Summary
HAMP domains are widespread prokaryotic signaling modules found as single domains or poly-HAMP chains in both transmembrane and soluble proteins. The crystal structure of a 3 unit poly-HAMP chain from the P. aeruginosa soluble receptor Aer2 defines a universal parallel four-helix bundle architecture for diverse HAMP domains. Two contiguous domains integrate to form a concatenated di-HAMP structure. The three HAMP domains display two distinct conformations that differ by changes in helical register, crossing angles, and rotation. These conformations are stabilized by different subsets of conserved residues. Known signals delivered to HAMP would be expected to switch the relative stability of the two conformations and the position of a coiled-coil phase stutter at the junction with downstream helices. We propose that the two conformations represent opposing HAMP signaling states and suggest a signaling mechanism whereby HAMP domains interconvert between the two states, which alternate down a poly-HAMP chain.
Introduction
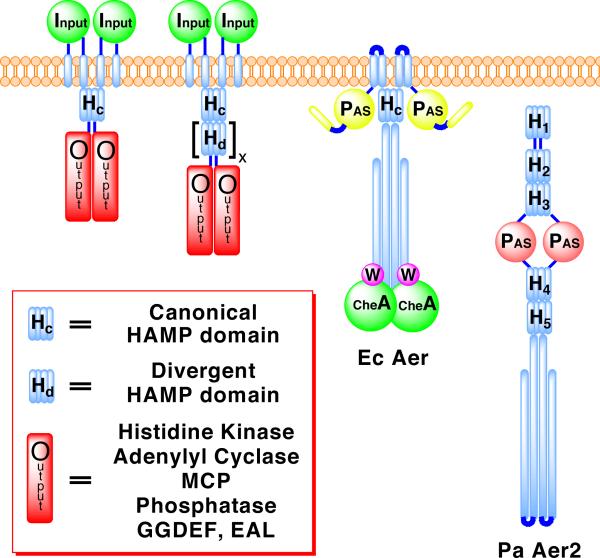
For their survival, prokaryotes depend on two-component signaling pathways to respond to changing environmental conditions (Szurmant et al., 2007). Many of the proteins that underlie these pathways contain HAMP domains, which function as signal relay modules that couple motions of transmembrane helices to the activity of a downstream cytoplasmic output domain (Hazelbauer et al., 2008). HAMP domains were named for the type of proteins in which they were originally identified: Histidine kinases, Adenylyl cyclases, Methyl-accepting chemotaxis proteins (MCPs), and Phosphatases (Aravind and Ponting, 1999). However, they also occur with other types of output modules: for example, diguanylate cyclases (GGDEF) and phosphodiesterases (EAL). The number and variety of signaling proteins in which they are featured highlight the importance and versatility of HAMP domains to prokaryotic signal transduction. The mechanism by which HAMP domains propagate conformational changes is of great interest for understanding how signals traverse membranes. In one of the best-studied examples, HAMP domains are an essential component of transmembrane chemoreceptors (MCPs), in which they connect the last transmembrane helix (TM2) to the cytoplasmic kinase interacting domain (Hazelbauer et al., 2008). Typically, transmembrane receptors, like MCPs, contain a canonical HAMP domain as a single unit (Figure 1). HAMP domains can be swapped interchangeably between different proteins without loss of function, which suggests a conserved mechanism for propagating signals (Appleman et al., 2003; Hulko et al., 2006; Zhu and Inouye, 2003).
Figure 1. Domain Architectures of Representative HAMP Containing Proteins.
Schematic depicting the location of canonical HAMP domains and poly-HAMP chains in transmembrane and soluble signaling proteins. Poly-HAMP chains, [Hd]x, can extend from 2 to 31 consecutive HAMP domains. EcAer is shown binding to the histidine kinase CheA and coupling protein CheW, proteins PaAer2 also likely interacts with.
HAMP subunits (~50 residues) contain two α-helices, AS1 and AS2, bridged by a flexible connector of approximately 14 residues (Butler and Falke, 1998). NMR studies of an archeal HAMP domain of unknown function (Af1503) demonstrated that the HAMP domain folds into a parallel four-helix bundle (Hulko et al., 2006). Each of the two helices are composed of a typical heptad repeat (a-g), with hydrophobic residues in positions a and d forming a buried hydrophobic core. The flexible connector spans the length of the four-helix bundle and contains a motif of three critical residues that is found in most HAMP domains (Ames et al., 2008; Hulko et al., 2006). Cross-linking studies of the aerotaxis receptor Aer (Watts et al., 2008) and the chemoreceptor Tar (Swain and Falke, 2007) confirmed the parallel four-helix bundle architecture for the HAMP domains of these well-studied proteins.
The mechanism of signal transduction by HAMP domains has been extensively studied in the bacterial chemotaxis system and with sensor histidine kinases. In the canonical HAMP systems of MCPs and the histidine kinase NarX, signal input is received from a transmembrane helix (TM2), attached to AS1, that undergoes a “piston-like” motion induced by ligand binding to a periplasmic sensing domain (Cheung and Hendrickson, 2009; Falke and Hazelbauer, 2001). Alternatively, the TM2's of the phototransducing element NpHtrII have been shown to undergo both a horizontal displacement and a 15° clockwise rotation after light excitation (Moukhametzianov et al., 2006).
A number of mechanisms have been proposed to describe the molecular motions of HAMP domains during signal transduction. These models, which are not necessarily mutually exclusive, encompass (i) a gearbox model involving a concerted rotation of helices (Hulko et al., 2006), (ii) a dynamic model of four-helix bundle stability (Zhou et al., 2009), and (iii) a scissor-like motion with a change in helix-helix crossing angles (Swain and Falke, 2007). Additionally, a model based on cryo-electron microscopy studies of chemoreceptor assemblies suggests HAMP domains interconvert between a compact trimer of HAMP dimers and an expanded form (Khursigara et al., 2008). In each case, HAMP signaling can be described by a two-state model in which modest structural changes separate states whose conformational stabilities are closely balanced in free energy.
Pseudomonas aeruginosa contains two transducers of aerotaxis: PaAer is homologous to the membrane bound E. coli aerotaxis receptor Aer, whereas the second, PaAer2, lacks transmembrane helices and is a soluble receptor (Figure 1) (Hong et al., 2004). Aer2 contains a heme-binding PAS domain, which can signal in response to diatomic gases (K. Watts, unpublished data). A total of five HAMP domains are predicted in the full-length protein, with three N-terminal and two C-terminal to the PAS domain. The Aer2 HAMPs belong to a group of divergent HAMPs recently identified in the gene sequences of a large number of diverse bacteria (Dunin-Horkawicz, submitted). Poly-HAMP chains (composed of repeating HAMP units) and soluble receptors, such as Aer2, are typically composed of divergent HAMP domains (Figure 1). The C-terminus of Aer2 contains a cytoplasmic signaling region, with a high degree of similarity to E. coli chemoreceptors, which likely couples the protein to the histidine kinase CheA and other components of the chemotaxis pathway (Hong et al., 2004).
Here, we present the crystal structure of the N-terminus of the soluble aerotaxis receptor Aer2 from P. aeruginosa, which contains three HAMP domains and represents the first structure of a di-HAMP unit. These three structures confirm the universality of the parallel four-helix bundle structure and provide a view of the diverse, low-energy conformational states available to HAMP domains. The two HAMP domains composing the di-HAMP share an extensive and interwoven interface to produce a concatenated signaling unit. The structure reveals the presence of two distinct conformations of HAMP domains, which are differentiated by changes in helical register, rotation, and crossing angle.
