Summary
During the course of a viral infection, viral proteins interact with an array of host proteins and pathways. Here we present a systematic strategy to elucidate the dynamic interactions between H1N1 influenza and its human host. A combination of yeast two hybrid analysis and genome-wide expression profiling implicated hundreds of human factors in mediating viral-host interactions. These factors were then examined functionally through depletion analyses in primary lung cells. The resulting data point to potential roles for some unanticipated host and viral proteins in viral infection and the host response, including a network of RNA binding proteins, components of WNT signaling and viral polymerase subunits. This multilayered approach provides a comprehensive and unbiased physical and regulatory model of influenza-host interactions, and demonstrates a general strategy for uncovering complex host-pathogen relationships.
Introduction
Mammalian cells have developed complex systems to detect and eliminate viral pathogens, while viruses have evolved mechanisms to co-opt host processes and suppress host defenses. For example, influenza A is a segmented, single-stranded, negative-sense RNA virus that has adapted to infect multiple species. Upon infection by influenza, host cells detect viral RNA through pathogen sensors, such as RIG-I, and induce Type I IFNs and an anti-viral program that is common to many RNA viruses (Figure 1A) (Takeuchi and Akira, 2009). At the same time, the 10 major (and 1 minor) gene products of influenza virus mediate the viral life cycle and modulate cellular processes. Most notably, the NS1 protein subverts host defenses through several mechanisms, including suppression of RIG-I/TRIM25-mediated sensing of viral RNA (Gack et al., 2009; Pichlmair et al., 2006), PKR antiviral activity (Li et al., 2006) and cellular mRNA processing (Krug et al., 2003). Immune regulatory functions for the other influenza proteins have yet to be defined, as their assigned roles have been limited to viral entry into cells, viral RNA trafficking, replication and transcription, as well as assembly of mature virions. Similarly, the function of the vast majority of host factors remains unexplored. Previous studies on viral and host factors have focused on specific interactions but have not produced global models of the viral-host relationship, with few exceptions (Brass et al., 2008; Bushman et al., 2009; Konig et al., 2008; Krishnan et al., 2008; Li et al., 2009).
Figure 1. Integrative strategy to generate a physical, regulatory and functional map of influenza-host interactions.
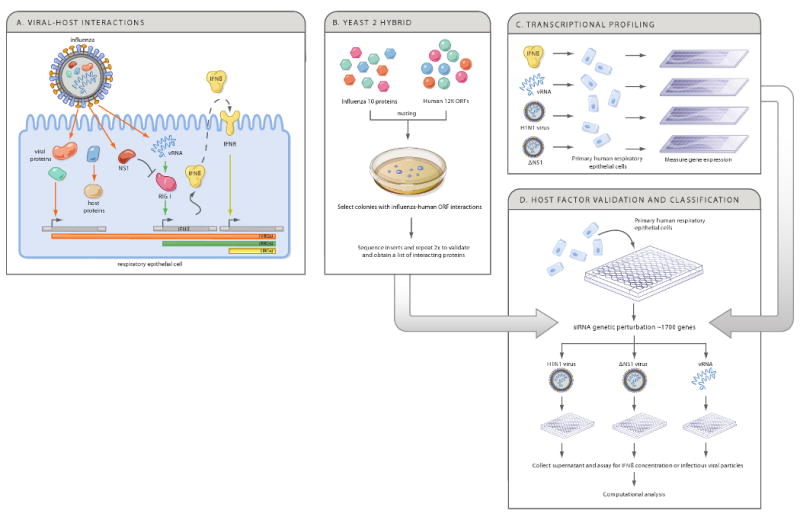
(A) When influenza infects host cells, viral components, including viral RNA (vRNA) and viral proteins interact with host proteins to induce changes in host gene expression or cellular functions. The RIG-I sensor detects virus-derived RNA and regulates host gene expression, including IFNβ, which in turn activates an anti-viral program through the interferon receptor (IFNR). We distinguish viral regulated genes (VRGs, orange), affected by infection, RNA regulated genes (RRGs, green), affected directly by vRNA, and interferon regulated genes (IRGs, yellow), affected directly by interferon treatment. NS1 is shown to inhibit the RIG-I response. (B-D) A genomic strategy to deconstruct influenza-host interactions. Host proteins that physically interact with each of the 10 viral proteins are identified using a systematic yeast-2-hybrid approach (B), and arrays are used to define the transcriptional responses of primary human bronchial epithelial cells (HBECs) to components of the virus and to virus infection (C). The physical and transcriptional maps were used to computationally predict human factors and pathways that affect the viral life cycle or host response. We tested these predictions by perturbing each gene and measuring the effect on IFN production and viral replication in primary HBECs (D).
Here, we use an integrative functional genomics strategy (Figure 1B-D) to generate a draft model of influenza-host interactions for the H1N1 strain A/PR/8/34 (‘PR8’). Our experimental and computational approach uncovers host networks contacted by viral proteins, cellular transcriptional responses to infection, and functional roles for candidate factors in influenza-infected primary lung epithelial cells. We integrate these datasets to generate a physical, regulatory and functional map that implicates hundreds of host factors in the influenza-human relationship.
Results
Identification of a human protein network that physically interacts with 10 viral proteins
To identify host factors that may participate in the pathogenesis of influenza infection, we first sought to identify those factors that are directly manipulated through physical associations with viral proteins. We used a yeast 2-hybrid (Y2H) approach to systematically identify direct binary contacts amongst the 10 major viral proteins of the H1N1 influenza strain A/PR/8/34 (‘PR8’), as well as between each viral protein and each of ∼12,000 human proteins available in the Human ORFeome v3.1 collection (Lamesch et al., 2007). We discovered 31 intra-viral interactions (out of 55 possible interactions, including homodimers) among the ten viral proteins (Figure 2A, Table S1A), and 135 pairwise interactions between the 10 viral proteins and 87 human proteins (‘H1’ genes, Figure 2B, Table S1B), 73 of which are expressed in primary human bronchial epithelial cells (HBECs). These included the previously reported association between NS1 and STAU1, interactions between NS1 and PRKRA and TARBP2 (regulators of PKR-mediated transcription; for a review of NS1-host interactions, see Hale et al., 2008), as well as interactions between influenza and eight proteins that are targeted by other viruses (Table S1B). Several known associations were not observed, either because the interacting protein was not among the 12,000 proteins in our assays (e.g. TRIM25 (Gack et al., 2009) and DDX58/RIG-I (Pichlmair et al., 2006), or for unknown reasons (e.g. we did not detect the PKR-NS1 interaction (Li et al., 2006), yet we identified the kinase that phosphorylates PKR).
Figure 2. A map of viral-human protein interactions identifies a dense interconnected network, coupled to key cellular signaling pathways. (A) Viral-viral interactions.
