 1.8 | 0.0028 | 2.8 | > 10 | 4.1 | > 10 | 1.4 | 0.28 | 0.89 | 0.032 | 0.35 | 0.02 |
| 10 | 0.78 | 0.017 | 10 | > 10 | 9.0 | > 10 | > 10 | > 10 | ND | ND | ND | ND |
PI-083, but not Bortezomib, inhibits the growth of human breast and lung tumors in a nude mouse model
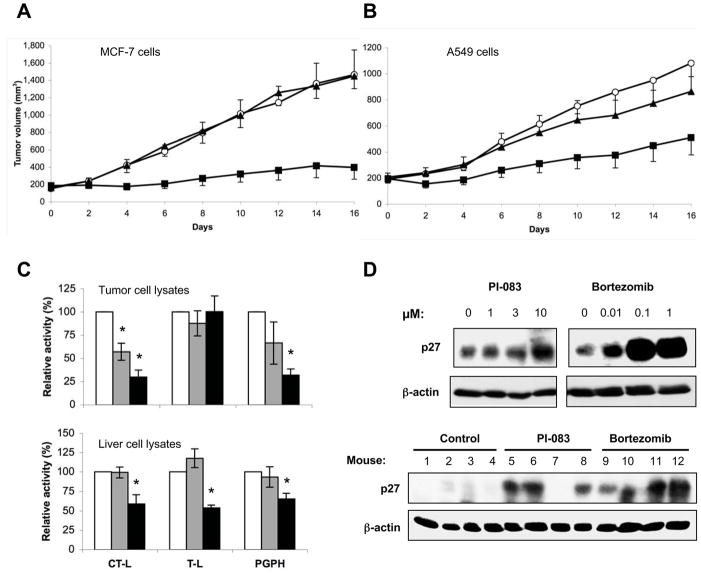
We next evaluated the anti-tumor activities of PI-083 and Bortezomib in a nude mouse xenograft model. To this end, we implanted MCF-7 and A549 cells s.c. in nude mice, and when tumors reached an average size of 200 mm3, the mice were treated 2x weekly either with vehicle, 1 mg/kg body weight (mpk) Bortezomib or 1 mpk PI-083. MCF-7 tumors from control and Bortezomib-treated animals grew to an average size of 1,465 ± 285, and 1,448 ± 145 mm3, respectively. In contrast, tumors from PI-083-treated animals grew to an average size of about 396 ± 137 mm3. Thus, while Bortezomib appeared to be ineffective, treatment with PI-083 resulted in a significant tumor growth inhibition of 84 % (Fig. 6A). We next determined the antitumor activity of PI-083 and Bortezomib in A549 xenografts. Fig. 6B shows that A549 tumors from control and Bortezomib-treated animals grew to an average size of 1,081 ± 103, and 864 ± 207 mm3, respectively. Tumors from PI-083-treated animals grew to an average size of about 511 ± 134 mm3, which corresponds to a tumor growth inhibition of 54 %. Using the Wilcoxon Signed Rank Test, we determined that the sizes of the PI-083-treated, but not the Bortezomib-treated tumors, were significantly different (p < 0.008) from the control animal tumors.
Figure 6. Effects of PI-083 and Bortezomib on tumor growth and the levels of the proteasomal substrate p27kip1 in vivo.
The growth of human tumors following the injection of (A) MCF-7 breast cancer and (B) A549 lung cancer cells into nude mice was determined as described in Materials and Methods. Mice were treated with DMSO (○), 1 mpk Bortezomib (▲) or 1 mpk PI-083 (□). Data represent the means ± standard error of one of three independent experiments with 4 to 6 animals in each group. Asterisks indicate statistical significance (p < 0.05). Proteasomal activities in tumor cell lysates (C, upper panel) or liver cell lysates (C, lower panel) following treatment with DMSO (n=4, white), PI-083 (n=4, gray) or Bortezomib (n=4, black). The asterisks indicate p values ≤ 0.006 for a comparison of experimental and DMSO-treated mice. (D, upper panel) A549 lung cancer cells were treated with 0.1 % DMSO (lane 1) or water (lane 5) or the indicated drug concentrations for 48 h. Cell lysates were then subjected to Western blot analyses with antibodies to p27Kip1 and β-actin as a loading control. (D, lower panel) p27Kip1 levels were determined in lysates prepared from vehicle- or drug-treated A549-tumors by Western blots, with β-actin serving as a loading control.
