Abstract
Several acute lymphoblastic and myelogenous leukemias are correlated with alterations in the human Mixed Lineage Leukemia protein-1 (MLL1) gene. MLL1 is a member of the evolutionarily conserved SET1 family of histone H3 lysine 4 (H3K4) methyltransferases, which are required for the regulation of distinct groups of developmentally regulated genes in metazoans. Despite the important biological role of SET1 family enzymes and their involvement in human leukemia’s, relatively little is understood about how these enzymes work. Here we review several recent structural and biochemical studies that are beginning to shed light on the molecular mechanisms for the regulation of H3K4 methylation by the human MLL1 enzyme.
Chromosomal translocations that disrupt the Mixed Lineage Leukemia protein-1 gene (MLL1, ALL1, HRX, Htrx)) are associated with a unique subset of acute lymphoblastic or myelogenous leukemias [1–4]. The product of MLL1 gene is a large protein that functions as a transcriptional co-activator required for the maintenance of Hox gene expression patterns during hematopoiesis and development [5–8]. The transcriptional co-activator activity of MLL1 is mediated in part by its histone H3 lysine 4 (H3K4) methyltransferase activity [6], an epigenetic mark correlated with transcriptionally active forms of chromatin [9, 10]. MLL1 complexes catalyze mono-, di-and trimethylation of H3K4, the regulation of which can have distinct functional consequences. MLL1 contains a number of conserved functional domains that work together for the assembly of multiprotein complexes that influence the appropriate targeting and regulation of the H3K4 methylation activity of MLL1. In this review, we summarize recent structural and functional studies that are beginning to provide a picture of how these domains are used to regulate the targeting, assembly and enzymatic activity of MLL1 complexes.
The MLL protein
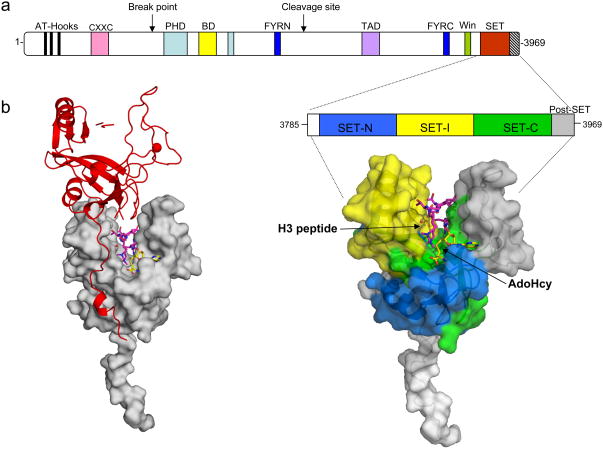
The MLL1 gene encodes a large protein of 3,969 amino acid residues and contains several conserved domains with functions implicated in chromatin mediated transcriptional regulation [11] (Figure 1). Domains include DNA binding AT hooks, a cysteine rich CXXC domain with homology to DNA methyltransferases, plant homeodomain (PHD) finger motifs, a Bromo domain (BD), a transactivation domain (TAD), a nuclear receptor interaction motif (NR box), a WDR5 interaction or Win motif, and a C-terminal SET domain, which is responsible for MLL1’s histone methyltransferase activity [6, 12, 13]. Upon normal expression of the MLL1 gene, the full-length protein is proteolytically processed into two fragments with opposite transcriptional properties; MLL-N and MLL-C, which associate to form a complex in vivo (Figure 1a) [14, 15]. The mature protein assembles with numerous regulatory proteins into multi-molecular complexes important for MLL1’s transcriptional co-activator activity [12, 16–21].
Figure 1.
Schematic representation showing the domain architecture of the MLL1 protein. a) The full-length MLL1 protein is rapidly processed by the Taspase 1 enzyme into MLL-N and MLL-C fragments, which reassociate through FYRN and FYRC motifs to form a stable complex. This mature protein then assembles with a number of proteins to form MLL1 complexes in the cell. b) Known three-dimensional structures of conserved MLL1 domains (colored green in each image). On the top from left-to-right is the CXXC domain (PDB code: 2j2s), and the c-terminal SET domain (PDB code: 2w5z). On the bottom from left to right is the MLL1 TAD domain (green) bound to the CBP:c-Myb complex (orange and blue, respectively) (PDB code: 2agh); and the MLL1 Win motif (green) bound to the WDR5 protein (purple) (PDB code: 3eg6).