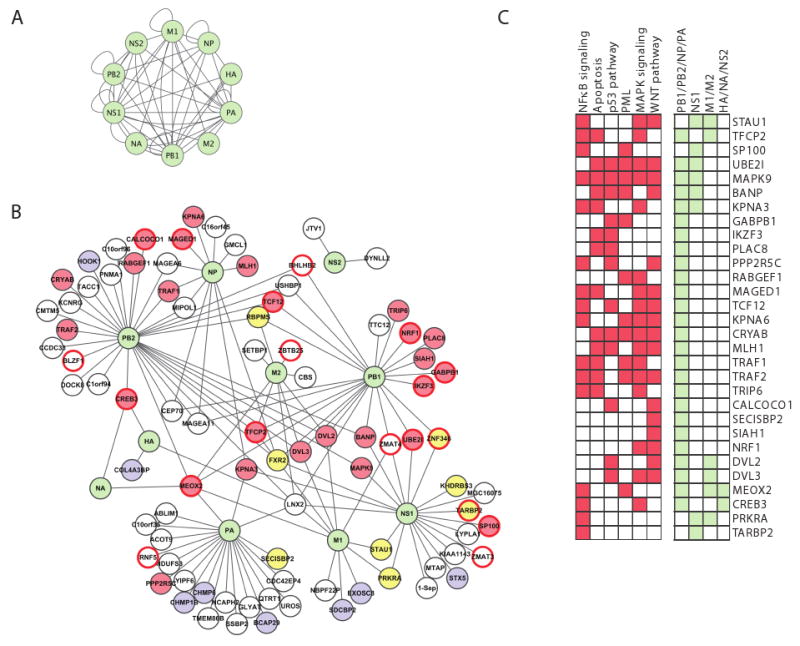
Green nodes, viral proteins; edges, direct physical interactions observed in a Y2H assay. (B) Influenza-human interactions. 134 interactions (edges) connect the ten influenza proteins (green nodes) to 87 ‘H1’ human proteins. Yellow fill, RNA-binding proteins; blue fill, protein transport; red border, transcription factors; red fill, 30 proteins that play a role in four major signaling pathways (NFκB, apoptosis, MAPK and WNT signaling); white fill, proteins with other functions. (C) 30 host interactor proteins (H1) are shown with their membership in specific pathways (red) or direct interactions with influenza proteins (light green). H1 proteins are either known components or established direct interactors with components of these pathways (see Experimental Procedures). Many of the 30 proteins are involved in multiple signaling pathways, and interact with polymerase subunits. Description of influenza A proteins: PB1, PB2, PA form the viral polymerase; NP complexes with viral RNA and is required for viral polymerase activity on RNA; HA and NA are membrane proteins involved in the fusion and release of viral particles; M1 is a matrix protein involved in export and assembly of RNA and viral particles; M2 modulates fusion through its ion channel activity; NS1 is a non-structural protein that regulates host pathways; NS2 is involved in RNA export.
The connectivity pattern of the intra-viral and viral-human network revealed three important principles. First, the influenza intra-viral network is extremely interconnected (Figure 2A, Table S2A), consistent with findings from other viruses (Bailer and Haas, 2009). This may be required for forming compact virions and functional viral complexes. Second, influenza proteins are linked on average with a significantly greater number of human proteins than expected from the human interaction network (13.5 vs. 6.5 expected, P<0.06, permutation test), even when compared to other viruses (Table S2A), or when comparing to the full 12,000 prey human network (data not shown). This may reflect the fact that a virus has to maximize the diversity of functions per protein. Third, some of the human proteins contact a greater than expected number of influenza proteins (24 human proteins interact with at least 2 flu proteins, P<10-5, permutation test, Figure 2B, Table S2B). These may be required for the formation of viral-host multi-protein complexes.
The H1 proteins form inter-connected hubs within the cellular protein network, suggesting that the virus targets proteins that play a central role in their respective cellular pathways. The 87 H1 proteins connect with each other through 51 interactions and with other human proteins (first neighbors; ‘H2 genes’) through 2717 interactions, a higher than expected connectivity (P<10-5, permutation test). The higher density of interactions is observed even when excluding highly connected H1 proteins, or when considering only the H1 proteins associated with individual viral proteins (Table 1).
Table 1. Network parameters of influenza and cellular protein interaction networks.
The parameters in this table quantify the numbers of interactions between a defined set of proteins (one set in each column) with the entire human interaction network (see Experimental Procedures). For example, there are 87 H1 proteins (i.e. direct interactors with the 10 PR8 viral proteins) that interact with 2768 human proteins with an average of 34.4 interactions per H1 protein (with standard deviation of 42.9). The H1H2 set includes the 87 H1 proteins and 566 H2 proteins that interact with them based on curated associations. X-H1 consists of the direct human interactors of the viral protein X.
| Network parameters | All-H1 | All-H1H2 | PB1-H1 | PB2-H1 | PA-H1 | NP-H1 | M1-H1 | M2-H1 | NS1-H1 | NS2-H1 | HA-H1 | NA-H1 | H. Sapiens |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nodes | 87 | 653 | 23 | 32 | 21 | 12 | 11 | 9 | 20 | 2 | 3 | 2 | 11624 |
| Total edges | 2768 | 17106 | 1312 | 1640 | 156 | 686 | 378 | 384 | 549 | 50 | 9 | 11 | 57206 |
| Average Degree | 34.4 | 30.6 | 60.0 | 55.4 | 8.2 | 57.3 | 37.8 | 48.3 | 32.5 | 9.9 | |||
| Stdev Degree | 42.9 | 43.6 | 50.3 | 49.6 | 7.7 | 36.2 | 40.6 | 48.7 | 47.1 | 19.9 | |||
| Median Degree | 17 | 15 | 53 | 47 | 5 | 59 | 31 | 32 | 17 | 4 |
Furthermore, we identified a core cellular sub-network that is enriched for H1 proteins (P<0.05-10-4, hypergeometric test; Experimental Procedures, Table S3). This sub-network contains six H1 proteins that bind at least three other H1 proteins (e.g. TRAF2, DVL2, FXR2) and 37 non-H1 proteins that fit the same criteria (e.g. p53, PKR, ILF3 and PSMF1, none of which contacts any viral protein directly). The network (Figure S1) consists of diverse proteins including RNA-binding proteins (see below for more details on functional assays), regulators of ubiquitination and sumoylation, transcription factors, mediators of apoptosis, and components of immune signaling pathways (Figures 2B,C, S1 and S2).
Our observations also hold in another influenza strain, the H3N2 A/Udorn/72 influenza virus (‘Udorn’). Using the same Y2H approach, we detected 81 interactions between 10 Udorn viral proteins and 66 human proteins (U-H1). Of these, 56 human proteins also interact with PR8 (P<10-10; hypergeometric test, Table S1C and Figure S3A), including most RNA binding proteins, regulators of transcription, protein transport and signaling (Figure S3B). For example, out of 30 signaling proteins and 19 transcription factors that directly interact with PR8 (PR8-H1) (Figure 2B,C), 28 and 16 proteins (respectively) also directly bind to Udorn proteins (Figure S3B). Most (63%) of the H1 proteins associated with PR8 polymerase subunits (PB1, PB2, PA), NP or NS1 were also found to interact with their counterparts in the Udorn strain (Figure S3C and Table S1B), reflecting conserved functions of viral proteins.
Viral proteins interact with the NF-κB, apoptosis and WNT pathways primarily through NS1 and polymerase subunits
To identify the key cellular pathways coupled to the virus, we considered the 87 H1 and 566 H2 cellular proteins from a manually curated database (IPA), and found that they are enriched for components of several signaling pathways (Table S4A, Figure 3D; see Experimental Procedures). 30 of the 87 H1 interactors couple the virus to six major pathways, including p53-, PML- and TNFR/Fas-mediated apoptosis, NF-κB and WNT/β-catenin (Figure 2C). The interactions with these pathways are conserved for the Udorn strain (Figure S3C), suggesting that the discovered interactions reflect a generalized strategy of influenza to manipulate the host. A role for these pathways in viral infections has been described (e.g. p53, Turpin et al., 2005), yet their direct physical association with influenza proteins was not previously reported.