Results
Structure Determination and Overall Description
Sequence analysis of the N-terminal region of Aer2 (1-172) predicts that this region contains three successive HAMP domains. We overexpressed this fragment of Aer2 in E. coli and obtained a soluble dimer that was amenable for crystallization. Crystals of Aer2 1-172 (space group P43212, a = b = 114.0 Å, c = 64.3 Å) grew in 1.1 M Li2SO4, 15-18% glycerol, 0.1 M Tris, pH 8.5, and diffracted to 2.64 Å. The structure was determined by multi-wavelength anomalous diffraction (MAD) using selenomethionine protein crystals and refined to an R factor of 0.237 and an Rfree of 0.270 (Table 1).
Table 1.
Data collection, MAD structure solution, and refinement statistics
| Data Collection | ||||
|---|---|---|---|---|
| Native | Se-Met Peak | Se-Met Inflection | Se-Met Remote | |
| Wavelength (Å) | 0.97918 | 0.97857 | 0.97914 | 0.95682 |
| Space group | P43212 | |||
| Cell parameters (Å) | a = b = 114.0, c = 64.3 | |||
| Resolution (Å) | 50-2.64 (2.69-2.64) | 50-3.15 (3.20-3.15) | 50-3.42 (3.48-3.42) | 50-3.25 (3.31-3.25) |
| No. of reflections | 90969 | 54421 | 43482 | 50373 |
| No. of unique reflections | 12812 | 13826 | 10963 | 12741 |
| Completeness (%) | 97.7 (99.2) | 97.2 (99.2) | 97.7 (100.0) | 97.8 (99.8) |
| Rsyma | 0.049 (0.411) | 0.099 (0.433) | 0.108 (0.433) | 0.084 (0.464) |
| I/σ(I) | 38.4 (4.5) | 12.1 (3.2) | 13.2 (3.1) | 16.3 (2.9) |
| MAD Structure Solution Statistics | ||||
|---|---|---|---|---|
| Resolution cutoff (Å) | 3.42 | |||
| No. of anomalous sites found | 4 (of 4) | |||
| Mean figure of merit (FOM) | 0.73 | |||
| Overall Z-score | 40.8 | |||
| Refinement statistics | ||||
|---|---|---|---|---|
| Resolution range | 40-2.64 Å | |||
| R factor, % | 23.7 (43.9) | |||
| Rfree, % | 27.0 (46.2) | |||
| Atoms (protein/solvent) | 1231, 44 | |||
| Mean B-values (Å2) | ||||
| Overall | 75.0 | |||
| Main chain | 63.2 | |||
| Side chain | 61.0 | |||
| Rmsd from ideal | ||||
| Bonds mainchain | 0.032 Å | |||
| Bonds sidechain | 0.281 Å | |||
| Angles mainchain | 0.6 deg | |||
| Angles sidechain | 1.2 deg | |||
| Ramachandran plot, % | ||||
| Most favored | 93.6 | |||
| Additionally allowed | 6.4 | |||
| Generously allowed | 0.0 | |||
| Disallowed | 0.0 | |||
| Missing residues | 158-172 | |||
Data for outermost resolution shell are given in parenthesis
Rsym = ΣΣj |Ij – <I>|/Σ<I>
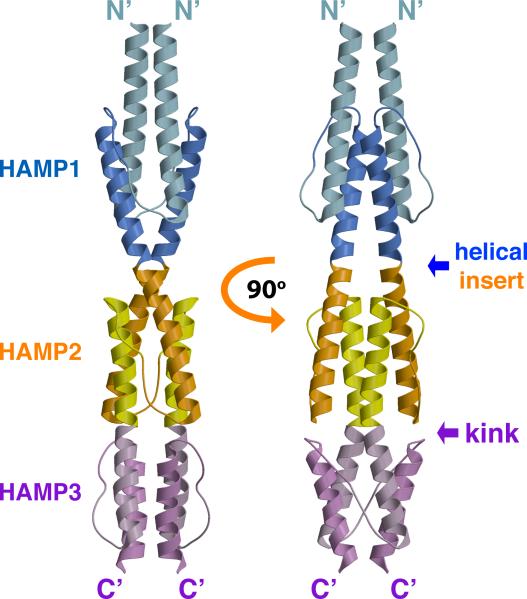
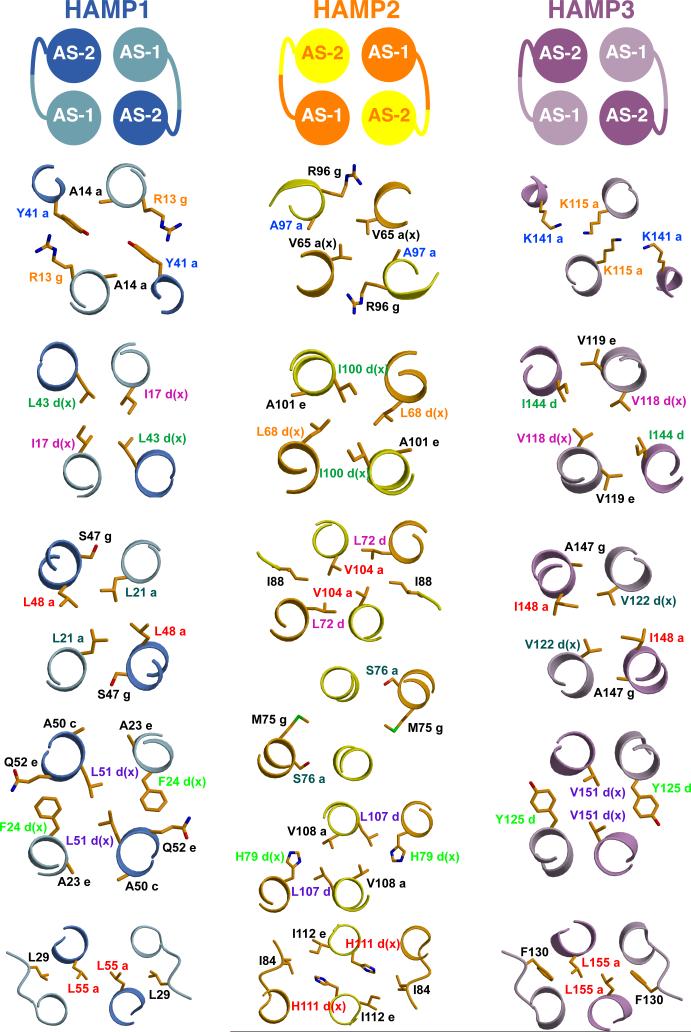
The structure of Aer2 1-172 is a symmetric dimer containing three HAMP domains (Figure 2), each similar to the NMR structure of Af1503. The basic construction of each HAMP domain consists of a monomeric unit of two parallel α-helices (AS1 and AS2) joined by an elongated connector of 12-14 residues (Figure 3). This unit dimerizes and coils around a central supercoil axis to form a parallel four-helix bundle. The residues of the heptad repeat point inward, forming a buried core, with the remaining residues exposed to the solvent to varying degrees. A helical insert separates HAMP1 and HAMP2, whereas HAMP2 and HAMP3 share a continuous helix, creating an interwoven, concatenated di-HAMP structure (Figure 2).
Figure 2. Crystal Structure of the Aer2 N-Terminal Domain Contains Three Successive and Interwoven HAMP Domains.
Ribbon presentation of the Aer2 (1-172) dimer with HAMP1 (AS1: light blue, AS2: blue), HAMP2 (AS1: orange, AS2: yellow), and HAMP3 (AS1: light purple, AS2: purple). HAMP2/3 forms a concatenated structure. AS2 of HAMP1 is contiguous with AS1 of HAMP2 and AS2 of HAMP2 is contiguous with AS1 of HAMP3. HAMP3 is rotated roughly 90 relative to HAMP1 and HAMP2.