Because of its large size, full-length MLL1 protein has thus far proven refractory to structural analysis. However, the modular nature of MLL1 has allowed structural analysis of some individual domains alone or in complex with functionally relevant ligands (Figure 1b). Structures that have been determined include the MLL1 CXXC domain [22], a portion of the MLL1 TAD domain bound to the KIX domain of the CREB binding protein (CBP) [23], a peptide from the Win motif of MLL1 bound to the WD-40 repeat protein, WDR5 [24, 25], and the C-terminal SET domain in the presence and absence of histone peptides and the cofactor product, s-adenosyl-homocysteine (AdoHcy) [26] (Figure 1b). These structures provide clues as to how MLL1 is targeted to MLL1 dependent genes and how MLL1’s enzymatic activity is regulated.
CXXC domain
The molecular mechanisms by which the MLL1 protein is recruited to specific target genes are poorly understood. The CXXC domain of MLL1 binds selectively to nonmethyl CpG DNA [27], and is essential for target gene recognition, transactivation and myeloid transformation in MLL1 fusion proteins [28]. Because the promoters of active genes in vertebrates are generally hypomethylated [29], the CXXC domain of MLL1 may play a role in targeting MLL1 to active genes. To identify the molecular basis of DNA recognition by the MLL1 CXXC domain, Allen et al., [30] determined the solution structure of the MLL1 CXXC domain consisting of amino acid residues 1146–1214, and used chemical shift mapping and site directed mutagenesis to identify residues involved in DNA recognition. The overall structure adopts an extended crescent-like shape that coordinates two zinc ions using the two conserved CGXCXXC motifs (Figure 2a). The zinc ions are required for the structural integrity of the protein, as mutation of any of the cysteine residues involved in zinc coordination result in protein unfolding [30]. The structure contains a positively charged surface groove containing a number of residues that were shown by chemical shift mapping and site directed mutagenesis to be important for DNA binding (Figure 2a). The MLL1 CXXC domain binds to unmethylated CpG DNA with a dissociation constant of ~4 μM as measured by Isothermal Titration Calorimetry (ITC) [30], but does not bind to similar DNA containing methyl-CpG dinucleotides- consistent with previous observations [27, 28]. These studies suggest a model in which the phospho-backbone of DNA binds to the positively charged groove on the CXXC domain, while residues from the extended loop insert into the major groove to interact with the CpG dinucleotide [30]. It is hypothesized that methylation of the CpG prevents the extended loop from interacting with the CpG dinucleotide, resulting in reduced affinity for DNA.
Figure 2.
The CXXC and TAD domains of MLL1 help recruit MLL1 to target loci. a) Transparent surface representation of the MLL1 CXXC domain (purple) determined by heteronuclear NMR spectroscopy (PDB code: 2j2s). A cartoon of the protein backbone is shown with zinc ions represented as spheres. The surfaces of amino acid residues perturbed by DNA binding in chemical shift and mutagenesis experiments are indicated in blue. The location of the extended loop is indicated with an arrow. b) The CBP-KIX domain:cMyb binary complex. The CBP-KIX domain is shown in orange and the c-Myb transactivation domain is shown in blue (drawn from PDB code: 1sb0). The positions of E665 and E555 of the CBP-KIX domain, and residues K291 and K294 of the c-Myb tranactivation domain are indicated. c) The CBP-KIX:cMyb:MLL TAD domain ternary complex (drawn form PDB code: 2AGH). The MLL1 TAD domain is shown in green and the colors for the CBP-KIX:cMyb are as in a). Upon formation of the ternary complex, residues E665 and E666 of the CBP-KIX domain become ordered and interact with the c-Myb transactivation domain (indicated with the arrow).
