Abstract
Signaling pathways usually activate transcriptional targets in a cell type-specific manner. Notable exceptions are pathway-specific feedback antagonists, which serve to restrict the range or duration of the signal. These factors are often activated by their respective pathways in a broad array of cell types. For example, the Wnt ligand Wingless (Wg) activates the naked cuticle (nkd) gene in all tissues examined throughout Drosophila development. How does the nkd gene respond in such an unrestricted manner to Wg signaling? Analysis in cell culture revealed regions of the nkd locus that contain Wg response elements (WREs) that are directly activated by the pathway via the transcription factor TCF. In flies, Wg signaling activates these WREs in multiple tissues, in distinct but overlapping patterns. These WREs are necessary and largely sufficient for nkd expression in late stage larval tissues, but only contribute to part of the embryonic expression pattern of nkd. These results demonstrate that nkd responsiveness to Wg signaling is achieved by several WREs which are broadly (but not universally) activated by the pathway. The existence of several WREs in the nkd locus may have been necessary to allow the Wg signaling-Nkd feedback circuit to remain intact as Wg expression diversified during animal evolution.
Keywords: naked cuticle, feedback antagonist, Wnt, TCF, Wingless
Introduction
Throughout development, some signaling pathways are used in a reiterated manner to achieve the proper regulation of gene expression. For example, the Drosophila gene wingless (wg), which encodes a member of the Wnt family of secreted signaling proteins, has numerous essential roles in embryogenesis and larval stages (Couso et al., 1993; Klingensmith and Nusse, 1994). Because Wg signaling so profoundly affects cell fate, its expression is tightly controlled both spatially and temporally (Couso et al., 1993; Sanson, 2001). To achieve this complex pattern of expression, the wg locus contains multiple enhancers, each active in different tissues and controlled by various signal and local inputs (Costas et al., 2004; Lessing and Nusse, 1998; Neumann and Cohen, 1996; Pereira et al., 2006).
The Wg ligand is the trigger for an evolutionarily conserved signaling cascade that promotes nuclear accumulation of the fly β-catenin, Armadillo (Arm). Nuclear Arm binds to the transcription factor TCF, converting it from a transcriptional repressor to an activator of Wg targets (Parker et al., 2007; Stadeli et al., 2006). Consistent with its multitude of functions, Wg signaling regulates gene expression in a cell and tissue-specific manner. For example, in the embryo, epidermal Wg activates Engrailed (En) and Hedgehog expression in the epidermis (Sanson, 2001) but mesodermal Wg regulates a different set of targets in the visceral and cardiac mesoderm (Bilder and Scott, 1998; Riese et al., 1997). In wing imaginal discs, Wg signaling regulates a different set of targets (Cadigan, 2002). The specificity of Wg activation is thought to occur through combinatorial regulation with other factors or signaling pathways. For example, the Wg Response Element (WRE) responsible for cardiac expression of even-skipped (eve) is also directly regulated by Decapentaplegic (Dpp) and RAS signaling (Halfon et al., 2000; Knirr and Frasch, 2001; Han et al., 2002). A high degree of specificity is also observed in vertebrate cell culture, where microarray analysis revealed minimal overlap between Wnt targets in different cell types (Vlad et al., 2008).
Many developmental signaling pathways activate the expression of feedback antagonists, which limit their range of action. These include naked cuticle (nkd) and notum/wingful for Wg signaling (Gerlitz and Basler, 2002; Giraldez et al., 2002; Zeng et al., 2000), as well as patched, daughters against dpp and argos for the Hedgehog, Dpp and Epidermal Growth Factor signaling pathways, respectively (Alexandre et al., 1996; Golembo et al., 1996; Tsuneizumi et al., 1997). In contrast to the cell-specific nature of most target genes, these feedback antagonists are ubiquitously activated by their respective signaling pathways regardless of cell type. The regulatory cis-acting sequences controlling these universal responders have not been characterized. Do universal enhancers activate expression of these antagonists in all cell types, or do these targets require multiple tissue-specific enhancers? To address this question, we examined the regulation of nkd by Wg signaling.
nkd has all the essential features of a signal-induced feedback antagonist. Its expression requires Wg signaling and it is often expressed in a slightly broader pattern than Wg, owing to the ability of secreted Wg to diffuse to neighboring cells (Zeng et al., 2000). nkd encodes an EF hand protein that is thought to antagonize Wg signaling through binding to Dishevelled, a protein which mediates Wg-dependent stabilization of Arm (Rousset et al., 2001). Loss of nkd in fly embryos results in elevated Arm levels and ectopic activation of Wg signaling, causing a dramatic reprogramming of epidermal cell fate (Waldrop et al., 2006; Zeng et al., 2000).
Our lab previously identified a WRE in the first intron of the nkd gene that is bound by TCF and activated by Wg signaling in cell culture (Fang et al., 2006; Li et al., 2007; Parker et al.,2008). In this report, we used chromatin immunoprecipitation (ChIP) to identify a region upstream of the nkd gene that is also bound by TCF. This region contains two overlapping WREs (UpE1 and UpE2). Mutagenesis of TCF binding sites in these WREs and the intronic WRE (IntE) demonstrate direct regulation by the pathway through TCF. Each of these WREs is activated by Wg signaling in patterns very similar to that of nkd transcripts. The WREs are active in broad, partially overlapping patterns in larval imaginal discs, and the sum of the three can largely account for the entire nkd pattern in late third instar larva. In contrast, these WREs only partially recapitulate the embryonic nkd pattern. When deleted from the endogenous nkd locus, loss of IntE has a minimal effect on nkd expression whereas loss of a region containing UpE1/UpE2 displays a dramatic reduction in imaginal disc expression. However, neither deletion affected embryonic expression of nkd. Our data demonstrate that multiple WREs are needed for nkd to respond to Wg signaling in all tissues. The overlapping specificity of these WREs may provide robustness to the Wg-Nkd feedback circuit in the various cells where it operates. In addition, multiple nkd WREs may have been required for the activation of nkd to be maintained as the expression of the Wg ligand became more elaborate during animal evolution.
Materials and Methods
Drosophila cell culture
Kc167 (Kc) and S2R+ cells were cultured in the Schneider’s Drosophila media (Invitrogen) containing 5% or 10% FBS at 25°C, respectively. Clone8 cells were cultured as described (http://flyrnai.org/RNAi_index.html).
RNAi, Wg-conditioned media (Wg-CM) treatment and qRT-PCR
Double-stranded RNA (dsRNA) corresponding to control, Axin, arm, and TCF was synthesized as described (Fang et al., 2006). The Wg pathway in cultured cells was activated by depleting Axin or adding Wg-CM as described (Chang et al., 2008; Fang et al., 2006; Li et al., 2007). Briefly, 1 × 106 Kc, S2R+ or Clone 8 cells were seeded in 12-well plates and 10 ug of Axin or control dsRNA were added to each well. When Axin dsRNA was combined with another RNA duplex, 10 ug of each dsRNA was used. Cultures were incubated with dsRNAs for 6 days before harvesting for qRT-PCR analysis.
Wg-CM was prepared using stable pTub-wg S2 cells, kindly provided by Dr Roel Nusse from Stanford University. Wg-CM was collected from dense cultures (typically 7–10 million cells/ml) lacking hygromycin. Usually 200 ul of unconcentrated Wg-CM was added to 600 ul cell suspensions containing 1–3 million cells. As a control, media collected from S2 cells was used. Cells were treated for 5 hours with control media or Wg-CM before harvesting for qRT-PCR analysis.
After the treated cells were collected, total RNA was isolated with Trizol reagent (Invitrogen) and cDNA synthesis was performed using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocols. Additional details of the qRT-PCR and the primer sequences used are described (Fang et al., 2006).
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed essentially as described (Fang et al., 2006) except that the dimethyl 3,30-dithio-bis(propionimidate) dihydrochloride treatment was eliminated. Typically 3 ×106 cells and 5–10 ul of TCF antibody were used per ChIP and all precipitated DNA samples were quantified by qPCR. Data are expressed as the percent of input DNA. The specific primer pairs for UpE, IntE and ORF correspond to #N1, #N5, and #N0 primer sets described previously (Fang et al., 2006).
