Abstract
DDB2 is an essential subunit of the damaged-DNA recognition factor DDB, which is involved in global genomic repair in human cells. Moreover, DDB2 is mutated in the repair-deficiency disease xeroderma pigmentosum (Group E). Expression of DDB2 in human cells is induced by P53, BRCA1 and by ionizing radiation. The DDB2 protein associates with transcriptional activator and coactivator proteins. In addition, DDB2 in conjunction with DDB1 associates with cullin 4A and the Cop9/signalosome. We generated a mouse strain deficient for DDB2 (DDB2−/−). Consistent with the human disease (XP-E), the DDB2−/− mice were susceptible to UV-induced skin carcinogenesis. We observed a significant difference in the initial rate of cyclobutane pyrimidine dimer (CPD)-removal from the skin following UV irradiation. Also, the DDB2-deficient mice exhibited a significantly reduced life span compared to their wild-type littermates. Moreover, unlike other XP-deficient mice, the DDB2-deficient mice developed spontaneous malignant tumors at a high rate between the ages of 20 and 25 months. The observations suggest that, in addition to DNA repair, the other interactions of DDB2 are significant in its tumor suppression function.
Keywords: DDB2, DNA repair, XP-E, UV-induced tumor, DDB2 knockout mice
Introduction
DDB2 is a subunit of the damaged-DNA recognition factor DDB, which binds to various forms of damaged-DNA, including UV-induced 6–4 photoproducts, cis-platin-modified DNA and bent DNA (Abramic et al., 1991; Hwang and Chu, 1993; Keeney et al., 1993; Hwang et al., 1998a; Fujiwara et al., 1999). Moreover, DDB2 has been shown to participate in global nucleotide excision repair (NER) (Hwang et al., 1998a; Tang et al., 2000; Wakasugi et al., 2002). The proteins involved in the NER pathway are mutated in the eight different complementation groups of the repair-deficiency disease xeroderma pigmentosum (XP: A–G and V) (Friedberg et al., 1995; Sancar, 1996; Cleaver et al., 1999). XP is a rare autosomal recessive disease characterized by sun-sensitivity and a high incidence of skin malignancies, and these phenotypes are believed to arise from a deficiency in the NER pathway of DNA repair. The DDB2 gene has been linked to XP-E because patients belonging to the XP-E group were shown to harbor mutations in the DDB2 gene (Nichols et al., 1996, 2000; Chu and Chang, 1988; Itoh et al., 2000, 2003). Despite these studies indicating an involvement of DDB2 in DNA repair, it is not clear how DDB2 participates nucleotide excision repair (NER) because the NER assays work efficiently on naked DNA without DDB2 (Reardon et al., 1993; Kazantsev et al., 1996). In addition to its role in DNA repair, DDB2 has been also implicated in transcription. DDB2 associates with the transcriptional activator protein E2F1 (Hayes et al., 1998). Moreover, DDB2 also associates with the transcriptional coactivator proteins CBP/p300 and the STAGA complex (Datta et al., 2001; Martinez et al., 2001). The association with CBP/p300 and the STAGA complex suggests a role of DDB2 in chromatin remodeling and that might explain its repair and transcription functions. It is noteworthy that DDB is a target of viral proteins (Lee et al., 1995; Lin and Lamb, 2000).
The human DDB2 gene is a downstream target of p53, and it is transcriptionally activated by p53 (Hwang et al., 1998b). It was shown that p53 could associate directly with the transcription-regulatory region of the human DDB2 gene and stimulate expression. In addition to p53-mediated activation, the DDB2 gene is transcriptionally activated by the tumor suppressor BRCA1 (Takimoto et al., 2002). It was shown that BRCA1 could activate expression of DDB2 independently of p53 (Hartman and Ford, 2002). Therefore, it is believed that the human DDB2 gene is a downstream mediator in the tumor suppression pathways of p53 and BRCA1. Recent studies provided evidence for a feedback loop in which DDB2 plays a role in the stabilization of p53 (Itoh et al., 2000, 2003). The XP-E cells were shown to be somewhat impaired in activating the levels of p53 following UV irradiation, indicating a role of the DDB2 gene in the stabilization of p53 (Itoh et al., 2003).
DDB2 has been shown to associate with cullin 4A, an E3 ubiquitin ligase, which is likely to be involved in regulating the protein level of DDB2. DDB2 is a cell cycle-regulated protein. It is undetectable in nondividing cells; the level of DDB2 protein increases in the mid-G1 phase and remains high until the G1/S boundary, following which the level drops significantly to an undetectable level in S phase (Nag et al., 2001a, b). The cell cycle-regulation appears to be mainly at the protein level, as the mRNA level does not change significantly (Nag et al., 2001a). Inhibitors of the 26S proteasome cause an accumulation of the DDB2 protein, suggesting a role of the ubiquitin–proteasome pathway-mediated proteolysis in the cell cycle regulation of DDB2. Moreover, overexpression of cullin 4A induces ubiquitination and increased decay of DDB2 (Chen et al., 2001; Nag et al., 2001a). However, the proteolysis of DDB2 may not be the only consequence of the DDB2/cullin 4A interaction because a stable complex of DDB1/DDB2/cullin 4A can be easily detected in cell extracts (Shiyanov et al., 1999; Groisman et al., 2003). Interestingly, it was shown that the DDB1/DDB2/cullin 4A complex remains associated with the Cop9/signalosome (Groisman et al., 2003). UV irradiation causes a dissociation of the interaction with the signalosome and the released DDB1/DDB2/cullin 4A complex associates with the UV-damaged chromatin (Groisman et al., 2003), which is consistent with our previous observation that the DDB1/DDB2/cullin 4A complex binds to UV-damaged DNA with a high affinity (Shiyanov et al., 1999). Therefore, it is possible that DDB2 participates in global nucleotide excision repair by recruiting ubiquitinating enzymes, such as cullin 4A. Interestingly, the transcription-coupled repair protein CSA, like DDB2, also associates with the DDB1-cullin 4A–signalosome complex (Groisman et al., 2003).
Recently, Itoh et al. (2004) generated a strain of DDB2−/− mice and demonstrated a role of DDB2 in preventing UV-induced skin carcinogenesis. We, independently, have generated a strain of DDB2-deficient mice. Consistent with the observations of Itoh et al. (2004), our DDB2-deficient mice were highly susceptible to skin carcinogenesis following UV irradiation. We observed that, while a significant difference in repair could be detected in the skin, the mouse embryonic fibroblasts (MEFs) from the DDB2-deficient mice exhibit only a marginal deficiency in the repair of cyclobutane pyrimidine dimers. Surprisingly, the DDB2-deficient mice developed spontaneous tumors at a high rate. We suggest that the DDB2 protein participates in tumor suppression through multiple pathways.