While the recognition of unmethylated CpG dinucleotides by the CXXC domain of MLL1 likely contributes to MLL1 targeting, as previously noted [7], several genes that are not regulated by MLL1 also contain unmethylated CpG dinucleotides in their promoters, indicating that other mechanisms contribute to target gene recognition by MLL1. A more recent structure of the TAD domain of MLL1 bound to the CBP protein describes one such additional mechanism that could also be involved in targeting MLL1 to specific loci.
TAD domain
The cyclic AMP response element-binding (CREB) binding protein (CBP) and its homolog p300 are general transcriptional co-activators that contain histone and transcription factor acetylation activities [31]. In addition, the CBP protein contains a number of protein binding domains that mediate transcription factor recruitment. The MLL1 TAD domain interaction with CBP was originally identified in a yeast three-hybrid screen using the CREB-CBP complex as bait [32], and was shown to be important for MLL1-mediated transcriptional activation [32]. Domain mapping experiments localize MLL1’s interaction to the KIX or CREB binding domain of CBP [32]. The KIX domain of CBP is a structural platform that is capable of binding several different families of transcriptional activators [31], and evidence indicates that the KIX domain has the ability to simultaneously interact with at least two different polypeptides in a cooperative manner [32, 33]. To identify the molecular basis of cooperative transcription factor binding by CBP, De Guzman et al., [23] determined the solution structure of a peptide derived from the MLL1 TAD bound to the KIX domain:c-MYB binary complex.
The overall structure of the c-Myb:KIX binary complex resembles a four-helix bundle where the c-Myb peptide adopts a helical conformation that binds to helices α1 and α3 of KIX (Figure 2b) [34]. When the MLL1 TAD peptide is added to the binary complex, the TAD peptide adopts a helical conformation where the conserved residues of the MLL1 TAD domain (residues 2845–2853) bind in a hydrophobic groove on the opposite side of the KIX domain between helices α2 and α3 (Figure 2c) [23]. No direct interaction between the c-Myb and MLL1 peptides are observed when bound to the KIX domain, suggesting that the mechanism of cooperative transcription factor binding is transmitted through subtle conformational changes in the KIX domain [23]. Consistent with allosteric binding, residues of the α3 helix of KIX that are disordered in the binary complex become ordered when MLL1 binds (see arrow in Figure 2c). This conformational change results in the placement of conserved KIX domain amino acids E665 and E666 into positions for optimal electrostatic interactions with conserved residues R294 and K291 of the c-Myb transactivation domain, respectively. Thermodynamic binding experiments show the interaction of MLL1 with the KIX domain increases CBP’s affinity for the c-MYB transactivation peptide by ~2-fold [33].
These experiments begin to provide a picture for how the recruitment of MLL1 can increase the binding of other important transcriptional activators that ultimately could result in the synergistic activation of gene transcription. In addition, cooperative transcription factor binding through CBP could provide a mechanism to help MLL1 recognize its target genes. MLL1 recruitment to chromatin results in the methylation of histone H3 at lysine 4 (H3K4) by the SET domain of MLL1, an activity that is regulated in part by a core complex of proteins that include WDR5, RbBP5, Ash2L and DPY-30 [26, 35–38]. H3K4 methylation is an epigenetic mark correlated with transcriptionally active forms of chromatin [10]. Several recent investigations have provided structural and functional information that describe how the histone methyltransferase activity of MLL1 is regulated.
SET domain
MLL1 contains an evolutionarily conserved SET domain which is found in a number of chromatin associated proteins with diverse transcriptional activities [39]. The SET domain is a histone methyltransferase motif named for its presence in Drosophila chromatin regulators SuVar3–9, E(z), and Trx [40]. SET domain proteins can be classified into several families that differ with respect to substrate specificity, processivity, and the presence of associated domains, and include the SUV39, SET1, SET2, E(z), Riz, SMYD and the SUV2–20 families [41]. MLL1 belongs to the SET1 family of SET domain proteins, which are found in conserved multisubunit complexes that regulate cellular H3K4 methylation levels [9, 42]. Because of the role of H3K4 methylation in diverse cellular processes ranging from stem cell differentiation to metazoan development and cancer, there has been an intense interest in understanding how SET1 family enzymes regulate H3K4 methylation.