Plasmids
Luciferase reporter constructs containing various nkd WREs were made by incorporating MluI/XmaI PCR fragments into a pGL3-Basic vector (Promega) containing the Drosophila hsp 70 minimal promoter. hsp 70 promoter was cloned into pGL3-Basic via PCR using 5’ ATCTCGAGCTCGAGATCTGAGCGCCGGAGT3’ (XhoI site is underlined) and 5’ATAAGCTTAAGCTTCCCAATTCCCTATTCAGAGTTCTC3’ (HindIII site is underlined) primers. The specific primers to amplify the UpE and IntE genomic fragments are the following: UpE, 5’TCCTACGCGTGGCTGGGCTCGATGCAGATAA3’ and 5’AATTCCCGGGGG GCCGCTGTCGGCCAACTG3’; UpE1, 5’TCCTACGCGTGGCTGGGCTCGATGCAGA TAA3’ and 5’GGTGCCCGGGTTTGTAGTTTGCGGTGGT3’; UpE2, 5’AATTACGCGTCAG GAGGTCTGCCAACTTAAGTAG3’ and 5’AATTCCCGGGGGGCCGCTGTCGGCCAAC TG3’; IntE (869 bp) 5’TTAGACGCGTGCTCTCGGGCCAC3’ and 5’CCAGCCCGGG TTCCTCAAAGCAACC3’; IntE (255 bp) 5’GCCACGCGTATAGTTTGTGTATAGTT3’ and 5’CCCAGCCCGGGTTCCTCAAAGCAACC3’. The MluI (ACGCGT) and XmaI sites (CCCGGG) are underlined. Deletions of the WREs (UpE #1, #2, #3, and IntE 558bp) were made by standard PCR cloning or subcloning.
TCF binding sites in the reporter constructs were destroyed using quick change site-directed mutagenesis (Stratagene). Base substitutions were A to C or T to G (or vice versa). For UpE1 and UpE2 all eight nucleotides of the TCF binding site (SSTTTGWW) were substituted while the 4th, 6th and 7th positions were altered in IntE.
For analysis in fly tissues, WREs were cloned into the pH-Pelican lacZ reporter (Barolo et al., 2000) and introduced into the fly genome by P-element transgenesis (Bestgene Inc.). The combined nkd WRE has a 1084 bp UpE fragment upstream of a 869 bp IntE fragment.
Transfection and reporter gene assays
Transient transfections and reporter assays were done essentially as previously described (Fang et al, 2006, Li et al., 2007). Briefly, a mixture of plasmids containing 100 ng luciferase reporter, 5 ng pAclacZ (Invitrogen) and 100 ng of pAc-Arm* (Fang et al., 2006) were co-transfected with 1 × 106 cells. The pAc-Arm* is a derivative of pAc5.1 expression vector (Invitrogen) encoding a constitutively active form of Arm which has Thr52, Ser56 substituted with Ala (Freeman and Bienz, 2001). The empty pAc5.1 vector was used to normalize the DNA content or as controls. Cells were harvested 3 days after transfection for further reporter assays.
Luciferase and β-galactosidase activities were assayed using the Tropix Luc-Screen and Galacto-Star kits (Applied Biosystems) and quantified with a Chameleon plate luminometer (Hidex Personal Life Science). Transfection efficiency was normalized using the pAclacZ β-galactosidase activities. When Wg–CM was used to activate Wg signaling instead of co-expression of Arm*, cells were transfected with the same amount of reporter and pAcLacZ and incubated for 2 days before the cells were treated with Wg-CM for 24 h prior to harvesting for the reporter assays.
Drosophila genetics
Fly stocks were maintained on standard medium at 25°C unless otherwise indicated. The P[En-Gal4] and P[Dpp-Gal4] are as described (Li et al., 2007). The dominant negative TCF transgene (P[UAS-TCFDN]) and constitutively active arm (P[UAS-armS10]) were obtained from M. Peifer (Pai et al., 1997; van de Wetering et al., 1997). wgCX4 is a molecular null (van den Heuvel et al., 1993). Df(3L)ED4782, a large (175 kb) deficiency lacking the entire nkd locus and a hypomorphic allele nkdl(3)4869 (Zeng et al., 2000) were obtained from Bloomington Stock Center. Homozygous Df(3L)ED4782 embryos and Df(3L)ED4782/nkdl(3)4869 transheterozygotes display loss of cuticular denticles characteristic of nkd loss-of-function. Experiments with En-Gal4 and Dpp-Gal4 were carried out at 25°C.
A 13 kb deletion lacking UpE (ΔUpE) was generated by mitotic recombination using hsFLP and PBac{RB}e00194 and P{XP}d09466 chromosomes (Parks et al., 2004). Two transposon insertions, PBac{RB}e00194 and P{XP}d09466 (obtained from the Exelixis stock center, Harvard Medical School) were outcrossed to w1118 flies for three generations before isogenization, removing at least one linked lethal from each line. In the dysgenic cross, males with darker eye color than any of single transposon line were obtained (see Supplemental Fig. 5) and molecular mapping with PCR confirmed the deletion. The ΔUpE allele is homozygous semi-lethal but ΔUpE/Df(3L)ED4782 transheterozygotes are viable and fertile. This indicates that the semi-lethality of homozygous ΔUpE is due to a linked mutation(s).
A 3 kb genomic deletion removing IntE was created by imprecise excision of the P[KG0529] transposon (obtained from Bloomington Stock Center) as described previously (Zhou et al., 2003). The P[KG0529] line was outcrossed to w1118 flies for three generations before isogenization. The deletion was characterized using PCR and the relevant PCR bands were sequenced to confirm the deletion breakpoints.
Immunostaining, in situ hybridization and microscopy
Immunostaining and in situ hybridization of fly embryos and imaginal discs were performed as described previously (Lin et al., 2004; Parker et al., 2002), using rabbit anti-LacZ (1:500) (Abcam Inc.), guinea-pig anti-Sens (1:500) (Fang et al., 2006) and mouse anti-Wg antisera (1:100) (Developmental Studies Hybridoma Bank at the University of Iowa). Cy3- and Alexa 488-conjugated secondary antibodies were from Jackson Immunochemicals and Molecular Probes, respectively. Samples were examined using Leica confocal microscope DM6000B-CS (Leica) and processed in Adobe Photoshop 8.0.
Probes for in situ hybridization of nkd transcripts were made by PCR of genomic DNA with the following oligos: 5’GAATTAATACGACTCACTATAGGGAGAGCTGCTGGTC AGCGAACGTGACAATAA3’ and 5’GAATTAATACGACTCACTATAGGGAGACAGACC CGTGGGCAACTTCTTCAGTTT3’. Underlined sequences are T7 promoter sites. Antisense dioxygenin probes were synthesized using the Ambion T7 Megascript kit with the Roche DIG RNA labeling mix. Samples for in situ analysis were photographed with a Nikon Eclipse800 compound microscope using DIC optics.
Quantification of nkd transcripts in wing imaginal discs
20 wing imaginal discs were collected from late 3rd instars of transheterozygotes +/Df(3L)ED4782, ΔUpE/Df(3L)ED4782, and ΔIntE/Df(3L)ED4782 flies. After pelleting, the discs were homogenized with 1.5 pellet pestles (South Jersey Precision Tool and Mold Inc.) in 200 µl Trizol reagent (Invitrogen). After addition of another 300 µl of Trizol, the samples were processed according to the manufacturer’s protocols. Total RNA was resuspended in 10 ul of RNAase-free water and 2 ug of total RNA was used to synthesize cDNA using Superscript III (Invitrogen) according to the manufacturer’s protocol. nkd and β-tubulin 56B transcripts were measured by qRT-PCR with primers used for cell culture experiments (Fang et al., 2006). The level of nkd transcript was normalized by the level of β-tubulin 56B. The value for nkd transcripts from +/Df(3L)ED4782 flies was normalized to 1 and the relative level of transcripts in ΔUpE or ΔIntE was determined. The error represents the standard deviations from four independent experiments.
Results
The nkd locus contains several WREs that are directly activated by Wg signaling in cultured cells
As previously reported (Fang et al., 2006; Li et al., 2007; Parker et al., 2008), expression of nkd is induced in Drosophila Kc167 (Kc) cells upon stimulation with Wg-conditioned media (Wg-CM) or RNAi depletion of Axin. RNAi knockdown of either TCF or arm significantly reduced this nkd induction (Fig. 1A, B). When the nkd transcription unit and surrounding DNA (see cartoon in Fig. 1C) were assayed for TCF binding via ChIP, TCF was highly enriched in the region containing an intronic WRE (IntE) and this binding was enhanced upon Wg-CM treatment (Fang et al., 2006; Parker et al., 2008). Thus, IntE is likely to be a major WRE in mediating Wg-dependent activation of nkd expression in Kc cells.
Fig. 1. TCF is recruited to two regions in or near the nkd transcription unit upon Wg stimulation of fly Kc cells.