Results
Targeted disruption of the mouse DDB2 gene
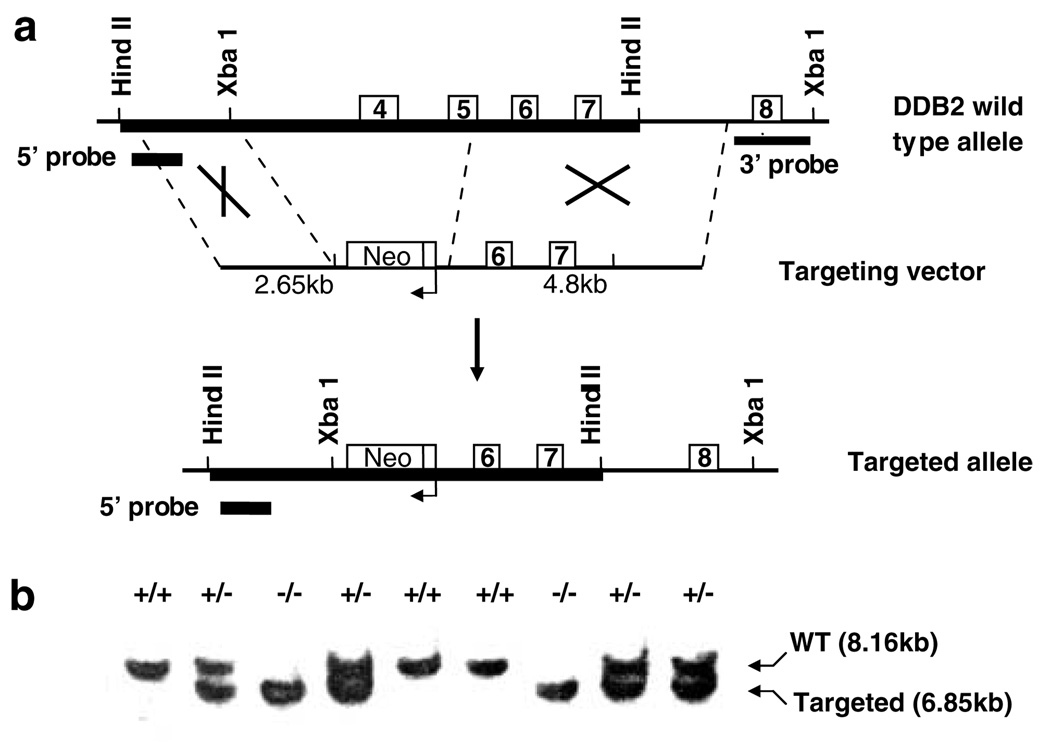
To investigate the in vivo function of DDB2, we sought to generate a strain of mice lacking expression of DDB2. The mouse DDB2-cDNA was characterized by the group of Linn (Zolezzi and Linn, 2000). It was shown that the mouse protein possesses 75% sequence identity and about 85% homology with the human DDB2. Linn’s group also carried out a partial characterization of the mouse DDB2 gene, which is identical to our data (not shown and see Zolezzi and Linn, 2000). To generate DDB2 null mutation, we deleted exons 4 and 5 through homologous recombination in ES cells (Figure 1a). Mouse chimeras were used to obtain stable heterozygotes, which were crossed to obtain the three genotypes. Southern blot experiments with genomic DNA from three genotypes of mice were performed to confirm the recombination. Probes corresponding to the 3′ and 5′ regions confirmed correct recombination. A Southern blot probed for the 5′ region is shown (Figure 1b). The available DDB2-antibody does not work well for mouse DDB2. Therefore, to detect expression of DDB2, total RNA from the MEFs and liver tissues of all three genotypes were analysed by a highly sensitive RT–PCR assay. The PCR primers corresponded to sequences present in both wild-type and mutated alleles. No DDB2-mRNA expression was detected in the MEFs or liver tissue of the DDB2−/− mice (Figure 2). The two PCR products in the wild-type and heterozygotes (Figure 2) corresponded to two alternatively spliced mRNAs: the shorter product corresponded to an mRNA lacking exon 4 (data not shown). The DDB2−/− mice appear to be devoid of developmental defect and are fertile. Analysis of hundreds of litters from crosses of the heterozygotes exhibited an expected Mendelian ratio of the three genotypes.
Figure 1.
Generation of DDB2−/− mice. (a) Targeted disruption strategy to delete the mouse DDB2 gene in embryonic stem (ES) cells. The thin line represents the intron sequences and boxes with numbers represent DDB2 exons. Shown from the top to bottom, the wild-type (WT) DDB2 allele, and the disrupted allele with neo cassette replacing exons 4 and 5. (b) Mouse genotyping by Southern blot analysis. Genotypes shown are the result of DDB2 heterozygous intercross mating. Mouse tail genomic DNA was digested with HindIII and subjected to Southern blot analysis using 5′ probe (833 bp, represented as thick lines on the 5′ side of wt and mutant allele) and also XbaI-digested DNA were probed with 3′ probe (1000 bp, represented as a thick line on the 3′ side of wild-type allele) to confirm correct recombination (data not shown). When probed with 5′ probe, expected 8.16 kb band was detected in the wild-type mice, and a 6.85 kb band was detected in homozygous DDB2 null mice. In heterozygotes, both bands were detected
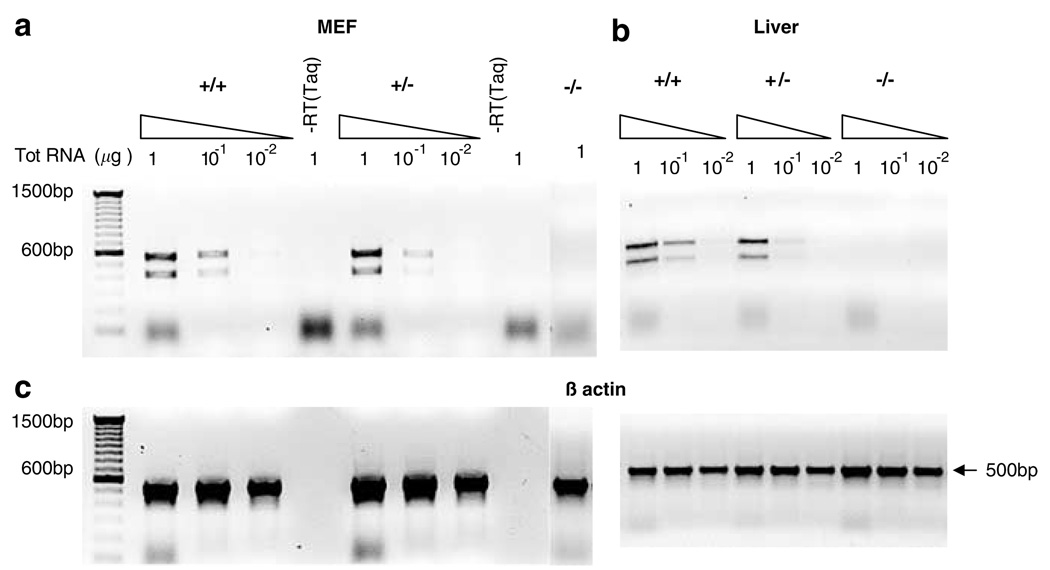
Figure 2.