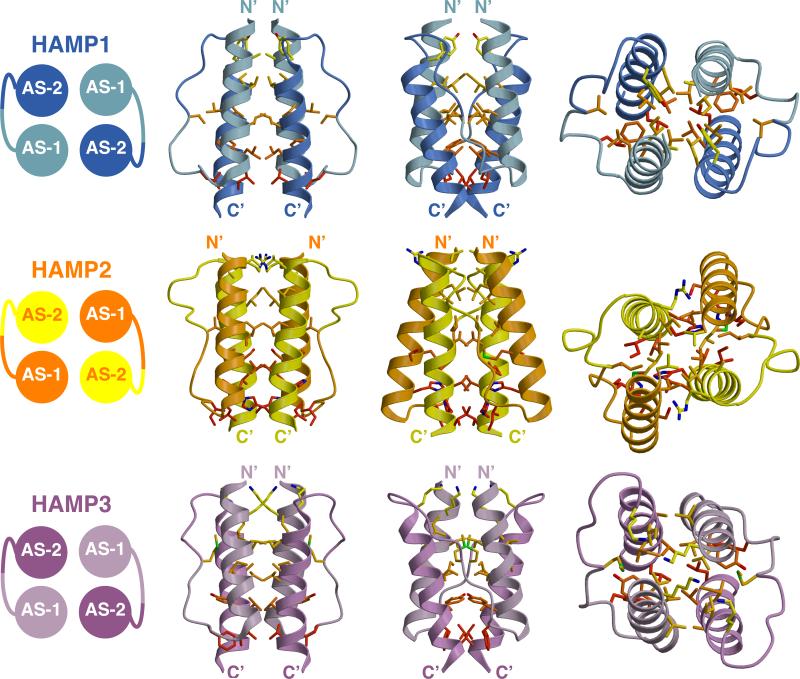
Figure 3. Structures of the Aer2 HAMP Domains.
Side and top views (90° rotation) of individual Aer2 HAMP domains with buried side chains (Carbon: yellow to red, Nitrogen: blue, Sulfur: green). HAMP1 and HAMP3 sidechains are in-register and HAMP2 sidechains are offset. HAMP2 has an unusual trapezoidal shape (as viewed from the side) and rhombic arrangement of helices in cross section. The position of the connector and hydrophobic residue 2 (HR2) correlates with changes in helical register, helix crossing angles, and helical rotation. I88 (HR2, HAMP2) inserts between AS1 and AS2, and V33 (HR2, HAMP1) and M134 (HR2, HAMP3) pack against the periphery of AS1 and AS2.
Two Distinct HAMP Conformations
The Aer2 HAMP domains each adopt a unique four-helix bundle. However, the three structures fall into two distinct structural conformations. Superposition shows that HAMP1 and HAMP3 adopt a conformation resembling Af1503, with only minor differences in helical tilt and orientation (Figure 4). In this conformation, the AS1 and AS2 helices are in-register, and the sidechains that form the hydrophobic core are positioned in the same plane to produce four layers of interacting residues (Figure 3).
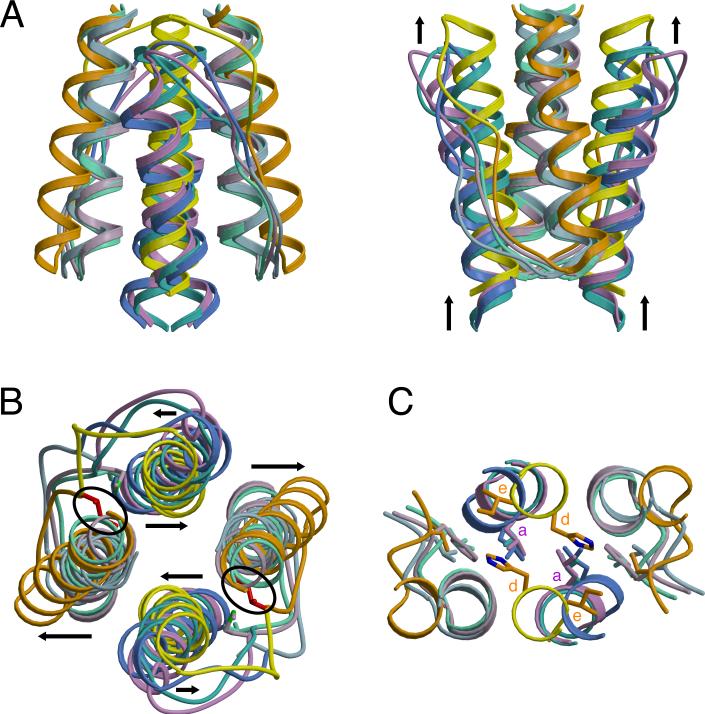
Figure 4. Superposition of HAMP Structures Reveals Two Distinct Conformations for Providing Different Signals to Downstream Domains.
Ribbon presentation of HAMP superpositions in (A) side and (B) top views displaying differences in helix orientation between overlapping HAMP1, HAMP3, and Af1503 (AS1: light green, AS2: turquoise) helices and distinct conformation of HAMP2 (AS1: orange, AS2: yellow) with offset helices, change in helix crossing angles (black arrows), and the inserted HR2 (I88: red) residue (black circle). Note the vertical displacement of AS2 relative to AS1. (C) Distal region of HAMP domains where signal output is directly linked to the conformation of AS2. His111 (HAMP2) occupies a typical “d” position, with the analogous residue in other HAMPs (L55: HAMP1, L155: HAMP3, L326: Af1503) holding an “a” position, in which they interact with hydrophobic residue 1 (HR1) of the connector.
A defining feature of coiled-coils is the position and direction of sidechain packing. Typically, coiled-coils make knobs-into-holes interactions in which the sidechains of one helix occupy a hole formed by a set of interacting residues on an adjacent helix. Af1503 contains unusual x-da packing, with one set of sidechains directed toward the supercoil axis, in an “x” position, and a second set forming a ring of interacting residues, in “da” positions. The HAMP1 and HAMP3 sidechains do not conserve the x-da packing mode and adopt a variety of packing arrangements, including knobs into holes (a-d), x-da, and x-x layers (Figure 5 and Figure 6). HAMP1, HAMP3, and Af1503 belong to different HAMP groups, based on sequence analysis (Dunin-Horkawicz, submitted), and each group may adopt a different packing mode to stabilize a similar four-helix bundle conformation. Overall, the variety of packing modes observed demonstrates that exclusive x-da packing is not a necessary structural feature of all HAMP domains.
Figure 5. Residue Conservation and Packing Modes of HAMP Domains.
Sequence alignment of representative HAMP domains: Aer2, Af1503, EcTsr, and EcAer, showing conservation of the buried core (blue) and connector motif (red) residues (top panel). The bottom panel denotes the heptad position of core residues (a, d, e, g, z = undefined, x = directed at supercoil axis), as defined by TWISTER, in a variety of packing modes: a-d, x-da (x-ga or x-de), and x-x. Sets of interacting residues are color-coded to highlight the sidechains of HAMP1, HAMP3, and Af1503 (in-register) and HAMP2 (offset) (bottom panel). “G” and “h” correspond to conserved residues of the connector motif and are also color coded with the layer of interacting residues. Position of the conserved proline residue (purple) in canonical HAMP domains corresponds to residues that occupy different heptad positions (peach) in alternative HAMP conformations. Aer2 HAMPs conserve Gly residues (peach) at the beginning of AS2, which allows close association of domains in a poly-HAMP chain, compared to the DExG motif of canonical HAMPs (purple).
Figure 6. Sidechain Interactions of Aer2 HAMP domains.
Cross-sections of the Aer2 HAMP domains displaying a variety of sidechain packing modes: a-d, x-da (x-ga or x-de), and x-x. Clockwise and counterclockwise rotation of AS1 and AS2 alters the corresponding sidechain positions (color-coded) in HAMP2 compared to HAMP1 and HAMP3. These changes generate a greater helix-helix separation that increases the overall solvent accessibility of HAMP2 residues at the inter-helix interface (Figure S3). Sidechains of HAMP2 do not occupy the same plane due to a change in helical register; they are shown with the nearest layer. Helices are viewed with the N-terminus pointing toward the viewer.