We next determined whether PI-083 and Bortezomib inhibited the proteasome activity in tumors and livers of mice that were injected i.p. with either PI-083 or Bortezomib. To this end, A549 tumors and livers were extracted 2 h after the last drug injection, and the lysates processed for CT-L, T-L and PGPH activities of the proteasome as described under Materials and Methods. Fig. 6C (upper panel) shows that PI-083 treatment resulted in 43, 12, and 34 % inhibition of tumor CT-L, T-L and PGPH activities, respectively, compared to those of tumors from vehicle-treated controls. On the other hand, Bortezomib treatment resulted in 70, 0, and 68 % inhibition of CT-L, T-L, and PGPH activities, respectively, compared to vehicle-treated controls.
We next determined the effects of PI-083 and Bortezomib on proteasome activities in livers of tumor-bearing mice. PI-083 did not inhibit liver proteasome activities, whereas Bortezomib significantly inhibited hepatic CT-L, T-L, and PGPH activities following in vivo treatments (Fig. 6C, lower panel). These results are consistent with our cell culture data where PI-083 inhibited the proteasome activity more potently in tumor cells compared to non-transformed “normal” cells.
Both PI-083 and Bortezomib increase the tumor levels of the cyclin-dependent kinase inhibitor p27Kip1, a proteasomal substrate
The ability of PI-083 and Bortezomib to inhibit CT-L activity in tumors following i.p. treatments of mice suggests that these drugs may be able to accumulate proteasomal substrates. We therefore examined the levels of p27Kip1 in cultured A549 lung cancer cells exposed to PI-083 and Bortezomib. As shown in Fig. 6D (upper panel), both of these drugs triggered a marked increase in p27Kip1 levels, although the increase produced by Bortezomib was more pronounced than PI-083. Furthermore, we examined the levels of p27Kip1 in tumor lysates derived from A549-bearing nude mice treated with PI-083 or Bortezomib as described in Fig. 6B. As shown in Fig. 6D (lower panel), 3 out of 4 PI-083-treated tumors, and 4 out of 4 Bortezomib-treated tumors demonstrated a dramatic upregulation of p27Kip1 protein as compared to vehicle-treated controls.
Discussion
In this manuscript, we describe the discovery of a novel proteasome inhibitor, PI-083. Our docking studies (based on the actual X-ray-determined structure of Bortezomib complexed to the β5/β6 subunit of the proteasome) suggest remarkable similarities between the binding modes of PI-083 and Bortezomib to the active site of the CT-L enzyme within the proteasome. Indeed, molecular modeling suggests that they engage identical amino acids to bind CT-L. For example, Asp114 binds to Bortezomib through its pyrazine nitrogen and to PI-083 through its pyridine nitrogen. In the case of PI-083 this hydrogen bond is apparently mediated by an intervening water molecule, whereas with Bortezomib there is a direct hydrogen bond between one of its pyrazine nitrogen atoms and protonated Asp114. Note that the pKa of protonated pyrazine is <1 and thus presumably Asp114 is protonated since in the X-ray structure the O-N distance is 2.9 Å, indicative of a strong hydrogen bond. The intervening water molecule is also hydrogen-bonded to one of the sulfonamide oxygen atoms in PI-083, and in Bortezomib it is hydrogen-bonded to the oxygen atom of the carbonyl group attached to the pyrazine ring. Similarly, Ala49 and Thr21 of CT-L form hydrogen bonds with PI-083 through its sulfonamide group and to Bortezomib through its two amide groups. Furthermore, Gly47 and Thr1 (active site) of CT-L are hydrogen-bonded to Bortezomib through its boronate hydroxyls, whereas PI-083 is hydrogen-bonded through one of the carbonyl groups of its naphthoquinone ring to Gly47. This carbonyl group is also possibly hydrogen-bonded to Thr1, but the distance between the Thr1 hydroxyl oxygen and the carbonyl oxygen is 3.4 Å in our computer model, which is a bit long for a strong hydrogen bond. The 100-fold difference in potency between Bortezomib and PI-083 is possibly due to the fact that Bortezomib forms a covalent bond to the β5 subunit through formation of a boronate complex with Thr1 that is further stabilized by hydrogen bonding between Thr1 and Gly47 with the boronate hydroxyl groups. Bortezomib is a potent inhibitor in spite of the fact that the P2 phenylalanine side chain does not make any appreciable interactions with the protein and, in fact, is observed to be oriented toward the solvent interface in the X-ray structure. It is worth noting that we cannot completely rule out covalent bond formation between Thr1 and PI-083 that could conceivably occur via displacement of the chloro substituent in PI-083 by the Thr1 hydroxyl group to form an ether. In our computer model, though, the distance between the threonine hydroxyl oxygen atom and carbon atom to which the chloro group is attached is 5.9 Å, which would mean that a rather significant conformational change in the protein and/or repositioning of the ligand would be required for the obligatory nucleophilic addition/elimination reaction to take place.