Mouse DDB2 expression analysis by RT–PCR. Total RNA isolated from primary embryonic fibroblasts (MEFs) and liver tissues were reverse transcribed and amplified by RT–PCR. RT–PCR using primers which are designed to amplify from exon 3 to exon 6 revealed expected 572 bp fragment and a smaller 420 bp fragment in the wild type and in the heterozygous mice-derived RNA but not in the DDB2−/− mice-derived RNA samples. The smaller 420 bp fragment was further analysed by sequencing and identified as an alternatively spliced mRNA lacking the exon 4. (a) Varying amount of total RNA (1, 0.1 and 0.01 µg) from MEFs was assayed. Lanes marked as ‘–RT (Taq)’ represent negative controls by using Taq DNA polymerase instead of reverse transcriptase to detect any possible genomic DNA contamination. (b) RT-PCR results from liver tissue-derived RNA samples. (c) Loading controls showing 500 bp β-actin band amplification. Same amount of total RNA was used for β-actin band amplification as the lanes above in (a) and (b)
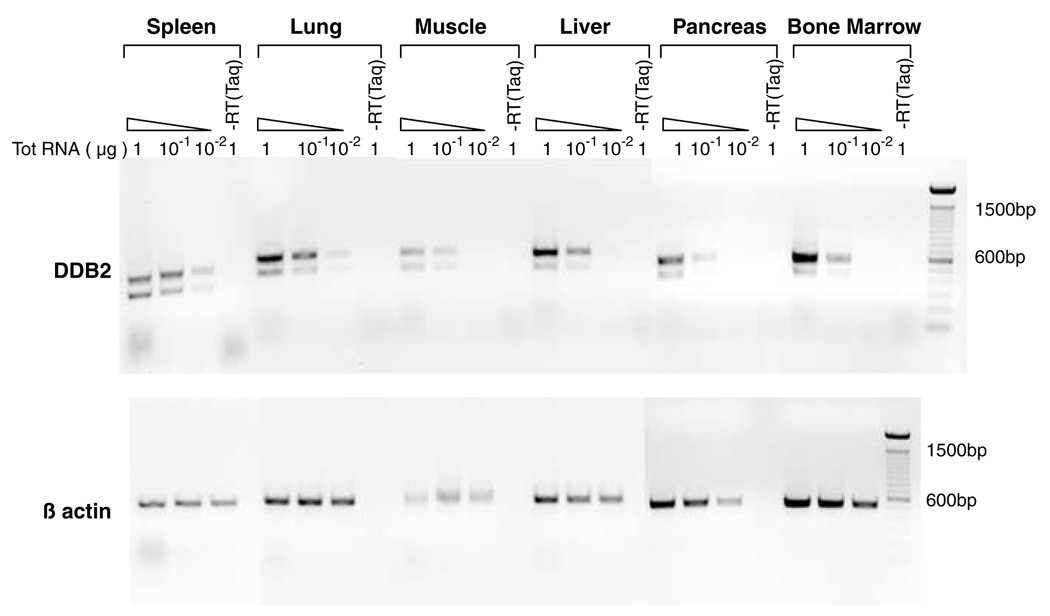
The human DDB2 gene exhibits a broad expression pattern based on RT–PCR assays of total RNA from various human tissues, including heart, lung, liver, brain and others (Inoki et al., 2004). To investigate the expression pattern of mouse DDB2, we analysed total RNA from muscle, lung, liver, pancreas, spleen and bone marrow. The expression was analysed using different levels of total RNA in RT–PCR assays. As shown in Figure 3, expression of DDB2-mRNA could be detected in all tissue-types tested. Those tissues also expressed the splice-variant lacking exon 4 only. Interestingly, that splice-variant was not observed in human tissues (Inoki et al., 2004). Thus, there are differences in the splicing patterns that generate the minor DDB2 splice-variants in humans and mice.
Figure 3.
Tissue distribution of DDB2. Total RNA isolated from various tissues of wild-type mice, including spleen, lung, muscle, liver, pancreas, and bone marrow were subjected to RT–PCR analysis using the same set of primers used for DDB2 expression analysis in Figure 2. Varying amount of total RNA (1, 0.1 and 0.01 µg) from different tissues was assayed to detect DDB2 within a linear range of amplification. Lanes marked as ‘–RT (Taq)’ represent negative controls by using Taq DNA polymerase instead of reverse transcriptase to detect any possible genomic DNA contamination. (Top) DDB2 expression in different tissues. (Bottom) Loading controls showing the 500 bp β-actin band amplification
DDB2−/− mice are susceptible to UV-induced skin cancers
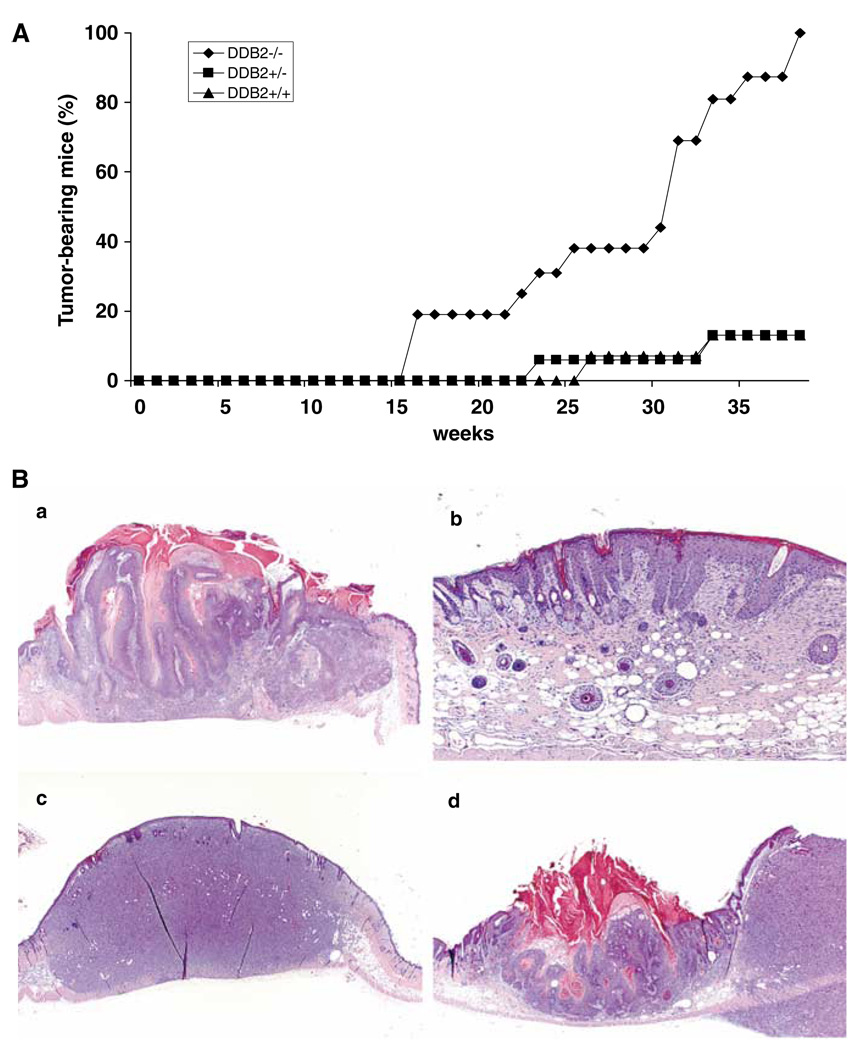
Mice harboring mutation in the XPA or XPC gene did not develop spontaneous tumors, however, they developed tumors at a high frequency upon UVB irradiation for a period of 20–30 weeks (Nakane et al., 1995; Sands et al., 1995). A recent study on DDB2−/− mice reported high incidences of UV-induced skin carcinogenesis (Itoh et al., 2004). In agreement with that study, we observed a high susceptibility to skin carcinogenesis by UV irradiation. In all, 15–16 mice of each genotype, 8–12 weeks of age, were subjected to UVB irradiation starting at 2 kJ/m2 3 days a week. The dosage of UVB irradiation was slowly increased to a maximum level of 6 kJ/m2 at the 33rd week. Irradiation was carried out only on a small shaved area on the dorsal side, and rest of the body was shielded from irradiation. By the end of the 38th week of irradiation, all mice (16 out of 16) with homozygous mutation in the DDB2 gene developed skin carcinomas, whereas only two out of 15 of the wild type or two out of 16 heterozygote mice developed skin carcinoma (Figure 4A). The tumors were confirmed by histological analysis (Figure 4B). Analysis of the tumor tissue sections from the DDB2−/− mice revealed several nonmelanoma types of skin cancer, including squamous cell carcinoma and soft-tissue sarcoma. The tumors were aggressive, as the tumor-bearing mice died within 3 weeks after tumor development, others were killed within 4 days after the appearance of the tumors. Our data on the heterozygotes are different from the previous report showing higher incidences of UV-induced carcinogenesis in the DDB2+/− mice (Itoh et al., 2004). We think that the difference resulted from the difference in the UV irradiation protocols.