To understand the structural basis of H3K4 methylation by the MLL1 SET domain, Southhall et al., [26] determined the X-ray crystal structures of a minimal MLL1 SET domain fragment in complex with its cofactor product S-adenosylhomocysteine (AdoHyc) in the presence and absence of a peptide mimicking the methylated histone H3 N-terminal tail (Figure 3). Much like other SET domains where the structures have been determined [43], the overall structure of the MLL1 SET domain contains two canonical conserved regions, SET-N and SET-C, that are separated by a less conserved insert region (SET-I) (expanded region in Figure 3a). The MLL1 SET domain is flanked on the C-terminus by a 22-amino acid post-SET domain, which provides several conserved residues that coordinate a zinc atom that is required for enzymatic activity (Patel and Cosgrove, unpublished observation). In the ternary complex, the histone H3 peptide binds in a deep channel that divides a pair of acidic lobes, one of which is composed of residues from the SET-I region and the other composed of residues from the SET-C and post-SET regions. Lysine 4 of histone H3 is inserted into a channel, at the end of which is the AdoHcy binding site, which is composed of residues from SET-N, SET-C and the post-SET domain (Figure 3a).
Figure 3.
X-ray crystal structure of the c-terminal MLL1 SET domain bound to s-adenosyl homocysteine (yellow) and histone H3 peptide (purple) (PDB code: 2W5Z). (a) At the top is a schematic representation of the full-length MLL1 protein and blown up is the construct used for crystallization of the MLL1 SET domain (residues 3785–3969). The SET-N, SET-I, and SET-C sub-domains are colored in blue, yellow and green, respectively. The post-SET domain is colored in grey, and the N-flanking region is colored white. The positions of histone H3 and AdoHcy are indicated. b) Crystal packing constrains the MLL1 SET domain into an open conformation. Surface representation of the MLL1 SET domain (grey) shown with a symmetry related molecule in red. The N-terminus of the symmetry related molecule interacts extensively with the SET-I region-constraining the MLL1 SET domain in an open conformation.
In published three-dimensional structures of other SET domain proteins that also contain the canonical post-SET domain [44–47], the formation of the ternary complex results in the ordering of the post-SET domain, so that the two lobes that flank the peptide binding site closes around the peptide, presumably to exclude solvent from the active site. However, comparison of the binary and ternary complexes of the MLL1 SET domain crystal structures reveals that the two lobes remain in a relatively open conformation, a conformation that is not optimal for catalysis [26]. It has been suggested on the basis of this observation that proteins that interact with the SET domain are required to induce the correct conformation of the active site [26], which is consistent with the poor catalytic activity of the isolated MLL1 SET domain. However, an analysis of crystal packing forces suggests that the SET-I lobe may be constrained in an unnatural conformation in the crystalline state by residues from the N-terminus of a symmetry related molecule (Figure 3b). It therefore remains to be determined to what extent the observed conformation of the isolated MLL1 SET domain in the crystal structure represents the range of possible conformations that may exist in solution.
Consistent with the conformational change hypothesis, SouthHall et al., [26] observed that the addition of other components of the MLL1 core complex, namely WDR5, RbBP5, Ash2L and DPY-30 increases H3K4 methylation by ~20-fold compared to that of the isolated MLL1 SET domain. However, the extent to which this 20-fold increase in enzymatic activity is due to a conformational change in the MLL1 SET domain is unclear at present. This is because the construct used to determine the structure of the MLL1 SET domain lacks the evolutionarily conserved Win motif in the region flanking the N-terminus of the SET domain [26], which has been shown to be essential for the assembly and dimethyltransferase activity of the MLL1 core-complex [24, 25, 37]. In addition, recent work from our laboratory indicates that the non-SET domain components of the MLL1 core complex possess a previously unrecognized H3K4 methyltransferase activity that is independent of the MLL1 SET domain [36] (see below). It is therefore possible that the observed increase in H3K4 methylation activity observed by Southhall et al., [26] is due at least in part to the independent activities of the MLL1 SET domain and the sub-complex containing WDR5, RbBP5, ASH2L, and DPY-30, which do not significantly interact in the absence of the MLL1 Win motif [25, 37].