One of the most striking and critical differences between PI-083 and Bortezomib is the ability of PI-083 to selectively inhibit the CT-L enzymatic activity in cancer cells over non-transformed “normal” cells. This was seen in both cultured cells (MCF-7/MCF-10A, C7-Kras/C7, and T80-Hras/T80) as well as in vivo (tumors vs. livers). Although we do not know why PI-083 is more selective for cancer cells, a plausible explanation could be that normal cells metabolically inactivate PI-083, but not Bortezomib. Analysis of the metabolic profile of PI-083 within cancer cells and their non-transformed counterparts as well as within tumors and livers in vivo could provide support for this possibility. Regardless of the mechanism of selectivity, the fact that PI-083, but not Bortezomib, inhibits the CT-L activity selectively in tumor cells but not normal cells may explain why PI-083 inhibits the growth and induces apoptosis selectively in cancer cells. This selectivity of PI-083 for cancer over “normal” cells should translate into less toxicity. This is consistent with the fact that no weight loss, change in appetite or activity occurred in mice treated with PI-083. Recently, an elegant study characterized another proteasome inhibitor argyrin A that is currently in preclinical testing and whose potent antitumor effects depend on accumulation of p27Kip1, which in turn triggers apoptosis 41. In addition, NPI-0052, a compound that is orally bioactive, has been reported to target NF-κB and act synergistically with Bortezomib to combat MM 42. However, neither of these studies rigorously addressed the issue of selectivity for tumor vs. non-tumor cells. In addition to its selectivity for cancer over “normal” cells, another potential advantage of PI-083 is its rapid action. Indeed, although both PI-083 and Bortezomib inhibit the CT-L activity of the proteasome within 24 h, PI-083, but not Bortezomib, inhibits growth and induces tumor cell death within 24 h in a wide variety of human cancer cells lines as well as fresh bone marrow aspirates from multiple myeloma patients. The fact that Bortezomib requires 48 to 72 h may necessitate its presence in the patient’s blood for long periods of time, and this, coupled to its ability to inhibit CT-L equally in tumor and normal cells may contribute to its toxicities in patients.
In addition to potentially being less toxic, PI-083 may also have a broader spectrum of anti-tumor activity. Indeed, in two animal xenograft models of solid tumors, MCF-7 human breast tumors and A549 non-small cell lung tumors were sensitive to PI-083, but not Bortezomib. The fact that Bortezomib is ineffective against MCF-7-derived tumors is consistent with the work of others 43. The resistance of A549 tumors to Bortezomib is also consistent with the work of others showing that even in combination with other agents, Bortezomib’s antitumor activity against A549 tumors is marginal 18. Thus, we have discovered the proteasome inhibitor PI-083, which presents major advantages in that it is selective for cancer over “normal” cells and has a potentially broader spectrum of antitumor activity. These observations warrant further advanced preclinical studies.
Materials and Methods
Materials
DMEM, RPMI-1640, DMEM/Ham’s F-12, horse serum, Keratinocyte-SFM penicillin and streptomycin were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum was from Atlanta Biologicals (Atlanta, GA). Purified 20S proteasome (rabbit), fluorogenic peptide substrates Suc-Leu-Leu-Val-Tyr-AMC (for the proteasomal CT-L activity), benzyloxy-carbonyl (Z-Leu-Leu-Glu-AMC (for the proteasomal PGPH activity) were purchased from Boston Biochem (Cambridge, MA). Fluorogenic peptide substrates Bz-Val-Gly-Arg-AMC (for the proteasomal T-L activity) were obtained from Biomol International (Plymouth Meeting, PA). Antibodies were obtained from the following suppliers: p27Kip1 (BD Biosciences, San Jose, CA), and β-actin (Sigma-Aldrich, St. Louis, MO). The APO-Direct Kit was from BD Biosciences (San Jose, CA). The proteasome inhibitor NSC-45382 (PI-083) was synthesized in-house as reported previously 44. All other reagents were from Sigma-Aldrich unless otherwise noted.