Figure 4.
Tumor formation in DDB2−/− mice by chronic UVB irradiation. (A) Mice with 8–12 weeks of age were subjected to UVB irradiation (Fisher Scientific FB-UVXL1000 UV crosslinker with UVB tubes) starting at 2kJ/m2 3 days a week for up to 38 weeks. The dosage was slowly increased up to 6kJ/m2 by the 33rd week. Mice were shaved once a week and only the shaved dorsal area was exposed to UVB irradiation. After 38 weeks, two out of 15 DDB2+/+ mice (13%), two out of 16 DDB2+/− mice (13%), and all 16 out of 16 DDB2−/− mice (100%) developed tumors (P<0.0001). The tumors were confirmed by histological analysis. (B) Histological analysis of UVB-induced tumors in DDB2−/− mice. (a) Squamous carcinoma in situ (1.2 × 1.2 magnification). (b) Early squamous papilloma showing severe epidermal hyperplasia (4 × 1.2). (c) Soft-tissue sarcoma/fibrosarcoma (1.2 × 1.2). (d) Squamous papilloma and adjacent soft tissue sarcoma (1.2 × 1.2)
Deficiency in CPD removal following UVB irradiation
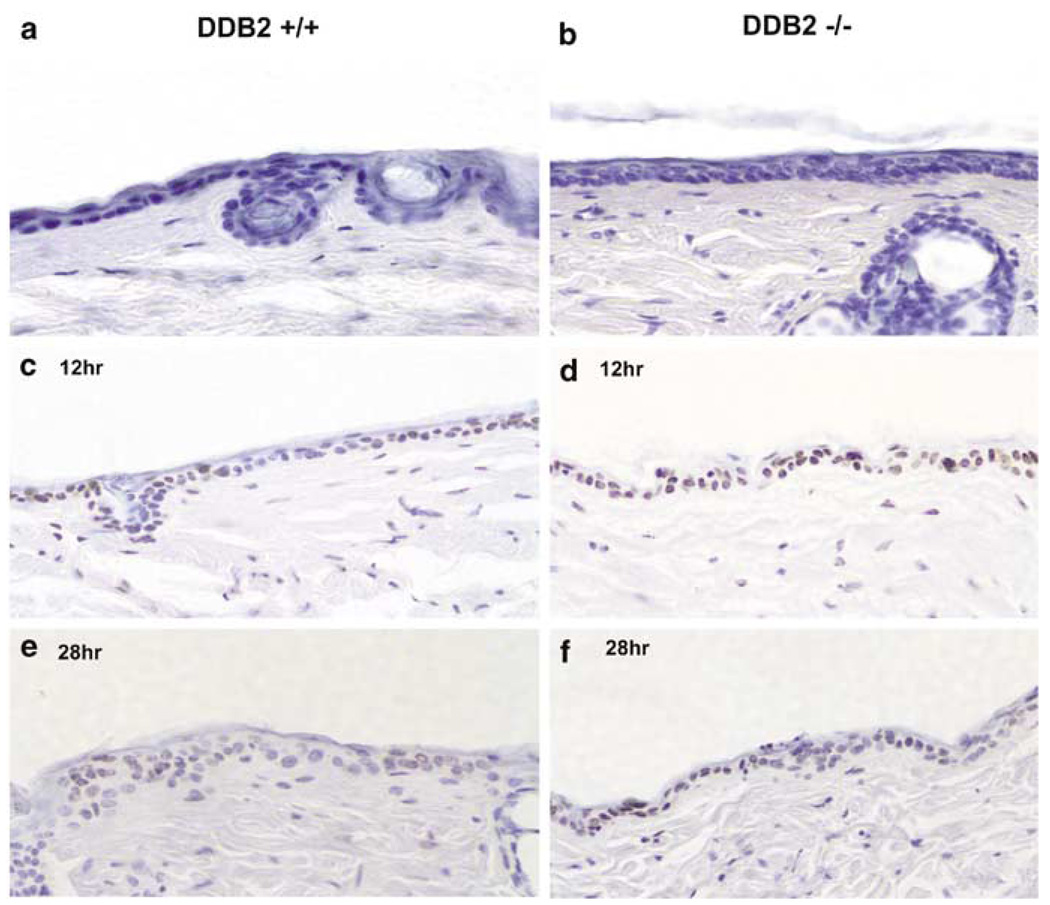
UVB irradiation damages DNA in several ways, including formation of thymine dimers or CPDs and 6–4 photoproducts. DDB2 was shown to stimulate removal of CPDs in an in vitro nucleotide excision repair assay (Wakasugi et al., 2002), as well as in cell culture-based assays (Iwao et al., 1999). We sought to investigate whether the DDB2−/− mice are capable of efficient repair of the CPDs. MEFs were shown to be deficient in nucleotide excision repair activities, which is believed to be the pathway stimulated by DDB2 (Tang et al., 2000). Therefore, we investigated repair of CPDs in the epidermal layers of the DDB2−/− mice. It was shown that the hairless SKH-1 strain of mice, within minutes following UVB treatment, generates CPDs, which are repaired between 12 and 36 h (Lu et al., 1999). To investigate repair of CPDs, wild-type and DDB2−/− mice (5–6 months old) were shaved in the dorsal region and the exposed skin regions were irradiated with UVB (2.5kJ/m2). At 12 and 28 h following irradiation, the mice were killed and skin samples containing epidermis and associated dermis were processed for immunohistochemical analysis. The CPDs in the epidermal cells were detected by a horse-radish peroxiodase-labeled thymine-dimer monoclonal antibody and visualized using a color (brown) assay (see Materials and methods). The sections were also counterstained with hematoxylin (blue) to visualize the cells. We analysed several skin sections from two mice for each time point for the CPDs using the monoclonal thymine-dimer antibody. We detected thymine dimers in both supra-basal and basal layers (Figure 5). Both at the 12 and 28 h time-points, we consistently detected a much higher percentage cells positive for thymine dimers in the skin sections of the DDB2−/− mice compared to the wild-type mice. The hematoxylin-stained cells and the thymine dimer-positive cells in 10 randomly chosen fields per section were counted (Table 1). At 12 h following UVB irradiation, 40 or 36% of the cells in the skin sections from the wild-type mice were positive for thymine dimers, whereas about 75 or 66% of the cells in the skin sections of DDB2−/− mice were positive for thymine dimers. A similar difference was also observed in the skin samples 28 h after UVB irradiation (see Table 1). In addition, the intensity of immunostaining for thymine dimers was much stronger in the skin sections of the DDB2−/− mice (compare panels c and d in Figure 5). These observations are consistent with the notion that the DDB2−/− mice are deficient in repairing thymine dimers generated following UVB irradiation. We also analysed the rate of CPD removal in the MEFs following UV irradiation following a previously described procedure that measures global genomic repair (Wakasugi et al., 2002). However, we failed to detect any significant difference (data not shown).