Win motif
The WD-40 repeat protein WDR5 is a conserved component of SET1 family complexes ranging from yeast to humans and has been shown to be important for H3K4 methylation and HOX gene expression in hematopoiesis and development [48]. Recent studies have shown that WDR5 interacts directly with MLL1 or other SET1 family members and functions to bridge interactions between MLL1 and other components of the MLL1 core complex [20, 49]. It has also been suggested that WDR5 functions within the MLL1 core complex as a histone binding module that presents histone H3 for further methylation by MLL1 [48, 50]. In an effort to identify the WDR5 binding surface in MLL1, two independent groups mapped the WDR5 binding site in MLL1 to a short 6-residue conserved sequence in the N-flanking region of the MLL1 SET domain [25, 37]. This sequence, called the Win or WDR5 interaction motif, is highly conserved among metazoan MLL1 orthologs and other SET1 family members [37]. To determine the structural basis for the interaction between MLL1 and WDR5, two groups independently determined high resolution crystal structures of WDR5 bound to peptides derived from the MLL1 Win motif [24, 25]. Surprisingly, the structures reveal that the MLL1 Win motif forms a 310-helix that binds to the central opening of WDR5, the same site that was previously suggested to bind histone H3 (Figure 4). Conserved arginine 3765 of the MLL1 Win motif inserts into the central opening and is stabilized by a number of hydrogen bond, cation-Pi, and Pi-Pi interactions with conserved residues from WDR5. Consistent with a central role for the MLL1 Win motif for the interaction by WDR5, substitution of arginine 3765 with alanine in MLL1 abolishes the interaction between MLL1 and WDR5 [25, 37]. Furthermore, the same amino acid substitution, or a synthetic peptide derived from the MLL1 Win motif abolishes the interaction between MLL1 and the WDR5-RbBP5-Ash2L sub-complex, which also results in the loss of the H3K4 dimethylation activity of the MLL1 core complex [37]. These results have led to a model in which the conserved Win motif of MLL1 and other metazoan SET1 family members functions to bind the WDR5 component of the WDR5-RbBP5-Ash2L sub-complex, which is required for the assembly and H3K4 dimethylation activity of the MLL1 core complex [37]. These results also suggest that Win motif peptides or related compounds could have therapeutic value as inhibitors of SET1 family complexes.
Figure 4.
X-ray crystal structure of the MLL1 Win motif peptide in complex with WDR5. At top the domain architecture of full-length MLL1 is shown. The blown up portion shows a cut-away view of the MLL1 Win motif (green) bound to the central opening of WDR5 (PDB code 3EG6). The position of the conserved Arg 3765 is indicated. On the left, a stick representation is used to show the position of the MLL1 Win motif residues 3762–3770 (green) bound to the central opening of WDR5. MLL Win motif residue numbers are indicated.
The binding of the MLL1 Win motif to the central arginine binding pocket of WDR5 raises questions about the proposed role of WDR5 in binding histone H3, at least while WDR5 is incorporated into the MLL1 core complex. This is because structure-function studies show that histone H3 and MLL1 compete for the same binding site on WDR5 (Reviewed in [49]). To reconcile these models, it has been suggested that the WDR5-MLL1 interaction in the MLL1 core complex may be displaced by the mono- or dimethylated H3K4 product of the MLL1 core complex in a potential feedback mechanism [25]. Indeed, it has been demonstrated that H3 peptides that are mono- or dimethylated at H3K4 more efficiently disrupt the interaction between MLL1 and WDR5 than similar peptides that are unmodified or trimethylated at H3K4 [25]. Since WDR5 is required for the assembly of the MLL1 core complex [35, 37], this model predicts that the mono- and dimethylated forms of H3K4 could potentially regulate the assembly of the MLL1 core complex at specific loci [49]. However, this hypothesis is difficult to reconcile with the high-affinity interaction between WDR5-MLL1 (estimated at 120 nM measured by analytical ultracentrifugation) [37], with the relatively weaker binding of the mono- and dimethyl H3K4 peptides to WDR5, for which a broad range of estimated dissociation constants have been reported in solution (~7–115 μM for H3K4me1 and ~5–77 μM for H3K4me2, as measured by ITC [51, 52]). It remains to be determined if the H3K4me1 and H3K4me2 peptides can displace the WDR5-MLL1 interaction within the context of the holo-MLL1 core complex.