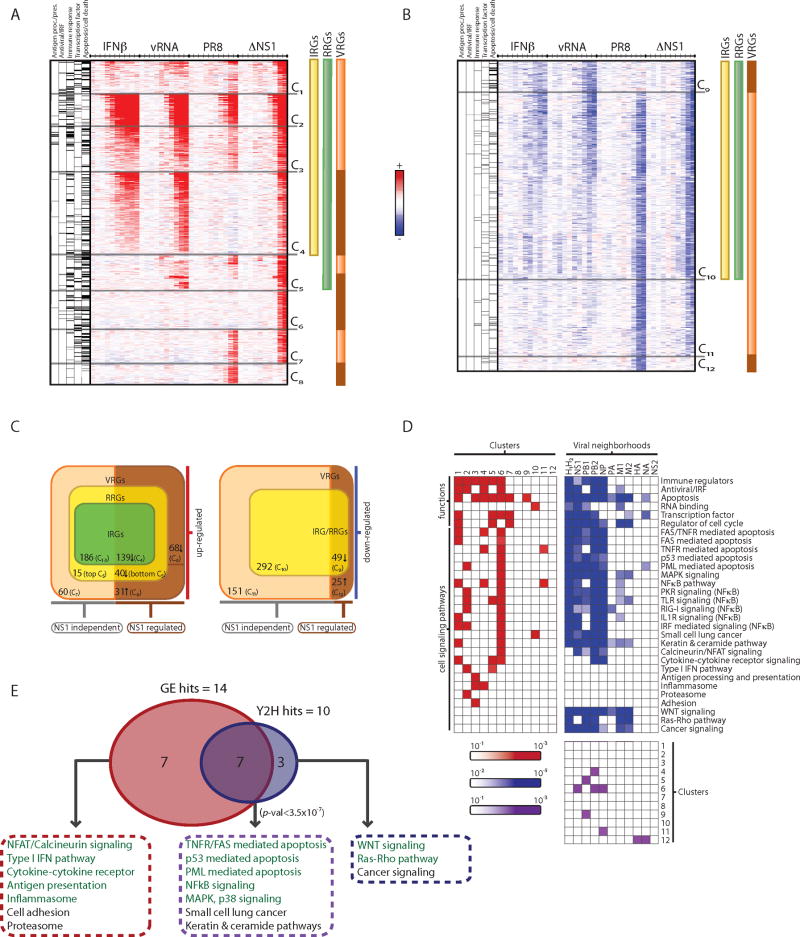
Figure 3. Functional decomposition of transcriptional responses to influenza infection identifies viral-specific gene regulation.
(A, B) Gene expression changes in HBECs in response to IFNβ (IRGs, yellow bar), vRNA (RRGs, green bar), wild type influenza (PR8) and mutant ΔNS1 virus (VRGs, orange bar; genes differentially regulated by NS1, brown bar) at 10 time points (.25, .5, 1, 1.5, 2, 4, 6, 8, 12, 18 hours, tick marks). Upregulated (A, red) and downregulated (B, blue) genes, relative to the expected level from mock-treated cells, were grouped into 12 clusters (C1-C12). Left columns denote gene membership in five major functional categories (black lines, category is enriched in cluster; grey lines, category is not enriched in cluster). (C) Venn diagrams indicate number of members in each class of regulated genes and their dependence on NS1 (bottom), within the subset of upregulated (left) and downregulated (right) genes. (D) Functional and pathway annotation of expression clusters and interaction neighborhoods. Shown are the functional categories and pathways (rows) enriched in each of the 12 expression clusters (red, left matrix) and interaction neighborhoods (H1 and H2) of each viral protein (blue, right matrix). Bottom matrix shows the significant overlaps (purple) between expression clusters (1-12, rows) and viral neighborhoods (columns). (E) Enrichment analysis identifies pathways that are over-represented in the influenza physical network (10 pathways, blue) and in transcriptional responses (14 pathways, red), with an overlap of 7 pathways enriched in both (purple, P <3.5×10-7). Pathways chosen for functional follow-up assays are colored in green.
While NS1 is considered the major viral protein to modulate host signaling, we found that 26 of the 30 H1 proteins associated with these pathways interact with viral polymerase subunits and NP, but only 8 interact with NS1 (Figure 2C). In particular, H1 and H2 interactors of PB1, PB2, NP are highly enriched (P≪ 10-10, Fisher's combined probability test) for the six key pathways, but those of PA are not (Figure 3D; this result also holds for Udorn, Figure S3C). This suggests that viral polymerase proteins may also act as direct modulators of host signaling pathways.
Expression profiling of the response to viral infection in primary human lung epithelial cells
We next defined the major transcriptional responses in primary human bronchial epithelial cells (HBECs) after either infection with influenza or treatment with relevant ligands. We used four different strategies, each highlighting distinct components of the response. (1) We infected cells with the wild-type PR8 influenza virus that can mount a complete replicative cycle. (2) We transfected cells with viral RNA (‘vRNA’) isolated from influenza particles. This does not result in the production of viral proteins or particles and identifies the effect of RNA-sensing pathways (e.g., RIG-I.). (3) We treated cells with interferon beta (IFNβ), to distinguish the portion of the response that is mediated through Type I IFNs. (4) We infected cells with a PR8 virus lacking the NS1 gene (‘ΔNS1’). The NS1 protein normally inhibits vRNA- or IFNβ-induced pathways, and its deletion can reveal an expanded response to infection. We could not assess the role of any other viral protein but NS1, since deletion strains cannot be propagated easily (Wressnigg et al., 2009). For each of the four stimuli, we profiled the cellular transcriptional response at ten time points (.25, .5, 1, 1.5, 2, 4, 6, 8, 12, and 18 hours) in duplicate experiments.
Transcription patterns decompose the response into interferon-, RNA- and virus-responsive genes
We found twelve major temporal and functional patterns of gene expression (C1-C12, Figure 3A,B), each associated with one or more stimulus, and covering 1056 genes that were all regulated in response to viral infection (VRGs, virus-regulated genes). Among these, we found 666 interferon-regulated genes (IRGs) that are affected by interferon directly (325 induced, C1-C4, and 341 repressed, C9-C10, Figure 3A-C). All of the IRGs are similarly affected by vRNA transfection and ΔNS1 virus but with an observed time delay, likely due to the induction of IFNβ by these stimuli. PR8 infection induces IRGs to a much lower level (with few exceptions, C2, 57 genes), and abrogates the downregulation of 49 IRGs (C9). This is consistent with the known role of NS1 in dampening RNA sensing and downstream interferon production (Pichlmair et al., 2006) (see Figure S4).
Next, we found 721 RNA-regulated genes (RRGs) that are directly modulated by transfected vRNA (380 induced, C1-C5; 341 repressed, C9-C10, Figure 3A-C). All of the RRGs are similarly affected by ΔNS1 virus infection, while 171 are regulated by PR8 infection. Thus, viral RNA present in the infecting virion and produced during modest viral replication (as with ΔNS1 virus) can induce a potent response. Induced RRGs (C1-5) were enriched for antigen presentation, apoptosis, NFκB and IRF signaling (P<10-4-10-17; hypergeometric test, Figure 3D).
Most of the induced RRGs are also induced by interferon treatment (C1-C4, 325 of 380), but a few IFNβ-independent RRGs (C5) were induced only by vRNA and ΔNS1 virus. These include important antiviral genes (e.g. IFNB1, IL7R, ING3, IRF2, PELI1) and are enriched for TLR pathway components, cytokines, chemokines, and cell cycle and apoptosis (P<10-3, Figure 3D). An IFNβ-independent mechanism (e.g. IRF3 based on promoter sequence analysis, data not shown) likely mediates the transcription of these genes.
Finally, we identified virus-specific response genes (VSRGs) that are transcriptionally regulated following PR8 or ΔNS1 virus infection, but not following vRNA transfection or IFNβ treatment (C6-8 and C11-12). 68 VSRGs are induced only by ΔNS1 (C6), and are enriched for regulators of apoptosis and NFκB (e.g. BCL10, TRAF6, NFKB1 and NFKBIE). NS1 may block their induction and dampen the NFκB pathway by an unknown, RNA- and IFN-independent mechanism. 60 VSRGs are induced by both PR8 and ΔNS1 (C7) and are enriched for regulators of apoptosis, cell cycle and transcription factors (P<10-3, Figure 3D). Finally, 31 VSRGs (C8) are induced only by PR8, possibly directly by NS1 or as the result of the higher burden of a replicating virus.