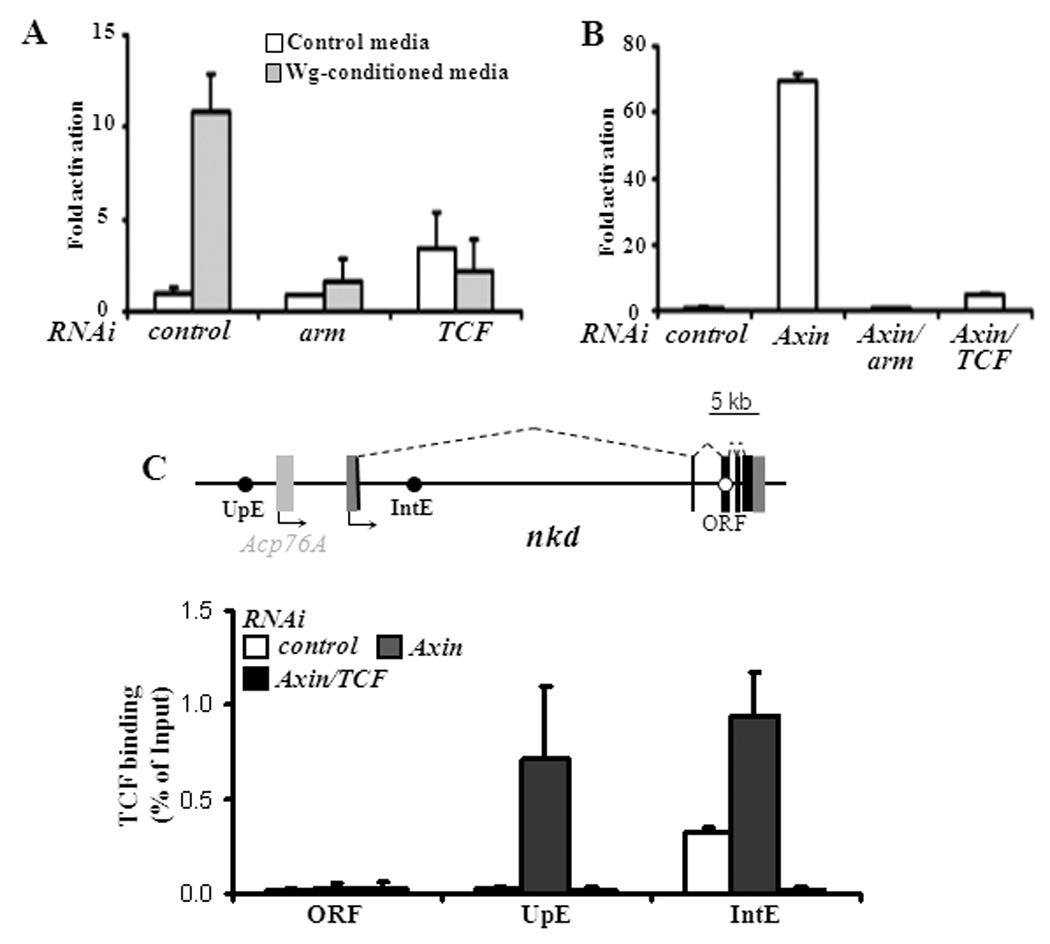
(A) Stimulation with Wg-CM for 5 hours resulted in elevated nkd expression, which was greatly reduced by RNAi depletion of arm or TCF. (B) Activation of Wg signaling by RNAi depletion of Axin for 6 days caused a huge increase in nkd transcript levels in an Arm and TCF-dependent manner. Transcript levels were determined by qRT-PCR as described in Materials and Methods. Fold activation is relative to nkd expression in cells treated with control RNAi and control media in panel A and control RNAi alone in panel B. Each bar is the mean of triplicates from cultures at each condition, with the standard deviation indicated. (C) Schematic of the nkd locus showing the location of three regions (UpE, IntE and ORF) assayed for TCF occupancy. Acp76A is not expressed at detectable levels in Kc cells. (D) Binding of TCF in Kc cells with the indicated RNAi treatments. In the absence of Wg signaling (control RNAi), TCF is bound to the region containing IntE but not UpE or ORF. In cells where the pathway is activated (Axin RNAi), there is strong binding to both UpE and IntE. TCF binding is dramatically lowered in Axin, TCF depleted cells, indicating that the ChIP signal is specific for TCF. Each bar represents the mean of duplicate ChIP samples with the standard error indicated. The data shown are representative examples from more than three separate experiments.
Occasionally, more modest TCF binding was also observed ~10 kb upstream of the nkd transcriptional start site (TSS) in Wg-CM treated Kc cells (Parker et al., 2008). TCF binding to this upstream region (UpE) was much more pronounced when cells were treated with Axin RNAi, reaching levels similar to those seen at IntE (Fig. 1D). Cells simultaneously depleted for Axin and TCF lost TCF binding at both locations, demonstrating the specificity of the TCF antisera (Fig. 1D). As observed previously (Fang et al., 2006; Li et al., 2007; Parker et al., 2008), no significant binding of TCF was found at the ORF (Fig. 1D).
To test whether UpE is a functional WRE, a genomic fragment (1084 bp) containing this region was cloned into a hsp 70 core promoter/luciferase reporter. Such reporters can be assayed for Wg responsiveness by cotransfection with a stabilized form of Arm (Arm*) (Fang et al., 2006; Parker et al., 2008). This UpE fragment was activated almost 1000-fold by co-expression with Arm* (Fig. 2A), indicating that it possesses a high level of WRE activity.
Fig. 2. Dissection of the UpE and IntE regions reveals WREs that contain functional TCF binding sites.
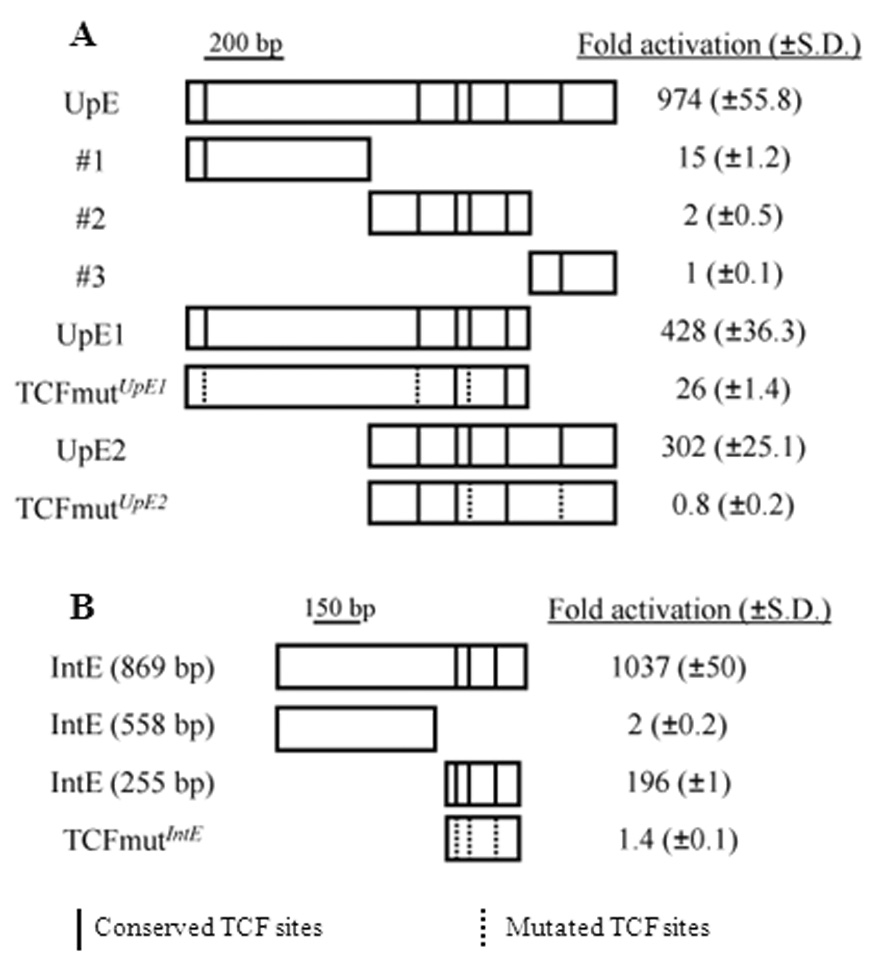
(A) When cloned upstream of a hsp70 core promoter/luciferase reporter, UpE activated luciferase expression when co-expressed with Arm*. UpE was divided into three fragments (#1 – 3), none of which were highly responsive to Arm*. However, two overlapping stretches (UpE1 and UpE2) possessed strong WRE activity. Vertical lines in the boxes represent the predicted TCF sites that are conserved in 12 Drosophila species, while the dotted lines denote mutated TCF sites. UpE1 and UpE2 both require a subset of the TCF sites for Arm* responsiveness. (B) The IntE genomic region contains a 255 bp WRE that requires three TCF binding sites for Arm* responsiveness. Fold activation is relative to luciferase expression without Arm* expression for each reporter construct. Each result is the mean of triplicate transfections, with the standard deviation indicated in parenthesis. The data shown are a representative example from more than three separate experiments.
To further localize the WRE activity in UpE, it was split into three parts and tested in reporter assays. None of the smaller fragments (#1, #2 or #3) had high WRE activity (Fig. 2A). However, regions containing the 5’ (UpE1) or 3’ (UpE2) two thirds of UpE were dramatically activated by Arm* (Fig. 2A). These data suggest that the UpE region contains two WREs, albeit with overlapping sequences.