Mechanism of Multiple Lysine methylation by MLL1
SET domain enzymes differ in their ability to add one, two, or three methyl groups to the epsilon amino group of a lysine side chain, a phenomenon that has been termed “product specificity” [45]. Structure-function studies have demonstrated that product specificity of SET domain enzymes is determined by the presence of a phenylalanine or a tyrosine at a key position in the SET domain active site, called the Phe/Tyr switch position [45, 53–56]. Enzymes with a Phe at the switch position have a relatively larger active site volume that can accommodate the addition of more than one methyl group to the lysine side chain. In contrast, SET domain enzymes with a Tyrosine at the switch position have a relatively smaller active site volume and are predominantly monomethyltransferases. While site directed mutagenesis studies have validated the Phe/Tyr switch hypothesis for a number of SET domain enzymes [45, 53], SET1 family enzymes appear to contradict this rule [53]. This is because SET1 family enzymes are predicted to be monomethyltransferases based on the presence of a conserved tyrosine at the Phe/Tyr switch position. However, mono-, di-, and trimethylation activities have been attributed to SET1 family complexes in vivo and in vitro [53]. To resolve this paradox, it has been proposed that the product specificity of SET1 family enzymes is regulated by proteins that bind to and alter the conformation of the SET domain active site [26, 57].
To test the conformational change hypothesis, we have developed an in vitro system to examine the enzymatic activity and product specificity of the MLL1 SET domain in the presence and absence of MLL1 interacting proteins WDR5, RbBP5, Ash2L and DPY-30 [36]. This analysis reveals that the isolated MLL1 SET domain is a relatively slow H3K4 monomethyltransferase, which is consistent with the predictions of the Phe/Tyr switch hypothesis [36]. Substitution of Tyrosine 3942 with phenylalanine in MLL1 converts MLL1 into a mono-, di- and trimethyltransferase [36], suggesting that Tyr 3942 largely limits the product specificity of wild type MLL1 to that of a monomethyltransferase. In contrast, when WDR5, RbBP5, Ash2L and DPY-30 are added to the MLL1 SET domain, enzymatic activity increases ~600 fold, but only to the dimethyl form of histone H3 [36], suggesting that the product specificity of the MLL1 core complex is that of a dimethyltransferase. Contrary to expectations, kinetic experiments suggest that the mechanism of multiple lysine methylation is distinct from that expected from a conformational change in the SET domain active site [36]. To test the alternative hypothesis that one of the other components of the MLL1 core complex catalyzes dimethylation of H3K4, we assembled the MLL1 core complex with a catalytically inactive MLL1 SET domain variant, and discovered that the non-SET domain components of the MLL1 core complex possess a previously unrecognized histone methyltransferase activity that catalyzes H3K4 dimethylation within the MLL1 core complex [36]. In addition, it was shown that the non-SET domain components of the MLL1 core complex (WDR5, RbBP5, Ash2L and DPY-30 (WRAD)) pos sesses an H3K4 monomethyltransferase activity in the absence of the MLL1 SET domain [36]. Since the WRAD components lack homology to a conserved SET or DOT1-like methyltransferase fold (Figure 5a), WRAD represents a new class of non-SET domain histone methyltransferases. These results suggest that the mechanism of multiple lysine methylation by the MLL1 core complex involves the sequential addition of two methyl groups at two distinct active sites within the same complex (Figure 5b).
Figure 5.
New model for the mechanism of multiple lysine methylation by the MLL1 core complex (adapted from [36]). a) The MLL1 core complex is composed of two distinct H3K4 methyltransferases each possessing H3K4 monomethylation activity on their own. The dashed oval on the WDR5-RbBP5-Ash2L-DPY-30 sub-complex indicates that the catalytic motif is presently unknown, and may be shared between subunits. b). WDR5’s recognition of the MLL1 Win motif results in the assembly of the MLL1 core complex, which possesses H3K4 dimethyltransferase activity. We suggest that the MLL1 SET domain catalyzes monomethylation of histone H3 at lysine 4, which is followed by transfer of the monomethylated histone H3 to a second active site on the WRAD sub-complex, where H3K4 dimethylation occurs. We propose that mechanisms that control the assembly of the MLL1 core complex will be important for the regulation of H3K4 methylation states in the cell.