The conformation of HAMP2 is distinct and represents a possible alternative signaling state of HAMP domains. The unique conformation of HAMP2 arises from a combination of structural differences that affect the intra and inter subunit sidechain interactions, helix-connector interactions, and the intra and inter helix-helix crossing angles. The most notable change is an offset of the helical register between the AS1 and AS2 helices by half a helical turn (~2-3 Å), which staggers the sidechains that form the buried core (Figure 3). The loss of interacting residue layers essentially leads to the formation of a two-stranded coiled-coil of the AS2 helices. The AS1 helices also supercoil around the same axis but they have limited interactions with each other.
The crossing angles of the HAMP2 helices change drastically compared to the other HAMP domains, with the AS1 helices flaring out away from the supercoil axis and packing against the AS2 helices in a ridges-into-grooves manner (Figure 3). In turn, the AS2 helices rotate into a parallel position. Although the different packing modes of the Aer2 HAMP domains preclude an exact quantification, a general clockwise rotation of AS1 and counterclockwise rotation of AS2 relates HAMP2 to HAMP1/3. The rotation of AS1 and AS2 in opposite directions turns the sidechains forming dimer contacts outward. This increases the solvent exposure of residues at the dimer interface in AS1, which flares out, but not those of AS2, which move inward toward the supercoil axis (Figure 6 and Figure S3).
These changes in HAMP2 combine to form a trapezoidal four-helix bundle (Figure 4A). This helical rearrangement is directly coupled to an increased integration of a connector residue, I88, into the hydrophobic core of HAMP2. To our knowledge, HAMP2 is a unique parallel four-helix bundle that displays an offset ridges-into-grooves interaction; however, the structure of HAMP2 bears some resemblance to the three stranded coiled-coil of spectrin (Yan et al., 1993).
Role of the Connector in Stabilizing Alternative Conformations
Additional structural variations are found in the position and interactions of the conserved HAMP connector motif. This motif, Gly-x-HR1-x-x-x-HR2, contains two hydrophobic residues (HR1 and HR2) and begins immediately after AS1 (Figure 5). An extensive mutagenesis analysis of the connector region of the E. coli serine chemoreceptor (Tsr) found that these residues (Gly, HR1, and HR2) were the only critical residues for function (Ames et al., 2008). Mutation of G235 in Tsr to nearly any other residue led to an altered or complete loss of chemotaxis in E. coli cells that contained Tsr as the only chemoreceptor. Similarly, mutation of L237 (HR1) and I241 (HR2) to hydrophilic or small hydrophobic residues also led to complete loss of function. These residues each perform a significant role in stabilizing the Aer2 HAMP structures but different subsets of these conserved HAMP residues play more prominent roles in stabilizing the two distinct conformational states.
In both conformations, the Gly residue located at the C-terminus of AS1 provides the flexibility to accomplish a sharp turn, with each Gly adopting ϕ/ψ conformations disallowed by other amino acids. In the similar conformations of HAMP1 and HAMP3, the first hydrophobic residue of the motif, HR1 (L29: HAMP1, F140: HAMP3), stabilizes each structure by packing against a conserved residue of the heptad repeat in an “a” position at the end of AS2 (L55: HAMP1, L155: HAMP3) (Figure 4C). In contrast, the HAMP2 AS2 helices rotate counterclockwise to place His111 (which occupies the conserved heptad position) in a “d” position where it cannot interact with I84 (HR1) of the same subunit. The counterclockwise rotation of AS2 instead brings I112 in close proximity to I84 (HR1). These structural changes are located at the C-terminal end of the HAMP domain, where the conformation of AS2 must relay signal output to a C-terminal domain.
Drastic conformational differences are observed in the position and interactions of HR2 among the three HAMPs (V33: HAMP1, I88: HAMP2, M134: HAMP3). These changes reflect the wide conformational variability and flexibility of the connector. Each interaction serves to stabilize the particular HAMP structure in a different way, which points to a concerted rearrangement of helical structure and connector interactions during conversion between HAMP signaling states. Variable extensions of the connector, combined with different helical tilts in the three conformations, allow HR2 to interact with different layers of the HAMP structure. In HAMP1, V33 (HR2) packs against the protein backbone and hydrophobic residues of layer 3: L21 and L48 (Figure 3). In HAMP3, a more extended conformation positions M134 (HR2) above layer 3, and the sidechain of Met134 points upward to make contact with V122 and I148 in layer2 (Figure 3). Again, HAMP2 adopts a more divergent conformation, fully inserting I88 (HR2) between AS1 and AS2, where it makes hydrophobic contacts with residues above and below (L68, A101, L72, V104, M75, and V108) (Figure 3). The position of I88 in HAMP2 reveals why HR2 has such a critical role for HAMP function and suggests that other HAMP domains may utilize a similar interaction.
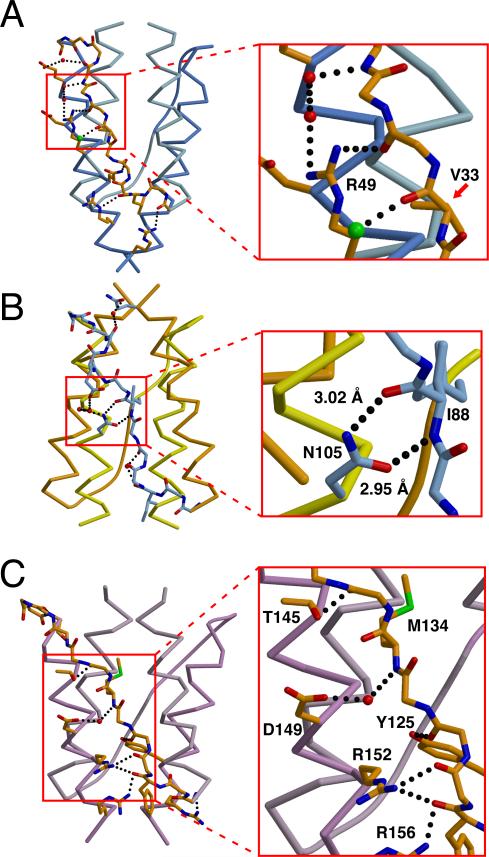
Complementary to the roles of HR1 and HR2, the connector stabilizes each HAMP domain through hydrogen bond interactions. These interactions do not involve the non-critical sidechains of the connector but rather occur primarily between the sidechains of AS2 and the connector peptide backbone. For HAMP1 and HAMP3, the resulting structural outcome is a close association of the connector with the AS2 helix (Figure 7). In contrast, the HAMP2 connector resides between AS1 and AS2 (N-terminal to HR2) and is closely associated with AS2 (C-terminal of HR2). At the center of this junction is N105 in AS2, which hydrogen bonds to the carbonyl oxygen and the amide nitrogen of the I88 (HR2) peptide backbone. The Asn sidechain is highly conserved in homologous Aer2 HAMP2-3 units (Figure S3) and is ideal to link conformational changes of AS2 with a rearrangement of the connector. This may explain why Asn is prevalently found in this position for many HAMP domains (Dunin-Horkawicz, submitted). The HAMP domains of MCPs are an exception, and other residues capable of hydrogen bonding (e.g. Arg or Ser) could perform a similar function.
Figure 7. Hydrogen Bonding Interactions Between the Connector and Helices Stabilize the Aer2 HAMP Domains.
Variable hydrogen bonding interactions and associations between the connector (peptide backbone) and helices (Cα trace) stabilize the different conformations of (A) HAMP1, (B) HAMP2, and (C) HAMP3. N105 hydrogen bonds (dotted line) with the peptide backbone to stabilize the position of the inserted I88 residue (hydrophobic residue 2: HR2) in HAMP2 (inset B). R49 forms a hydrogen bonding network with the connector peptide backbone and two water molecules (red). Peptide backbone of V33 (HR2) in HAMP1 forms a salt bridge with a chlorine atom (green) (inset A). D149 of HAMP3 forms a salt bridge with a water molecule (red) to the amide nitrogen of M134 (HR2) (inset C).