Figure 5.
Evidence for repair deficiency of UVB-induced cyclobutane pyrimidine dimers (CPDs) in the epidermis of DDB2−/− mice. Five to six months old DDB2+/+ (a, c, and e) and DDB2−/− (b, d, and f) mice were irradiated with a single dose of UVB (2.5 kJ/m2), killed after 12 h (c and d), and 28 h (e and f) later and their skins were fixed in 10% formalin, processed, and embedded in paraffin for sectioning. Prepared skin section slides (4–5 µm) were then subjected to immunohistochemical analysis using HRP (horse radish peroxidase)-labeled anti-thymine dimer monoclonal antibody (Kamiya Biochemical Co., Seattle, WA, USA). In the epidermis, thymine dimer-positive nuclei are stained as dark brown and nonpositive cells are counterstained as blue color by Mayer’s hematoxylin (Sigma Chemical Co., St Louis, MO, USA). (a) and (b) are skin sections prepared from nonirradiated DDB2+/+ and DDB2−/− mice respectively
Table 1.
CPD repair in epidermis of DDB2+/+ and DDB2−/− mice
| % thymine dimer-positive cells (# of CPD-positive cells/total # of cells) |
||
|---|---|---|
| Hours after UV | DDB2+/+ | DDB2−/− |
| 12 | 40 | 75 |
| 36 | 66 | |
| 28 | 38 | 56 |
| 29 | 49 | |
Percentage of thymine dimer-positive cells was calculated from the number of brown color-stained CPD-positive cells per total cells counted in 10 randomly chosen fields (400-fold magnification, 60–150 epidermal cells per field) from each mouse. For each genotype, two mice were used per time point
DDB2-deficient mice develop spontaneous tumors
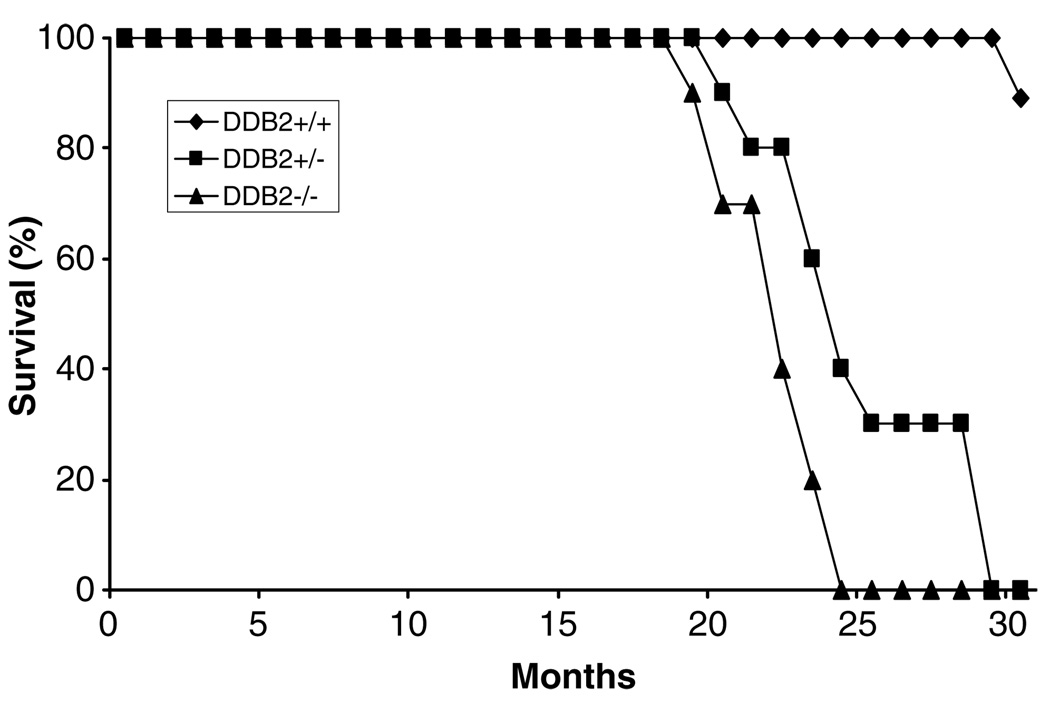
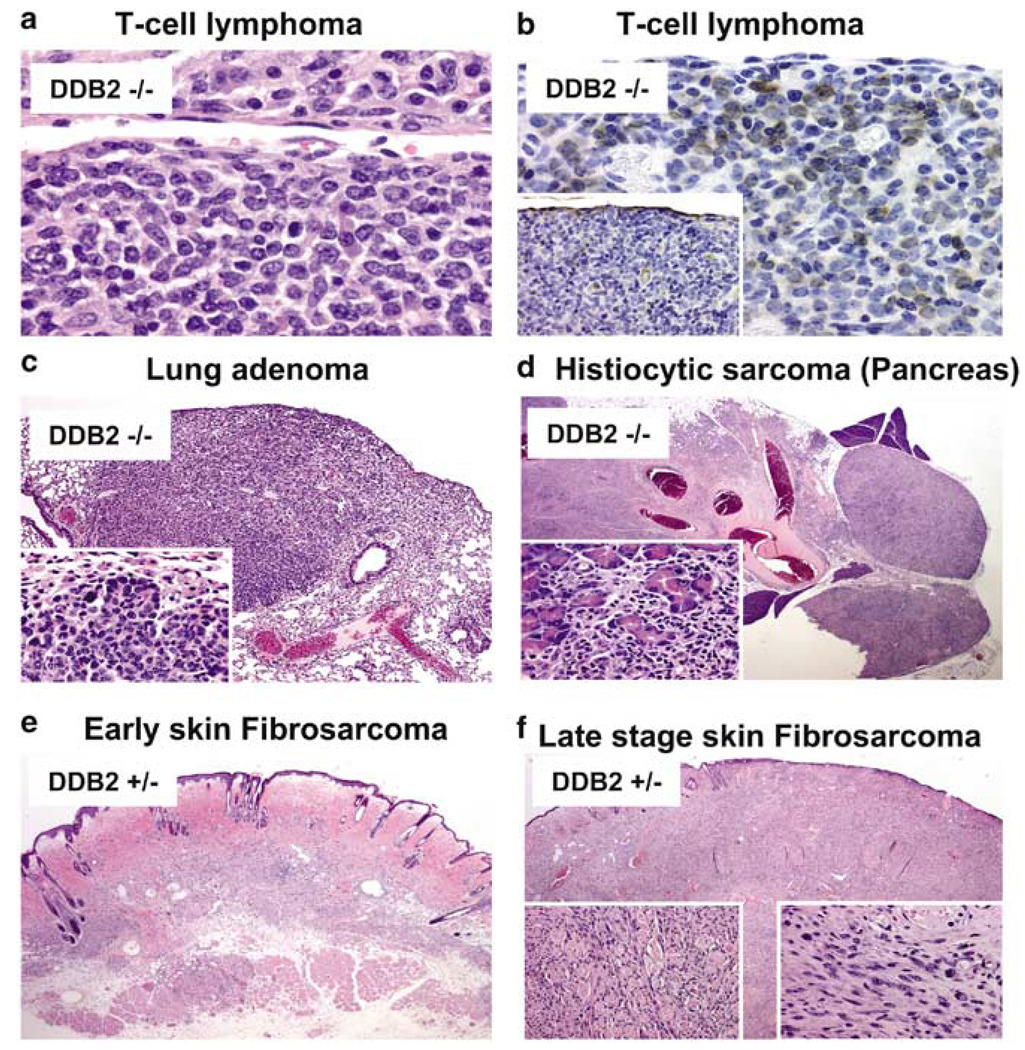
To investigate whether DDB2-deficiency has any effect on the life span and age-related diseases, we maintained and observed a small group of mice of each genotype for over 30 months. The wild-type littermates lived disease-free for at least 30 months. On the other hand, high percentages of the heterozygous (+/−) and the homozygous (−/−) mice died between the ages of 20 and 25 months. Seven out of the 10 DDB2+/− (70%) and 10 out of the 10 DDB2−/− (100%) died between the ages of 20 and 26 months (Figure 6). Three of the seven DDB2+/− mice could be analysed for tumors. One developed dermal fibrosarcoma, another sebaceous carcinoma and the third one suffered from bronchioloalveolar adenoma as well as sebaceous adenoma (Table 2). Histological analyses of an early stage and a late stage fibrosarcoma in a heterozygous mouse are shown in Figure 7 (bottom panels). It is unclear at this time whether the heterozygous mice developed tumors as a result of loss of heterozygosity at the DDB2 loci. Six of the DDB2−/− mice could be analysed at their moribund stage or immediately after they had died. Those mice developed broad spectrum of tumors with a bias towards lymphoid and hematopoietic neoplasms (Table 2). One of the mice had T-cell lymphoma involving spleen, kidney and lymph nodes. Histological staining of a mesenteric node along with immunohistochemical staining for T-cell marker is shown in Figure 7 (upper two panels). The neoplastic lymphocytes were negative for the B-cell marker CD79 Figure 7 (inset, upper right panel). Another DDB2−/− mouse had hematopoietic neoplasms involving spleen, and it also developed phaeochromocytoma. A fifth mouse that was analysed in detail developed bronchioloalveolar adenoma, sebaceous cystadenoma and histiocytic sarcoma involving the pancreas. Histological analyses of the bronchioloalveolar adenoma and histiocytic sarcoma are shown in Figure 7 (middle panels). The sixth mouse developed pulmonary carcinoma. A seventh mouse was diagnosed to have developed mammary adenoma and tumor in the cervical region based on external examination by a veterinarian, but detailed histological analyses were not done for that animal. The high percentages of tumors in the DDB2-deficient mice are clearly consistent with a role of DDB2 in inhibiting age-dependent development of tumors.
Figure 6.