Surprising is the lack of H3K4 trimethylation by the in vitro assembled MLL1 core complex [36]. This observation is in contrast to previous results suggesting that an insect cell immunoprecipitated complex containing MLL1, WDR5, RbBP5, and Ash2L represent the minimal complex required for H3K4 trimethylation activity [35, 38]. A possible reason for this discrepancy could be due to the different assays used to quantitate the degree of H3K4 methylation in enzymatic reactions [36]. In previous investigations [35, 38], the degree of H3K4 methylation was monitored with methylation state specific antibodies, which can sometimes provide misleading results due to antibody cross-reactivity [58]. Indeed, we and others [59] have observed significant cross-reactivity of α-H3K4me3 antibodies with H3K4me2 epitopes in enzymatic assays. In contrast, in the investigation by Patel et al., [36], MALDI TOF mass spectrometry was used to quantitate the degree of H3K4 methylation, which shows an accumulation of the dimethyl from of H3K4 with little evidence for H3K4 trimethylation under the assayed conditions. These results suggest that an additional unidentified protein or posttranslational modification may be required for H3K4 trimethylation by the MLL1 core complex [36]. The possibility that an additional enzyme is required for H3K4 trimethylation is strengthened by the existence of a SET domain enzyme (PRDM9 (Meisetz)) that can only trimethylate H3K4, but not mono- or dimethylate H3K4 [60]. Further experimentation with more quantitative techniques to assess the degree of H3K4 methylation will be required to understand how H3K4 trimethylation is regulated by the MLL1 core complex.
Future Prospects
The regulatory mechanisms in the pathways that control eukaryotic transcription remain poorly understood. The analysis of the molecular mechanisms that regulate the enzymes that introduce covalent modifications into histones is expected lead to a deeper understanding of how transcription initiation, elongation, and termination are controlled in the context of chromatin. It is likely that the key enzymes in these pathways have evolved to integrate cellular, extracellular and feedback signals in mechanisms that result in exquisite control over enzymatic activity. Defects in this process are expected to be highly detrimental for development of an organism or in the specification of cell fate. Leukemias associated with loss-of-function or gain-of-function variants of MLL1 are prime examples of the importance of maintaining the enzymatic activity of MLL1 under tight control. Identifying the protein structural features that account for the enzymatic activity of MLL1 and other SET1 family members will be essential for understanding how the regulation of H3K4 methylation is integrated into eukaryotic transcriptional circuits. A future challenge is defining from a structure-function perspective how MLL1 interacts with and is regulated by other proto-oncoproteins including the Multiple Endocrine Neoplasia type 1 (Men1) [61] or the Integrase Interactor 1 (Ini/hSNF5) [62] tumor suppressor proteins. An additional challenge is in understanding the molecular basis for how all the different MLL fusion proteins disrupt normal MLL functioning and contribute to cellular transformation. In addition, important questions that remain unanswered include how does MLL1 regulate the trimethyl form of histone H3? Does the regulation of H3K4 methylation involve posttranslational modifications in MLL1 or other proteins that regulate the assembly of the MLL1 core complex? How does MLL1 discriminate among potential target genes? It is expected that such knowledge will be valuable for the development of new therapeutic strategies for the treatment of some forms leukemia and other aggressive cancers.
Acknowledgments
This work is supported in part by a Research Scholar Grant (RSC-09-245-01-DMC) from the American Cancer Society and by NIH grant number R01CA140522 from the National Cancer Institute (to M.S.C.). We thank Venkat Dharmarajan for a critical reading of this manuscript. We would also like to dedicate this manuscript to the memory of Warren DeLano, the creator of the molecular graphics program PyMol, which was used to create the figures in this manuscript.