Threaded HAMP Sequences
The differences in helix orientation and sidechain packing between the Aer2 and Af1503 HAMP domains prompted us to test if each HAMP sequence was specific for only one HAMP conformation. The Rosetta program is a powerful tool that has been used to evaluate the compatibility of protein sequences with different 3-dimensional structures (Ambroggio and Kuhlman, 2006; Das and Baker, 2008). Using Rosetta Design, which applies a potential function incorporating terms for stereochemistry, sterics, solvation, and electrostatics (see Methods for a more detailed description), each sequence was threaded onto all four known HAMP conformations to generate one native and three altered conformations. The threaded structure was given a score, in Rosetta energy units (REUs), that represents the compatibility of each sequence and associated sidechain conformations with the experimentally determined fixed backbone structure.
The HAMP1, HAMP3, and Af1503 structures, which all share the in-register conformation but contain different packing modes (e.g. x-da, x-x), had comparable REUs for their native sequence but had much higher scores when threaded with the other sequences (Table 2). This indicates that each HAMP sequence favors a specific packing mode to stabilize a common conformational state. In contrast, the HAMP2 sequence was compatible with the out-of-register structure of HAMP2 and the in-register structure of HAMP1. This suggests that HAMP2 could adopt a conformation similar to HAMP1 during signal transduction. When the HAMP3 and Af1503 sequences were threaded onto the HAMP2 structure, the resulting REUs were only slightly higher than with their native structures. This suggests that a HAMP2 conformation would be accessible for the HAMP3 and Af1503 sequences. In contrast, HAMP1 did not thread well onto the distorted four-helix bundle of HAMP2, which may indicate that HAMP1 could adopt a HAMP2-like structure, but only with an altered backbone conformation and packing mode.
Table 2. Relative Threading Scores for HAMP Sequences onto Known Conformations.
Numbers refer to the Rosetta Design Score, given in Rosetta Energy Units (REUs), with lower scores being more stable. The lowest stable score is highlighted in grey for each conformation in our two-state model.
| Protein Sequences | Conformations | |||
|---|---|---|---|---|
| HAMP1 | HAMP2 | HAMP3 | Af1503 | |
| Hamp1 | 38 | 2047 | 1347 | 5510 |
| Hamp2 | 358 | -74 | 2792 | 1001 |
| Hamp3 | 1555 | 564 | 145 | 2599 |
| Af1503 | 1085 | 304 | 1667 | 148 |
| Aer | 2016 | 613 | 1615 | 6773 |
| Mt Rv3645 | 212 | 506 | 937 | 4616 |
| NarX | 1757 | 416 | 490 | 1298 |
| Tar | 1985 | 486 | 431 | 3000 |
| Tsr | 1606 | 459 | 255 | 885 |
We then threaded the sequences of other canonical HAMP domains of unknown structure onto each known structure. Again, each sequence was most compatible with only one of either the HAMP1, HAMP3, or Af1503 structures, supporting the hypothesis that each HAMP domain sequence favors a distinct packing mode (Table 2). Interestingly, all the sequences threaded well onto the HAMP2 structure, with REUs close in value to the lowest score obtained from HAMP1, HAMP3, or Af1503. This may be a reflection of the more expanded conformation of HAMP2, which allows for a greater degree of freedom in sidechain size. For these reasons, one could expect a HAMP2-like structure to be more dynamic than a HAMP1, HAMP3, or Af1503 conformation. Overall, the threading analysis suggests that both canonical and divergent HAMP sequences are compatible with both an in-register conformation and a more dynamic, out-of-register, HAMP2-like conformation.
The Concatenated Di-HAMP 2-3 Unit
Concatenated HAMP2-3 gives the first view of a di-HAMP structure and provides insight into signal transduction by poly-HAMP systems. HAMP2 and HAMP3 not only share a contiguous helix, but their interface is also highly interwoven. The sidechains of K115 and K141 in HAMP3 are directed upward and insert into the HAMP2 structure to form a polar pocket with H79 and H111 (Figure S1). Sequence analysis of homologous Aer2 HAMP2-3 units reveals a high degree of residue conservation at this interface (Figure S2). Two conserved Gly residues at both the beginning and end of the connector are necessary to achieve the close proximity between HAMP2 and HAMP3. A number of hydrogen bonds are formed between HAMP2 and HAMP3, including one between the connector peptide backbones of G82 and K140, and others between various sidechains (Figure S1). Structurally, these two HAMP domains are highly integrated and may function as a single unit containing two opposing conformations.
Unlike the interwoven HAMP2-3 unit, a helical insert separates HAMP1 and HAMP2 of Aer2. However, analysis of 132 sequences homologous to Aer2 1-156 (identified using pBLAST) found that Aer2 was the only protein containing this helical linker. Thus, this element is unlikely to be functionally important. Altogether, signal transduction through poly-HAMP systems is most likely to involve concerted structural changes that propagate through concatenated, repeating units.
Output Mechanism Involving “Stutter Compensation”
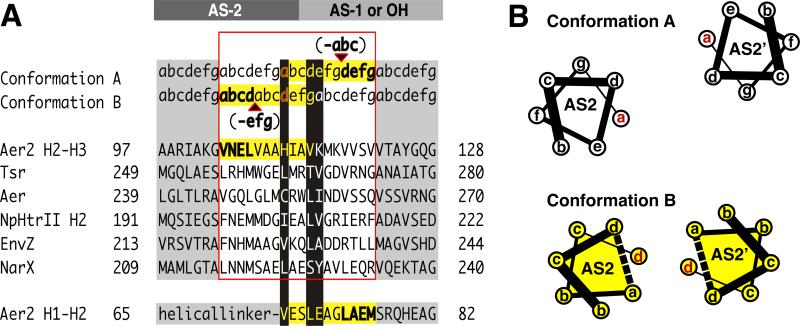
Insight into how HAMP domains may propagate signals downstream is gained by examining the similar heptad discontinuities between continuous AS2 and AS1 helices of the concatenated HAMP2/3 unit and between canonical AS2 helices and downstream output helices (OHs) (Figure 8). As discussed in other HAMP sequences (Stewart and Chen, 2009; Zhou et al., 2009), at both of these junctions the heptad repeat of continuous helices contains a “stutter” (Lupas and Gruber, 2005), differing by the insertion of four (or deletion of 3) residues. In Aer2 HAMP2, this stutter occurs at the end of AS2. It alters the heptad position of H111 to a “d” position (Figure 4) and aligns the heptad repeat to match that of AS1 of HAMP3 (Figure 8). However in HAMP1/3, the same residue (L55/L155) is in an “a” position and will thus offset the heptad repeat of a continuous coiled-coil from ideal packing. To compensate for the offset, the stutter could move to AS1 of a downstream HAMP, as observed in HAMP2 (Figure 8), or output helix (OH). Critical residues at these junctions may alternately occupy a core heptad position (a or d) to an e or g position.
Figure 8. Output Mechanism Involving “Stutter Compensation”.
(A) Model for signal transfer that shifts the position of a stutter on either side of the AS2 junction with AS1 (poly-HAMPs) or output helices (OH) (canonical HAMPs). Helical junctions are shown for representative HAMP domains and Aer2 HAMP 2-3. Stutters (bold) are defined as either an insertion of four residues or a deletion of three residues in the heptad repeat (a-g) and effect adjacent residues (yellow highlight). Residues at the junction switch heptad positions (black highlight) and are compatible with different heptad positions. This model predicts that Aer2 HAMP2 contains a stutter (shown in bold) in AS1 (bottom sequence). Whereas the heptad assignments in HAMP2 AS-1 are as shown, the splayed helices of AS-1 do not generate a true hendecad repeat but do contain an elongated i to i+4 (E66-A70) hydrogen bond distance (3.5 Å) typical of stutters. (B) Helical wheel diagrams of AS2 helices in conformation A (HAMP1/3) and B (HAMP2) highlighting the helical rotation and translation associated with a stutter (dotted line) in AS2.