NF-κB, MAPK and apoptosis pathways are regulated through both transcriptional and physical interactions
Some of the host systems affected at the transcriptional level by viral infection may be linked to the virus through physical interactions. Indeed, we found that the cellular network of direct interactors (H1) and first neighbors (H2) is enriched for genes that are transcriptionally regulated upon viral infection (70 of 1056 VRGs, P<4×10-4, hypergeometric test). For example, the NS1 neighborhood is enriched in C6 (P<0.01), a VSRG cluster induced only in response to infection with ΔNS1 virus (e.g. NFKB1, BCL10). Similarly, the neighborhood of the polymerase subunit PB2 and NP is also enriched in C6 (P<7×10-4), further supporting the potential role of the viral polymerase in modulating host pathways in concert with NS1.
While some cellular pathways are uniquely associated with either the physical network (e.g. WNT, Ras/Rho) or transcriptional responses (e.g. Type I IFN and antigen presentation), many are enriched for both (P<3.5×10-7, hypergeometric test, Figure 3D,E). These include p53-mediated apoptosis, PML, NFκB, MAPK and p38 signaling. These pathways are mostly associated with rapidly and highly-induced IRGs (C2) or VSRGs that are inhibited by NS1 (C6). Thus, the virus physically engages critical pathways while inducing transcriptional changes in their components.
Functional interrogation of viral interactors and transcriptionally responsive genes
The physical interactions, transcriptional responses and associated pathways together identified 1745 candidate genes that could impact influenza infection. These included 1056 genes that were transcriptionally regulated, 259 direct interactors and their first neighbors (H1/H2) (67 of them are also transcriptionally regulated), and 504 further candidates predicted from our analyses (e.g. pathway members) that are expressed in HBECs.
To test the functional contribution of these genes to viral replication and type I IFN production, we measured the effect of perturbing each gene using targeted siRNA pools in three functional assays. In the viral replication assay, we infected siRNA-transfected primary HBECs with PR8 virus and measured virus production after 48 hours using a cellular reporter system that is analogous to conventional plaque assays (Experimental Procedures). In two independent assays, we used a reporter cell line to measure levels of IFNβ in siRNA-transfected HBECs in response to ΔNS1 virus infection or vRNA transfection.
We determined the relative effect of each of the 1745 siRNA pools in each assay using a statistical scoring approach (Experimental Procedures) that identifies significant changes in phenotypes relative to the background of all tested genes. Since we selected a focused set of candidates for functional testing, this scoring approach is highly conservative. Furthermore, because cell number impacts production of IFN, we used AlamarBlue to determine cellular viability following siRNA knockdown and to effectively normalize IFN values to the number of cells in each well. We used a 2-fold threshold (see Experimental Procedures) to identify genes whose perturbation significantly impacted the phenotypes evaluated in each of the three assays, distinguishing positive and negative regulators of each phenotype.
616 of the 1745 candidate genes affected at least one of the phenotypes significantly. The number of genes with two or more significant phenotypes is substantially higher than expected by chance (P<10-4, permutation test, Figure S5). These included all the major sources of candidate genes, including 361 transcriptionally responsive genes, 88 direct interactors (H1) and first neighbors (H2), and 174 additional members of identified pathways. This suggests that many of the transcriptional and physical target pathways play an important role in infection.
Distinct functional signatures for regulators of IFN production and viral replication
We divided the 616 validated genes into 20 ‘phenoclusters’ based on the combinatorial behavior of each gene across the three functional assays (Figure 4A). vRNA-dependent regulators of IFNβ production are members of phenoclusters in which IFNβ levels changed in response to vRNA (211 positive regulators, P1-6; 145 negative regulators, P7-12). These genes correctly include many well-known regulators of Type I IFN, both activators (e.g., VISA, IRF3, RELA, IκBKγ, IκBKε, IκBKβ, and IRF9) and repressors (e.g., PTPN6, IRF2). Some of these genes did not affect PR8 replication (P1,2,7,8. e.g., IFNβ in P1), likely because the NS1 protein already ensures low levels of IFNβ post-infection. Others (P3,4,9,10, e.g. IRF3, IRF2) had opposing effects on IFN levels and viral replication, including genes (P4,10) that were essential for IFNβ-dependent anti-viral effects even in NS1-inhibited cells. Genes that were not previously known to affect IFN production included a potential ubiquitin ligase complex (CUL1, FBXO34), regulators of vesicle trafficking (e.g. CHMP6, ARL4A), peroxisomal components (PEX14), WNT pathway genes (below), and genes known to be involved in the life cycle of other viruses (e.g., TMF1 binding to the HIV TATA element (Table S5; Garcia et al., 1992).
Figure 4. Functional interrogation and classification of candidate genes identified through integrative analysis of influenza-human interactions.
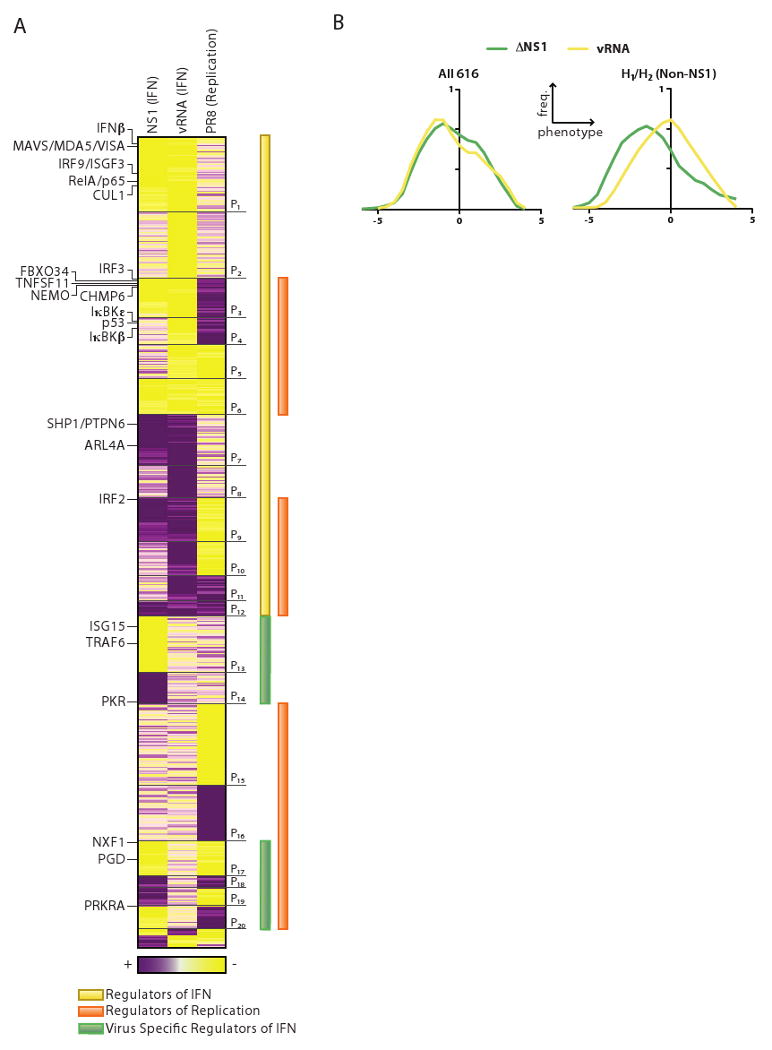
(A) Classification of phenotypes resulting from siRNA-mediated knockdown of 616 genes. Heat map shows phenotype scores corresponding to 3 functional assays (columns) performed on HBECs following transfection with siRNAs (rows). Gene phenotypes are hierarchically clustered, resulting in 20 major phenoclusters (P1-20). NS1 (IFN), assay for production of interferon following infection with ΔNS1 virus. vRNA (IFN), assay for production of interferon following transfection with viral RNA. PR8 (Replication), assay for infectious virion production following infection with PR8 virus. Yellow, positive regulator (lower IFN or virus titer). Purple, negative regulator (higher IFN or virus titer). Selected genes (also referred to in the text) are marked (left). (B) Distribution of phenotype scores for direct physical interactors and their first neighbors. Right panel: H1/H2 interactors (excluding NS1) have a significantly higher number of positive regulators of interferon production following infection with ΔNS1 (green) compared to vRNA transfection (yellow). There is no such shift in the distribution for all 616 genes shown in the phenocluster heatmap (left panel).