UpE1, UpE2 and IntE all contain multiple predicted TCF binding sites (Fig. 2) that are conserved among the sequenced Drosophila species, including the distantly related D. virilis (Supplemental Fig. 1B, C). Examination of the nine conserved TCF sites reveals a consensus of SCTTTGW (S = G or C; W = A or T) very similar to the preferred binding site of fly TCF (CCTTTGAT) (van de Wetering et al., 1997). In the 869 bp IntE previously identified (Fang et al., 2006), all three sites are clustered at the 3’ end of the fragment (Fig. 2B). A 255 bp fragment containing all three TCF sites still possessed a high level of WRE activity, though less than that of the 869 bp fragment (Fig. 2B).
To determine whether the conserved TCF sites in these WREs are functional, they were destroyed by site-directed mutagenesis. Individual mutation of the five TCF sites in UpE2 demonstrated that three of the five contributed to WRE activity (Supplemental Fig. 2A). Individual mutations in any of the three TCF sites in IntE (255 bp) reduced WRE activity (Supplemental Fig. 2B). Simultaneous mutation of three TCF sites in UpE1 resulted in a large reduction in Wg responsiveness (Fig. 2A). More emphatically, mutation of two TCF sites in UpE2 or all three sites in IntE completely abolished the activity of these WREs (Fig. 2A, B). Together with the ChIP data, the mutagenesis results demonstrate that these WREs are directly activated by TCF-Arm in Kc cells.
UpE1, UpE2 and IntE are highly responsive to Arm* in Kc cells (Fig. 2), which are derived from embryonic hemocytes (Goto et al., 2001). These WREs are also activated by Arm* in two other fly cell lines, S2R+ and Clone8 (Supplemental Table 1), derived from embryonic hemocytes and wing imaginal disc epithelia cells, respectively (Peel et al., 1990; Yanagawa et al., 1998). In addition, these WREs were also highly activated by Wg-CM treatment (Supplemental Table 1).
The TCF binding and reporter gene data suggest that the UpE1 and UpE2 act as WREs for the nkd locus but it is also possible that they activate other nearby genes. To explore this, three genes that are upstream of the nkd TSS (mkp3, CG3797 and Acp76A) and two downstream of the 3’ end of the gene (CG18136 and CG3808) were tested for Wg responsiveness. The only gene whose expression was altered by Axin RNAi was CG3808, which showed a 1.8 fold increase (nkd was activated 56 fold in the same experiment; data not shown). This suggests that UpE1 and UpE2 mediate transcriptional activation of the nkd gene by Wg signaling.
The nkd-WREs identified from cultured cells are also directly activated in vivo by Wg signaling
Wg is expressed in many embryonic tissues (e.g., Fig. 3A, F) and larval imaginal discs (e.g., Fig. 3K, P, U). In all tissues examined, nkd transcripts are found in patterns similar to that of Wg (e.g., Fig. 3E, J, O, T, Y). In some tissues the nkd pattern is broader than that of Wg (compare Fig. 3A and U with Fig. 3E and Y), consistent with non-autonomous activation by the secreted Wg ligand (Zeng et al., 2000).
Fig. 3. nkd-WRE reporter transgenes are expressed in partially overlapping patterns similar to nkd transcript distribution.
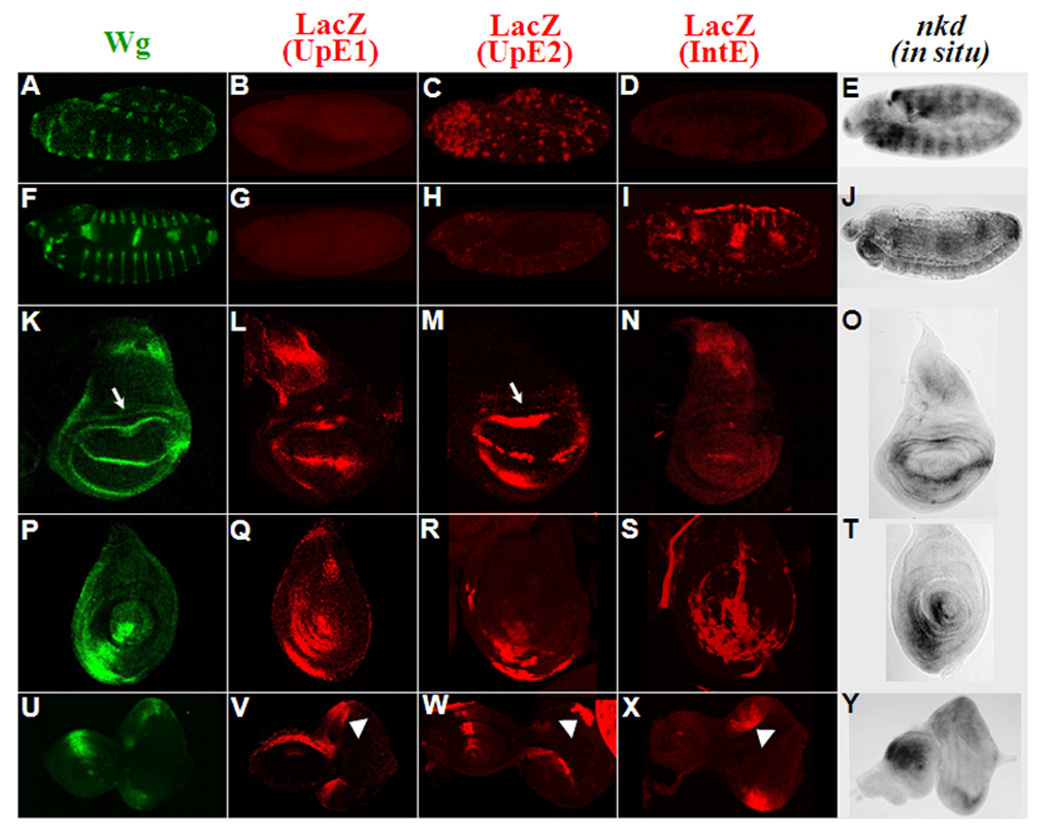
The patterns of Wg protein are shown in green, the LacZ patterns of UpE1, UpE2 and IntE reporter transgenes shown in red while nkd transcripts are in grey. Patterns are shown for embryos at stage 11 (A–E), and stage 14 (F–J). Late third instar wing (K–O), leg (Q–S) and eye-antennal (U–Y) imaginal discs are also shown. Arrows indicate the proximal ring of Wg expression (K) and faint lacZ expression in the same region of the UpE2 reporter (M). LacZ expression in eye imaginal discs is marked with arrowheads (V–X). The WRE reporters show overlapping expression domains containing a subset of the endogenous nkd pattern. Three independent lines of each WRE reporter showed similar results.
To determine whether the WREs from the nkd locus reflect the endogenous nkd pattern in fly tissues, nkd-UpE1, UpE2 and IntE (the 869 bp fragment) were cloned into the pH-Pelican lacZ reporter (Barolo et al., 2000) and introduced into the fly genome by P element transgenesis. All three reporters were expressed in patterns reminiscent of nkd transcript distribution. UpE1 was active in multiple imaginal discs (Fig. 3L, Q, V) but displayed no activity during embryogenesis (Fig. 3B, G). UpE2 partially recapitulates the epidermal striped nkd pattern during germband extension (Fig. 3C) but was not expressed in older embryos (Fig. 3H). UpE2 also has activity in several imaginal discs (Fig. 3M, R, W). IntE did not express LacZ reporter at germband extension (Fig. 3D), but was active in the mesoderm and endoderm in older embryos (Fig. 3I). This reporter also displayed activity in late third instar imaginal discs (Fig. 3N, S, X). In summary, each reporter recapitulated part of the endogenous nkd pattern.
The expression patterns of the nkd-WREs in various fly tissues are summarized in Table 1. In the larval eye disc, the activities of all three reporters are very similar (Fig. 3V–X). However, in all other tissues the WREs display a fair degree of specificity. While all three WREs were active in the leg discs, UpE1 activation was largely restricted to the columnar epithelia of the disc (Fig. 3Q) while IntE was mostly active in the peripodial membrane (Fig. 3S). UpE2 was active in both cell types (Fig. 3R). In the wing pouch, hinge region and antennae primordia, UpE1 and UpE2 are active (though UpE2 is significantly stronger) while IntE is very weak or not detected (Fig. 3L–N, V–X). Conversely, in older embryos, IntE is active in several mesodermal and endodermal tissues (Fig. 3I) similar to nkd (Fig. 3J) but UpE1 and UpE2 have no activity (Fig. 3G, H). In the notum, UpE1 is most active (Fig. 3L), IntE has intermediate expression (Fig. 3N) while UpE2 is not active (Fig. 3M). These data demonstrate that these WREs have partially overlapping patterns, but that they are also selectively used in many tissues.
Table 1.