Despite the AS2 stutter realigning the heptad repeat, it may not stabilize packing of downstream helices because accommodation of the stutter couples both helix rotation and translation to the formation of a kink at the junction of HAMP2/3 (Figure 2). Such a kink could alter OH packing and is consistent with the recently proposed “Yin-Yang” mechanism for MCP signaling, where adjacent coiled-coil domains reciprocally influence their helix-helix packing stability (Swain et al., 2009). Notably, in MCPs, a functionally critical Pro residue at the input junction may facilitate a similar helix bending through loss of a mainchain hydrogen bond (Figure 5).
Discussion
A Common Signaling Mechanism
Support for a common signaling mechanism among HAMP domains comes from the fact that HAMP domains from various proteins can be swapped interchangeably to produce functional chimeras (Appleman et al., 2003; Hulko et al., 2006; Zhu and Inouye, 2003). However, signal input into HAMP domains seems to involve several alternative modes of input. In MCPs, which contain a single canonical HAMP domain, ligand binding to a perisplasmic sensing domain generates a piston-like motion in the C-terminal helix that continues through the connecting transmembrane helix (TM2) to the HAMP domain (Falke and Hazelbauer, 2001). For native Tar, ligand binding is negatively cooperative and produces an asymmetric, downward motion of TM2 from one subunit. Nevertheless, Tar does not require an asymmetric signal and functions when the input signal is contrived to be symmetric (Draheim et al., 2005; Miller and Falke, 2004). For the histidine kinase NarX, nitrate binding occurs between the two perisplasmic sensing domains resulting in a symmetric 1 Å, upward shift of the periplasmic C-terminal helices, presumably pulling TM2 upwards (Cheung and Hendrickson, 2009). Domain swapping of the periplasmic domains of Tar and NarX results in a reverse output, consistent with the opposite displacement of TM2 (Ward et al., 2002).
Signal input to HAMP can also occur in other ways. The TM2 helices of NpHtrII, a phototransducing module from Natronomonas pharaonis, undergo a horizontal displacement and rotation upon light excitation (Moukhametzianov et al., 2006; Wegener et al., 2001), and the HAMP domain of Aer receives signal input from side-on interactions with an adjacent PAS domain (Taylor, 2007). In poly-HAMPs, signal input has not been biochemically characterized but most likely occurs through connecting HAMP subunits due to the highly interwoven nature of concatenated HAMPs. In our view, proposed HAMP signaling mechanisms should be consistent with the variable modes of input received by HAMPs.
A Model for HAMP Signal Transduction
The alternate structure of HAMP2 suggests a mode of signal transduction involving a change from an in-register four-helix bundle to a distorted, HAMP2-like structure. Helical displacement and rotation are two known signal inputs into HAMP that could readily shift the equilibrium between states. Both inputs merge into a screw-like motion required to interconvert HAMP conformations. The compatibility of HAMP domains with different signal inputs could be derived from a combination of structural rearrangements between opposing signaling states. The altered heptad pattern at the distal end of AS2 indicates that a HAMP2-like state would deliver a different conformational signal to a C-terminal output domain compared to HAMP1 or HAMP3. The identical heptad discontinuity at the C-terminus of canonical and poly-HAMP domains suggests a shared output mechanism and mode of signal transduction. We propose that canonical and poly-HAMP systems utilize a similar signaling mechanism and convert between the similar conformations of HAMP1, HAMP3, and Af1503 to a conformation resembling HAMP2.
Poly-HAMP Chains
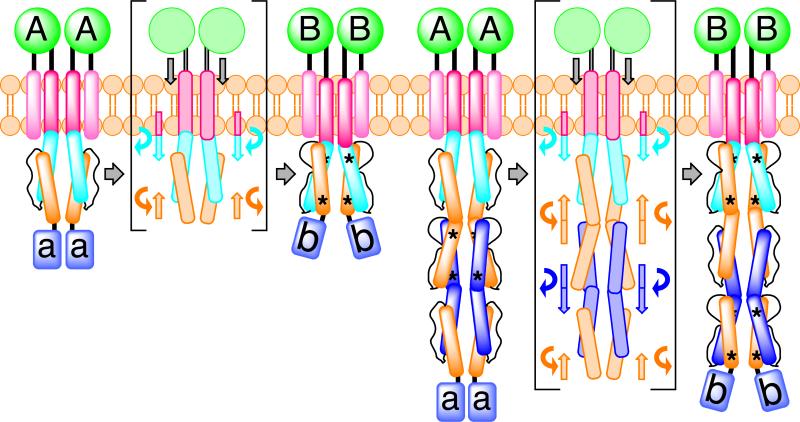
The structure of concatenated HAMPs, as revealed here, raises the interesting mechanistic question of how a signal is propagated through a poly-HAMP chain. Concatenation creates a heptad discontinuity at the junction of HAMP2/3 and forms the basis for a proposed “stutter compensation” mechanism. This model is consistent with the stutters that cause a heptad shift in both AS1 and AS2 of HAMP2. Assuming that, under physiological conditions, HAMPs exhibit a two-state switching mechanism, each HAMP domain would be capable of visiting one of two states: A or B. This model predicts that in poly-HAMP chains, repeating HAMP domains would assume alternating conformations (A-B-A-...) and interconvert in a binary fashion (B-A-B-...) (Figure 9). Two additional points support this mechanism. First, the HAMP domains of Aer2 abide by this pattern with HAMP2 and HAMP3 assuming distinct conformations. Second, amplification of poly-HAMPs typically occurs in sets of two (Dunin-Horkawicz, submitted). An alternating mechanism would require the addition of an even number of units to maintain the sign of the signal output. If this is true, any two concatenated HAMP domains would occupy opposing signaling states at a given moment. Consequently, the di-HAMP structure of HAMP2/HAMP3 may provide a snapshot of two HAMP signaling states with reversed signal output.
Figure 9. Model for HAMP Domain Signal Transduction.
Schematic of a ligand dependent, two-state (A and B) HAMP domain signal transduction model highlighting the movement and rotation of helices in a canonical HAMP system and a poly-HAMP system. In the poly-HAMP system successive AS1 helices are contiguous with the preceding AS2 helices. HAMP domain structural rearrangements correspond to a change in helical register, helix-helix crossing angle, and helix rotation. Asterisks denote stutters within HAMP domains. Signal input is shown as a symmetric and downward piston-like displacement of transmembrane helix 2 (TM2), but both an asymmetric displacement or a helical rotation of TM2 would likely lead to the same conformational changes within HAMP domains.
The shared helix and extensive contacts between HAMP2/3 suggest poly-HAMP chains propagate signals through concerted rearrangements. Interconversion between the alternative conformations in our structure results in a displacement and rotation of AS1 and AS2 in opposite directions, consistent with a concerted motion in poly-HAMPs (Figure 9) and not unlike the proposed “gearbox” model (Hulko et al., 2006). For homologous Aer2 HAMP2-3 units, the most conserved residues were those involved in interdomain contacts. The presence of Gly residues at the beginning and end of the connector allows for the HAMP2-3 subunits to come close enough together to hydrogen bond. Structurally, these Gly residues differentiate the Aer2 HAMPs from canonical HAMPs (which contain a DExG motif at the beginning of AS2) (Figure 5) and have been identified as common feature of most divergent HAMPs (Dunin-Horkawicz, submitted).
The Gearbox Model
The “gearbox” model is based on the observed x-da packing of Af1503, which has been proposed to convert into knobs into holes packing by a concerted 26 rotation of the helices (Hulko et al., 2006). However, the structures of HAMP1 and HAMP3 do not adhere to the uniform x-da packing modes upon which this model is based, although HAMP1 and HAMP3 produce the same position of the AS2 helices as seen in Af1503. In addition, sequence threading of other HAMP domains indicates that each HAMP sequence would favor a different packing mode to stabilize the same general conformation. Rotation of AS1 and AS2 in HAMP2 compared to HAMP1/3 does reflect an important feature of the gearbox model; however, large helix translations accompany the rotations.