Higher frequency of spontaneous tumor formation in DDB2-null mice. A group of DDB2−/− (triangle, n = 10), DDB2+/− (square, n = 10), and DDB2+/+ (diamond, n = 9) mice were observed without any treatment for 30 months. Moribund or recently died mice were subjected to detailed histological analysis. Tumors arising from these mice are described in Table 2 and Figure 7
Table 2.
Moribund or recently died mice were injected with 10% formalin into their abdominal and thoracic cavities and subjected to histological examination
| Genotype & case no. |
Months lived |
Type of tumor |
|---|---|---|
| DDB2−/− | ||
| K1 | 25 | Lymphoid hyperplasia |
| K2 | 20 | T-cell lymphoma involving spleen, kidney, lymph nodes |
| K3 | 21 | Bronchioloalveolar adenoma, sebaceous cystadenoma, histiocytic sarcoma of pancreas |
| K4 | 25 | Hematopoietic neoplasm involving spleen, phaeochromocytoma |
| K5 | 21 | Hematopoietic neoplasm involving spleen and bone marrow |
| K6 | 23 | Pulmonary carcinoma, locally extensive epidermal hyperplasia |
| K7 | 23 | Not analysed |
| K8 | 23 | Not analysed |
| K9 | 24 | Breast and cervical cancer (not analysed by immunohistochemistry) |
| K10 | 24 | Not analysed |
| DDB2+/− | ||
| H1 | 25 | Dermal fibrosarcoma |
| H2 | 25 | Sebaceous carcinoma |
| H3 | 24 | Bronchioloalveolar adenoma, sebaceous adenoma |
| H4–10 | 21–30 | Not analysed |
Figure 7.
Tumor histology. Tumor tissue section from DDB2−/− (a, b, c, and d), and DDB2+/− (e, f) mice. (a) T-cell lymphoma at mesenteric lymph node. (b) With immunohistochemical staining for the T-cell marker (CD3), cells with morphology typical of neoplastic population are strongly and clearly labeled at mesenteric node. On staining with a B-cell marker (CD79), the neoplastic cells are consistently negative with very rare lymphocytes present marking as B cells (inset). (c) A bronchioloalveolar adenoma at × 1.2 and at × 40 (inset). (d) Pancreas with histiocytic sarcoma × 1.2. The oval structures to the right represent adjacent lymph nodes that have become completely infiltrated by the advancing neoplasm. The detail of interface between residual pancreatic cells that are being irregularly infiltrated by the advancing neoplasm (inset). (e) An architectural view of an area of early sarcoma formation within the deep dermis. (f) Skin × 1.2. An architectural view of skin involved by fibrosarcoma at a much later stage of involvement. The rounded structures surrounded by cells in longitudinal orientation represent residual skeletal muscle fibers of the cutaneous skeletal muscle that are heavily infiltrated by the stromal neoplasm and are being progressively destroyed (left inset). Typical area of fibrosarcoma from the smaller lesion at an earlier stage of development (right inset)
Discussion
DDB is a multifunctional protein. It was identified as a damaged-DNA binding protein involved in global nucleotide excision repair. Recent studies suggest that DDB is a functional partner of cullin 4A and functions as an E3 ligase in the ubiquitin–proteasome pathway. DDB binds to cullin 4A and the DDB-cullin 4A complex associates with UV-damaged DNA. Therefore, it is possible that the E3 ligase function of DDB plays a role in DNA repair through ubiquitination of certain target proteins in the vicinity of the UV-damaged chromatin. Other studies suggested a role of DDB in recruiting chromatin-remodeling proteins onto UV-damaged chromatin. DDB has been shown to associate with the CBP/p300 family of histone acetyl transferase (Datta et al., 2001). In this regard, DDB is similar to the base excision repair protein thymine DNA glycosylase, TDG, which was shown to associate with CBP/p300 (Tini et al., 2002). The interaction between TDG and CBP/p300 is believed to play roles both in repair and transcription. Itoh et al. (2004) studied the function of the DDB2 subunit of DDB using a knockout strain of mice; and demonstrated that the DDB2−/− mice developed skin malignancies at a higher rate compared to the wild-type littermates following UV irradiation. Also, the MEFs lacking DDB2 were somewhat deficient in activating the level of p53 after UV irradiation. Consistent with that study, we observed that the DDB2-deficient mice are susceptible to UV-induced skin carcinogenesis.