Determination of proteolytic activity
In the high-throughput screen, we used fluorogenic peptides as substrates to assay 3,229 compounds of the NCI Diversity, Natural Product, Challenge and Mechanistic Sets (at 10 μM) for inhibitory activity against the proteolytic activities of the purified rabbit 20S proteasome, resulting in the identification of PI-083. Briefly, 70 ng of purified 20S proteasome was incubated with 20 μM Suc-Leu-Leu-Val-Tyr-AMC for the CT-L activity, Bz-Val-Gly-Arg-AMC for the T-L activity, and benzyloxycarbonyl Z-Leu-Leu-Glu-AMC for the PGPH activity for 1 h at 37°C in 100 μl of assay buffer (50 mM Tris-HCl, pH 7.6) with or without PI-083 and Bortezomib. After incubation, production of hydrolyzed 7-amido-4-methyl-coumarin (AMC) groups was measured using a WALLAC Victor2 1420 Multilabel Counter with an excitation filter of 355 nm and an emission filter of 460 nm (Perkin Elmer Life Sciences, Turku, Finland).
To determine proteasome activity in whole cell extracts from cultured cells (5 μg) or tumor and liver tissue extracts (30 μg) from nude mice, the same assay was used except that the buffer was changed to 20 mM HEPES, 0.5 mM EDTA, pH 8.0.
Cell culture and extract preparation
Human MCF-7 breast cancer and DU-145 prostate cancer cells were cultured in DMEM, and LNCaP prostate cancer as well as U266 and RPMI-8226 multiple myeloma cells were cultured in RPMI-1640 medium containing 10% fetal calf serum (FCS). Normal immortalized MCF-10A breast cells were cultured in DMEM/Ham’s F-12 containing 5% horse serum, 20 ng/ml epidermal growth factor (EGF), 100 ng/ml cholera toxin, 500 ng/ml hydrocortisone and 0.01 mg/ml insulin. Human lung carcinoma cell lines A549 and CaLu-1 were cultured in F-12 Kaighn’s and McCoy’s 5A medium, respectively, with 10% FCS. T80-Hras cells (an Hras-V12-transformed human ovarian epithelial cell line) and their normal/immortalized counterpart T80 cells (a generous gift from J. Liu and R. Bast 39) were cultured in Medium 199/MCDB 105 with 10% fetal calf serum. Normal/immortalized pancreatic duct epithelial cells HPDE6-C7 and their mutated K-ras derivatives HPDE6-C7K-ras (kindly provided by M.S. Tsao 40) were cultured in Keratinocyte-SFM supplemented with EGF and bovine pituitary extract. All media were supplemented with 100 units/ml of penicillin and 100 μg/ml of streptomycin. All cells were maintained at 37°C in a humidified incubator in an atmosphere of 5% CO2.
Whole cell extracts were prepared as follows: Cells were harvested, washed with PBS twice, and homogenized in a lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 0.5% NP-40) for 30 min at 4°C. Cell extracts from tumors and livers of nude mice were prepared in 50 mM HEPES, 0.5 mM EDTA, pH 8.0. Cell lysates were centrifuged at 12,000 g for 15 min, and the supernatants were collected as whole cell extracts.
Preparation of bone marrow samples from MM patients
MM patients’ bone marrow samples were collected from our Liquid Tissue Bank facility. Freshly isolated bone marrow samples were fractionated by Ficoll-Paque (Pharmacia Biotech, Piscataway, NJ) sedimentation. The mononuclear cellular layer was then resuspended in RPMI-1640 containing 10% heat-inactivated fetal bovine serum, 10 mM HEPES (pH 7.4), sodium pyruvate, L-glutamine, and 1% penicillin-streptomycin. The mononuclear cells were then treated with different concentrations of PI-083 or Bortezomib for indicated periods.
Trypan Blue exclusion assay
Adherent cells were harvested using trypsinization and pooled with suspension cells from media supernatant by pelleting at 300 g for 5 min at 4°C. The cells were then resuspended in an appropriate volume of media by pipetting gently up and down. Two 20 μl aliquots were removed and combined with an equal volume of 0.4% Trypan Blue and allowed to mix for 1 min. A 10 μl volume was loaded onto a hemacytometer and cells exluding Trypan Blue were scored as alive, whereas Trypan Blue-positive cells were considered dead/dying, and the percentage of dead/dying cells was calculated. To calculate the percentage of proliferation, the number of live cells in the treated samples was divided by the number of live cells in the untreated vehicle control.