Evidence for a HAMP2 Conformation in Canonical HAMP Domains
Helix-connector interactions distinguish HAMP domains from other four-helix bundles and play important roles in all known HAMP structures. A recent study identified the hydrophobic residues (HR1 and HR2) of the connector motif as critical to HAMP function (Ames et al., 2008). Different sets of these critical residues have prominent roles in stabilizing the alternative structures of our two-state model. In HAMP2, the conserved connector residue HR2 (I88) is the central residue in a hydrophobic pocket formed between the off-register AS1 and AS2 helices. In comparison, the corresponding residue in HAMP1 (V33), HAMP3 (M134), and Af1503 (V303) plays a less prominent role and packs weakly against the four-helix bundle. Thus, the high conservation of HR2 is more easily understood from the role it has in HAMP2, suggesting that other HAMP domains may adopt a conformation similar to HAMP2. In contrast, HR1 plays a more prominent role in stabilizing the in-register HAMP conformation and directly interacts with an output residue in AS2 whose heptad position may correlate with signal output.
Biochemical studies also provide evidence for a HAMP2-like conformation in canonical HAMP domains. A study aimed at obtaining secondary and tertiary structure information used a library of single cysteine residue substitutions to assay residue proximity in the HAMP domain of the aspartate chemoreceptor Tar (Butler and Falke, 1998). Symmetric disulfide bonds were formed at a significantly faster rate between residues of the AS2 heptad repeat. HAMP domains are highly dynamic (Doebber et al., 2008), and the rapid degree of cross-linking suggests that structural oscillations bring the AS2 helices, but not the AS1 helices, into close proximity, as observed in the HAMP2 conformation. EPR studies of NpHtrII found that the HAMP domain oscillates between two conformations: one resembling the compact Af1503 structure and another corresponding to a more-expanded, solvent exposed conformation (Doebber et al., 2008). The most dynamic region of NpHtrII was the C-terminal region of AS1, an observation consistent with the separation of AS1 from the bundle in the highly trapezoidal structure of HAMP2.
Sequence Divergence
Although canonical and divergent HAMP domains conserve residue types in positions important for bundle formation (hydrophobic residues of the heptad repeat) and connector interactions (G-x-HR1-x-x-x-HR2), sensitive HMM-HMM (Hidden Markov-Models) are needed to recognize many divergent HAMPs (Dunin-Horkawicz, submitted). Thus, canonical and divergent HAMPs may adopt distinct subsets of conformations, and a HAMP2 conformation may only be accessible to divergent HAMPs. Additionally, protein conformations can change due to constraints of the crystal lattices. However, the similar conformations observed between the NMR structure of Af1503 and the crystal structures of HAMP1 and HAMP3 suggest, at a minimum, that the in-register four-helix bundle is an important signaling state accessible to all HAMP domains. Furthermore, the functional importance of HR2 is best rationalized in the context of the HAMP2 conformation. Also, similar heptad discontinuities at the output helices of canonical and poly-HAMPs suggest that these modules function similarly. It follows that the distorted HAMP2 conformation would represent the opposing low-energy signaling state. Although extrapolation of the Aer2 structure into the functional dynamics of HAMP domains is at this point conjectural, the range of structural oscillation may lie somewhere near or between the two observed conformations.
Global Restructuring
As mentioned before, not all inputs to HAMP need necessarily involve a vertical helix displacement. The rearrangements that relate HAMP2 to HAMP1/3 can best be thought of as a global restructuring of the domain in which vertical helix displacements are coupled to helix rotations, tilts, and repacking of the connector. Any perturbation or interaction that favors the helix positions in one conformation over the other could, in principle, shift the equilibrium between states. In this context, structural perturbations delivered to HAMP could be asymmetric or symmetric, provided they preferentially stabilized one of the signaling states. The extensive subunit contacts displayed by HAMP domains ensures that asymmetric signals convert to a symmetric restructuring of the dimer. These inputs could include varying degrees of helix shifts and rotations, as well as direct side-on interactions from other domains, such as PAS, which is found in Aer-type sensors.
Although not definite, we believe that the HAMP2 conformation is most consistent with the kinase off state in MCPs because: (i) vertical displacement of AS1 would couple to a piston motion of TM2; (ii) the stutter in AS2 would kink and destabilize the helices of the adaption region, which correlates with a kinase off state (Swain et al., 2009); and (iii) replacement of I241 (HR2) in Tsr with Glycine or a charged residue (D, E, K, R), changes that would destabilize a HAMP2-like state, resulted in a CW-locked (kinase on) phenotype (Ames et al., 2008).
HAMP signaling is known to be bi-directional; compatible with adaptation responses through the methylation of chemoreceptor output bundles (Hazelbauer et al., 2008), as well as the inside-out signaling mechanism of LapD (Newell et al., 2009). The HAMP domains presented here are N-terminal to the PAS domain and not in the linear path from the PAS domain to the downstream signaling domain. However, preliminary evidence indicates that HAMP2 and HAMP3, but not HAMP1, are required for proper Aer2 function (K. Watts, unpublished data). We are actively pursuing this area of research to elucidate HAMP domain function and signaling in the context of soluble receptors.
Summary
The structure of an N-terminal fragment of the soluble receptor Aer2 has expanded the known examples of accessible HAMP conformations from one to four. HAMP2 represents a novel four-helix bundle and connector arrangement whose unique structure provides the basis for a two-state model of HAMP signaling that involves a complicated set of structural rearrangements. The two conformations we implicate in signaling are distinguished by changes in helical register, crossing angle, rotation, and likely dynamics. The conformational differences are greatest at the C-terminal output region, where the rhombic distortion of HAMP2 is most dramatic. Interconversion would shift a stutter on either side of connecting helical junctions, and could serve as an output mechanism. Moreover, a vertical displacement or rotation of AS1 caused by like movements in preceding N-terminal domains could readily induce the conformational switch. Overall, complex remodeling of the HAMP domain could be exploited to send signals to C-terminal output domains or through concatenated poly-HAMP chains.
Materials and Methods
Protein Expression and Purification
HAMP1-3 comprising residues 1-172 of the gene encoding Pseudomonas aeruginosa PAO1 Aer2 was cloned into the pET28a vector between NdeI and XhoI restriction sites, which added a cleavable N-terminal His-tag. For overexpression the plasmid was transformed into BL21 (DE3) cells, grown at 37° C in Luria Broth (LB) to an OD600 = 0.6 and incubated with 100 M IPTG at 23° C for 6 hrs before harvesting cells. Protein was purified using a Ni-NTA column following the manufacturers recommended protocol (Qiagen). After thrombin digestion His-tag free protein was applied to a Superdex 75 26-60 Hi-Prep Column equilibrated with 20 mM Tris, pH 7.5, 50 mM NaCl. Concentrated protein was aliquoted, flash frozen, and stored at -80°C.
To generate Se-Met protein, the Aer2 plasmid was transformed into B834 (DE3) cells, which are auxotrophic for methionine. An overnight culture in LB was spun down and washed twice with autoclaved water, then added to M9 minimal media supplemented with 19 standard amino acids and L-selenomethionine (50 mg/L). All Se-Met purification buffers contained 10 mM dithiothreitol (DTT) and Se-Met protein was purified otherwise as described for the native protein.