To investigate the basis for the susceptibility to UV-induced skin cancers, we analysed the repair activities in the skin of the mice or in the MEFs following UV irradiation. The observations in the skin and in the cultured fibroblasts were quite different. The MEFs from the DDB2+/+ and DDB2−/− did not exhibit any significant difference in the rates of CPD removal. By contrast, a higher percentage of the CPD-positive cells were detected in the skin sections following 12 and 24 h of UV irradiation. Many possibilities exist and further work will be necessary to explain the difference observed in the two systems. It is likely that another damaged-DNA binding protein substitutes for DDB2 in repairing CPDs in the MEFs. It is noteworthy that the XPC/HR23B heterodimer is able to recognize UV-damaged DNA. Moreover, it was suggested that in hamster cells XPC/HR23B could completely substitute for DDB (Tang et al., 2000). Therefore, it is possible that, in the MEFs, XPC/HR23B can substitute for the damaged-DNA recognition function of DDB2. In addition, consistent with the previous observations (Itoh et al., 2003, 2004), we did not see any difference in the survival between the wild-type and the DDB2-deficient MEFs following UV irradiation (data not shown). Nevertheless, we think that the difference in the CPD-positive cells in the skin sections is consistent with the UV-induced skin cancers observed in the DDB2-deficient mice.
We observed that the DDB2-deficient mice developed tumors in the absence of added carcinogen. The tumor spectrum was quite broad and consistent with the broad expression pattern of DDB2. The high rate of spontaneous tumors in the DDB2-deficient mice is interesting because the XPA−/− or XPC−/− mice (with similar genetic background), which lack nucleotide excision repair, rarely developed spontaneous tumors (de Vries et al., 1995; Wijnhoven et al., 2000). Therefore, it is possible that the spontaneous tumors in the DDB2-deficient mice might be unrelated to DDB2’s role in nucleotide excision repair. In that regard, it is noteworthy that DDB2 associates with the cell cycle transcription factor E2F1. It is possible that a deregulation of the E2F1 function in the DDB2-deficient mice is responsible for the spontaneous tumors. However, other possibilities involving a potential ubiquitination function of DDB2 would be important in determining the basis for spontaneous tumors in the DDB2-deficient mice.
The DDB1 subunit of DDB associates with cullin 4A. The DDB1-cullin 4A complex has been shown to be a part of larger E3 ligase complexes involving the V protein of the paramyxovirus SV5 or the human hDET1/hCOP1 proteins. The V protein of SV5 allows the DDB1-cullin 4A ligase to target the STAT proteins for ubiquitination and proteolysis by the 26S proteasome (Ulane and Horvath, 2002). hDET1 and hCOP1 form a heteromeric complex that functions as a substrate adaptor linking the c-jun protein to DDB1-cullin 4A (Wertz et al., 2004). Authors of these studies named the two E3 ligase complexes as VDC and DCXhDET1-hCOP1, respectively. Interestingly, the VDC or the DCXhDET1-hCOP1 complex was not shown to contain DDB2. Therefore, it is likely that DDB2-DDB1-cullin 4A is a distinct E3 ligase complex that might be involved in targeting other proteins for ubiquitination and proteolysis. It was shown that DDB2 could be ubiquitinated by cullin 4A (Chen et al., 2001; Nag et al., 2001a). However, the DDB2-DDB1-cullin 4A complex is quite stable inside the cell, as a stable complex can be easily detected in the cell extracts (Shiyanov et al., 1999). Therefore, it is possible that DDB2 functions as a substrate adaptor for ubiquitination. By analogy with the VDC or DCXhDET1-hCOP1 complex, we suspected that DDB2 might be involved in linking a substrate to the DDB1-cullin 4A ligase. Identification of those substrates will be valuable in understanding the tumor suppression function of DDB2.
Materials and methods
Generation of DDB2-deficient mice
Mouse DDB2 genomic clones were isolated by screening 129 mouse genomic ƛ library with DDB2 cDNA probes generated from mouse EST (GenBank Accession number AA756513 and AA516636). The pPNT vector (Tybulewicz et al., 1991) was used to generate the DDB2 targeting construct (see Figure 1a, legend). The 5′ homology region (2.65 kb) was amplified by PCR using primers (Sph1F–Kpn1 5′-CCCGGTACCGG-CATGCATGTGGTACACATG, Sph1R–Xba1 5′-AGGGTCTCTAGATAGATAGCGCTG), which contained Kpn1, and Xba1 restriction sites that were also present in targeting vector. For the 3′ homology region (4.8 kb) insertion, a DDB2 genomic DNA fragment in pGEM3Zf (+) was digested with Xba1 and BamH1 to release a 4.8kb 3′ homology region, which was blunt ended with T4 DNA polymerase. The 3′ homology region then was inserted into the targeting vector that was predigested with Xho1, blunt ended and dephosphorylated. The orientations of inserted homology regions were confirmed by restriction digestion and also by DNA sequencing. The targeting construct was electroporated into ES cells (Genomesystems) and clones were selected by incubating in selection medium containing G418 (150 µg/ml) and 2 µM ganciclovir for 7–10 days. Selected DDB2+/− ES clones were identified by Southern blot analysis. The selected ES cell clones were subjected to karyotyping to confirm correct chromosome copy number. The selected DDB2+/− ES clones were then microinjected into the C57BL/6 blastocysts. Male chimeras were mated with C57BL/6 females and tail DNA isolated from their offspring was subjected to Southern blot analysis to confirm germline transmission. Finally, the DDB2−/− mice were derived from DDB2 heterozygous intercross mating. The genotypes were confirmed by Southern blot and by Genomic PCR. For the Southern blot genotyping, mouse tail genomic DNA was digested with HindIII and subjected to Southern blot analysis (see Figure 1b, legend) using a 5′ probe (833 bp), and a XbaI-digested genomic DNA was probed with a 3′ probe to confirm correct recombination (data not shown). When probed with 5′ probe, one 8.16 kb band was detected in the wild-type mice, and a 6.85 kb band was detected in homozygous DDB2 null mice. In heterozygotes, both bands were detected.
DDB2 expression analysis by RT-PCR assay
Total RNA from various tissues was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RT-PCR was carried out using Superscript One-Step RT–PCR with Platinum Taq (Invitrogen) according to the manufacturer’s instructions. Varying amount of total RNA (1, 0.1 and 0.01 µg) from different tissues was assayed to detect the linear range of amplification. Primers (RT-F1 and RT-B1 respectively: 5′-TGCAGAAGTCCTTTTTGCACTCTC, 5′-GCTGTCTTTCCCTTTAATTTGGCG) were designed from mouse DDB2 cDNA to amplify the sequences flanking the region between exons 3 and 6. As a negative control, Taq DNA polymerase instead of reverse transcriptase was used to detect any possible genomic DNA contamination. β-actin was used as a loading control.
UVB-induced tumorigenesis
In all, 15–16 mice each from all three genotypes (DDB2+/+, +/−, and −/−) were subjected to UVB irradiation (Fisher Scientific FB-UVXL1000 UV crosslinker with UVB tubes) starting at 2kJ/m2 3 days a week for up to 38 weeks. The dosage was slowly increased up to 6kJ/m2 by the 33rd week. Mice were shaved once a week and only the shaved dorsal area was exposed to UVB irradiation by keeping the mouse in plasticshield tubing with square opening exposing only the shaved dorsal area. Mice with tumors were killed and the tumors were confirmed by histological analysis.
Tumor histology and immunohistochemistry
Moribund mice were humanely terminated by CO2 inhalation. Moribund or recently dead mice were injected with 10% formalin into their abdominal and thoracic cavities and around the tumor mass and then tissues were collected for histological examination. Formalin-fixed paraffin-embedded tissues were analysed by light microscopy and immunohistochemistry. Lesions were analysed according to the descriptions in the text, Pathology of the Mouse by RR Maronpot (Cashe River Press, 2003).