MTT (3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide) metabolism assay
Cells were plated in 96-well plates in 100 μl medium and allowed to attach overnight. Cells were then incubated for 24 to 72 h with varying concentrations of PI-083, Bortezomib or appropriate control. Media was aspirated and replaced with 100 μl complete media containing 1 mg/ml MTT and incubated for three hours at 37°C in 5% CO2 humidified incubator. Media was then aspirated and DMSO was added. Cells were incubated for 10 min at room temperature while shaking, and the absorbance was determined at 540 nm using a μQuant spectrophotometric plate reader (Bio-TEK, Winooski, VT).
TUNEL assay
Terminal deoxynucleotidyl transferase-mediated nick-end labeling (TUNEL) was used to determine the extent of DNA strand breaks 45. The assay was performed with the APO-Direct Kit following the manufacturer’s instructions. In brief, the harvested cells were fixed in 1% paraformaldehyde for 45 min on ice, washed twice with PBS, and then fixed again in 70% ethanol at −20° C overnight. The cells were then incubated in DNA labeling solution (containing terminal deoxynucleotidyl transferase (TdT) enzyme, fluorescence-conjugated dUTP and reaction buffer) for 60 min at 37° C. After rinsing the cells to remove the DNA labeling solution, the cells were incubated with the propidium iodide/RNase A solution, incubated for 30 min at room temperature in the dark, and analyzed by flow cytometry within 3 h of staining.
Antitumor studies of human tumor xenografts in nude mice
Nude mice (Charles River Laboratories, Wilmington, MA) were maintained and treated in accordance with the Institutional Animal Care and Use Committee procedures and guidelines. Seven days before inoculation with MCF-7 cells, the animals were fully anesthetized for subcutaneous implantation of estradiol pellets (0.25 mg per pellet, 60-day release; Innovative Research of America, Sarasota, FL) on the dorsal surface of mice. Exponentially growing MCF-7 and A549 cells were harvested via trypsinization, pelleted at 300 g for 5 min, resuspended in sterile PBS (Invitrogen) at 107 cells per 100 μl, and injected into each flank of mice. The tumor xenografts were monitored with an electronic caliper every other day for 16 days. Tumor volume was calculated using the formula V=W2L, where width is the largest diameter and length is the smallest diameter. When the tumors reached ~ 200 mm3, the animals were randomized and treatment schedules were implemented. Treatments consisted of intraperitoneal (i.p.) injections of PI-083 or Bortezomib at 1.0 mg/kg body weight (mpk) (twice per week) or vehicle control (100% DMSO). 2 h after the last injections animals were sacrificed via CO2 inhalation and then tumors and livers were harvested and snap frozen in liquid N2.
Western blot analysis
Cell lysates (50 μg) were separated by SDS-PAGE and transferred to a nitrocellulose membrane, probed with p27Kip1 and β-actin antibodies, and signals were visualized by enhanced chemoluminescence (ECL, Amersham, Piscataway, NJ) according to the manufacturer’s protocol.
Acknowledgments
This work was supported by NIH grant P01-CA118210 (to S.M.S.). G.M.S. is an Amos Fellow of the Robert Wood Johnson Foundation and is supported by NIH-3U19-CA067771-13S1. We are grateful to Drs. Kenyon Daniel, Shen-Shu Sung, and Wesley Brooks of the HTS core facility for their assistance with the molecular docking studies.