Summary of lacZ expression of the UpE1, UpE2 and IntE nkd-WRE reporters. The relative strength of expression in several tissues is indicated. All imaginal discs are from late third instar larva. See text for more details.
| nkd-WREs | |||
|---|---|---|---|
| Tissues | |||
| UpE1 | UpE2 | IntE | |
| Early embryos (stage 10–12) | − | + | − |
| Late embryos (stage 13–14) | − | − | + |
| Eye imaginal discs | + | + | + |
| Antennal imaginal discs | + | ++ | − |
| Leg imaginal discs (epithelia) | ++ | + | +/− |
| Leg disc (peripodal membrane) | +/− | + | ++ |
| Wing disc (notum) | + | − | + |
| Wing disc (D/V boundary) | ++ | ++ | + |
| Wing disc (hinge-distal ring) | + | ++ | − |
All the lacZ expression patterns described for the nkd-WREs are consistent with activation by Wg signaling. In several cases this was demonstrated experimentally. When the IntE reporter was crossed into a wg null mutant background, the expression pattern was lost in several tissues (compare Fig. 4B with 4A), except for the activity at the leading edge of the migrating dorsal epithelia (arrow in Fig. 4B). Wg regulation of these reporters in the wing imaginal disc was tested by expressing a dominant-negative form of TCF (TCFDN) in the posterior part of the wing pouch, via en-Gal4. TCFDN is known to potently inhibit Wg signaling (van de Wetering et al., 1997), as exemplified by inhibition of Sens (arrowheads in Fig. 4L; compare to Fig. 4K), a known Wg target (Parker et al., 2002). UpE1 and UpE2 expression was markedly reduced by TCFDN (arrowheads in Fig. 4F and 4I). Conversely, expression of an active form of Arm(ArmS10) (Pai et al., 1997) via dpp-Gal4 caused a dramatic increase in UpE1 expression in the wing pouch (arrows in Fig. 4G). Similar activation by ArmS10 was observed for UpE2 and IntE (data not shown). In all cases examined, loss of Wg signaling dramatically reduced nkd-WRE activity while activation of the pathway increased reporter expression.
Fig. 4. The nkd-WRE reporters are positively regulated by Wg signaling.
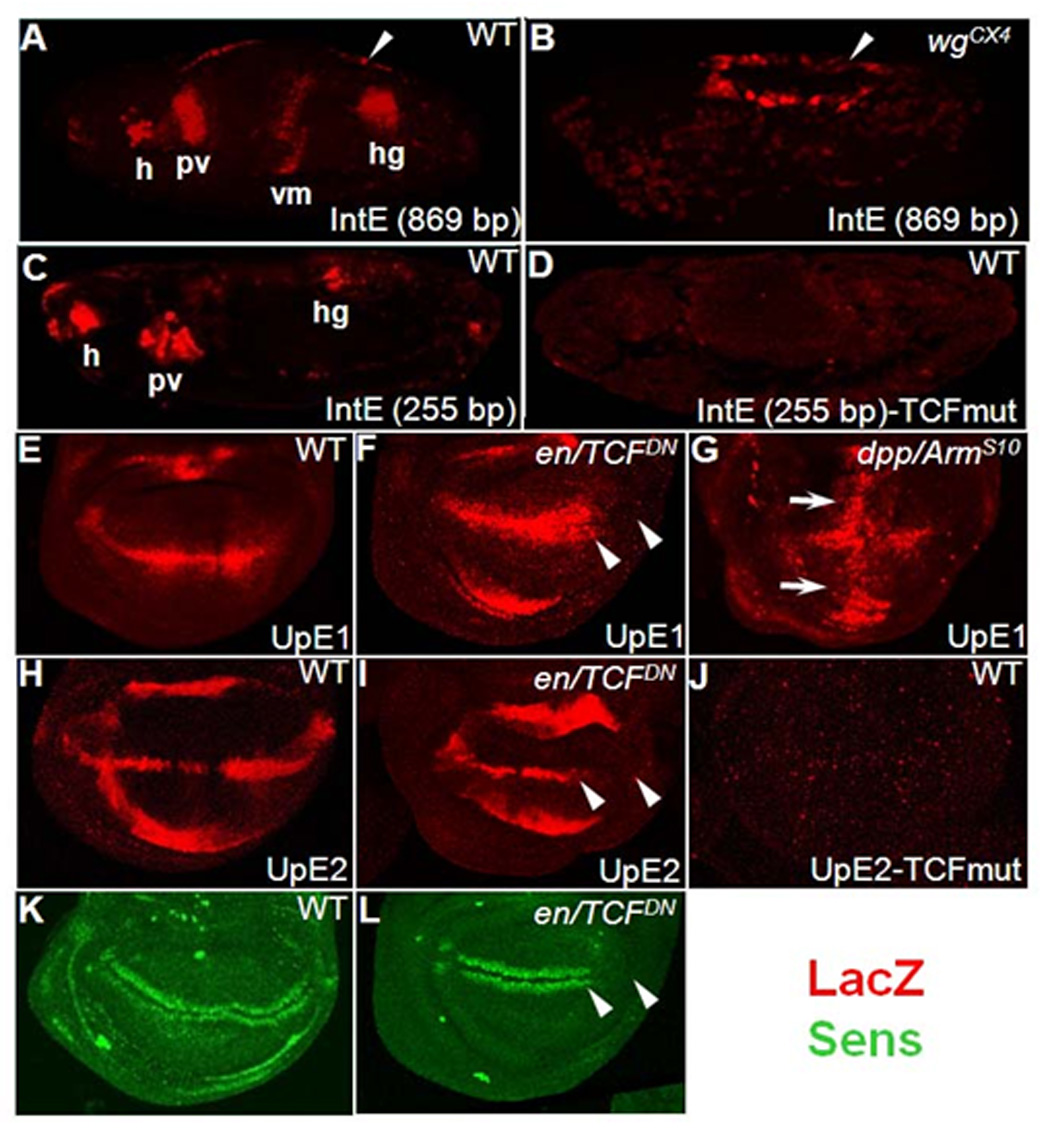
(A) Stage 14 embryo containing the nkd-IntE reporter stained for LacZ (red). Several expression domains consistent with positive regulation by Wg signaling are evident, including regions of the head (h), proventriculus (pv), visceral mesoderm (vm) and hindgut (hg). (B) IntE pattern in a wg mutant embryo. Most of the pattern is absent, except for the dorsal domain indicated by white arrowheads. (C) Stage 14 embryo with a smaller (255 bp) IntE reporter shows staining in a subset of the larger fragment. (D) IntE (255 bp) staining is abolished when the three TCF binding sites indicated in Fig. 2B are mutated. (E–L) The nkd-UpE1 and UpE2 WREs require Wg signaling in late third instar wing imaginal discs. (F, I, L) Expression of a dominant negative form of TCF (TCFDN) in the posterior compartment of the wing pouch (via En-Gal4; marked with arrowheads). TCFDN inhibits expression of UpE1 (F), UpE2 (I) and the Wg readout Sens (L). The broader expression of lacZ in the anterior compartment of UpE1 discs (F) is likely due to distortion of disc morphology caused by TCFDN expression. The slightly elevated expression of lacZ in the posterior compartment of UpE2 discs (H) is not always observed (see Fig. 3M). (G) Expression of a stable form of Arm (ArmS10) along the anterior/posterior boundary of the wing pouch (via Dpp-Gal4; white arrows) results in marked expansion of nkd-UpE1 expression. The decrease in lacZ expression at the dorsal/ventral boundary is likely due to distortion of patterning in the Dpp/ArmS10 discs. (J) Mutation of the two TCF binding sites in UpE2 (the same ones indicated in Fig. 2A) abolishes reporter expression in the pouch and hinge regions of the wing imaginal discs. For the TCF mutant constructs, three independent lines were examined with identical results as those shown.
To demonstrate that UpE2 and IntE are directly regulated by TCF in fly tissues, the functional TCF sites identified in cell culture (Fig. 2A, B) were destroyed in the WRE-lacZ reporters. In the case of IntE, a shorter (255 bp) transgene was the starting point. IntE (255 bp) has an identical pattern to the longer IntE (869 bp) but the expression is less robust in most tissues (Fig. 4C and data not shown). Mutation of the three TCF sites in IntE (255 bp) abolished the expression of LacZ in stage 13 embryos (Fig. 4D) and various imaginal discs (data not shown). In the case of UpE2, altering two TCF sites drastically reduced lacZ expression in the wing disc (Fig. 4J) and all other imaginal discs examined (data not shown). Thus, IntE and UpE2 are directly activated by Wg signaling in several fly tissues.
UpE and IntE are not sufficient to recapitulate the embryonic nkd pattern
The patterns of UpE1, UpE2 and IntE appear to cover most of the endogenous nkd pattern in the imaginal discs of late third instar larva. However, the sum of these WREs accounts for only part of the endogenous embryonic pattern of nkd (Fig. 3). To test whether the UpE region and IntE might act synergistically in the embryo, transgenic animals containing the entire UpE (1084 bp) and IntE (869 bp) cloned into the pH-Pelican vector (Fig. 5A) were created and monitored for lacZ expression.