Virus-dependent, vRNA independent, regulators of IFNβ production (137 genes, P13-14, P17-20) affect ΔNS1-induced, but not vRNA-induced, IFN production. These are subdivided into genes that do not affect (P13,14), inversely affect (P19,20) or concordantly affect PR8 replication (P17,18; we cannot rule out the possibility that these genes affect IFN production as a consequence of their effect on replication). These genes include PRKRA, a known regulator of PKR (in P20) and TRAF6 (in P13); known essential regulators of PR8 replication such as NXF1 (P17) and PGD (P17) (Hao et al., 2008; Satterly et al., 2007); and inflammasome-associated components (NOD2 and NLRP9, P18), consistent with recent findings (Sabbah et al., 2009). We also observe candidates such as TTC12 (P18) that associates physically with PB1 (Figure 2B). PKR, a known repressor of viral replication, is also a member of this group (P14) and shows no effect on PR8 replication in our assay. This likely reflects masking of PKR activity by the NS1 protein (Li et al., 2006), suggesting that other regulators of viral replication masked by NS1 are members of this class.
IFN-independent regulators of viral replication (107 genes, P15-P16) are genes that affected PR8 replication, but did not affect IFN production. These include PML, an ‘H2’ gene and a well-established negative regulator of viral replication (Everett and Chelbi-Alix, 2007), and an unappreciated negative regulator, USHPB1 that interacts physically with PB1 and PB2 (Figure 2B and Table S1B). P15 includes several candidate positive regulators of replication, including RIOK3 that is induced only by PR8 virus (C8) and ZMAT4 which interacts directly with M1/PB1/NS1 (Figure 2B).
We next determined the relative contribution of transcriptionally regulated genes to each of the phenoclusters. Virus-specific regulated genes (VSRGs) that are induced by both PR8 and ΔNS1 infection (C7) included 30 genes with effects in our functional assays. This class of genes was enriched in phenotypes (P<0.007) with (mostly positive) regulators of IFN production in ΔNS1. The observation that a wild type virus and a virus lacking NS1 could induce anti-viral (i.e. pro-IFN) genes suggests that influenza virus may possess NS1-independent mechanisms to bypass this anti-viral response.
A subnetwork of RNA binding proteins affects IFNβ production during infection
The functional assays revealed the importance of a number of densely connected areas of the influenza-cellular interaction network (Figure S1). One of these areas was enriched for RNA binding proteins (P < 10-4, Figure S1), including the known regulator PKR, and RBPMS, ILF3, FMR1, DHX9, ZNF346 and HNRPC. ILF3 is phosphorylated by PKR and associates with XBP-1 (both are key mediators of the stress response; Patel et al., 1999), and ILF3 in turn interacts with DHX9 and HNRPC (Reichman et al., 2003). Since influenza is an RNA virus, this enrichment may reflect direct regulation of the influenza life cycle. Indeed, we found that shRNA-mediated depletion of four of these genes significantly affected interferon production following ΔNS1 infection (Figure 5A). Two are negative regulators (PKR and ILF3) and two as positive regulators (DHX9 and HNRPC).
Figure 5. Functional roles of an RNA-binding protein subnetwork, the WNT pathway and the viral polymerase.
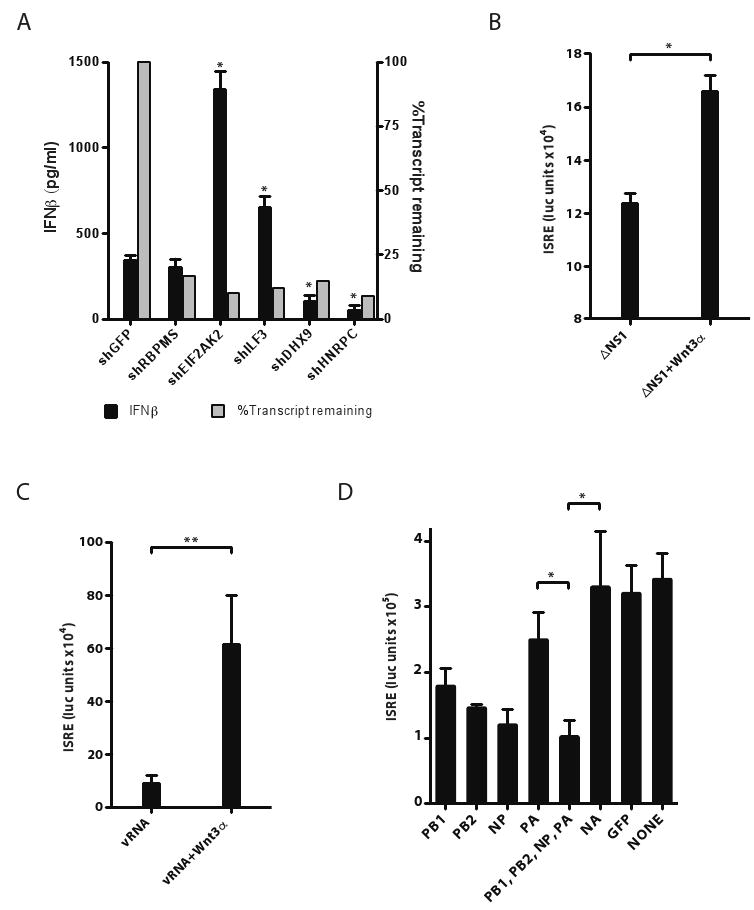
(A) RNA binding proteins play a role in regulating interferon production in HBECs infected with ΔNS1 virus. HBECs were infected with lentiviral shRNAs to knockdown each of 5 candidate RNA-binding proteins. Cells were selected in puromycin for 5 days and then stimulated with ΔNS1 virus. Supernatants were collected 24 hours post-infection, and IFNβ protein levels were quantified by ELISA (black bars, left Y-axis). In the same experiment, the efficacy of knockdown by each shRNA on its target mRNA was quantified (grey bars, right Y-axis, measured by qPCR relative to GAPDH). (n=3). (B,C) WNT protein potentiates ISRE responses in epithelial cells. 293T-ISRE-luciferase reporter cells were treated with WNT3α for 24 hrs and then infected with ΔNS1 virus or transfected with vRNA for 18 hours. Luciferase reporter activity (Y-axis) was quantified in response to (B) ΔNS1 infection or (C) transfected vRNA (purified from PR8 virions). (n=6). (D) Overexpression of viral polymerase subunits or NP and their effects on ISRE-inducing activity following vRNA transfection. 293T-ISRE-luciferase reporter cells were transfected with an expression plasmid encoding each influenza polymerase subunit or with combinations of plasmids (bottom panel) and then stimulated with transfected vRNA. ISRE responses to transfected vRNA were quantified for each of PB1, PB2, NP, PA, NA and control GFP, and their combinations. Similar results were obtained when cells were infected with ΔNS1 virus (data not shown). Significant effects of overexpression were found only for PB1, PB2, NP, PB1/PB2/NP/PA vs. NA (but only two are marked for clarity). (n=6). Error bars represent the standard deviation of the replicates. *, p<0.05 (t-test).
WNT pathway components modulate cellular responses to infection
Another highly enriched sub-network involved components of the WNT signaling pathway. There is a significant number of interactions between influenza proteins and members of the WNT/β-catenin pathway, and deletion of WNT pathway components significantly impacts influenza replication and interferon production (Figure S6). Consistently, recent studies have implicated the WNT pathway in the modulation of immune function (Staal et al., 2008), and in regulating cell survival and proliferation in EBV infected B-cells (Hayward et al., 2006). To test the involvement of the WNT pathway in influenza pathogenesis, we measured the effect of WNT3α treatment on interferon production following influenza infection or vRNA transfection. We found that direct treatment of cells with WNT3α increased IFN production in both assays (Figure 5B,C). The mechanism of action is yet to be defined.