References
- 1.Ziemin-van der Poel S, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci U S A. 1991;88(23):10735–9. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leegte B, et al. 11q- syndrome: three cases and a review of the literature. Genet Couns. 1999;10(3):305–13. [PubMed] [Google Scholar]
- 3.Canaani E, et al. ALL-1/MLL1, a homologue of Drosophila TRITHORAX, modifies chromatin and is directly involved in infant acute leukaemia. Br J Cancer. 2004;90(4):756–60. doi: 10.1038/sj.bjc.6601639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marschalek R. Mixed Lineage Leukemia: roles in human malignancies and potential therapy. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07608.x. [DOI] [PubMed] [Google Scholar]
- 5.Yu BD, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378(6556):505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 6.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10(5):1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 7.Milne TA, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005;102(41):14765–70. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terranova R, et al. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103(17):6629–34. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik S, Bhaumik SR. Mixed Lineage Leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahl BD, et al. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci U S A. 1999;96(26):14967–72. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasio D, et al. Complete exon structure of the ALL1 gene. Cancer Res. 1996;56(8):1766–9. [PubMed] [Google Scholar]
- 12.Nakamura T, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10(5):1119–28. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 13.Ansari KI, Mandal SS. Mixed Lineage Leukemia: Role in gene expression, hormone signaling and mRNA processing. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07606.x. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama A, et al. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100(10):3710–8. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh JJ, et al. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23(1):186–94. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121(6):873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24(13):5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wysocka J, et al. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17(7):896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, et al. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282(18):13419–28. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 20.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13(4):587–97. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 22.Allen MD, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. Embo J. 2006;25(19):4503–12. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Guzman RN, et al. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355(5):1005–13. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 24.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to Mixed Lineage Leukemia Protein-1 peptide. J Biol Chem. 2008;283(47):32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 25.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283(50):35258–64. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southhall SM, et al. Structural basis for the recruitment of additional factors for MLL1 SET domain actvity and recognition of epigenetic marks. Molecular Cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 27.Birke M, et al. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30(4):958–65. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24(23):10470–8. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5(3):309–14. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 30.Allen MD, GC, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006 Oct 4;125(19):4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14(13):1553–77. [PubMed] [Google Scholar]
- 32.Ernst P, et al. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21(7):2249–58. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goto NK, et al. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J Biol Chem. 2002;277(45):43168–74. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 34.Zor T, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J Mol Biol. 2004;337(3):521–34. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 35.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 36.Patel A, et al. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284(36):24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel A, et al. A conserved arginine containing motif crucial for the assembly and enzymatic activity of the Mixed Lineage Leukemia protein-1 core complex. J Biol Chem. 2008;283(47):32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 38.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13(9):852–4. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 39.Jenuwein T, et al. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54(1):80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 41.Dillon SC, et al. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6(8):227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20(3):341–8. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao B, Wilson JR, Gamblin SJ. SET domains and histone methylation. Curr Opin Struct Biol. 2003;13(6):699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, et al. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111(1):117–27. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12(1):177–85. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Y, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009;16(3):312–7. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min J, et al. Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol. 2002;9(11):828–32. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- 48.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 49.Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16(7):678–80. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- 50.Ruthenburg AJ, et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13(8):704–12. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13(8):698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 52.Schuetz A, et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5. Embo J. 2006;25(18):4245–52. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins RE, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280(7):5563–70. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian C, et al. Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J Mol Biol. 2006;359(1):86–96. doi: 10.1016/j.jmb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 55.Trievel RC, et al. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol. 2003;10(7):545–52. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 56.Xiao B, et al. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19(12):1444–54. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi YH, et al. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol. 2009;29(13):3478–86. doi: 10.1128/MCB.00013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheung P. Generation and characterization of antibodies directed against di-modified histones, and comments on antibody and epitope recognition. Methods Enzymol. 2004;376:221–34. doi: 10.1016/S0076-6879(03)76015-7. [DOI] [PubMed] [Google Scholar]
- 59.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2(7):E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438(7066):374–8. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 61.Yokoyama A, et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–18. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 62.Rozenblatt-Rosen O, et al. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci U S A. 1998;95(8):4152–7. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]