Abbreviations
- CT-L
chymotrypsin-like activity
- PGPH
peptidyl glutamyl peptide hydrolase
- T-L
trypsin-like
References
- 1.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–21. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 2.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–89. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 3.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Cancer. 2006;5:596–612. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–23. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 6.Wilk S, Orlowski M. Cation-sensitive neutral endopeptidase: isolation and specificity of the bovine pituitary enzyme. J Neurochem. 1980;35:1172–82. doi: 10.1111/j.1471-4159.1980.tb07873.x. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, Heller H, Elias S, Ciechanover A. Components of the ubiquitin-protein ligase system. J Biol Chem. 1983;258:8206–14. [PubMed] [Google Scholar]
- 8.Arrigo AP, Tanaka K, Goldberg AL, Welch WJ. Identity of the 19S ‘prosome’ particle with the large multifunctional protease complex of mammalian cells (the proteasome) Nature. 1988;331:192–4. doi: 10.1038/331192a0. [DOI] [PubMed] [Google Scholar]
- 9.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–47. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 10.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 11.Adams J. The proteasome: A suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–60. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 12.Raff MC. Social controls on cell survivial and cell death. Nature. 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 13.Downward J. Targeting ras signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 14.Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: Therapeutic implications. Eur J Cancer. 2004;40:2217–29. doi: 10.1016/j.ejca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Proteasome inhibition in the treatment of cancer. Cell Cycle. 2005;4:290–6. [PubMed] [Google Scholar]
- 16.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–6. [PubMed] [Google Scholar]
- 17.Ling Y-h, Liebes L, Zou Y, Perez-Soler R. Reactive Oxygen Species Generation and Mitochondrial Dysfunction in the Apoptotic Response to Bortezomib, a Novel Proteasome Inhibitor, in Human H460 Non-small Cell Lung Cancer Cells. J Biol Chem. 2003;278:33714–23. doi: 10.1074/jbc.M302559200. [DOI] [PubMed] [Google Scholar]
- 18.Mortenson MM, Schlieman MG, Virudachalam S, Bold RJ. Effects of the proteasome inhibitor bortezomib alone and in combination with chemotherapy in the A549 non-small-cell lung cancer cell line. Cancer Chemother Pharmacol. 2004;54:343–53. doi: 10.1007/s00280-004-0811-4. [DOI] [PubMed] [Google Scholar]
- 19.Williams S, Pettaway C, Song R, Papandreou C, Logothetis C, MCConkey DJ. Differential effects of the proteasome inhibitor bortezomib on apoptosis and angiogenesis in human prostate tumor xenografts. Mol Cancer Ther. 2003;2:835–43. [PubMed] [Google Scholar]
- 20.Ikezoe T, Yang Y, Saito T, Koeffler HP, Taguchi H. Proteasome inhibitor PS-341 down-regulates prostate-specific antigen (PSA) and induces growth arrest and apoptosis of androgen-dependent human prostate cancer LNCaP cells. Cancer Sci. 2004;95:271–5. doi: 10.1111/j.1349-7006.2004.tb02215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Proteasome Inhibitors: A Novel Class of Potent and Effective Antitumor Agents. Cancer Res. 1999;59:2615–22. [PubMed] [Google Scholar]
- 22.Sunwoo JB, Chen Z, Dong G, Yeh N, Crowl Bancroft C, Sausville E, Adams J, Elliott P, Van Waes C. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-kappa B, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clin Cancer Res. 2001;7:1419–28. [PubMed] [Google Scholar]
- 23.Bazzaro M, Lee MK, Zoso A, Stirling WL, Santillan A, Shih Ie M, Roden RB. Ubiquitin-proteasome system stress sensitizes ovarian cancer to proteasome inhibitor-induced apoptosis. Cancer Res. 2006;66:3754–63. doi: 10.1158/0008-5472.CAN-05-2321. [DOI] [PubMed] [Google Scholar]
- 24.Bold RJ, Virudachalam S, McConkey DJ. Chemosensitization of Pancreatic Cancer by Inhibition of the 26S Proteasome. J Surg Res. 2001;100:11–7. doi: 10.1006/jsre.2001.6194. [DOI] [PubMed] [Google Scholar]
- 25.Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS-341): a novel, first-in-class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control. 2003;10:361–9. doi: 10.1177/107327480301000502. [DOI] [PubMed] [Google Scholar]