The viral polymerase may mediate a non-NS1 effect on IFNβ production
The non-NS1 physical interactors and their direct neighbors (non-NS1 H1/H2) have a higher number of positive regulators of interferon production in the ΔNS1 assay than in the vRNA transfection assay (P<0.001, KS test, Figure 4B, right). This distinction is in marked contrast to the overall similarity in phenotypic effects of all the remaining genes (i.e. 616 non-H1/H2 genes) on IFNβ production in both assays (Figure 4B, left). This suggested that non-NS1 viral proteins participate in the manipulation of the IFN production in response to viral RNA.
To identify candidate non-NS1 mechanisms that mediate the effect on IFN production, we ranked each of the viral proteins based on their neighborhood enrichment for cellular pathways. The most prominently enriched neighborhoods of non-NS1 proteins were for two of the three viral polymerase subunits (PB1, PB2) and NP (Table 1 Figure 3D). These neighborhoods are also conserved in the Udorn strain (P<10-10, hypergeometric test), and are enriched for VSRGs (C6) whose expression is induced only in ΔNS1 infection. We thus hypothesized that the viral polymerase may play a previously unappreciated, NS1-independent, role in modulating interferon production.
To test this hypothesis, we measured the effect of over-expressing viral-polymerase subunits and NP on cellular production of IFN. Indeed, we found that over-expression of PB1, PB2 and NP, individually and in combination, was sufficient to inhibit cellular interferon responses to either vRNA transfection or ΔNS1 infection (Figure 5D). This disruptive effect is more prominent for PB1, PB2 and NP than for PA. This is consistent with the lower enrichment of the PA neighborhoods across immune functions and pathways and with the lower connectivity of PA in the PR8 and Udorn interaction networks (Figure 3D, Table 1). Taken together, our results illustrate the functional relevance of the Y2H interactions, and implicate non-NS1 viral proteins in cooperating with NS1 to modulate host responses.
Discussion
A physical, regulatory and functional map of influenza-host interactions
Influenza-host interactions have evolved over countless infections across diverse host species. To capture this complex relationship, we have constructed a first comprehensive map representing the physical and regulatory interactions between influenza virus and its primary human host cell. First, we assembled a physical map of binary associations between human and viral proteins. Second, we defined the regulatory responses of the host using genome-wide mRNA profiling of HBECs exposed to wild type virus, ΔNS1 virus, viral RNA or type I IFN. Third, we overlaid these maps within the context of known cellular human networks, expanding them to a neighborhood of the human interaction network and annotated cellular pathways. This led us to identify 1745 candidate genes that may play a role in influenza replication or the host response to infection. To validate the function of these candidate genes, we assessed the loss-of-function effects of each of the candidates in three independent in vivo assays in HBECs and classified each gene within a specific phenocluster. These extensive experimental and computational analyses allowed us to assign each to candidate gene a physical, transcriptional, and phenotypic ‘signature’ that reflects its specialized role in the host-pathogen network (Table S5).
Our results must be interpreted and validated with care. For example, Y2H interactions should be validated in vivo during infection, e.g. with appropriate pull-down assays. Similarly, we conservatively scored our functional assays, and may have masked relevant false negatives. Finally, analyses across other influenza strains may further generalize our findings. Nevertheless, by integrating across three sources of information, we were able to reveal new roles for viral and host proteins in manipulating the cellular machinery during infection.
Effects of the NS1 protein may be mediated by the p53 and WNT pathways
We rediscovered many of the known roles and interactions of the well-studied PR8 and Udorn NS1 proteins. For example, these included NS1-dependent dampening (Figure 3A) of the RIG-I-mediated transcriptional response to RNA (Pichlmair et al., 2006), and the direct interaction between NS1 and proteins involved in RIG-I/NFκB, PKR and mRNA processing functions (Figure 2B).
We also expanded the physical and regulatory scope of NS1 in the context of viral infection. We suggest a regulatory role for NS1 in the induction of 45 RNA/IFN-independent VSRGs (C8,12, Figure 3); 24 of which impact viral replication or the host IFN response in our assays (Table S5). We found previously unknown physical associations between NS1 and multiple proteins of the p53 and WNT pathways (Figure 2B,C). Components of these pathways affect the host response to virus in our assays (Table S5), and direct treatment with recombinant WNT protein increases cellular production of IFN in response to vRNA and viral infection (Figure 5B,C). While further experiments are required to understand the mechanistic basis of these findings, we propose that NS1 protein has an even broader impact on cellular processes than was previously appreciated.
A potential role for the viral polymerase in host-pathogen interactions
We discovered a large number of proteins and pathways that physically associate with the two viral polymerase subunits PB1, PB2 and NP of the PR8 and Udorn strains (Figures 2B,C), including the NF-κB, p53, apoptosis and WNT pathways. In addition, these host components are enriched in VSRGs, suggesting that the virus has also evolved to indirectly regulate mRNA levels of genes in these networks. Consistently, over-expression of one or more of the viral polymerase proteins (or NP) inhibits the IFN response to virus or vRNA in epithelial cells. Further experiments, including disruption of individual virus-host protein interactions (e.g. PB2-TRAF2 vs. PB2-DVL3) through point mutations, are needed to establish these mechanisms and to exclude non-specific effects.
Virus Specific Regulated Genes (VSRGs) play an important role in host defense
Transcriptional profiling of cells exposed to viruses or intermediate components of infection led us to discriminate a group of genes (VSRGs, C6-8,11-12) that respond only to virus but not to vRNA or IFN treatment. Many of these have not been previously described in this context. VSRGs include major components of the signaling pathways identified in the physical network, as well as regulators of IFN production and viral replication (Figure 3D; Table S5). How VSRGs are regulated and whether they are specific to influenza virus or respond to a broader class of viruses remains to be determined.
Towards an integrated model of host-influenza interactions
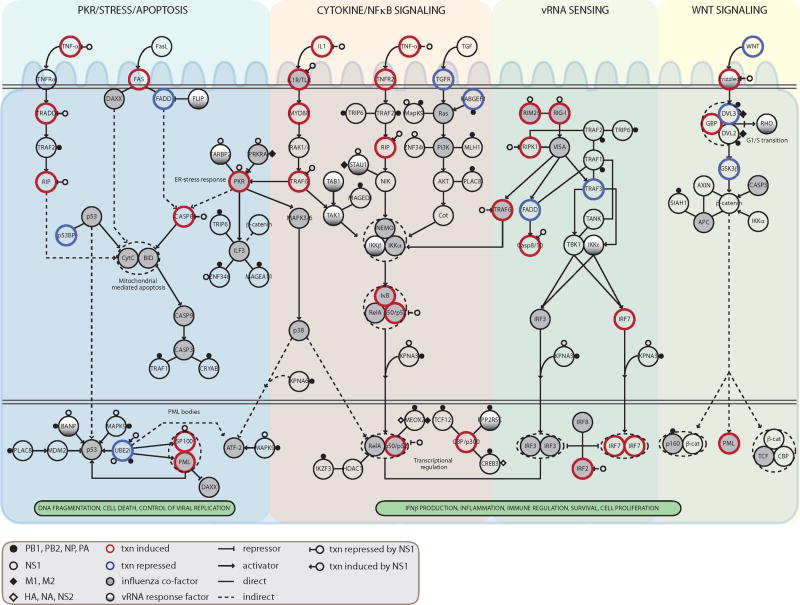
To build a model of the core networks targeted by the virus (Figure 6), we focused on the four main pathways identified in our analysis as key targets of the virus both physically and transcriptionally: NFκB (including RIGI and PKR), apoptosis, MAPK, and WNT. We incorporated all the H1 proteins that directly associate with these pathways into the model (see Experimental Procedures). We annotated each protein in the model with its mode of regulation (physical or transcriptional), the specific viral protein with which it associates, and the functional consequences on IFN and replication upon its knockdown.