Fig. 5. The combination of UpE and IntE largely recapitulates the endogenous nkd pattern in late larval third instar wing imaginal discs but not in embryos.
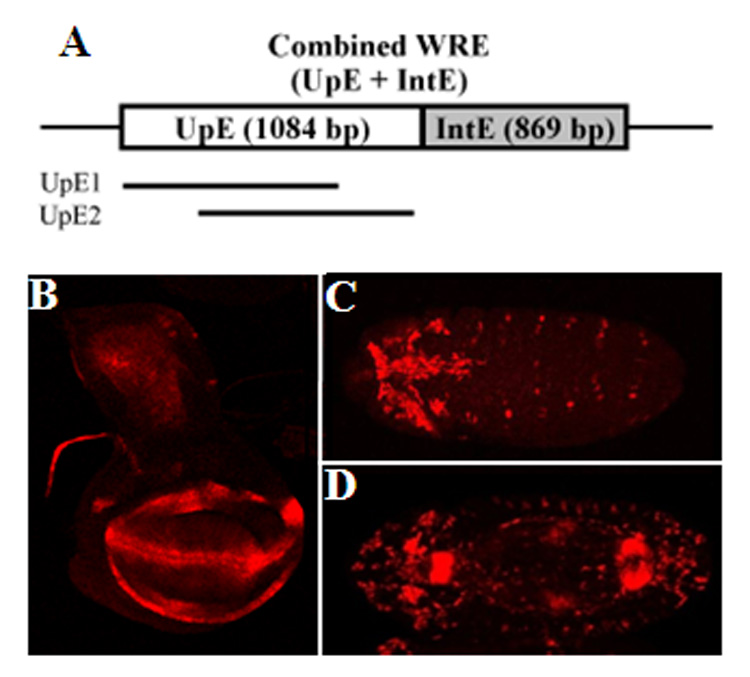
(A) The combined WRE contains the overlapping regions of UpE1 and UpE2 upstream of the 869 bp IntE in the pH-Pelican lacZ vector. (B) The expression of the combined WRE reporter in the wing disc is very similar to the endogenous nkd pattern (Fig. 3O) except for the proximal ring in the hinge (arrow in Fig. 6C). The combined reporter appears to be the sum of the three individual WRE reporters in regard to spatial pattern (compare with Fig. 3L –N). In regard to expression level, the combined reporter is greater in the wing hinge and pouch. The decrease gain of the confocal lasar in this image makes the notum staining less apparent (compare Fig. 5B to Fig 3L). (C, D) Ventral views of stage 11 (C) and stage 14 (D) embryos. The pattern is additive of the UpE2 and IntE WREs (Fig. 3C, I and Fig. 4A) and does not fully recapitulate the endogenous nkd pattern (Fig. 3E, J).
In the wing imaginal discs, the combined WRE reporter was active in all the locations observed with the individual UpE1, UpE2 and IntE reporters (Fig. 5B). However, the pattern was slightly more than the sum of the three individual WREs. For example, the combined reporter had elevated lacZ expression on either side of the Wg stripe at the dorsal/ventral boundary (Fig. 5B). In addition, the level of expression in the wing pouch and hinge region was consistently elevated in the combined reporter. Because of the elevated lacZ expression in this portion of the wing disc, the expression in the notum appears weaker in the combined reporter compared to the UpE1 reporter (compare Fig. 5B to Fig 3L). However, the level of expression of both reporters in the notum appear roughly similar (data not shown). The expression of the combined WRE reporter in the eye/antennal and leg imaginal discs is similar to that of endogenous nkd and appears to be the sum of the three individual WRE reporters (data not shown).
In embryos, the combined WRE reporter was active in weak stripes at germband extension (Fig. 5C), a pattern similar to UpE2 (Fig. 3C). In older embryos, the pattern of the combined WRE reporter (Fig. 5D) was similar to that found in IntE embryos (Fig. 3I). The additive nature of the expression patterns indicates no detectable cooperative interactions between UpE and IntE, suggesting that at least one other WRE exists in the nkd locus which might account for the missing embryonic pattern.
The data obtained from reporter assays strongly support the model that UpE1, UpE2 and IntE are bona fide WREs of nkd. To determine whether UpE or IntE were required for expression of endogenous nkd, two deletions in the gene were created. Using transposons inserted in the locus that contain Flp recombinase recognition sites (FRTs) (Parks et al., 2004), a deletion removing approximately 13 kb of sequence upstream of the nkd TSS (ΔUpE; Fig. 6A) was engineered (see Materials and Methods and Supplemental Fig. 3 for more details). Imprecise excision of a P-element in IntE was used to generate the ΔIntE allele, which lacks approximately 3 kb of intronic sequence including IntE (Fig. 6B).
Fig. 6. Deletion of IntE does not affect nkd expression, but a large deletion removing UpE reduces nkd expression in wing and leg imaginal discs.
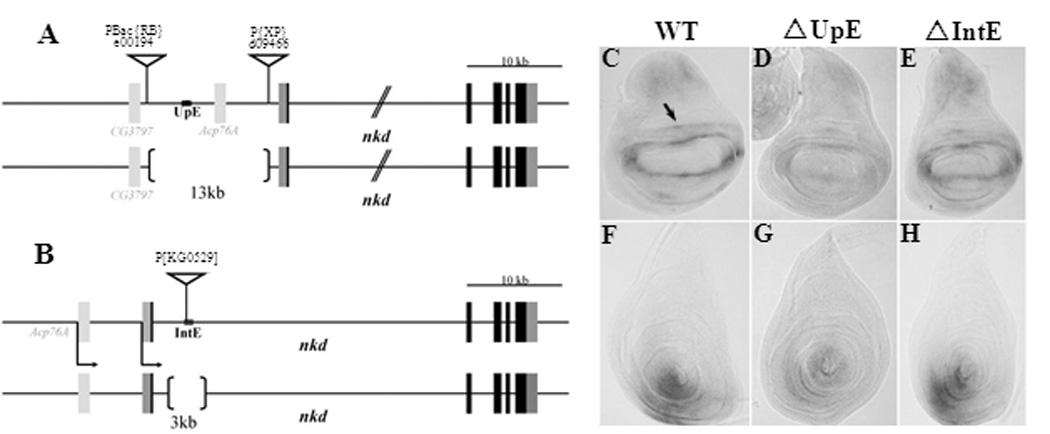
(A, B) Cartoon of the ΔUpE and ΔIntE deletions. See Materials and Methods and Supplemental Fig. 3 for details of the deletion construction. (C–H) nkd transcripts in wing (C–E) and leg (F–H) imaginal discs from +/Df(3L)ED4782 (C, F) ΔUpE/Df(3L)ED4782 (D, G) and ΔIntE/Df(3L)ED4782 (E, H) transheterozygotes. Df(3L)ED4782 is a large deficiency removing the entire nkd locus. No noticeable decrease of nkd expression is detected from imaginal discs (E, H) or embryos (data not shown) when IntE is deleted. ΔUpE/Df(3L)ED4782 transheterozygotes showed no decrease in nkd expression in the embryo (data not shown) but displayed a marked reduction in the wing and leg imaginal discs (D, G). This reduction was consistently observed in three independent in situ analyses and was also confirmed by qRT-PCR (see text).
To test the effect of the ΔUpE and ΔIntE alleles on nkd expression, they were placed over Df(3L)ED4782, a large (175 kb) deficiency removing nkd and several surrounding genes. These transheterozygous backgrounds produced viable fertile adults at about the same frequency as +/Df(3L)ED4782 individuals (data not shown). Since mutations in the nkd gene are embryonic lethal (Zeng et al., 2000), the viability of ΔUpE/Df(3L)ED4782 and ΔIntE/Df(3L)ED4782 indicates that UpE and IntE are dispensable for nkd expression in the embryo. Consistent with this, no detectable reduction in nkd transcript level was detected from ΔUpE/Df(3L)ED4782 and ΔIntE/Df(3L)ED4782 embryos as judged by in situ hybridization (data not shown).
Unlike the situation in the embryo, ΔUpE did affect nkd expression in larval tissues. In situ hybridization revealed that ΔUpE/Df(3L)ED4782 larvae had a marked reduction in nkd transcript levels compared to +/Df(3L)ED4782 controls in the wing and leg imaginal discs (Fig. 6C, D, F, G). A similar reduction was observed in the eye-antennal discs (data not shown). qRT-PCR quantification revealed that ΔUpE/Df(3L)ED4782 wing discs had 41% (S.D. ± 26%) of the nkd mRNA found in control wing discs. In contrast to ΔUpE, loss of IntE did not detectably change nkd expression in the imaginal discs, as judged by in situ hybridization (Fig. 6E, H) and qRT-PCR of RNA from wing imaginal discs (data not shown). With the important caveat that the ΔUpE allele deletes almost 12 kb of sequence besides UpE, the data obtained are consistent with the idea that UpE is a bona fide WRE of nkd. IntE, on the other hand is dispensable for expression of endogenous nkd in the tissues that were examined.