In situ skin CPD detection assay
Five- to six-month-old DDB2+/+ and DDB2−/− mice were irradiated with a single dose of UVB (2.5 kJ/m2), killed after 12 and 28 h and their skins were fixed in 10% formalin at 4°C for 14–18 h, processed, and embedded in paraffin for sectioning by standard procedure. Prepared skin section slides (4–5 µm) were then subjected to immunohistochemical analysis using HRP (horse radish peroxidase)-labeled anti-thymine dimer monoclonal antibody (Kamiya Biochemical). In the epidermis, thymine dimer-positive nuclei are stained as dark brown and nonpositive cells are counterstained as blue color by Mayer’s hematoxylin (Sigma Chemical). In brief, The skin sections were deparaffinized in xylene (2 × 5 min), rehydrated in grades of ethanol (100, 95, and 80%) twice 2min each and finally with distilled water. Endogenous peroxidase was blocked by incubating the section in 3% hydrogen peroxide in methanol for 10 min at room temperature. The slides were then treated with 0.125% trypsin for 10 min at 37°C in a moist chamber and rinsed with distilled water, incubated at room temperature for 30 min with 1 n HCl, incubated with goat serum for 10 min at room temperature. After that, the slides were covered with mouse anti-CPD monoclonal antibody (1 : 50) at room temperature for 90 min. Sections were rinsed with PBS and incubated with DAB substrate (Vector laboratories) for approximately 5 min at room temperature and washed in water for 5 min. Finally, the slides were counterstained with Mayer’s hematoxylin (Sigma), washed with tap water for 5 min and dehydrated in ascending concentrations of ethanol (80, 95, 100%, 2 × 2 min), cleared in xylene and mounted with a coverslip. Percentage of thymine dimer-positive cells was calculated from the number of brown color-stained CPD-positive cells per total cells counted in 10 randomly chosen fields (400-fold magnification, 100–150 epidermal cells per field) from each mouse. For each genotype, two mice were used per time point.
Acknowledgements
We thank Dr S Linn (UC, Berkeley) for helping us with reagents. We thank the UIC Transgenic Facility for blastocyst injections. HK is supported by a grant from the NCI (1R01CA100204) and a grant from the DOD (DAMD 17-02-1-0413). This work was supported by grants from the NCI (CA 77637 and CA 88863) to PR.
References
- Abramic M, Levine AS, Protic M. J. Biol. Chem. 1991;266:22439–22500. [PubMed] [Google Scholar]
- Chen X, Zhang Y, Douglas L, Zhou P. J. Biol. Chem. 2001;276:48175–48182. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- Chu G, Chang E. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Thompson LH, Richardson AS, States JC. Hum. Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, Raychaudhuri P. Mutat. Res. 2001;486:89–97. doi: 10.1016/s0921-8777(01)00082-9. [DOI] [PubMed] [Google Scholar]
- de Vries A, van Oostrom CThM, Hofhuls FMA, Dortant PM, Berg RJW, de Gruiji FR, Wester PW, van Kreiji CF, Capel PJA, van Steeg H, Verbeek SJ. Nature. 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington DC: ASM Press; 1995. [Google Scholar]
- Fujiwara Y, Masutani C, Mizukoshi T, Kondo J, Hanaoka F, Iwai S. J. Biol. Chem. 1999;274:20027–20033. doi: 10.1074/jbc.274.28.20027. [DOI] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Tanaka K, Nakatani Y. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Hayes S, Shiyanov P, Chen X, Raychaudhuri P. Mol. Cell Biol. 1998;18:240–249. doi: 10.1128/mcb.18.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AR, Ford JM. Nat. Genet. 2002;32:180–184. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Chu G. Biochemistry. 1993;32:1657–1666. doi: 10.1021/bi00057a033. [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Toering S, Francke U, Chu G. Mol. Cell Biol. 1998a;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang BJ, Ford JM, Hanawalt PC, Chu G. Proc. Natl. Acad. Sci. USA. 1998b;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki T, Yamagami S, Inoki Y, Tsuru T, Hamamoto T, Kagawa Y, Mori T, Endo H. Biochem. Biophys. Res. Commun. 2004;314:1036–1043. doi: 10.1016/j.bbrc.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Iwao K, Kawasaki H, Taira H, Yokoyama KK. Nucl. Acid. Symp. Ser. 1999;42:207–208. doi: 10.1093/nass/42.1.207. [DOI] [PubMed] [Google Scholar]
- Itoh T, Linn S, Ono T, Yamaizumi M. J. Invest. Dermatol. 2000;114:1022–1029. doi: 10.1046/j.1523-1747.2000.00952.x. [DOI] [PubMed] [Google Scholar]
- Itoh T, O’Shea C, Linn S. Mol. Cell Biol. 2003;23:7540–7553. doi: 10.1128/MCB.23.21.7540-7553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Cado D, Kamide R, Linn S. Proc. Natl. Acad. Sci. USA. 2004;101:2052–2057. doi: 10.1073/pnas.0306551101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A, Mu D, Nichols AF, Zhao X, Linn S, Sancar A. Proc. Natl. Acad. Sci. USA. 1996;93:5014–5018. doi: 10.1073/pnas.93.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Chang GJ, Linn S. J. Biol. Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- Lee T-H, Elledge SJ, Butel JS. J. Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GY, Lamb RA. J. Virol. 2000;74:9152–9166. doi: 10.1128/jvi.74.19.9152-9166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-P, Lou Y-R, Yen P, Mitchel D, Huang M-T, Conney AH. Cancer Res. 1999;59:4591–4602. [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Mol. Cell Biol. 2001;20:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Bondar T, Shiv S, Raychaudhuri P. Mol. Cell Biol. 2001a;21:6738–6747. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Datta A, Yoo K, Bhattacharyya D, Chakrabortty A, Wang X, Slagle B, Costa R, Raychaudhuri P. J. Virol. 2001b;75:10383–10392. doi: 10.1128/JVI.75.21.10383-10392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, Kato Y, Tsunoda Y, Miyauchi H, Horio T, Tokunaga T, Matsunaga T, Nikaido O, Nishimune Y, Okada Y, Tanaka K. Nature. 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- Nichols AF, Ong P, Linn S. J. Biol. Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, Linn S. J. Biol. Chem. 2000;275:21422–21428. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A. J. Biol. Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- Sancar A. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- Sands AT, Abulin A, Sanchez A, Conti CJ, Bradley A. Nature. 1995;377:162–165. doi: 10.1038/377162a0. [DOI] [PubMed] [Google Scholar]
- Shiyanov P, Nag A, Raychaudhuri P. J. Biol. Chem. 1999;274:35309–35312. doi: 10.1074/jbc.274.50.35309. [DOI] [PubMed] [Google Scholar]
- Takimoto R, Maclachlan TK, Dicker DT, Niitsu Y, Mori T, El-Deiry WS. Cancer Biol. Ther. 2002;1:177–186. doi: 10.4161/cbt.65. [DOI] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Mol. Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- Ulane CM, Horvath CM. Virology. 2002;304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. J. Biol. Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Science. 2004;303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- Wijnhoven SW, Kool HJ, Mullenders LH, van Zeeland AA, Friedberg EC, van der Horst GT, van Steeg H, Vrieling H. Oncogene. 2000;19:5034–5037. doi: 10.1038/sj.onc.1203844. [DOI] [PubMed] [Google Scholar]
- Zolezzi F, Linn S. Gene. 2000;245:151–159. doi: 10.1016/s0378-1119(00)00022-6. [DOI] [PubMed] [Google Scholar]