Abstract
The cp27 gene is a highly conserved and unique gene with important roles related to craniofacial organogenesis. The present study is a first analysis of the CP27 promoter and its regulation. Here, we have cloned the promoter of the mouse cp27 gene, examined its transcriptional activity, and identified transcription factor binding sites in the proximal promoter region. Two major transcription start sites were mapped adjacent to exon 1. Promoter function analysis of the 5′ flanking region by progressive 5′ deletion mutations localized transcription repression elements between −1993 bp to −969 bp and several positive elements between −968 bp and the preferred transcription start site. EMSA and functional studies indicated two function-cooperative CCAAT boxes and identified the NF-Y transcription factor as the CCAAT activator controlling transactivation of the CP27 promoter. In addition, this study demonstrated that for its effective binding and function, NF-Y required not only the minimal DNA segment length identified by deletion studies, but also a defined nucleotide sequence in the distal 3′ flanking region of the CP27 proximal promoter CCAAT box. These results provide a basis for our understanding of the specific regulation of the cp27 gene in the NF-Y-mediated gene transcription network.
Keywords: CP27, promoter structure, CCAAT boxes, NF-Y
1. Introduction
Craniofacial development occurs in a complex signaling environment in which growth factors, transcription factors and structural genes of the extracellular matrix maintain signal-response cascades that ultimately result in the formation of the vertebrate head (Davidson, 1993; Slavkin and Diekwisch, 1996; Thesleff and Sharpe 1997). These signaling cascades involve a continuous communication between epithelial and mesenchymal components of adjacent tissues (Thesleff, 1995; Thesleff and Sharpe, 1997). One such gene that is expressed at several crucial sites in the epithelial-mesenchymal interface during craniofacial development is CP27 (Diekwisch et al., 1999; Diekwisch et al., 2002).
CP27 is a unique gene that is highly conserved in many species such as human, mouse, bovine, deer, goat, sheep, giraffe, and pig (Nobukuni et al., 1997; Diekwisch et al., 2002). Sequence analysis has also revealed significant homologues in zebrafish (Danio rerio) and yeast (Saccharomyces cerevisiae) (Diekwisch et al., 2002; unpublished observation). Originally, CP27 was cloned from an E11 early embryonic library (Nobukuni et al., 1997; Diekwisch and Marches, 1997; Diekwisch et al., 2002). Northern blot analysis of RNA from multiple mouse tissues demonstrated high levels of expression in developing mouse teeth, heart, lungs, and liver. