Abstract
Milestoning is a method for calculating kinetics and thermodynamics of long time processes typically not accessible for straightforward Molecular Dynamics (MD) simulation. In the Milestoning approach, the system of interest is partitioned into cells by dividing hypersurfaces (Milestones) and transitions are computed between nearby hypersurfaces. Kinetics and thermodynamics are derived from the statistics of these transitions. The original Milestoning work concentrated on systems in which a one-dimensional reaction coordinate or an order parameter could be identified. In many biomolecular processes the reaction proceeds via multiple channels or following more than a single order parameter. A description based on a one-dimensional reaction coordinate may be insufficient. In the present paper we introduce a variation that overcomes this limitation.
Following the ideas of Vanden-Eijnden and Venturoli on Voronoi cells that avoid the use of an order parameter (J. Chem. Phys. 2009, 130, 194101), we describe another way to “Milestone” systems without a reaction coordinate. We examine the assumptions of the Milestoning calculations of mean first passage times (MFPT) and describe strategies to weaken these assumptions. The method described in this paper, Directional Milestoning, arranges hypersurfaces in higher dimensions that “tag” trajectories such that efficient calculations can be done and at the same time the assumptions required for exact calculations of MFPTs are satisfied approximately.
In the original Milestoning papers trajectories are initiated from an equilibrium set of conformations. Here a more accurate distribution, that mimics the first hitting point distribution, is used. We demonstrate the usage of Directional Milestoning in conformational transitions of alanine dipeptide (in vacuum and in aqueous solution) and compare the correctness, efficiency, and statistical stability of the method with exact MD and with a related method.
1. Introduction
Milestoning is a method to calculate kinetics and thermodynamics of molecular systems that evolve on long time scales typically not accessible for straightforward Molecular Dynamics (MD) simulation.1–8
Straightforward Molecular dynamics can be used to compute rate of reactions. In these applications coordinates and velocities are initiated in the reactant state and the equations of motion are integrated until the product state is reached. While considerably promising, there are caveats: (i) the numerical integration of a typical biomolecular process is computationally demanding and may not be feasible; (ii) actual realizations of reactive trajectories are noisy, making their analysis difficult and may require significant filtering to recover useful signals.
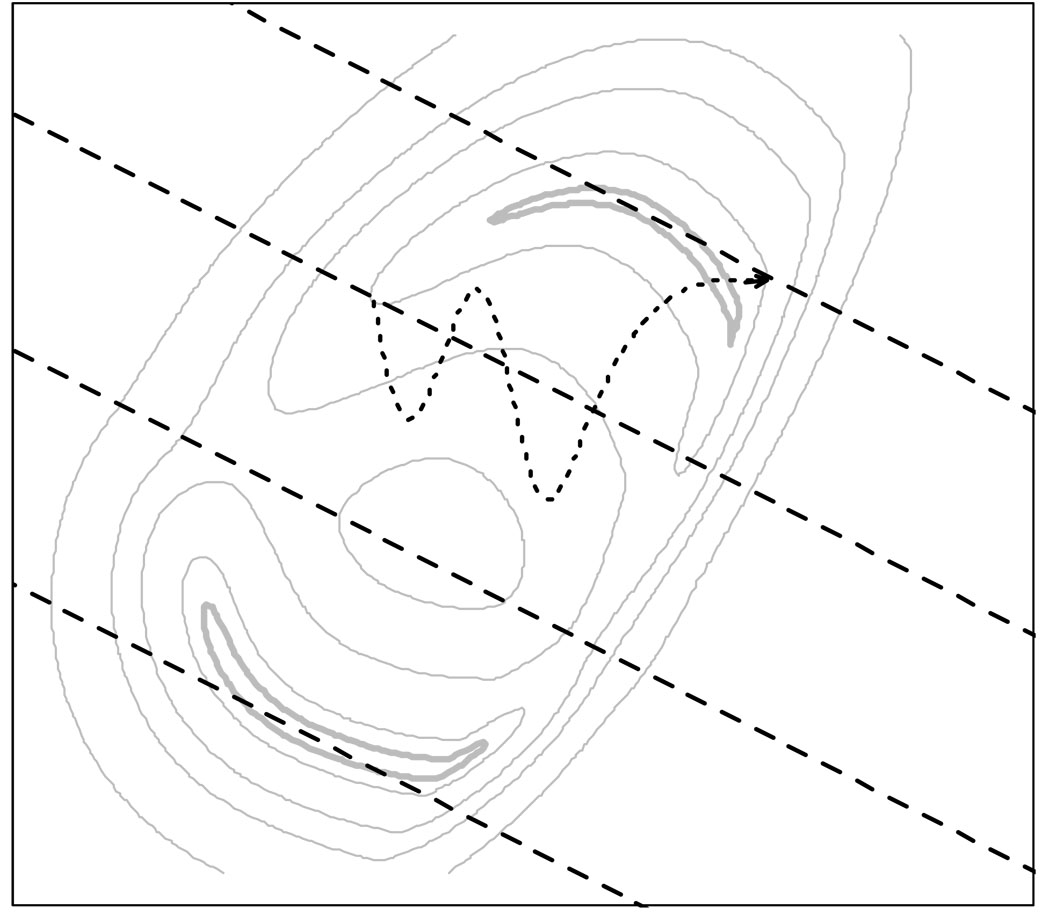
In Milestoning, the conformational space between the reactant and the product is partitioned by a set of dividing hypersurfaces called Milestones (Fig. 1). An ensemble of initial conditions is prepared at each Milestone and trajectories are simulated from each initial point until another nearby Milestone is reached. These trajectories are significantly shorter and trivially parallelized compared to a reactive trajectory of the overall process. The efficiency of the algorithm is discussed in 1.
Figure 1.
A schematic arangement of Milestones (dashed lines) in a two-well potential. Also shown is a trajectory (dotted line) starting on a second Milestone and terminating on the first one.
In the original milestoning papers,1,3 a theory that relates the statistical properties of the short trajectories initiated on each Milestone and the overall rate was developed. In the present work we consider a variant of the Markovian limit of Milestoning,1,2 a method that uses only the first moments of local first passage time (LFPT) distributions. The advantage of the Markovian limit of Milestoning is that it is easier to implement and is statistically more stable. As we will show in Section 2.1 it calculates the overall mean first passage times (MFPT) accurately, given that certain assumptions are met. Milestoning in its complete settings (non-Markovian) provides a useful alternative if more detailed understanding of the reaction process is desired, for example if the reaction is non-exponential in time.
Vanden-Eijnden et. al.,4 considered reaction dynamics with overdamped Langevin. It was shown that if Milestones are chosen as isocommittor surfaces, i.e. surfaces for which the probability of reaching the product state before the reactant is constant, then Milestoning calculation of the MFPT using Brownian dynamics is exact. However, determination of exact isocommittor surfaces can be very difficult in practice.
Other limits in which Milestoning is expected to be accurate are available for systems near equilibrium. As outlined in the original Milestoning papers,1,3, even when other surfaces are used (surfaces that are not isocommittors) Milestoning can still work well. If successive crossing events of Milestones are sufficiently separated in time to “lose” velocity memory Milestoning was illustrated to provide accurate results. This assumption is achieved in practice by placing Milestones sufficiently far from each other such that the average termination time of trajectories is at least a few hundred femtoseconds.1
In Section 2 we propose a variant of Milestoning in the Markovian limit which we call Directional Milestoning (DiM) – the dividing hypersurfaces are redefined in more than one dimension to capture features of the reaction (e.g. multiple reaction channels or multiple collective variables) that at the same time maintain the concept of Milestone separation, e.g. trajectories initiated on any Milestone have time to “lose memory” before terminating on other Milestones.
The original Milestoning approach approximates the initial ensemble on each hypersurface by equilibrium distribution. To be exact the initial distribution at a Milestone must be the first hitting point distribution (FHPD). A first hitting point is a phase space point on the Milestone crossed for the first time by a trajectory arriving from a nearby hypersurface. The distribution of these phase space points is complex and a closed form of it is known only for overdamped Langevin dynamics in low dimensions.4
In recent work,7 Vanden-Eijnden and Venturoli proposed a modification of Milestoning that avoids a generation of initial ensembles on each of the dividing surfaces. As we discuss later their approach is more accurate compared to the original Milestoning for the generation of the FHPD. Memory loss, however, is harder to control in the new approach. To improve the accuracy of the original Milestoning approach while retaining some of its advantages we propose in Section 2.4 another way to approximate FHPD, which is better than the original Milestoning.
In Section 3 we illustrate the Directional Milestoning (DiM) for the calculation of MFPT of a conformational transition of alanine dipeptide, both in vacuum and in water. We compare Directional Milestoning with exact Molecular Dynamics and with the related method Markovian Milestoning with Voronoi Tessellation (MMVT).7 We illustrate that as the complexity of the underlying energy surface increases, DiM becomes more effective. Discussions and conclusions are in presented Section 4.
2. Directional Milestoning – theory
2.1 Definition of Milestones in higher dimensions
We discuss below an extension of Milestoning that avoids the use of a reaction coordinate. Instead of placing hypersurfaces orthogonal to a one-dimensional curve as introduced in the original papers,1,3 we define the interfaces (Milestones) based on a set of coordinates (images) that sample the conformational space of the biophysical process under consideration. (Two of the images define the reactant and the product state.) These images may be obtained from long time simulations, high temperature trajectories, replica exchange simulations, etc., as discussed later in examples in the article. Having N images X1,…, XN placed in the conformational space, we intuitively want to arrange Milestones as interfaces between the images, which is the approach taken in the Voronoi Tessellation of Markovian Milestoning.7 However, we aim to place the Milestones in conformational space in such a way that a trajectory initiated on any Milestone has time and space to “lose memory” of its starting point before terminating at a different Milestone. A formal definition of “losing memory” will be given in the following section. For each pair of images Xi and Xj we define the Milestone Mi → j as a set of conformational points on which a trajectory enters the region of image Xj from the region of image Xi. Formally, the above intuitive requirements on Milestone placement can be accomplished in several different ways. We define a Milestone Mi → j as follows
| (1) |
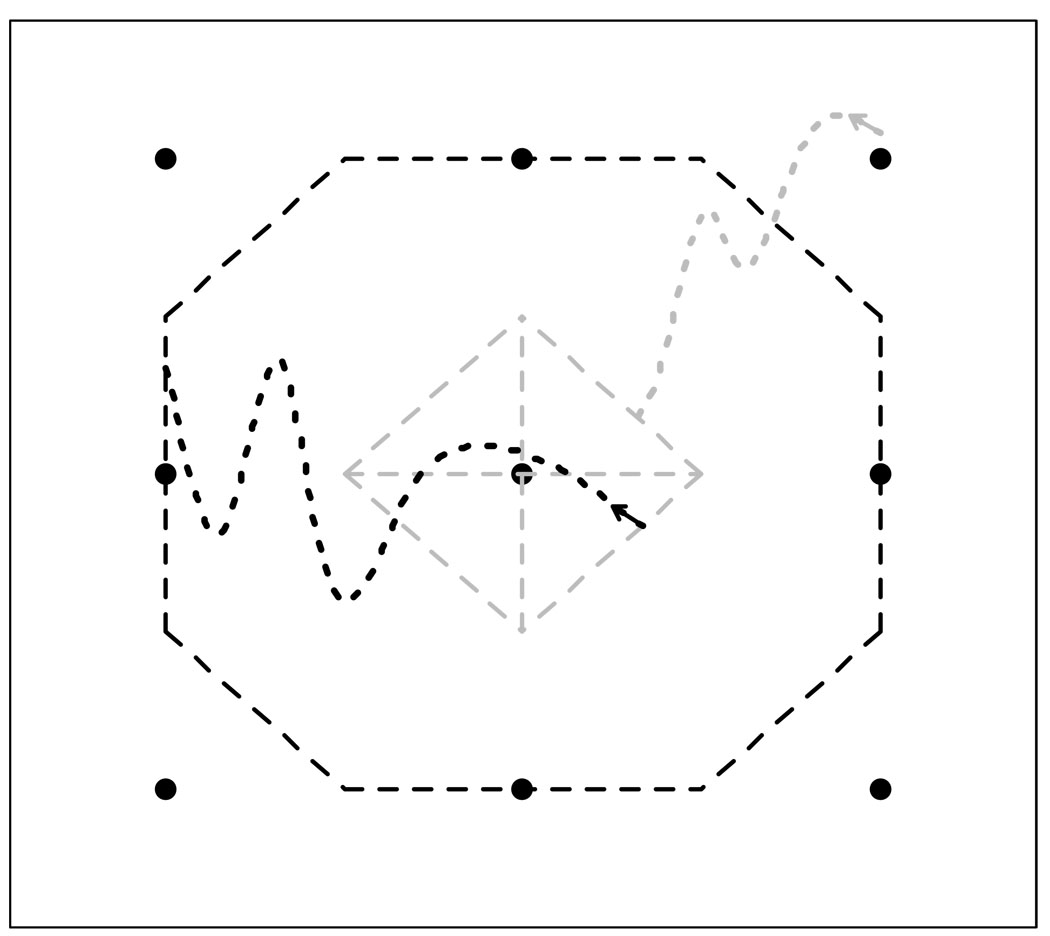
where d(X, Y) is a distance function of images X and Y and Δi = minj ≠ id(Xi, Xj). The arrangement (1) has a few important properties discussed in detail in Section 2.3. We name some of the properties here, referring the formal proofs to Section 2.3: A Milestone Mi → j is located in the region between the images Xi and Xj and is always closer to the image Xj. The Milestone Mi → j does not intersect any of Mi → l Milestones (for l ≠ j) and there is a finite separation in conformational space between the Milestones Mi → j and Ml → i. See Fig. 2 for an example of the proposed arrangement. As shown in the figure, the outgoing (black) Milestones bound the region of the central image and all the incoming (gray) Milestones are located within this region with a minimal distance to any of the outgoing Milestones.
Figure 2.
Example of Milestones according to definition (1). Conformational images are represented as black dots, Milestones related to the central image are displayed as dashed lines. A trajectory coming to the central region (gray, dotted) terminates on one of the gray Milestones (depending on the previously assigned region). A trajectory re-initiated on any of the gray (incoming) Milestones leaves the region through one of the black (outgoing) Milestones.
The proper selection of the conformational images X1,…, XN will be explained in more detail in Section 2.3; for now we assume their arbitrary placement. If Δi were omitted in the above definition (Δi ≠ 0) then the set of Milestones Mi → j is reduced to the Voronoi tessellation proposed in 7,8; we refer to this arrangement as Markovian Milestoning with Voronoi Tessellation [MMVT] throughout this paper. In the MMVT arrangement, the Milestone Mi → j is equivalent to the Milestone Mj → i and the only information they preserve is the identity of last crossed Milestone, not the direction of such a crossing. (In a private communication Vanden-Eijnden disclosed an extension of MMVT to make the Milestones velocity dependent).
It is important to emphasize that the proposed placements of Milestones is not a tessellation. In accord with the definition of the original Milestoning, a trajectory is identified by the last Milestone that it passes and not by its actual current position. A memory is carried out in time until the trajectory crosses another interface (Milestone).
Trajectories from Xi to Xj can be fundamentally different from trajectories from Xj to Xi. To exploit this observation it is useful to make the Milestones dependent on the direction. We therefore call Milestones defined according to eq. (1) Directional Milestones. The role of the additional flexibility offered by Δi is to avoid counting rapid transitions between interfaces due to spatial proximity of Milestones. As a result, the Milestones defined by eq. (1) depend on more than the coordinates alone. This is consistent with the notion of a Milestone Mi → j (Mk → j) as a state of a trajectory that arrives from the region Xi(Xk) to the region of image Xj. Hence the definition of a Milestone is extended to include information about the previous assignment of the trajectory. If the system is assigned to a region Xi0 at time 0 then by following a trajectory of the system one can deterministically identify the sequence of Milestones the trajectory has passed through Mi0 → i1, Mi1 → i2, Mi2 → i3, …, Mik − 1 → ik.
2.2 Calculation of Mean First Passage Times
In the rest of the manuscript we will use Roman subscripts do denote image index (as was done in the previous section) and Greek letters to denote Milestones. Consider the mean first passage time (MFPT) from any Milestone α to a given target Milestone β. We define it as follows: a trajectory is assigned to a Milestone α if the last Milestone it has passed through is α. One-step transition from a Milestone α to a Milestone β(β ≠ α) is a change of assignment of a trajectory from α to β. This step is clearly on a coarse Milestoning level and does not mean a single Molecular Dynamics step, which we will call a time-step. If such an event is possible we say that α connects to β. Note that by the definition given in equations (1), if α connects to β, the second index of α (e.g. Mi → j) must be equal to the first index of β (Mj → k). The first hitting point distribution on β, ρβ(p), is the distribution of phase space points (denoted by p) at which an equilibrium trajectory passes through β numerous times while the previous Milestone it passes through was not β. In further discussion only the relative weight of trajectories that pass through β is important so we can choose to normalize ρβ(p) such that ∫ ρβ(p) dp = 1. We denote by 〈ταβ(p)〉 the mean time of all trajectories that start from the phase space point p in α and terminate on Milestone β (possibly crossing other Milestones on the way). Integrating the last entity over p, weighting it by the probability that p is a phase space point at which an equilibrium trajectory hits β for the first time, ∫ 〈ταβ(p)〉 ρα(p) dp ≡ 〈ταβ〉, we obtain the MFPT from α to β.
Let the distribution of one-step transitions from α to β be Tαβ(p, q, t), where p is the phase space point at which a trajectory starts in α and q is the phase space point at which the trajectory changes its assignment to β after time t. Tαβ(p, q, t) is normalized in such a way that if we integrate over t and q we get conditional probability of a trajectory reaching β in one step given that it originates from p in α : ∫∫Tαβ(p, q, t) dqdt = P(β | α, p), or alternatively ∑β∫∫Tαβ(p, q, t) dqdt = 1. Note that by the definition of trajectory assignment, Tαα(p, q, t) = 0 for all p and q (since a trajectory cannot change its assignment from α to α).
Assuming that the phase space point p(t + dt) can be determined from p(t) only, as is true for most microscopic dynamics (e.g. Newtonian, or Langevin dynamics, but not Generalized Langevin dynamics) we make the following argument: The MFPT from α to β, 〈ταβ〉, is defined as the weighted average of termination times of trajectories from α to β. Each trajectory, starting at p in α jumps in one step to some other Milestone γ(γ ≠ α) at phase point q and then in multiple steps (possibly 0, if γ = β) continues to β. Consider all the trajectories that jump in one-step from p in α to q in γ exactly in time t and then eventually reach β (in potentially different total time). Since the microscopic dynamics is Markovian we can replace the contribution of these trajectories to 〈ταβ〉 by (t + 〈τγβ(q)〉) weighted by sum of the weights of all of them (which is ρα(p)Tαγ(p, q, t)). By doing this for all possible combinations γ of and q we get the following equation:
| (2) |
The first term of the above equation can be reduced as
| (3) |
where 〈tαγ(p)〉,〈tα(p)〉, and 〈tα〉 are average times of one-step transitions from p ∈ α to γ, from p ∈ α to any other Milestone, and from α to any other Milestone (averaged over p), respectively. In the second term of equation (2) the average time from q ∈ γ to β is weighed by a factor depending on the phase space point p ∈ α ! To overcome this problem we use the following assumption: The distribution at which any Milestone γ is hit does not depend on the Milestone to which the trajectory was assigned before the hit:
| (4) |
It is easier to illustrate the properties of equation (4) if we consider a one-dimensional arrangement of Milestones in which the forward and the backward Milestones occupy the same spatial coordinates. Consider a Milestone α that is pointing forward and is therefore denoted for the clarity of this discussion by α +. There are two Milestones that initiate trajectories that may terminate at α +. They are (α − 1)+ and (α − 1)−. Hence they occupy the same place in space but have their velocities pointing in the opposite directions. The assumption of equation (4) states that it does not matter if we start at (α − 1)+ or at (α − 1)−, both Milestones will generate the same hitting point distribution on α+. If the initial direction of the velocity de-correlates quickly there should be no difference in the results from Milestone (α − 1)+ and (α − 1)−. In this case the assumption formulated in equation (4) will be satisfied. Indeed, we observed empirically in reference 9 that even the usual Milestoning works well when the velocity de-correlates. This empirical formulation is now formulated mathematically. In higher dimension we will also require spatial de-correlation.
The multiplicative factor in the above equation is determined by the fact that if both sides of equation (4) are integrated over q, the left side equals to 1 and the right side to P(γ|α); the conditional probability that if a trajectory changes its assignment from α it changes to γ. Therefore using the above assumption the second term of equation (2) reduces to ∑γP(γ|α) 〈ταβ〉 and we obtain the final form for the MFPT:
| (5) |
The set of equation (5) is extended by boundary conditions 〈τββ〉 = 0, 〈τβ〉 = 0, and ∀γ P(γ|β) = 0. It is a set of linear equations for all the 〈ταβ〉 that can be solved by any standard linear solver. The size of the problem (the number of Milestones) never exceeded a few hundreds in our hands. Equation (5) can be directly generalized for considering more than a single target Milestone (e.g. all incoming interfaces to the folded state of a peptide). Alternative equations equivalent to equation (5) were derived in 1,4. These equations are independent of the type of microscopic dynamics that we use (e.g. overdamped Langevin or Newtonian). The system of linear equations (5) relates the overall rate (τ ‘s) with the local kinetics information (〈tα〉 and P(γ|α)). Milestoning collects this local information in a more effective way than running an ensemble of trajectories from α to β. On each Milestone α, Nα phase space points are sampled from the FHPD ρα (see Section 2.4 for details). As a second step, each of the sampled phase space points is propagated in time until a connected Milestone is reached. The termination times of these trajectories are typically several orders of magnitude shorter than the overall MFPT of the system. Furthermore the trajectories between Milestones are independent of each other and thus can be run in parallel. For each Milestone γ connected to α we record Nαγ- the number of trajectories that are initiated on α and terminated on γ. We also record t̄α, the mean termination time of all Nα trajectories regardless of their terminal Milestone. The collected information {Nα,t̄α} is used to estimate the required entities for equation (5) as
| (6) |
In practice instead of using equation (6) we employ Bayesian inference on the collected data to calculate the MFPT supported by the data as well as an estimate of the statistical error due to the finite size of collected data. This procedure is described in detail in Appendix B.
2.3 Properties of Directional Milestones
The use of equation (5) for calculating MFPT depends on validity of the assumption expressed in equation (4). It has been shown in 4 that the assumption formulated in equation (4) holds if overdamped Langevin dynamics is used and the Milestones are chosen as isocommittor surfaces. To our knowledge there is no efficient algorithm that identifies exact isocommittor surfaces and scales moderately with system size. However, there are other ways of satisfying equation (4). Instead we base our strategy on selecting Milestones according to equation (1), making sure that Milestones are sufficiently separated to allow for a memory loss of trajectories as outlined in the arguments of reference.1 Consider a pair of connected Milestones Mi → j, Mj → k (defined by coordinate images Xi, Xj, and Xk). Let Sjk be a hyperplane perpendicular to the line segment Xi − Xk and passing through its midpoint. From equation (1) that defines Mi → j we know that each point on Mi → j is closer to Xj than to Xk. Thus the Milestone Mi → j lies on the Xj’s side of Sjk. It follows from Lemmas A.1 and A.2 in Appendix A that Sjk and Mj → k are parallel, Mj → k lies on the Xk’s side of Sjk, and that . Therefore . This minimal separation of connected Milestones is a property of Directional Milestoning that allows for some velocity relaxation to at least approximately satisfy the assumption described in equation (4). Note that the lower bound for the distance d(Mi → j, Mj → k) is a function of distances between the images that we place at will. Minimal separation of any two images places a lower bound on Δj‘s; additionally if one guarantees for each connected pair Mi → j, Mj → k that d(Xj, Xk)is about Δj then .
2.4 Sampling of the First Hitting Point Distribution
The first step of Milestoning is to sample the initial conditions on each Milestone α from the first hitting point distribution ρα(p). An analytical expression for ρα(p) is in general unknown. In 4 the authors provided the formula ρα(x) ∝ e−βV(x)|∇q(x)| for the case of overdamped Langevin dynamics with Milestones being placed as isosurfaces of the committor function q(x). The last formula includes the gradient of committor function ∇q(x) which is difficult to get in high dimensions.
Instead of computing ρα(p) exactly (no exact expression is available for Newtonian dynamic), we approximate it. First, phase space points are sampled from the equilibrium distribution at Milestone Mi → j. It can be done either by running an MD simulation constrained to the Milestone,1,3 or by employing the Umbrella Sampling technique (see appendix C and 10). The second step involves filtering each of the sampled phase points to determine those that are indeed first hitting events of Mi → j. Exact verification tracks each of the sampled phase space points p back in time and tests termination on one of the incoming Milestones to the cell Xi(Mk → i) before the trajectory intersects any of Mi → l. (If Mi → j itself is crossed before any of Mk → i, p is not the first hitting event of Mi → j, it is at least a second hit of Mi → j; if Mi → l, l ≠ j, is crossed before any of Mk → i then the trajectory must have entered the cell of Xl before reaching p – therefore p cannot be the first hitting event of Mi → j). Tracking the trajectory back in time to any of the Milestones Mk → i is similar in spirit to Transition Interface Sampling,11–13 (TIS), the difference is that a TIS trajectory is propagated back in time until the reactant or the product state is hit. In DiM we perform significantly shorter backward verification, applicable only for equilibrium processes. TIS is exact, however it is more expensive since in Milestoning we still exploit the use of trajectory fragments. Trajectory fragments are easier to parallelize and they can lead to implicit long time trajectories while in TIS long time individual trajectories need to be computed explicitly.
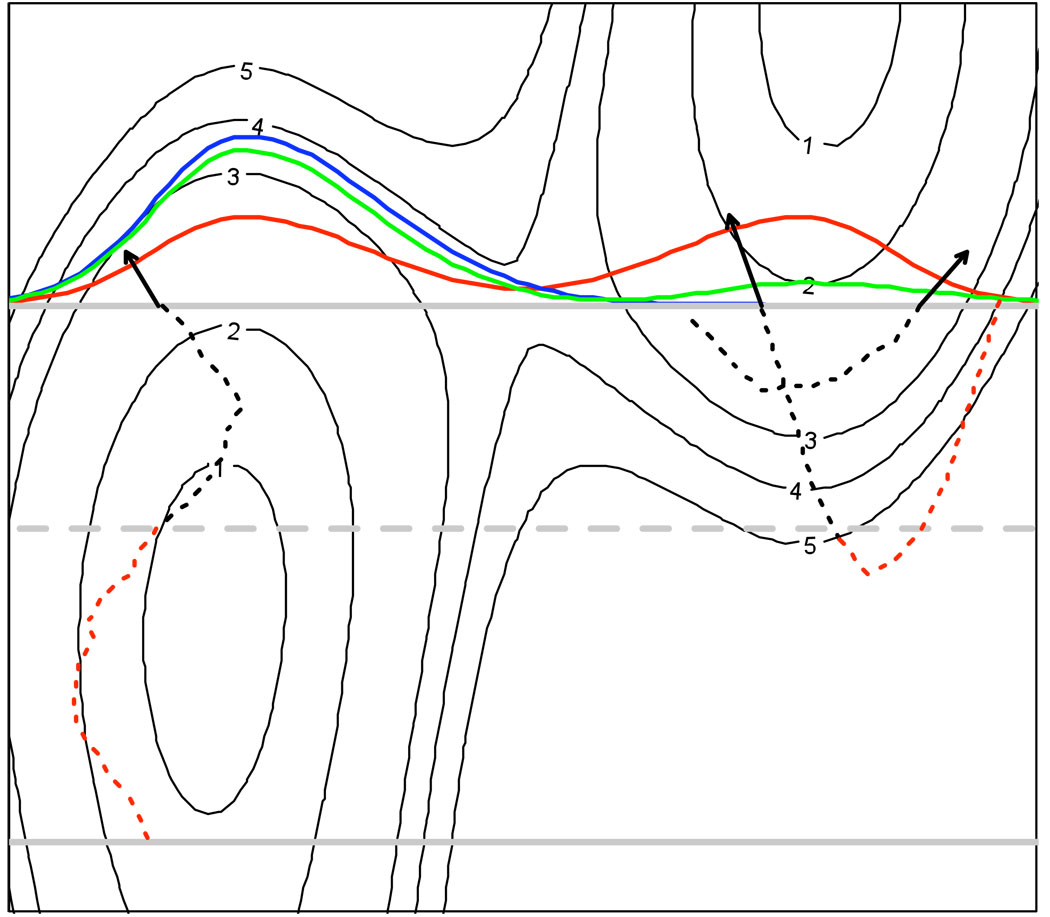
To retain high efficiency we track the trajectory back in time only until it reaches an empirical test boundary that is placed at a distance d on the Xi’s side of the target Milestone Mi → j (d being smaller than or equal to the minimal distance to any of Mk → i from Mi → j). If the trajectory reaches the checking boundary without re-crossing any other Milestone Mi → l, we assume that p is a first hitting event. Otherwise we reject it. The procedure is schematically illustrated on Fig. 3.
Figure 3.
Illustration of sampling of the first hitting point distribution of trajectories initiated on the lower gray Milestone and terminating on the top (target) Milestone. The FHPD on the target Milestone (blue) is centered in the left basin, which is different from the equilibrium distribution (red). The FHPD is approximated by sampling phase space points from the equilibrium distribution and following each of them back in time until it hits the target Milestone on which it was initiated (the point is rejected) or the test boundary shown as a dashed gray line (it is accepted). Tracking of three phase space points is shown; the algorithm tracks only the black parts of the trajectories. Two of the points are accepted; one of them, however, is accepted by a mistake. The point is accepted because the test boundary was reached, however if the trajectory were checked further on (the red part) it would have been detected that the trajectory turns back and is not coming from the lower Milestone. Because of these false positive samples the resulting distribution (green) only approximates ρα(p) (blue). As the test boundary approaches the originating Milestone (lower gray) the sampled distribution approaches the true FHPD.
In principle we can follow the trajectory back in time until one of the incoming Milestones to Xi (Mk → i) or any of the outgoing Milestones from Xi (Mi → l) is hit (a comment by Giovanni Ciccotti). By performing this complete verification the prepared ensemble on each Milestone would be the exact first hitting point distribution. However, the complete verification of each of the sampled phase points roughly doubles the overall computational cost (assuming reasonable acceptance ratio). The result of the more expensive exact verification will be reported elsewhere; in this paper we report results and analysis of the more efficient (but approximate) checking protocol.
3. Applications of Directional Milestoning
3.1 Alanine dipeptide solvated in water
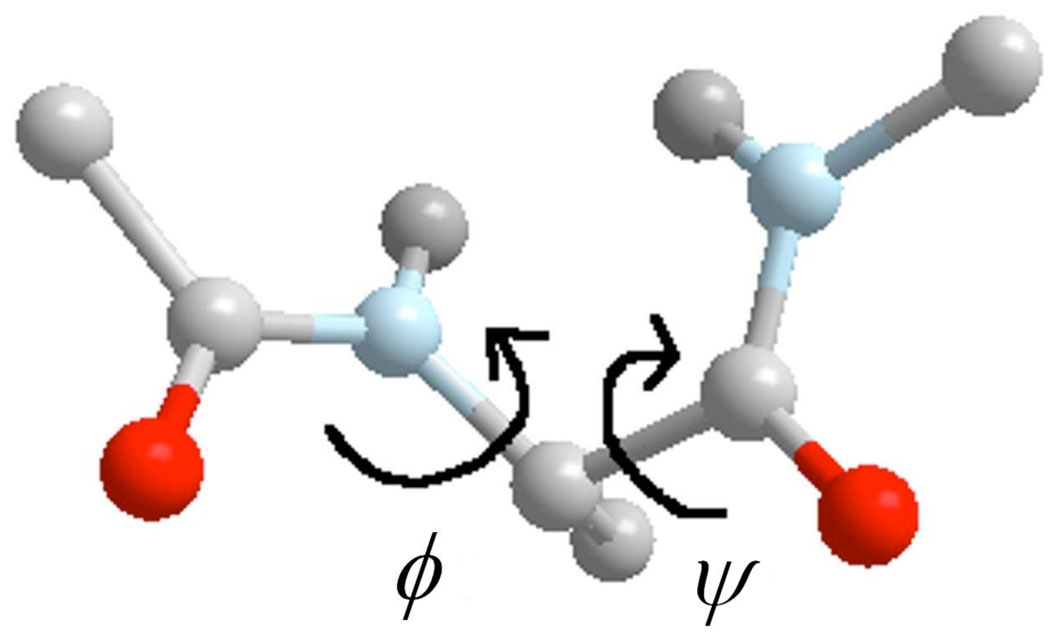
To demonstrate an application of Directional Milestoning we compute MFPT of the transition between α helix and β sheet conformations in solvated alanine dipeptide (Fig. 4). The thermodynamics and kinetics of alanine dipeptide has been investigated in several studies 1,8,14–16. In aqueous solution two dihedral angles, ϕ and ψ, shown in Fig. 4, are adequate coarse variables for the dynamics of the peptide. We therefore use a 2-norm distance in the reduced space of ϕ and ψ as the distance metric in the definition of Milestones (periodicity of the angles was taken into account in the calculation of a distance between two torsion angles).
Figure 4.
Alanine dipeptide.
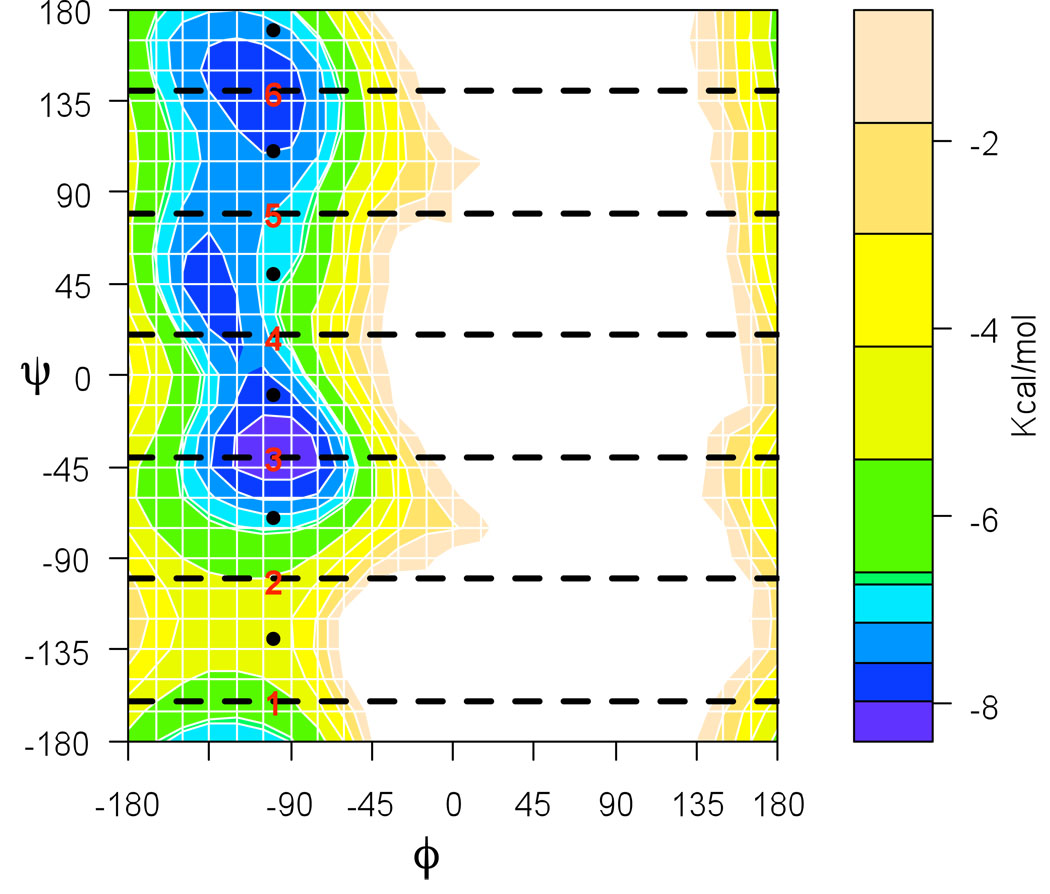
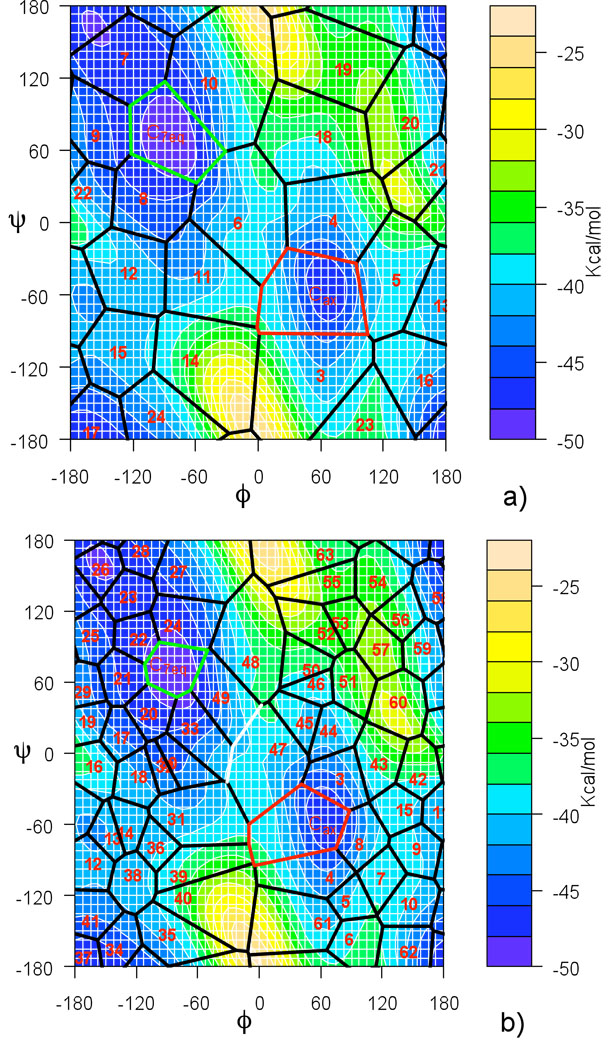
The new module for Directional Milestoning was created in the program MOIL,17 and is available at https://wiki.ices.utexas.edu/clsb/wiki. The peptide molecule is solvated in a periodic box (20 Å)3 of 248 TIP3P water molecules. The OPLS force field,18 is used with electrostatics real space cutoff of 9 Å augmented with Particle Mesh Ewald summation. Van der Waals interactions are cut at a distance of 8 Å. All calculations were run in NVT ensemble at a temperature of 303 K by employing a weak Andersen thermostat that acts only on the center-of-mass motion of the water molecules.19 The probability of velocity re-sampling was set to 5 · 10−4 per fs. For a water box of this size an average of 13 water molecules had their velocities re-sampled in a 100 fs interval. This weak coupling does not change the transition rate obtained from NVE (Newtonian) simulations (with initial conditions sampled from the NVT ensemble). The free energy surface as a function of the two dihedral angles (ϕ, ψ) is shown in Fig. 5. It was calculated from statistics of a 340 ns long MD simulation. The white region of the map was not visited by the trajectory. There are two local free energy minima corresponding to an α helix conformation (ϕ, ψ = −100, −40) and to a β sheet conformation (ϕ, ψ = −100, −140).
Figure 5.
Free energy profile of alanine dipeptide as a function of the two dihedral angles ϕ and ψ. It was calculated from statistics of a 340 ns long MD simulation. Images for DiM calculations are placed at the positions of the red numbers and for MMVT calculation at the location of the black points. Both algorithms with these placement of images infer the Milestones in the positions of the dashed lines, in DiM, however there are two directional Milestones for each line.
The height of the free energy barrier between the two metastable regions at 303K is less than 2kBT and the transitions between the metastable states are rapid on the trajectory time scale so the MFPTs can be estimated from straightforward MD simulations directly. We have performed five independent MD simulations of 68 ns. In each of the simulations more than 1000 transitions between the metastable regions occurred. The MFPT of α → β transition is 66.4 ps (± 2.7 ps) and that of the opposite transition is 53.8 ps (± 4.6 ps). We set up the Milestoning calculation by placing six images in the conformational space in the positions ϕi, ψi = −100°, −240°+60°i, (i = 1, …, 6). The positions of the images were not optimized. They were placed equidistantly in the region of conformation space that is accessible to the molecule. Table 1 shows the results of the Milestoning calculations for this system; it also includes the results of Markovian Milestoning with Voronoi Tessellation method.7 The MMVT calculation was performed with the same settings as for DiM, with the exception of the image placement; images for MMVT calculation were placed at (for) ϕi', ψi'=−100°,−210°+60°i (for i = 1, 2, …, 6) so that the Milestones are placed in the same positions as in Directional Milestoning.
Table 1.
Results of the MFPT calculations on alanine dipeptide solvated in water with 6 cells placed as shown on Fig. 5. Exact MFPTs were calculated by running five 68 ns long MD trajectories. The standard deviation of predicted MFPT of DiM and MD calculations are given in the brackets. For DiM, standard deviation was calculated from a single execution by using Bayesian inference (details in Appendix B). The total cost for DiM is given as a sum of the simulation time of all trajectories and the simulation time used for preparation of initial distributions.
| Method | MEPT [ps], (sd [ps]) α → β/β → α |
total cost [ns] |
|---|---|---|
| straightforward MD | 66.4 (2.7)/53.8 (4.6) | 68 |
| DiM, 100 trajectories/Milestone | 66.5 (11.1)/39.0 (4.6) | 5.0 + 0.6 = 5.6 |
| DiM, 250 trajectories/Milestone | 57.7 (5.4)/46.5 (3.6) | 12.5 + 1.0=13.5 |
| DiM, 500 trajectories/Milestone | 61.2 (4.2)/46.8 (2.6) | 22.8 + 2.0=24.8 |
| DiM, 1000 trajectories/Milestone | 57.0 (2.7)/45.2 (1.8) | 46.1 + 3.9=50.0 |
| DiM, 5000 trajectories/Milestone | 59.5 (1.3)/44.2(0.8) | 230 + 10.1=240.1 |
| MMVT, 0.4 ns /cell | 60.2/43.9 | 2.4 |
| MMVT, 0.8 ns/cell | 57.2/43.7 | 4.8 |
| MMVT, 1.6 ns/cell | 63.2/41.2 | 9.6 |
| MMVT, 3.4 ns/cell | 63.4/53.2 | 20.4 |
| MMVT, 12 ns/cell | 62.4/48.3 | 72.0 |
Note that the employed dynamics is almost deterministic and thus a trajectory reflected from an interface (procedure required in MMVT) would approximately track itself back in time. Therefore we have slightly modified the MMVT protocol in a way suggested by Vanden-Eijnden in a private communication: instead of reversing the velocities of all the degrees of freedom at a cell interface, only the velocities of peptide atoms are reversed. This modification should not influence the statistics of observed fluxes through the interfaces since only the peptide degrees of freedom are used in the definition of cell boundaries.
Both methods, DiM and MMVT, perform well in this scenario, though MMVT is more efficient for this simple system. If enough sampling is done, both techniques provide reasonable estimates of MFPTs between the metastable regions, the systematic error is lower for MMVT (6 % and 10%) as compared to our method (10 % and 18 %). Analysis of MMVT on the same system was performed recently.8 A different force field was employed in [8] and the MFPT reported differs by a factor of two from our calculations; however the relative error of MMVT for the reported α → β transition is about 6%, which is comparable to our result. Results of β → α transition were not reported in 8. Table 1 shows that MMVT needs about 2–3 times less CPU time compared to DiM to converge. DiM requires more computations in these setting since each interface of MMVT is effectively doubled for the two different directions. Furthermore, additional computation is needed in DiM to sample initial phase space points on each interface. In this one-dimensional set-up of Milestones with relatively large separation between Milestones and low free energy barrier MMVT is more efficient and as accurate as DiM. However, we will show below that with smaller separation between the interfaces, multi-dimensional arrangement of milestones, and rougher energy landscapes, DiM is better.
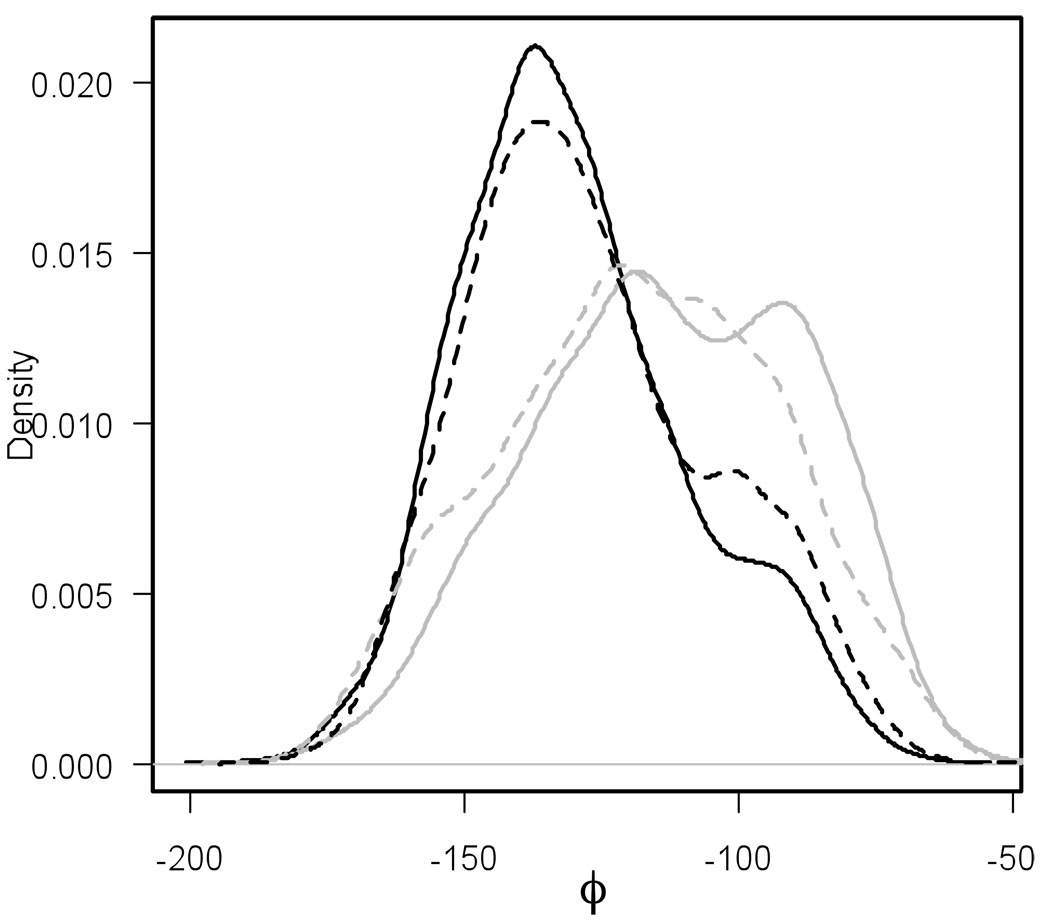
Even though previous Milestoning studies calculated accurately MFPTs on alanine dipeptide, memory effects in the system are not negligible. First hitting point distributions (in terms of ϕ angles) for the Milestones M4→5 and M6→5 are shown on Fig. 6. There is a noticeable difference between distributions of first hitting points on the Milestone M4→5 and on the Milestone M6→5. As shown on the figure, the approximate sampling described in Section 2.4 distinguishes the first hitting point distributions arriving from different directions to the region of image X5 reasonably well.
Figure 6.
Distributions of ϕ angle of the first hitting point conformations of the region of image X5 (located at ψ = 80°): distributions observed in a long MD simulation for conformations arriving to the hypersurface at X5 from the hypersurface of X4 (black solid), or from that of X6 (gray solid). Distributions sampled on the Milestone M4→5 (black dashed) and the Milestone M6→5 (gray dashed).
In Table 2, we examine the use of directional Milestones on this system. The table shows that transitions between the six Milestones (if direction is not part of the description) are not Markovian. If no memory effects were present in the system then the probability of transiting to Milestone i+1 from Milestone i would not depend on the Milestone visited before i, i.e. the second and the forth columns of Table 2 would be the same within the error bars. We however see differences of up to 21% (for i=5) or by a factor of up to 2.2 (for i=1). One can see that the values of these relative probabilities estimated by Directional Milestoning (columns 3 and 5 in Table 2) are in good agreement with the true values.
Table 2.
This table shows that dynamics of the alanine dipeptide system is not fully reducible to a Markov jump process between six hypersurfaces shown on Fig 5. The probability of jumping to the Milestone i + 1 from the Milestone i depends on the Milestone visited before i. Probabilities (from a long MD trajectory) of jumping from i to i + 1 if the Milestone i − 1 (i + 1) was visited before the hypersurface i are listed in the second (fourth) column. The third and fifth columns list these probabilities as measured by DiM calculation by starting 1000 trajectories from each Milestone. Note that in contrast to DiM, the original Milestoning assumes that P(i → i +1|i − 1 → i) = P(i → i +1|i + 1 → i).
| i | P(i → i + 1|i − 1 → i) | NMi−1→iMi→i+1/NMi−1→i | P(i → i + 1|i + 1 → i) | NMi+1→iMi→i+1/NMi+1→i |
|---|---|---|---|---|
| 1 | 3.9 | 3.6 | 8.6 | 8.3 |
| 2 | 82.4 | 84.8 | 89.4 | 92.0 |
| 3 | 84.9 | 88.1 | 91.0 | 88.0 |
| 4 | 39.0 | 37.5 | 49.0 | 50.0 |
| 5 | 39.2 | 41.4 | 60.6 | 50.5 |
| 6 | 26.3 | 32.0 | 35.0 | 34.1 |
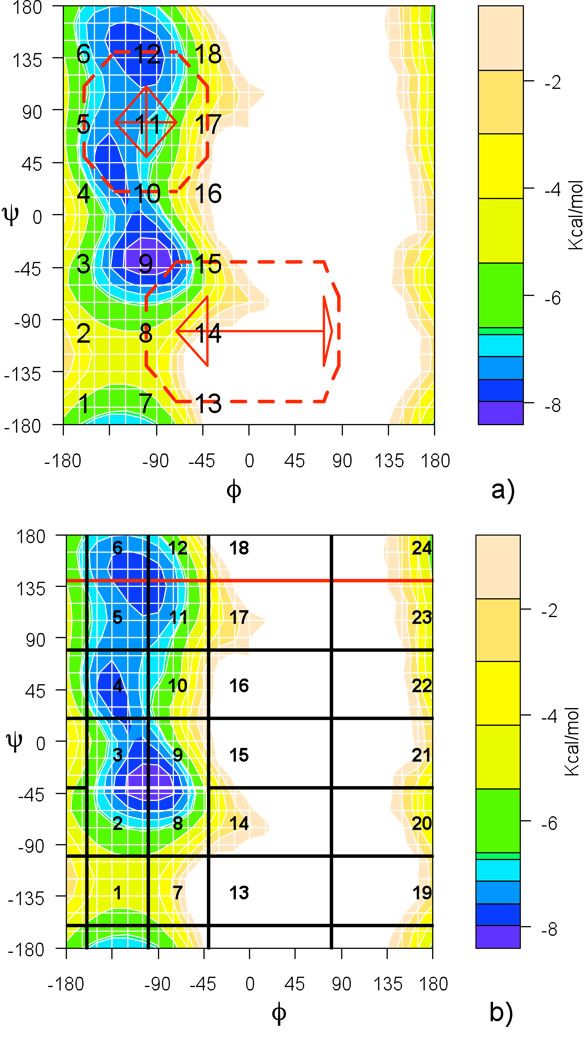
In the second experiment we examine both methods (DiM and MMVT) on the same system with Milestones in more than one dimension. This experiment is performed to empirically illustrate that placing Milestones in a non-linear arrangement does not compromise accuracy of DiM calculations. Images are placed in a two-dimensional grid covering the accessible space at the target temperature (conformations with torsional angle ϕ < 0). For DiM, 18 images are placed in the positions marked on 1, …, 18 on Fig. 7a). Each image has 8 incoming Milestones and 8 outgoing Milestones (displayed in solid and dashed on Fig. 7a) respectively). We calculated the MFPT from M12→11 (or M10→11) to the union of M10→9 and M8→9 for the β → α transition. The MFPTs from these two Milestones differ from each other by about 0.3 ps and we report their average in Table 3. The opposite transition (α → β) was defined in the equivalent way.
Figure 7.
Placement of images on a two dimensional grid. (a) DiM settings: total of 18 images, located at position of numbers in the plot, are placed in a two dimensional grid. For two of the images, X11 and X14, the outgoing (dashed) and incoming (solid) Milestones are shown. (b) Arrangment for MMVT. 24 images are placed in the conformational space so the resulting milestones are in the positions equivalent to DiM. Average of MFPT from the two white Milestones to any of the red Milestones is reported in the results for α → β transition.
Table 3.
Results of the MFPT calculations on alanine dipeptide solvated in water with 18 cells placed as on Fig. 7a. Standard deviations are in the brackets. Total cost for DiM is given as a sum of the simulation time of all trajectories and the simulation time used for preparation of the initial ensemble on each Milestone.
| Method | MEPT [ps], (sd [ps]) α → β/β → α |
total cost [ns] |
|---|---|---|
| straightforward MD | 66.4 (2.7)/53.8 (4.6) | 68 |
| DiM, 100 trajectories/Milestone | 68.2 (10.0)/56.9 (8.9) | 10.0 + 2.6 = 12.6 |
| DiM, 300 trajectories/Milestone | 63.5 (4.9)/56.6 (4.1) | 31.1 + 8.7= 39.8 |
| DiM, 1000 trajectories/Milestone | 62.8 (2.5)/53.2 (1.6) | 103 + 26=129 |
| DiM, 2000 trajectories/Milestone | 65.7 (1.6)/52.2 (1.1) | 207 + 52=259 |
| MMVT, 5 ns/cell | 48.6/37.0 | 120 |
| MMVT, 10 ns/cell | 44.3/37.1 | 240 |
For MMVT the images were placed in slightly different positions than for DiM (see Fig. 7b) such that the Milestones inferred by the Voronoi Tessellation are in equivalent positions to those used in Directional Milestoning. For the α → β transitions, we calculated the MFPT of trajectories starting from the two white Milestones in Fig. 7b (M2↔3 and M8↔9) and terminating at the union of the red Milestones. MFPT of the transitions from these two starting points differ by less than 0.2 ps so only their average is reported in Table 3. The β → α calculation was performed in the equivalent way (from the two central Milestones in the β sheet conformation (ψ = 140°) to the union of all the Milestones with ψ = −40°).
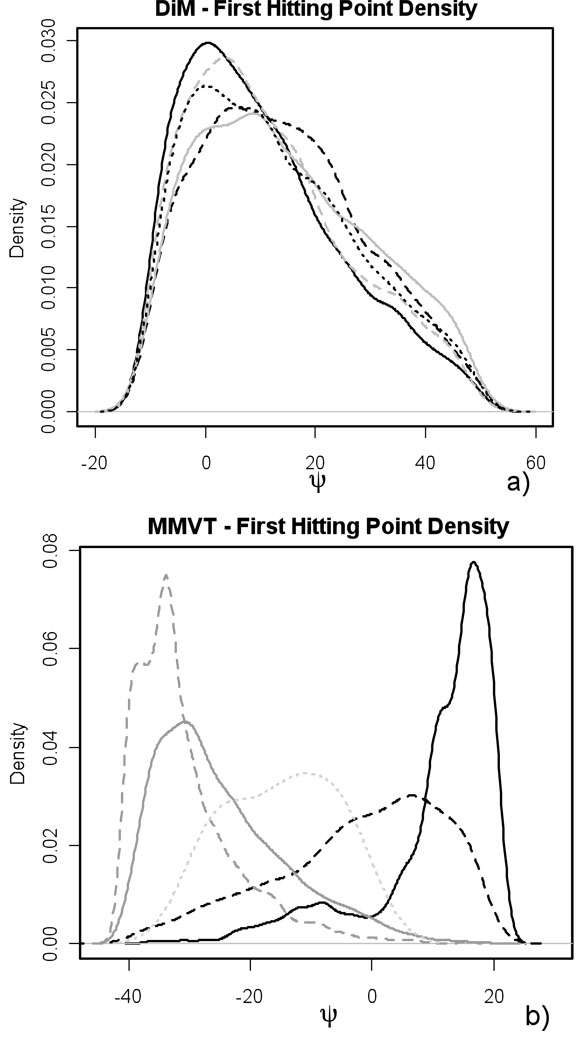
The results of both methods are listed in Table 3. The accuracy of Directional Milestoning is not compromised by multidimensionality; hence DiM works well for higher dimensions or higher connectivity of Milestones. The relative error of the MMVT method increased to 33 % (31 %). We think that this is mainly due to the corners between Milestones in the MMVT arrangement that cause rapid termination times between nearby Milestones and unwanted correlations between touching Milestones. Evidence of this can be seen in Fig. 8.
Figure 8.
First hitting point distributions. (a) For DiM, distribution of ψ torsional angle of conformations arriving to the Milestone M4→10 from the Milestones M9→4 (black, solid), M11→4 (black, dashed), M10→4 (gray, solid), M3→4 (gray, dashed), and M5→4 (black, dotted). (b) For MMVT, distribution of ψ torsional angle of conformations arriving to the Milestone M3↔9 from the Milestones M10↔9 (black, solid), M4↔3 (black, dashed), M8↔9 (gray, solid), M2↔3(gray, dashed), and M15↔9 (gray, dotted). Note that the target Milestone M3↔9 in the MMVT arrangement is in the same position as M4↔10 in the DiM arrangement, only shifted by 30° in ψ direction.
3.2 Alanine dipeptide in vacuum
In vacuum there are two stable conformers C7eq and Cax of alanine dipeptide (Fig. 9). The state C7eq is further split into two sub-states denoted by C7eq and (located at X26 in Fig. 9b) separated by a small barrier. We calculate the MFPT of transition from C7eq to Cax at two different temperatures, 400 K and 350 K, using Langevin dynamics. This is performed by calculating MFPT starting from each of the incoming Milestones to C7eq region (green on Fig. 9) and considering union of the incoming Milestones to the region Cax (red on Fig. 9) as the final state. The MFPT is not sensitive to exact identity of the starting Milestone (variation of less than 2%) therefore an average MFPT from all green Milestones is considered. The friction constant of Langevin dynamics was set to 30 ps−1.
Figure 9.
The shown landscape is an adiabatic ϕ, ψ energy map. The energy is minimized while constraining the ϕ and ψ dihedrals to specified values. Placement of (a) 24 images, (b) 63 images in the conformational space based on the algorithm described in Subsection 3.1.1 is shown. Also displayed is the Voronoi Tessellation based on the periodic Euclidean metric in the reduced space of ϕ and ψ torsions.
3.2.1 Image and cell generation
The images were generated by the following expansion. We start with the set of images S = {X1, X2}, where X1 is a conformation located at Cax and X2 at C7eq. Then we iteratively pick an image X from the set S and “expand” it: We launch trajectories starting from X with randomly initiated velocities and run each of these trajectories until it departs at least a pre-specified distance δ from X. Then we cluster the set of end points of these trajectories to existing images in S and potentially add new images to the set S if there are end points that are farther than δ from all images of S. We repeat this process until no new images are generated, i.e. we have tried launching trajectories from all images in S and all end coordinates are in S. There are three parameters in this algorithm: (i) the distance cutoff δ, (ii) the number of expanding trajectories Ne, and (iii) the clustering algorithm employed. For alanine dipeptide we have used expectation-maximization as a clustering algorithm,20 with Ne set to 400 and two different values of δ, δ1 = 0.6 Å and δ2 = 0.4 Å. The root mean squared distance after optimal overlap21 (RMSD) is the distance metric (the RMSD between X1 and X2 is 1.25 Å) for the purposes of clustering as well as the distance function in the definition of Milestones (1).
3.2.2 Results for alanine dipeptide in vacuum
By using different values for δ we obtained sets of images of size 24 (for δ1) and 63 (for δ2); both are shown on Fig. 9. The tessellations shown in black in this figure are only approximate since they are based on the Euclidean distance in (ϕ, ψ) space, where the real interfaces (Milestones) are defined using the RMSD distance. The MFPT of the transitions between the metastable conformations are significantly longer than those in the solvated peptide due to higher free energy barriers. Table 4 and Table 5 summarize the results of the Milestoning calculations in this system. At the high temperature (400 K) both methods, DiM and MMVT, predict accurate MFPT from C7eq to Cax (with systematic error of about 10%). MMVT needs to run about 1.5 µs MD simulations to obtain converged results, while DiM requires about 2.5 µs. Both of them provide significant speed up against straightforward MD simulation, even though a rough estimate of MFPT of the C7eq to Cax transition can be obtained by running about 11 independent MD simulations (equivalent to 4 µs of the total simulation time); however, both MMVT and DiM can be trivially parallelized to thousands of CPUs, shortening the actual time to perform the calculation.
Table 4.
Results of the MFPT calculations on alanine dipeptide in vacuum with 24 cells placed as on Fig. 9a) at temperature 400 K. Standard deviations are in the brackets. Estimation of the exact MFPT was performed by launching five groups of 400 trajectories from C7eq state and running them until Cax state is reached (the MFPT reported in the table is calculated as the MFPT of all 2000 trajectories; the error is estimated by standard deviation of MFPTs calculated from each of the five groups). Total cost for DiM is given as a sum of the simulation time of all trajectories and the simulation time used for preparation of the initial ensemble on each Milestone.
| method | MFPT [ns] | total cost [µs] |
|---|---|---|
| straightforward MD at T = 400 K | 375 (16) | 150 |
| DiM, 500 trajectories/Milestone | 630 (299) | 0.13 + 0.09 = 0.22 |
| DiM, 1K trajectories/Milestone | 217 (103) | 0.26 + 0.18 = 0.46 |
| DiM, 3K trajectories/Milestone | 306 (76) | 0.78 + 0.47 = 1.25 |
| DiM, 10K trajectories/Milestone | 344 (37) | 2.6 + 1.6 = 4.2 |
| DiM, 20K trajectories/Milestone | 387 (34) | 5.2 + 3.1 = 8.3 |
| DiM, 30K trajectories/Milestone | 352 (31) | 7.8 + 4.7 = 12.5 |
| MMVT, 10 ns /cell | 135 | 0.24 |
| MMVT, 20 ns /cell | 289 | 0.48 |
| MMVT, 40 ns /cell | 322 | 0.96 |
| MMVT, 60 ns /cell | 359 | 1.5 |
| MMVT, 130 ns /cell | 351 | 3.1 |
| MMVT, 400 ns /cell | 336 | 9.6 |
Table 5.
Results of the MFPT calculations on alanine dipeptide in vacuum with cells placed as on Fig. 9a/b) at temperature 350 K. DiM was performed with 24 cells, MMVT in two different settings: 24 and 63 cells. Standard deviations are in the brackets. Estimation of the exact MFPT was performed by launching five groups of 200 trajectories from C7eq state and running them until Cax state is reached. Standard deviation and average of the MFPT calculated from each group are reported in the table. Total cost for DiM is given as a sum of the simulation time of all trajectories and the simulation time used for preparation of the initial ensemble on each Milestone.
| method | MEFP [µs] | total cost [µa] |
|---|---|---|
| straightforward MD at T = 350 K | 2.05 (0.3) | 410 |
| DiM, 5K trajectories/Milestone | 2.78 (0.65) | 2.3 + 1.4 = 3.7 |
| DiM, 10K trajectories/Milestone | 1.74 (0.40) | 4.7 + 2.8 = 7.5 |
| DiM, 20K trajectories/Milestone | 1.75 (0.33) | 9.4 + 5.6 = 15.0 |
| DiM, 60K trajectories/Milestone | 1.77 (0.20) | 28 + 16.8 = 44.8 |
| MMVT, 24 cells, 2.00 µs /cell | 69.7 | 48 |
| MMVT, 63 cells, 0.75 µs /cell | 3798 | 47 |
| MMVT, 63 cells, 2.25 µs /cell | 855 | 142 |
When the temperature is lowered to 350 K (see Table 5) the C7eq to Cax transition is slower with MFPT of about 2.0 µs. As listed in Table 5, Directional Milestoning calculates the MFPT with systematic error of about 15% with as few as 7.5 µs of total simulation time. That is significant speedup compared to straightforward MD since DiM can be easily parallelized on thousands of processors. MMVT fails to calculate the MFPT accurately. The main reason for this failure is poor statistics. An important difference between DiM and MMVT is that DiM allocates computational resources to each Milestone, where MMVT allocates the computational resources to a cell. If a transition between two specific interfaces in a cell is needed to describe the reaction and the transition is significantly less likely than transitions between other interfaces of the cell, then sampling this transition using MMVT is inefficient. A simple realization of this effect is the existence of a barrier in the middle of the cell. In that case MMVT trajectory is likely to be confined to a one minimum, to collide with the same interface many times (hits that do not count for the statistics) and to record only a few successful transitions to the other minimum. In contrast DiM launches a large number of short trajectories. These trajectories terminate quickly, and contribute to the statistics.
In DiM, sampling is done (extensively) at the interfaces, so the probability of observing a transition between interfaces of interest is greatly enhanced, since at least one end of the transitional event is sampled extensively. A potential problem in DiM is a large number of interfaces that may make sampling expensive. To avoid sampling irrelevant interfaces (at a given temperature) trajectories are initiated at few initial interfaces and only interfaces that are hit at least once during the DiM calculation are sampled and launched. We stop the DiM calculation when the process converges (i.e. no new interfaces besides those already sampled are reached). For the MMVT calculation with 24 images, many cells cover a relatively large part of the conformational space with a rough energy landscape (see for example cell X6 on Fig. 9a). This arrangement may cause poor statistics for those regions since the trajectories spend most of their time in low free energy regions, rarely visiting interfaces higher in free energy. To increase the probability of having a double hit at the two desired surfaces, we run the same calculation with 63 images as well. But even when 63 images are used, the allocation of computational resources is highly unbalanced. For example, we consider the frequency of hitting the interfaces 49 → 47 and 33 → 47 (displayed in white on Fig. 9b) that are both important for the overall MFPT. In both, 49 and 33 cells, confined simulations of total time of 2.25 µs hit a cell boundary more than 2 · 107 times. However, the interface 33 → 47 is hit only 17 times and the interface 49 → 47 only 7 times. In contrast DiM allocates an equal number of starting trajectories to each of the Milestones and transitions from Milestones located near the transition states are sampled as well as other Milestones. We have not experimented with any selection criterions for allocation of computational resources to different cells (in MMVT) or to different Milestones (in DiM) but both methods may benefit from selective allocation of resources to “important regions” of conformational space.
4. Discussions and conclusions
In this paper, we proposed a method to compute dynamics in high dimensions called Directional Milestoning. We have shown that the mean first passage times between Milestones can be calculated accurately given that the distribution at which a Milestone is hit does not depend on the previously assigned Milestone (the assumption formulated in equation (4)). Directional Milestoning arranges dividing hypersufaces in a special way, aiming to satisfy the above assumption: i) Milestones in DiM are made directional, so the local progress of the reaction (going from the region of Xi to Xj as opposed to being at the interface between Xi and Xj) is made part of the description, ii) the arrangement of Milestones guarantees a lower bound on spatial separation of any connected pair of Milestones so trajectories initiated on a Milestone have space and time to “lose memory” before terminating on a different Milestone.
The algorithm, while based on the trajectory fragments of Milestoning, is a step in the direction of Transition Interface Sampling 11–13 (TIS) and the Forward Flux Sampling (FFS) methods22,23 compared to the original Milestoning. Here we use some trajectory tracking. The main difference between these methods and Directional Milestoning is that TIS and FFS are tracking trajectories all the way back to the reactant state. This tracking has the advantage of not relying on any assumption about the initial ensemble on an interface like is done in Milestoning. On the other hand, sampling of trajectories in TIS and FFS is computationally more expensive than in Milestoning because every attempted trajectory in these methods is tracked back to the reactant state where in (Directional) Milestoning a trajectory is tracked only until it reaches a different Milestone. Computations of trajectory fragments can be done in Milestoning in a massively parallel way. The PPTIS method uses a conceptually similar approach of trajectory fragments 24.
An important distinction of Directional Milestoning compared to TIS, FFS, and the original Milestoning is that it allows for arbitrary arrangement of Milestones in conformational space, not necessarily following a linear arrangement along an order parameter or a reaction coordinate. A similar (arbitrary) arrangement of interfaces is used in the MMVT method,7 nonequilibrium umbrella sampling method,25,26 and Trajectory Parallelization and Tilting method.27 The last two techniques are using short trajectories in cells and balance the fluxes between cells. Recently the non-equilibrium umbrella sampling26 was illustrated to be more efficient than FFS 28 The Weighted Ensemble approach was also shown recently to work without a reaction coordinate 29.
We have compared DiM with MMVT and showed that the performance of MMVT (in terms of effectiveness and correctness) is comparable to that of DiM in some of the examples, but that the correctness and/or effectiveness of MMVT can be compromised in systems with high free energy barriers, or in cells with two interfaces that are hard to reach. Another problem for straightforward implementation of MMVT is the existence of corners between Milestones along more than one dimension that contribute to termination times that are too short. So while DiM is in general somewhat slower than MMVT it provides reliable results more consistently, including cases in which MMVT fails.
We also would like to comment on the similarities (and the differences) of our approach to the Markov State Model (MSM -- for a recent study see 30). In the applications of MSM that we are aware of, long to very long Molecular Dynamics trajectories at normal conditions are used to estimate transition times and population of different cells. MMVT and DiM are designed to avoid such long trajectories (at the cost of approximate matching of probability densities at the interfaces). Once a sample of conformational space is available (which can be done in numerous ways, reaction path calculations, replica exchange simulations, or high temperature trajectories) only very short Molecular Dynamics trajectories are required to estimate the local kinetics. These short trajectories can be trivially parallelized providing profound computational saving compared to straightforward Molecular Dynamics simulations. While significant progress has been made in parallelizing a single trajectory 31, overhead still remains and special hardware that is frequently used is more expensive to buy and to maintain
Acknowledgements
This research was supported in part by NIH grant GM059796 and NSF grant 0833162 to RE.
Appendix A
Lemmas regarding the Milestones geometry
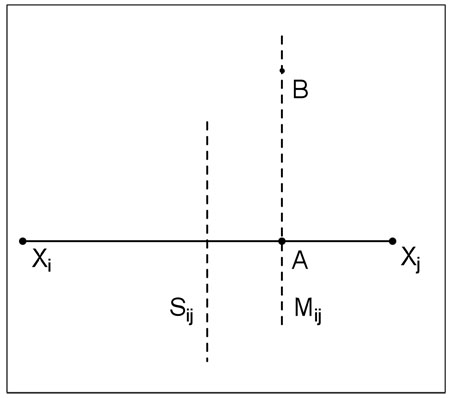
Lemma A.1
Let Xi and Xj be two images in conformation space such that Mi → j exists. Let A be an intersection of the line segment XiXj with Mi → j. Then a point B on the hyperplane perpendicular to XiXj and passing through A belongs to Mi → j iff ∀k d(Xk, B) ≥ d(Xj, B).
Proof of Lemma A.1: From def. (1) of Mi → j:
By using the Pythagoras theorem for triangles and XiAB and XjAB:
Consequence of Lemma 1: Mi → j is a hyperplane segment perpendicular to XiXj.
Lemma 2
Let Sij be the hyperplane perpendicular to the line segment XiXj and passing though its midpoint. Then .
Proof of Lemma 2: Since both Sij and Mi → j are perpendicular XiXj to the distance d(Sij, Mi → j) is equal to the distance of the XiXj midpoint, Pij, and the intersect of Mi → j with Xij, A. Thus:
Appendix B
Statistical reasoning
We describe an estimate of the statistical error of a milestoning calculation from a single set of collected data using Bayesian reasoning. As shown in Section 2, equation (5), repeated here as (B.1),
| (B.1) |
relates MFPTs (〈ταβ〉) and local kinetics entities (〈tα〉 and P(γ|α)). Milestoning aims to estimate 〈tα〉 and P(γ|α) by launching Nα trajectories from each Milestone α. Nαγ of them terminate on the Milestone γ and the mean incubation time (time to termination) of all trajectories is t̄α. In Bayesian inference a statistical model of the transitions among Milestones is needed. We closely follow and extend notation used in the analysis of Markovian Milestoning with Voronoi Tesselations; for more details consult 7. The same kinetic formulas (with different notation) are also available from 9. We assume a continuous Markov jump process between the Milestones controlled by a transition matrix Q defined in the following way: let the probability distribution of the system over all the Milestones be, ρ = (ρ1,…,ρN) where ρα is the probability that the system is assigned to a Milestone α. Under continuous Markov jump process, ρ behaves as:
| (B.2) |
For transition matrix Q, by definition qαα = −∑α ≠ βqαβ and it can be shown by simple algebra that P(β|α) = qβα/∑γ ≠ αqγα and 〈tα〉 =1/∑γ ≠ α qγα (for derivation see for example 1,2,7). By plugging the last three identities to the linear system (B.1) it reduces to
| (B.3) |
where 〈τ〉 is the row vector (〈τ1β〉, …, 〈τβ−1β〉, 〈τβ+1β〉, …, 〈τNβ〉)T and Q' is a (N − 1) × (N − 1) matrix created from Q by skipping the row and the column related to the Milestone β. In order to infer 〈τ〉 from the collected data, {Nαγ, t̄α}, using Eq. (B.3), a relation between {Nαγ, t̄α} and Q' is needed. Following the derivations from ref. 7: for a system ruled by (B.2) the probability of staying in a state α for time t and then jumping to a state β in the time interval <t,t + dt> is e−∑γ≠αqαγtqαβ dt. Using this equality,s the likelihood of observing the collected data, L({Nαγ, t̄α}|Q), is
| (B.4) |
By using the Bayes’ rule the likelihood that the true transition matrix is Q given the collected data, L(Q|{Nαγ, t̄α}), is:
| (B.5) |
where P(Q) is the prior probability distribution of Q without seeing any data (typically this is set to uniform if we do not have any prior knowledge about the system). Eq. (B.5) is typically used in maximum likelihood estimators, e.g. one estimates unknown entity Q with Q*, the matrix that maximizes likelihood L(Q|{Nαγ, t̄α}). In this particular case, Q* has form what is in agreement with estimators given in Eq. (6) in the main text. Instead of using purely Q* for calculations of MFPTs we can examine whole distribution of transition matrices according to Eq. (B.5) and understand what is the distribution of MFPTs consistent with the data collected. Therefore we typically sample a number of (typically 300) transition matrices from distribution (B.5) and look at the variance of MFPTs predicted by them. If standard deviation of MFPTs is large it suggests that more data about the system shall be collected. We report standard deviation obtained by this algorithm in the results of Section 3.
Appendix C
Sampling equilibrium distribution on a Milestone using Umbrella Sampling
As described in Section 2.4 the equilibrium ensemble from a Milestone Mi → j is used to sample the first hitting point distribution on the Milestone Mi → j. The Milestone Mi → j is defined in equation (1) as , where {X1,…,XK} is a set of images in the conformational space. In practice we work with the following approximation of Mi → j:
| (1.1) |
Clearly as λ→0, converges to Mi → j. We have used λ = 0.5° or λ = 0.01 Å for the calculations on alanine dipeptide.
To sample conformations in from equilibrium distribution the following Umbrella Sampling protocol is employed. We run NVT trajectory of the system (using Andersen thermostat) with a modified potential function U and examine a conformation every few steps (every 100 – 400 fs for examples described in this paper). If an examined conformation belongs to it is saved; otherwise it is discarded. If conformation is saved, corresponding velocities are sampled from Boltzmann distribution. The potential function U is modified to bias the system towards the region in the following way:
By definition for , U'(X) = U(X) and therefore saved points from are sampled with the true equilibrium probabilities. If on the other hand NVT trajectory of the system is outside of the region , the terms and/or force the system to return back to , the strength of this bias is controlled by force constants K1 and K2 (both are set to 103 Kcal mol−1 rad−2 or 104 Kcal mol−1 Å−2 for alanine dipeptide system).
REFERENCES
- 1.West AMA, Elber R, Shalloway D. The Journal of Chemical Physics. 2007;126:145104. doi: 10.1063/1.2716389. [DOI] [PubMed] [Google Scholar]
- 2.Shalloway D, Faradjian AK. The Journal of Chemical Physics. 2006;124:054112. doi: 10.1063/1.2161211. [DOI] [PubMed] [Google Scholar]
- 3.Faradjian AK, Elber R. The Journal of Chemical Physics. 2004;120:10880. doi: 10.1063/1.1738640. [DOI] [PubMed] [Google Scholar]
- 4.Vanden-Eijnden E, Venturoli M, Ciccotti G, Elber R. The Journal of Chemical Physics. 2008;129:174102. doi: 10.1063/1.2996509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elber R. 2007;92:L85. doi: 10.1529/biophysj.106.101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuczera K, Jas GS, Elber R. The Journal of Physical Chemistry A. 2009;113:7461. doi: 10.1021/jp900407w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanden-Eijnden E, Venturoli M. The Journal of Chemical Physics. 2009;130:194101. doi: 10.1063/1.3129843. [DOI] [PubMed] [Google Scholar]
- 8.Maragliano L, Vanden-Eijnden E, Roux B. Journal of Chemical Theory and Computation. 2009;5:2589. doi: 10.1021/ct900279z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West AMA, Elber R, Shalloway D. Journal of Chemical Physics. 2007;126 doi: 10.1063/1.2716389. [DOI] [PubMed] [Google Scholar]
- 10.Torrie GM, Valleau JP. Journal of Computational Physics. 1977;23:187. [Google Scholar]
- 11.van Erp TS, Bolhuis PG. Journal of Computational Physics. 2005;205:157. [Google Scholar]
- 12.Moroni D, van Erp TS, Bolhuis PG. Physica A: Statistical Mechanics and its Applications. 2004;340:395. [Google Scholar]
- 13.Moroni D, Bolhuis PG, van Erp TS. The Journal of Chemical Physics. 2004;120:4055. doi: 10.1063/1.1644537. [DOI] [PubMed] [Google Scholar]
- 14.Ren W, Vanden-Eijnden E, Maragakis P, E W. The Journal of Chemical Physics. 2005;123:134109. doi: 10.1063/1.2013256. [DOI] [PubMed] [Google Scholar]
- 15.Maragliano L, Vanden-Eijnden E. The Journal of Chemical Physics. 2008;128:184110. doi: 10.1063/1.2907241. [DOI] [PubMed] [Google Scholar]
- 16.Ensing B, De Vivo M, Liu Z, Moore P, Klein ML. Accounts of Chemical Research. 2005;39:73. doi: 10.1021/ar040198i. [DOI] [PubMed] [Google Scholar]
- 17.Elber R, Roitberg A, Simmerling C, Goldstein R, Li H, Verkhivker G, Keasar C, Zhang J, Ulitsky A. Computer Physics Communications. 1995;91:159. [Google Scholar]
- 18.Jorgensen WL, Tirado-Rives J. Journal of the American Chemical Society. 2002;110:1657. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 19.Juraszek J, Bolhuis PG. 2008;95:4246. doi: 10.1529/biophysj.108.136267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartley H. Biometrics. 1958;14:174. [Google Scholar]
- 21.Kabsch W. Acta Crystallographica Section A. 1976;32:922. [Google Scholar]
- 22.Valeriani C, Allen RJ, Morelli MJ, Frenkel D, ten Wolde PR. The Journal of Chemical Physics. 2007;127:114109. doi: 10.1063/1.2767625. [DOI] [PubMed] [Google Scholar]
- 23.Allen RJ, Frenkel D, ten Wolde PR. The Journal of Chemical Physics. 2006;124:024102. doi: 10.1063/1.2140273. [DOI] [PubMed] [Google Scholar]
- 24.Moroni D, Bolhuis PG, van Erp TS. Journal of Chemical Physics. 2004;120:4055. doi: 10.1063/1.1644537. [DOI] [PubMed] [Google Scholar]
- 25.Warmflash A, Bhimalapuram P, Dinner AR. The Journal of Chemical Physics. 2007;127:154112. doi: 10.1063/1.2784118. [DOI] [PubMed] [Google Scholar]
- 26.Dickson A, Warmflash A, Dinner AR. The Journal of Chemical Physics. 2009;130:074104. doi: 10.1063/1.3070677. [DOI] [PubMed] [Google Scholar]
- 27.Vanden-Eijnden E, Venturoli M. The Journal of Chemical Physics. 2009;131:044120. doi: 10.1063/1.3180821. [DOI] [PubMed] [Google Scholar]
- 28.Allen RJ, Frenkel D, ten Wolde PR. Journal of Chemical Physics. 2006;124:17. doi: 10.1063/1.2140273. [DOI] [PubMed] [Google Scholar]
- 29.Zhang BW, Jasnow D, Zuckerman DM. Journal of Chemical Physics. 2010;132 doi: 10.1063/1.3306345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noe F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19011. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, Eastwood MP, Gagliardo J, Grossman JP, Ho CR, Ierardi DJ, Kolossvary I, Klepeis JL, Layman T, McLeavey C, Moraes MA, Mueller R, Priest EC, Shan YB, Spengler J, Theobald M, Towles B, Wang SC. Communications of the Acm. 2008;51:91. [Google Scholar]