Figure 6. An integrated model of the key signaling pathways modulated by influenza-host interactions.
For each host component within computationally selected pathways (see text), we show its mode of regulation and its functional role: (i) direct physical contact with viral proteins (small circles and diamonds); NS1 interactions with PKR, RIG-I, TRIM25 were added manually based on previous reports; (ii) transcriptional regulation in response to influenza infection in HBECs (increase in gene expression, thick red border; decrease, thick blue border); (iii) transcriptional regulation by the NS1 protein (open circle with inhibitory edge or activating edge); (iv) role in modulation of IFN production or PR8 replication (filled, gray); or role only in the IFN response to vRNA (filled, gradient). Txn, transcriptional. Influenza co-factor indicates significant change in PR8 replication upon siRNA knockdown of that gene. vRNA response factor indicates a gene whose knockdown caused significant change only in the vRNA-induced interferon assay.
We find that the virus targets diverse signaling pathways by affecting multiple components in each pathway through both physical and regulatory interactions. While the same pathway is often targeted by both mechanisms, the direct effect can be mediated through distinct genes. This observation emphasizes the virus' capacity for combinatorial regulation of cellular processes, and the importance of an integrated analysis approach for revealing a more complete picture of the viral-human relationship.
Many unrelated viruses (e.g. HSV, HIV, EBV, KSHV, Table S4B) also target the same pathways or components as influenza (Brander and Walker, 2000; Hiscott et al., 2006). For example, HIV targets NFKB1, p53 and β–catenin; EBV targets TRAF1/2 (Luftig et al., 2004) and SP100. Also consistent with other viruses (Konig et al., 2008), perturbation of H1 proteins does not typically result in a phenotypic change. Nevertheless, for influenza, perturbation of a subset of the H1 proteins (e.g. TARBP2, BANP, STAU1, PPP2R5C; see Figure 6) does affect IFN production in response to vRNA, suggesting that the virus may inhibit their function by direct binding.
Our analysis identified the involvement of several pathways whose role in the host response had not been fully appreciated. For example, we found that a group of inflammasome-related sensors are important in modulating IFN production (NLRP14, NOD2, NLRP9, NLRP10) and virus replication (NOD2). The WNT, p53, ER stress, apoptosis and PML pathways also impact IFN and replication. Finally, we identified host proteins which were not previously associated with influenza, each of which interacts with multiple viral proteins and is essential for the control of PR8 viral replication (e.g. USHBP1, ZMAT4 and MAGEA11). Such proteins are located at the host-pathogen interface and are likely to mediate essential viral and host activities. Among these are several RNA binding proteins, whose function we validated in an independent assay (Figure 5A).
With rapid advances in comprehensive measurement and perturbation technologies, we will be able to produce increasingly detailed maps of the physical and regulatory interactions underlying viral-host relationships. Such models could provide a basis for interrogating host circuitry across human individuals to help identify host susceptibility factors and causal genetic polymorphisms. Indeed, there is a significant overlap between the proteins in our model and the genes induced in patients during influenza infection (data not shown). Comprehensive models could also help explain differences in the host response to influenza infection, when the virus is transmitted from one species to another. While some physical and regulatory interactions and their effects on cell function are likely conserved across birds, pigs and humans, others may be host-specific and could help account for the differences in virulence across pathogenic strains.
Experimental Procedures
Primary cell cultures and virus strains
Primary human bronchial epithelial cells (HBEC; Lonza, Basel, Switzerland) derived from normal human bronchial epithelium, were maintained in vented T225 tissue culture flasks and grown in bronchial epithelial cell basal medium (Lonza, Supplementary Information). All experiments were performed with low passage (P) cells (P2-P5). Both PR8 and ΔNS1 viral strains were grown in Vero cells (which allow efficient growth of the ΔNS1 virus) in serum-free DMEM with 10% BSA and 1μg/ml TPCK trypsin. Viral titers were determined by standard MDCK plaque assays.
Yeast 2 hybrid assay
Stringent Y2H assays were carried out as described (Venkatesan et al., 2009) with ORFs from PR8 and Udorn strains as DB-ORFs in MATα Y8930 yeast and against the Human ORFeome v3.1 as AD-prey. Each primary screen was done twice, and all initial positive pairs from the two primary screens were individually retested three times using fresh stocks of DB-Flu and AD-Human yeast strains. The final datasets contain those interaction pairs that successfully retested at least two times without exhibiting autoactivation of the yeast HIS3 reporter gene (Rual et al., 2005; Venkatesan et al., 2009).
Human interaction network
We generated a comprehensive human interaction network by combining information from the interaction databases BioGRID, BIND and INTACT. To maximize coverage across the network, we included direct binary interactions, co-complexes, and protein modifications. In total, the network contained 57,206 interactions among 11,624 human proteins. To analyze the first neighbors of H1 proteins (H2), we used both curated and non-curated resources. Non-curated neighbors consisted of 1923 human proteins that physically interact with H1 proteins based on the human interaction network. Curated first neighbors consisted of 653 human proteins that interact directly with H1 proteins based on the Ingenuity Pathway Analysis (IPA) Interactions Knowledge Base (Ingenuity, Redwood City, CA), when including only direct relationships among proteins (i.e., excluding transcription regulation, protein-DNA interactions and protein-chemical relations). The statistical analysis of connection degrees around viral host proteins was calculated by permutation testing (n=10000). All reported p-values for network connections were adjusted for multiple testing using an FDR correction (p<0.05).
Functional annotations
All gene lists for 36 functional categories were downloaded from the IPA database. We also tested all 29 relevant categories specified by the GO Slim generic list (Ashburner et al., 2000) and augmented it with an additional seven immune-related Ingenuity categories. All p-values for functional enrichments were adjusted for multiple testing using an FDR correction (p<0.05).
Pathway analyses
We collected 646 cellular pathway gene sets, including the MSigDB (Subramanian et al., 2005) ‘canonical pathways’ collection and viral-response pathway gene sets from IPA., We grouped these into 212 distinct pathway groups (Figure S7) to avoid redundancies. We tested whether the H1 proteins, their curated first neighbors (H2), and each expression cluster were over-represented in the pathway gene set (a hyper-geometric enrichment test; the same result is obtained when using the non-curated H2 set, Figure S8). All p-values were adjusted for multiple testing using an FDR correction (p<0.05).
mRNA expression profiling
HBECs were stimulated with a 15 minute pulse of 1000U/ml IFNβ (PBL, Piscataway, NJ), 100ng/ml vRNA (purified directly from PR8 virus) with LTX transfection reagent (Invitrogen; Carlsbad, California), wild type H1N1 influenza (A/PR/8/34) or ΔNS1 virus (PR8 with a deleted NS1 gene, gift from Dr. Garcia-Sastre). Viruses were used at a multiplicity of infection (moi) of 5. Control samples were incubated with media or LTX under the same conditions. Cells were washed, supplemented with warm media and harvested at 11 timepoints (0, .25, .5, 1, 1.5, 2, 4, 6, 8, 12, and 18 hours post-treatment).
Array hybridizations and pre-processing
All array hybridizations and pre-processing were done using Affymetrix HT Human Genome U133 Arrays (see Supplementary Information). We defined the fold change of each gene in a stimulated sample as the average intensity of its probe set in two biological replicates, normalized by its expected intensity at the same time point without treatment. To calculate the expected intensities, genes were clustered based only on their intensities in the mock treatment (PCluster (Segal et al., 2003), n=30). For each time point, all genes within a cluster were compared to the same expected intensities (i.e. the average intensities of the particular mock time point across the entire cluster). To identify genes whose expression is affected by stimulation, we selected genes with ≥1.6-fold change in 2 consecutive time points (or ≥2-fold for each of the single time points, 12h or 18h) (Table S6). At this threshold, we expect a 10% error rate, estimated by the number of genes that cross the cutoff in mock LTX (false positives) vs. vRNA+LTX transfection (true positives; data not shown).