Crystallization and Data Collection
Crystals of native Aer2 1-172 protein (20 mg/mL) were grown by vapor diffusion against a reservoir containing 1.1 M Li2SO4, 15-18% glycerol, and 0.1 M Tris, pH 8.5 overnight at room temperature. Se-Met Aer2 1-172 protein (60 mg/mL) crystallized in the same space group (P43212) against a reservoir of 1.35 M (NH4)2SO4, 12-15% glycerol, 0.1 M Tris, pH 8.5. Solutions containing 1.25 M Li2SO4 or 1.5 M (NH4)2SO4, 18% glycerol, 0.1 M Tris, pH 8.5 were used as cryoprotectants. Native diffraction data were collected at the APS NE-CAT 24-ID-E beamline on an ADSC Quantum 315 CCD. MAD data were collected at the Cornell High Energy Synchrotron Source (CHESS) F2 beamline on an ADSC Quantum 210 CCD. Data were processed with HKL2000 (Otwinowski and Minor, 1997).
Structure Determination and Refinement
Diffraction data for Se-Met protein were processed with SOLVE (Terwilliger and Berendzen, 1999) and RESOLVE (Terwilliger, 2000) to generate initial electron density maps based on anomalous diffraction from Se-Met sites (figure of merit = 0.73, resolution cutoff = 3.42 Å). The structure was built into initial maps with XFIT (McRee, 1999) and then used for refinement against the high resolution native data set (Brunger et al., 1998). Structure refinement was carried out using CNS, amidst cycles of manual model building, minimization, B-factor refinement, and solvent molecule placement to produce the final model (R-factor = 23.7 %, Rfree = 27.0%) (Table 1).
Superposition of HAMP structures
Superpositions of the Aer2 and Af1503 HAMP domains were carried out using (i) Cα atoms of AS1, AS1’, AS2, and AS2’ helices or (ii) Cα atoms of AS1 and AS1’ with both giving similar results. For optimal clarity, all figures are shown from the superposition of AS1 and AS1’ Cα atoms.
Threading, Energy Minimization, and Energy Determination
The Rosetta Design program was used to generate and calculate all threaded sequences and their corresponding energy scores. The conformation of each sidechain was determined by sampling all rotamer conformations on a fixed backbone and calculating the corresponding energies of all combinations. The rotamer combination with the lowest score was taken as the final model and represents the final overall score reported. The Rosetta Design energy function calculates threading scores by evaluating a number of factors. These factors include attractive and repulsive Lennard-Jones potentials, solvation (Lazardis-Karplus solvation model), internal energy of side chain rotamers (from Dunbrack's statistics), intra-residue clashes, salt bridges, phi and psi angles (Ramachandran preferences), hydrogen bonding interactions and void volume of residues (tightness of packing).
Sequence Analysis
Sequence alignment of Aer2, Af1503, Tar, and Aer HAMP domains was generated using ClustalW and manually adjusted as presented. Homologs of Aer2 1-156 (HAMP1-3) and 63-156 (HAMP2-3) were identified using pBLAST against the nr database (non-redundant protein sequences) (Altschul et al., 1997) and returned 132 homologous protein sequences. An alignment of the top 100 sequences homologous to Aer2 63-156 (E-value cutoff = 4 × 10-5) was generated using ClustalW and used to visualize HAMP2-3 sequence conservation in a WebLogo (Crooks et al., 2004).
Sequence alignment of the AS2/AS1 and AS2/OH junctions of various HAMP proteins was generated using ClustalW. Heptad positions were determined using the program TWISTER (Strelkov and Burkhard, 2002). TWISTER first assigns the heptad positions “a” and “d” based on orientation relative to the supercoil axis. Remaining positions are determined based on these initial assignments.
Solvent Exposure
Solvent exposures of each HAMP domain were calculated using the CCP4 program AREAIMOL, which determines the solvent accessibility of all atoms in a protein structure based on proximity to neighboring atoms and summarizes the data per residue.
Supplementary Material
Acknowledgements
The authors thank Andrei Lupas and Stanislaw Dunin-Horkawicz (Max Planck Institute) for sharing the results of their bioinformatic analysis prior to publication and for useful discussions. Special thanks to John S. Parkinson for helpful discussions of HAMP signaling mechanisms. We also thank Joachim Schultz and Murray Cole for useful discussions; David Neau at 24ID-E beamline (APS) and Irina Kriksunov at F2 station (CHESS) for assistance during data collection; Abiola Pollard for assistance with MAD data sets; and Crane lab members for their help and support. This work was supported by the NIH grant GM066775 to B.R.C., NIH MBTG training grant GM08267 to M.V.A, and NIH grant GM029481 to Barry Taylor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroggio XI, Kuhlman B. Design of protein conformational switches. Current Opinion in Structural Biology. 2006;16:525–530. doi: 10.1016/j.sbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. Journal of Bacteriology. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleman JA, Chen LL, Stewart V. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. Journal of Bacteriology. 2003;185:4872–4882. doi: 10.1128/JB.185.16.4872-4882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. Fems Microbiology Letters. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Hendrickson WA. Structural Analysis of Ligand Stimulation of the Histidine Kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with Rosetta. Annual Review of Biochemistry. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Doebber M, Bordignon E, Klare JP, Holterhues J, Martell S, Mennes N, Li L, Engelhard M, Steinhoff HJ. Salt-driven equilibrium between two conformations in the HAMP domain from Natronomonas pharaonis - The language of signal transfer? Journal of Biological Chemistry. 2008;283:28691–28701. doi: 10.1074/jbc.M801931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draheim RR, Bormans AF, Lai RZ, Manson MD. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry. 2005;44:1268–1277. doi: 10.1021/bi048969d. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends in Biochemical Sciences. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends in Biochemical Sciences. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. Fems Microbiology Letters. 2004;231:247–252. doi: 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Khursigara CM, Wu XW, Zhang PJ, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. 2005;70:37–+. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- McRee DE. XtalView Xfit - A versatile program for manipulating atomic coordinates and electron density. Journal of Structural Biology. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Miller AS, Falke JJ. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moukhametzianov R, Klare JP, Efremov R, Baeken C, Goppner A, Labahn J, Engelhard M, Buldt G, Gordeliy VI. Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature. 2006;440:115–119. doi: 10.1038/nature04520. [DOI] [PubMed] [Google Scholar]
- Newell PD, Monds RD, O'Toole GA. LapD is a bis-(3 ',5 ')-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography. 1997;276(Pt A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Stewart V, Chen LL. The S helix mediates signal transmission as a HAMP domain coiled-coil extension in the NarX nitrate sensor from Escherichia coil K-12. Journal of Bacteriology. 2009 doi: 10.1128/JB.00172-09. Published online 4 December 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. Journal of Structural Biology. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: A disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain KE, Gonzalez MA, Falke JJ. Engineered Socket Study of Signaling through a Four-Helix Bundle: Evidence for a Yin-Yang Mechanism in the Kinase Control Module of the Aspartate Receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, White RA, Hoch JA. Sensor complexes regulating two-component signal transduction. Current Opinion in Structural Biology. 2007;17:706–715. doi: 10.1016/j.sbi.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Molecular Microbiology. 2007;65:1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallographica Section D-Biological Crystallography. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallographica Section D-Biological Crystallography. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Delgado A, Gunsalus RP, Manson MD. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Molecular Microbiology. 2002;44:709–719. doi: 10.1046/j.1365-2958.2002.02902.x. [DOI] [PubMed] [Google Scholar]
- Watts KJ, Johnson MS, Taylor BL. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. Journal of Bacteriology. 2008;190:2118–2127. doi: 10.1128/JB.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener AA, Klare JP, Engelhard M, Steinhoff HJ. Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. Embo Journal. 2001;20:5312–5319. doi: 10.1093/emboj/20.19.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Winograd E, Viel A, Cronin T, Harrison SC, Branton D. Crystal-Structure of teh Repetitive Segments of Spectrin. Science. 1993;262:2027–2030. doi: 10.1126/science.8266097. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Inouye M. Analysis of the role of the EnvZ linker region in signal transduction using a chimeric Tar/EnvZ receptor protein, Tez1. Journal of Biological Chemistry. 2003;278:22812–22819. doi: 10.1074/jbc.M300916200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.