Discussion
We previously identified binding of TCF to a region in the nkd intron that corresponds to IntE (Fang et al., 2006; Parker et al., 2008). In this report, we find that TCF is also highly enriched on the chromatin about 10 kb upstream of the nkd TSS (UpE) when the Wg pathway is chronically activated by Axin RNAi (Fig. 1). Wg signaling also increases the binding of TCF to the region containing IntE (Fig. 1), but this effect occurs within a few hours of pathway stimulation (Fang et al., 2006; Parker et al., 2008). The increase in TCF-binding in the UpE region is not due to increased TCF, since the expression of TCF and its nuclear localization are not affected by Axin RNAi (Chang et al., 2008). Rather, we postulate that widespread histone acetylation at the nkd locus upon Wg stimulation (Parker et al., 2008) allows subsequent recruitment of TCF to the UpE region in Kc cells.
Further analysis of the UpE region with reporter gene assays revealed the presence of two overlapping stretches of DNA (UpE1 and UpE2) that confer a high degree of responsiveness to Wg signaling (Fig. 2A). Mutagenesis of two to three predicted TCF binding sites in UpE1, UpE2 and IntE largely abolished their ability to respond to Wg signaling (Fig. 2A,B). While this suggests that TCF sites within a WRE act in a redundant manner, mutation of individual sites indicates that some sites contribute more than others. For example, in UpE2, the TCF 2 and TCF 3 sites share the same sequence (Supplemental Fig. 1A), but mutation of TCF 2 reduces Wg activation while mutation of TCF 3 does not (Supplemental Fig. 2A). These data suggests that the exact sequence of the TCF binding site is likely not as important as the context in which they are located within the WRE.
When tested in flies, UpE1, UpE2 and IntE are all activated by Wg signaling in several tissues in patterns that partially recapitulate that of the endogenous nkd gene (Fig. 3, Fig. 4 & Table 1). In the leg and eye imaginal discs, all three WREs are active. In addition, each WRE has unique tissue-specific activities. For example, in later embryogenesis IntE is the only WRE that is active. Even though UpE1 and UpE2 share more than 400 bp of sequence (Fig. 2A), only UpE2 is active in the embryonic epidermis. UpE1 and UpE2 are both active in the wing and antennal imaginal discs, (IntE shows no or minimal expression in these tissues), but UpE1 is expressed strongly in the notum whereas UpE2 is not. Each WRE is active in multiple tissues but also contains information that confers tissue-specific Wg responsiveness.
The basis for the tissue specificity of the various nkd-WREs is not clear at present. It could be that different TCF sites within each WRE are utilized in different tissues. However, our data in cell culture argue that multiple TCF sites are required in each WRE in a partially redundant manner (Fig. 2; Supplemental Fig. 2). In addition, the same TCF sites that are required for UpE2 and IntE activity in Kc cells are required for WRE activation in all tissues examined (Fig. 4D, J; data not shown). Therefore, we favor the view that the tissue specificity for the different WREs is derived from the presence of other cis-acting elements that work with the TCF sites to allow activation by Wg signaling.
When all three WREs are placed within a single reporter construct the resulting pattern largely recapitulates that of the endogenous nkd gene in imaginal discs at the late third larval instar stage (Fig. 5B; data not shown). However, we have not examined the regulation of our WRE reporters at earlier larval stages, where Wg is also expressed (Williams et al., 1993; Neumann and Cohen, 1996). Even at the late larval stage examined, the pattern of the reporters does not completely match that of endogenous nkd in the wing disc. Wg is expressed in a double ring pattern in the hinge region (Fig. 3K; the proximal ring indicated by the arrow) and nkd transcripts are also found in a double ring (Fig. 6C; arrow). However, expression of WRE reporters in the proximal ring is weak (Fig. 3M) and often not present (Fig. 5B). This suggests the existence of at least one other WRE for the wing imaginal disc.
In the embryo, it is even more obvious that additional regulatory information for nkd expression remains to be identified. In the embryonic epidermis the pattern of the combined WRE construct is only a subset of the endogenous nkd pattern and is equal to the sum of the IntE and UpE2 WREs (Fig. 5C, D; data not shown). This suggests the presence of at least one other WRE that is active in the embryonic epidermis. Consistent with this, deletions of genomic fragments containing IntE or UpE do not affect the expression of nkd in embryos or the viability of the animals when heterozygous with a nkd deficiency (data not shown).
In the wing and leg imaginal discs, loss of UpE results in a significant decrease in nkd transcript levels (Fig. 6D, G). In contrast, the IntE deletion had nkd expression in the normal range (Fig. 6E, H). Even with the large UpE deletion, there is still significant nkd expression in the wing disc (41% of wild type; see Results). These data could be evidence for redundancy between IntE and UpE in these tissues. It is also possible that additional WREs exist that contribute to imaginal disc expression, which are still present in the IntE and UpE deletions.
Our data indicate that nkd does not contain a universally responding WRE that is activated by Wg signaling in all tissues. Rather there are at least several WREs that can respond to the pathway in multiple, overlapping tissues. It appears that only limited multi-tissue responsiveness can be obtained with any individual WRE. In the absence of a universal WRE, the strategy of having multiple WREs responding to Wg signaling in each tissue may be required to ensure the robustness of the Wg-Nkd feedback circuit. Whether this is the case for the regulation of other Wnt feedback antagonists or those acting in other signaling pathways remains to be determined.
The finding that the nkd locus does not contain a universal WRE raises the question of how the Wg-Nkd relationship could remain intact during animal evolution as the Wg expression pattern became more elaborate. We postulate that the existence of several WREs with broad tissue specificity could have ensured that when an enhancer evolved that expressed Wg in a new location, at least one of the existing nkd WREs would be able to respond to the pathway in that tissue. This precludes the need to have a tissue-by-tissue de novo synthesis of nkd WREs every time Wg was expressed in a new pattern. Retaining the feedback inhibition of Wg signaling by Nkd may have allowed Wg to be used more readily during the diversification of animal body plans.
Supplementary Material
The sequences and conservation of the TCF sites in the nkd-WREs. Predicted TCF sites that fit the consensus (SSTTTGWW) with one substitution allowed and are conserved in 12 Drosophila species (alignment was done via http://genome.ucsc.edu/) are shown. (A) Six TCF sites from UpE and three from IntE (255 bp) were identified. TCF sites in red represent TCF sites that were functional in the reporter assays (Supplemental Fig. 2). “(rev)” indicates the site is on the reverse strand. (B, C) Alignments of TCF sites and flanking sequences between D. melanogaster and D. virilis. We found there is a high correlation between phylogenetic conservation and functionality of the TCF sites. While 3 non-conserved TCF sites in UpE or IntE did not contribute to WRE activity (data not shown), 7 of 9 conserved TCF sites are required for UpE or IntE to be fully activated by Wg signaling. Despite their high similarity to the consensus TCF sequence, the conserved T3 and T5 sites in UpE are dispensable for activation in the reporter assays.
Not every TCF binding site is functional or contributes to WRE activity equally. The indicated reporter constructs were activated by Arm* in Kc cells. The fold activation of UpE1, UpE2 and IntE (255 bp) was normalized to 100. (A) Compared to the UpE1 reporter, the TCF1mut construct showed a 36% reduction in the Wg signaling-responsiveness. The functionality of other TCF sites was tested in the context of UpE2. While the TCF2mut, 4mut and 6mut constructs had reduced WRE activity to different degrees, TCF3mut or 5mut were similar to wild type UpE2. Despite the different contributions of individual TCF sites to WRE function, there is no significant difference in their sequences or conservation (Supplemental Fig. 1). (B) In the IntE (255 bp) WRE, three TCF sites contribute to Wg signaling-responsiveness. Individual mutation of these TCF sites reduces the activation of IntE by Arm* and mutation in all three TCF sites completely abolishes the response All experiments shown here were done with triplicate samples (± S.D.) and the results were reproducible in more than three separate experiments.
Schematic showing the generation of the ΔUpE deletion. (A) Two insertions, PBac{RB}e00194 and P{XP}d09466, were used to generate ΔUpE. (B) The FRT site present in PBac{RB}e00194 was recombined to one of the two FRTs present in P{XP}d09466. As a consequence, the intervening sequences having UpE and other flanking sequences were removed from one chromosome and the UpE region and Acp76A gene were duplicated on the other chromosome. Flies having the ΔUpE chromosome were selected based on the enhanced eye color and the mutation was confirmed by PCR. To test whether the P{XP}d09466 transposon insertion influences nkd transcription, nkd transcripts in wild type and in P{XP}d09466 were compared via in situ hybridization and no noticeable variation was detected (data not shown).