Clustering of expression data
We clustered the time-series gene expression data by a modification of the PCluster algorithm. We first applied PCluster (k=30) as previously described (Segal et al., 2003) and then iteratively improved the partition. There are two steps at each iteration: (1) splitting and (2) merging of clusters. The splitting step learns the best partition of each cluster into two clusters, through a search over every pair of consecutive time points. The query that best partitions the gene expression at two consecutive time points into two distinct distributions is chosen until no significant split exists (t-test p-value cutoff of 10-23). In the merging step, we merge pairs of clusters that are not significantly distinct in any two consecutive time points (same t-test cutoff). This fits our sparse temporal information, when many genes are only regulated in a few consecutive time points (e.g., C6,8). The 35 resulting clusters were grouped manually into 12 categories called ‘expression clusters’ (Figure 3A,B C1-C12). Expression cluster 1 (C1) includes those clusters that overlap but do not clearly fall into any of these categories.
siRNA transfection and stimulation of primary HBECs
3.5×103 HBECs (filtered through a 0.4μm filter) were seeded in wells of triplicate 96-well plates. 24 hours later, 25nM (final concentration) of siRNA duplexes (SMARTpools, Dharmacon) were transfected (HiPerFect, Qiagen) and incubated at 37°C for 3 hours, followed by a media change. Cells were incubated for 3 days with one media change at 24h post-transfection. Following knock-down, we used AlamarBlue (Invitrogen, Carlsbad, CA) to determine live cell numbers in all wells (in some replicate plates). Cells were then washed twice with complete media. To assess the effects of siRNAs on influenza virus replication, cells were inoculated with PR8 virus at a moi of 1. At 48 hours post-infection, HBEC supernatants were harvested and frozen with 5μg/ml TPCK trypsin. To assess effects on IFN production in response to vRNA and ΔNS1, cells were transfected with 100ng/ml vRNA or infected with ΔNS1virus at a moi 5. At 24 hours post-transfection or infection, HBEC supernatants were harvested for IFN assays.
Virus titering of HBEC supernatant
2×106 293T cells (filtered through a 0.4μm filter) were seeded in 10cm dishes and transfected with a vRNA luciferase reporter plasmid based on prior design (Lutz et al., 2005) using Transit-LT1. Cells were trypsinized at 24h post-transfection and 104 transfected reporter cells were re-seeded in white Costar plates. Supernatants (frozen with trypsin) from PR8-infected HBECs were added to reporter cells and incubated for 24 hours. Reporter activity was measured with firefly luciferase substrate (Steady-Glo, Promega, Madison, WI). Luminescence in 96-well plates was quantified using the Envision Multilabel reader (Perkin Elmer, Waltham, MA) fitted with an automated plate stacker.
Determining interferon production from HBEC supernatants
Cells were infected with a lentivirus containing ISRE-luciferase (Cignal Lenti ISRE Reporter, SA Biosystems, Frederick, MD). Following selection with puromycin, stably infected cells were cloned by limiting dilution and tested for responsiveness to human IFNβ (PBL, Piscataway, NJ). A clone with high signal to background ratio was selected and found to be sensitive to low levels of IFNβ (<1U/mL) with a >100× dynamic range. To measure IFNβ in supernatants from experimental assays, ISRE-Luc reporter cells were seeded in flat bottom white Costar plates at 2×104/well. 24 hours later, supernatants were added and assayed for ISRE-Luc inducing activity with firefly luciferase substrate.
Scoring of functional assays and identification of phenoclusters
For each assay, luminescence values were quantified using the Envision reader for ISRE-luc and vRNA-luc reporters. These values were normalized using robust Z-score normalization (RNAeyes program, A. Derr, Broad Institute) and averaged across three replicates. Robust Z-scores were further normalized to cell number per well by calculating the distance of each robust Z-score from the running average of robust Z-scores vs. AlamarBlue values. These cell number-normalized Z-scores, referred to as phenotype scores, reduce the impact of cell number variation on assay measurements and allow comparisons across wells and plates. Genes with a phenotype score equivalent to a 2-fold or more change in IFN or replication (compared to the median score for the relevant assay; see Supplementary Information) in each assay were analyzed further and clustered into 20 ‘phenoclusters’ with single-linkage hierarchical clustering using the Cluster software (Eisen et al., 1998). Statistical enrichments of phenotype scores in expression clusters or pathway gene sets were evaluated by permutation tests (n=10,000).
Supplementary Material
Acknowledgments
We thank D. Lieber and K. Maciag for discussions and help with data analysis. We thank C. Shamu and S. Chiang at ICCB (HMS) for the siRNA library and expert advice, S. Gupta and the Broad's Genetic Analysis Platform for microarray processing, H. Le, A. Derr, B. Wong and the staff at the Broad RNAi Platform for assistance with RNAi studies and analysis, R. Cadagan, A. Garcia-Sastre for ΔNS1 virus, E. Fodor for PR8 plasmids, R. Lamb for Udorn plasmids, and Sigrid Hart for assistance with artwork. Supported by NIH U01 AI074575 (NH, DR, DH), NERCE NIH program, and the NIH New Innovator Award (NH), Ford Foundation Predoctoral Fellowship (MG), EMBO Postdoctoral Fellowship (IGV), The Ellison Foundation, Boston, MA and NIH grant R01 HG001715 and P50 HG004233 (DH), the Howard Hughes Medical Institute, a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, the NIH Pioneer Award, and the Sloan Foundation (AR). Complete microarray data sets available at the Gene Expression Omnibus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer SM, Haas J. Connecting viral with cellular interactomes. Curr Opin Microbiol. 2009;12:453–459. doi: 10.1016/j.mib.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brander C, Walker BD. Modulation of host immune responses by clinically relevant human DNA and RNA viruses. Curr Opin Microbiol. 2000;3:379–386. doi: 10.1016/s1369-5274(00)00108-9. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JA, Ou SH, Wu F, Lusis AJ, Sparkes RS, Gaynor RB. Cloning and chromosomal mapping of a human immunodeficiency virus 1 “TATA” element modulatory factor. Proc Natl Acad Sci U S A. 1992;89:9372–9376. doi: 10.1073/pnas.89.20.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward SD, Liu J, Fujimuro M. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci STKE. 2006;2006 doi: 10.1126/stke.3352006re4. re4. [DOI] [PubMed] [Google Scholar]
- Hiscott J, Nguyen TL, Arguello M, Nakhaei P, Paz S. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene. 2006;25:6844–6867. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Min JY, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E, Seed B, Kieff E. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc Natl Acad Sci U S A. 2004;101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz A, Dyall J, Olivo PD, Pekosz A. Virus-inducible reporter genes as a tool for detecting and quantifying influenza A virus replication. J Virol Methods. 2005;126:13–20. doi: 10.1016/j.jviromet.2005.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RC, Vestal DJ, Xu Z, Bandyopadhyay S, Guo W, Erme SM, Williams BR, Sen GC. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Reichman TW, Parrott AM, Fierro-Monti I, Caron DJ, Kao PN, Lee CG, Li H, Mathews MB. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J Mol Biol. 2003;332:85–98. doi: 10.1016/s0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin E, Luke K, Jones J, Tumpey T, Konan K, Schultz-Cherry S. Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J Virol. 2005;79:8802–8811. doi: 10.1128/JVI.79.14.8802-8811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wressnigg N, Shurygina AP, Wolff T, Redlberger-Fritz M, Popow-Kraupp T, Muster T, Egorov A, Kittel C. Influenza B mutant viruses with truncated NS1 proteins grow efficiently in Vero cells and are immunogenic in mice. J Gen Virol. 2009;90:366–374. doi: 10.1099/vir.0.006122-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.