- 26.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) Cancer Investig. 2004;22:304–11. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 27.Jagannath S, Durie BG, Wolf J, Camacho E, Irwin D, Lutzky J, McKinley M, Gabayan E, Mazumder A, Schenkein D, Crowley J. Bortezomib therapy alone and in combination with dexamethasone for previously untreated symptomatic multiple myeloma. Br J Haematol. 2005;129:776–83. doi: 10.1111/j.1365-2141.2005.05540.x. [DOI] [PubMed] [Google Scholar]
- 28.Oakervee HE, Popat R, Curry N, Smith P, Morris C, Drake M, Agrawal S, Stec J, Schenkein D, Esseltine DL, Cavenagh JD. PAD combination therapy (PS-341/bortezomib, doxorubicin and dexamethasone) for previously untreated patients with multiple myeloma. Br J Haematol. 2005;129:755–62. doi: 10.1111/j.1365-2141.2005.05519.x. [DOI] [PubMed] [Google Scholar]
- 29.Aghajanian C, Soignet S, Dizon DS, Pien CS, Adams J, Elliott PJ, Sabbatini P, Miller V, Hensley ML, Pezzulli S, Canales C, Daud A, Spriggs DR. A phase I trial of the novel proteasome inhibitor PS341 in advanced solid tumor malignancies. Clin Cancer Res. 2002;8:2505–11. [PubMed] [Google Scholar]
- 30.Scagliotti G. Proteasome inhibitors in lung cancer. Crit Rev Oncol Hematol. 2006;58:177–89. doi: 10.1016/j.critrevonc.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Papandreou CN, Logothetis CJ. Bortezomib as a potential treatment for prostate cancer. Cancer Res. 2004;64:5036–43. doi: 10.1158/0008-5472.CAN-03-2707. [DOI] [PubMed] [Google Scholar]
- 32.Bang SM, Lee JH, Yoon SS, Park S, Min CK, Kim CC, Suh C, Sohn SK, Min YH, Lee JJ, Kim K, Seong CM, Yoon HJ, Cho KS, Jo DY, Lee KH, Lee NR, Kim CS. A multicenter retrospective analysis of adverse events in Korean patients using bortezomib for multiple myeloma. Int J Hematol. 2006;83:309–13. doi: 10.1532/IJH97.A30512. [DOI] [PubMed] [Google Scholar]
- 33.Groll M, Koguchi Y, Huber R, Kohno J. Crystal structure of the 20S proteasome:TMC-95A complex: A non-covalent proteasome inhibitor. J Mol Biol. 2001;311:543–8. doi: 10.1006/jmbi.2001.4869. [DOI] [PubMed] [Google Scholar]
- 34.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of the 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–71. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 35.Kisselev AF, Goldberg AL. Proteasome inhibitors: From research tools to drug candidates. Chem Biol. 2001;8:739–58. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 36.Groll M, Berkers CR, Ploegh HL, Ovaa H. Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure. 2006;14:451–6. doi: 10.1016/j.str.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat Rev Cancer. 2002;2:331–41. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 38.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–83. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, Patterson A, Marquez RT, Auersperg N, Yu Y, Hahn WC, Mills GB, Bast RC., Jr A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–63. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 40.Qian J, Niu J, Li M, Chiao PJ, MST In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K-ras oncogenic activation in pancreatic carcinogenesis. Cancer Res. 2005;65:5045–53. doi: 10.1158/0008-5472.CAN-04-3208. [DOI] [PubMed] [Google Scholar]
- 41.Nickeleit I, Zender S, Sasse F, Geffers R, Brandes G, Sörensen I, Steinmetz H, Kubicka S, Carlamagno T, Menche D, Gütgemann I, Buer J, Gossler A, Manns MP, Kalesse M, Frank R, Malek NP. Argyrin A reveals a critical role for the tumor suppressor protein p27Kip1 in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008;14:23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, Mitsiades C, Mitsiades N, Yasui H, Letai A, Ovaa H, Berkers C, Nicholson B, Chao T-H, Neuteboom STC, Richardson P, Palladino MA, Anderson KC. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;8:407–19. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Codony-Servat J, Tapia MA, Bosch M, Oliva C, Domingo-Domenech J, Mellado B, Rolfe M, Ross JS, Gascon P, Rovira A, Albanell J. Differential cellular and molecular efffects of bortezomib, a proteasome inhibitor, in human breast cancer cells. Mol Cancer Ther. 2006;5:665–75. doi: 10.1158/1535-7163.MCT-05-0147. [DOI] [PubMed] [Google Scholar]
- 44.Prescott B. Potential antimalarial agents. Derivatives of 2-chloro-1,4-naphthoquinone. J Med Chem. 1969;12:181–2. doi: 10.1021/jm00301a053. [DOI] [PubMed] [Google Scholar]
- 45.Kazi A, Wang Z, Kumar N, Falsetti SC, Chan TH, Dou QP. Structure-activity relationships of synthetic analogs of (−)-epigallocatechin-3-gallate as proteasome inhibitors. Anticancer Res. 2004;24:943–54. [PubMed] [Google Scholar]