Acknowledegments
We thank members of the Cadigan lab for helpful discussions and Chandan Bhambhani in particular for careful reading of the manuscript. This work was supported by NIH Grant R01CA95869 to K.M.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandre C, Jacinto A, Ingham PW. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Bilder D, Scott MP. Hedgehog and wingless induce metameric pattern in the Drosophila visceral mesoderm. Dev. Biol. 1998;201:43–56. doi: 10.1006/dbio.1998.8953. [DOI] [PubMed] [Google Scholar]
- Cadigan KM. Regulating morphogen gradients in the Drosophila wing. Semin. Cell Dev.Biol. 2002;13:83–90. doi: 10.1016/s1084-9521(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Chang JL, Lin HV, Blauwkamp TA, Cadigan KM. Spenito and Split ends act redundantly to promote Wingless signaling. Dev. Biol. 2008;314:100–111. doi: 10.1016/j.ydbio.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Costas J, Pereira PS, Vieira CP, Pinho S, Vieira J, Casares F. Dynamics and function of intron sequences of the wingless gene during the evolution of the Drosophila genus. Evol. Dev. 2004;6:325–335. doi: 10.1111/j.1525-142X.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- Couso JP, Bate M, Martinez-Arias A. A wingless-dependent polar coordinate system in Drosophila imaginal discs. Science. 1993;259:484–489. doi: 10.1126/science.8424170. [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. Embo J. 2006;25:2735–2745. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M, Bienz M. EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep. 2001;2:157–162. doi: 10.1093/embo-reports/kve019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz O, Basler K. Wingful, an extracellular feedback inhibitor of Wingless. Genes Dev. 2002;16:1055–1059. doi: 10.1101/gad.991802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Copley RR, Cohen SM. HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev. Cell. 2002;2:667–676. doi: 10.1016/s1534-5807(02)00180-6. [DOI] [PubMed] [Google Scholar]
- Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122:223–230. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, Kadowaki T, Beck K, Kitagawa Y. A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J. 2001;359:99–108. doi: 10.1042/0264-6021:3590099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Han Z, Fujioka M, Su M, Liu M, Jaynes JB, Bodmer R. Transcriptional integration of competence modulated by mutual repression generates cell-type specificity within the cardiogenic mesoderm. Dev. Biol. 2002;252:225–240. doi: 10.1006/dbio.2002.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingensmith J, Nusse R. Signaling by wingless in Drosophila. Dev. Biol. 1994;166:396–414. doi: 10.1006/dbio.1994.1325. [DOI] [PubMed] [Google Scholar]
- Knirr S, Frasch M. Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev. Biol. 2001;238:13–26. doi: 10.1006/dbio.2001.0397. [DOI] [PubMed] [Google Scholar]
- Lessing D, Nusse R. Expression of wingless in the Drosophila embryo: a conserved cis-acting element lacking conserved Ci-binding sites is required for patched-mediated repression. Development. 1998;125:1469–1476. doi: 10.1242/dev.125.8.1469. [DOI] [PubMed] [Google Scholar]
- Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM. CBP/p300 are bimodal regulators of Wnt signaling. Embo J. 2007;26:2284–2294. doi: 10.1038/sj.emboj.7601667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HV, Rogulja A, Cadigan KM. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development. 2004;131:2409–2418. doi: 10.1242/dev.01104. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Sternopleural is a regulatory mutation of wingless with both dominant and recessive effects on larval development of Drosophila melanogaster. Genetics. 1996;142:1147–1155. doi: 10.1093/genetics/142.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development. 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- Parker DS, Blauwkamp T, Cadigan KM. Wnt/□-catenin-mediated transcriptional regulation. In: Sokol S, Wassarman PM, editors. Wnt Signaling in Embryonic Development, Advances in Developmental Biology. Vol. 17. San Diego: Elsevier; 2007. pp. 1–61. [Google Scholar]
- Parker DS, Jemison J, Cadigan KM. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development. 2002;129:2565–2576. doi: 10.1242/dev.129.11.2565. [DOI] [PubMed] [Google Scholar]
- Parker DS, Ni YY, Chang JL, Li J, Cadigan KM. Wingless signaling induces widespread chromatin remodeling of target loci. Mol. Cell. Biol. 2008;28:1815–1828. doi: 10.1128/MCB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Peel DJ, Johnson SA, Milner MJ. The ultrastructure of imaginal disc cells in primary cultures and during cell aggregation in continuous cell lines. Tissue Cell. 1990;22:749–758. doi: 10.1016/0040-8166(90)90069-l. [DOI] [PubMed] [Google Scholar]
- Pereira PS, Pinho S, Johnson K, Couso JP, Casares F. A 3' cis-regulatory region controls wingless expression in the Drosophila eye and leg primordia. Dev. Dyn. 2006;235:225–234. doi: 10.1002/dvdy.20606. [DOI] [PubMed] [Google Scholar]
- Riese J, Yu X, Munnerlyn A, Eresh S, Hsu SC, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- Rousset R, Mack JA, Wharton KA, Jr, Axelrod JD, Cadigan KM, Fish MP, Nusse R, Scott MP. Naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev. 2001;15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson B. Generating patterns from fields of cells. Examples from Drosophila segmentation. EMBO Rep. 2001;2:1083–1088. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadeli R, Hoffmans R, Basler K. Transcription under the control of nuclear Arm/beta-catenin. Curr. Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Harryman-Samos C, Klingensmith J, Perrimon N, Nusse R. Mutations in the segment polarity genes wingless and porcupine impair secretion of the wingless protein. Embo J. 1993;12:5293–5302. doi: 10.1002/j.1460-2075.1993.tb06225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad A, Rohrs S, Klein-Hitpass L, Muller O. The first five years of the Wnt targetome. Cell Signal. 2008;20:795–802. doi: 10.1016/j.cellsig.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Waldrop S, Chan CC, Cagatay T, Zhang S, Rousset R, Mack J, Zeng W, Fish M, Zhang M, Amanai M, Wharton KA., Jr An unconventional nuclear localization motif is crucial for function of the Drosophila Wnt/wingless antagonist Naked cuticle. Genetics. 2006;174:331–348. doi: 10.1534/genetics.106.061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Paddock SB, Carroll SB. Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development. 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- Yanagawa S, Lee JS, Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J. Biol. Chem. 1998;273:32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- Zeng W, Wharton KA, Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- Zhou H, Cadigan KM, Thiele DJ. A copper-regulated transporter required for copper acquisition, pigmentation, and specific stages of development in Drosophila melanogaster. J. Biol. Chem. 2003;278:48210–48218. doi: 10.1074/jbc.M309820200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences and conservation of the TCF sites in the nkd-WREs. Predicted TCF sites that fit the consensus (SSTTTGWW) with one substitution allowed and are conserved in 12 Drosophila species (alignment was done via http://genome.ucsc.edu/) are shown. (A) Six TCF sites from UpE and three from IntE (255 bp) were identified. TCF sites in red represent TCF sites that were functional in the reporter assays (Supplemental Fig. 2). “(rev)” indicates the site is on the reverse strand. (B, C) Alignments of TCF sites and flanking sequences between D. melanogaster and D. virilis. We found there is a high correlation between phylogenetic conservation and functionality of the TCF sites. While 3 non-conserved TCF sites in UpE or IntE did not contribute to WRE activity (data not shown), 7 of 9 conserved TCF sites are required for UpE or IntE to be fully activated by Wg signaling. Despite their high similarity to the consensus TCF sequence, the conserved T3 and T5 sites in UpE are dispensable for activation in the reporter assays.
Not every TCF binding site is functional or contributes to WRE activity equally. The indicated reporter constructs were activated by Arm* in Kc cells. The fold activation of UpE1, UpE2 and IntE (255 bp) was normalized to 100. (A) Compared to the UpE1 reporter, the TCF1mut construct showed a 36% reduction in the Wg signaling-responsiveness. The functionality of other TCF sites was tested in the context of UpE2. While the TCF2mut, 4mut and 6mut constructs had reduced WRE activity to different degrees, TCF3mut or 5mut were similar to wild type UpE2. Despite the different contributions of individual TCF sites to WRE function, there is no significant difference in their sequences or conservation (Supplemental Fig. 1). (B) In the IntE (255 bp) WRE, three TCF sites contribute to Wg signaling-responsiveness. Individual mutation of these TCF sites reduces the activation of IntE by Arm* and mutation in all three TCF sites completely abolishes the response All experiments shown here were done with triplicate samples (± S.D.) and the results were reproducible in more than three separate experiments.
Schematic showing the generation of the ΔUpE deletion. (A) Two insertions, PBac{RB}e00194 and P{XP}d09466, were used to generate ΔUpE. (B) The FRT site present in PBac{RB}e00194 was recombined to one of the two FRTs present in P{XP}d09466. As a consequence, the intervening sequences having UpE and other flanking sequences were removed from one chromosome and the UpE region and Acp76A gene were duplicated on the other chromosome. Flies having the ΔUpE chromosome were selected based on the enhanced eye color and the mutation was confirmed by PCR. To test whether the P{XP}d09466 transposon insertion influences nkd transcription, nkd transcripts in wild type and in P{XP}d09466 were compared via in situ hybridization and no noticeable variation was detected (data not shown).


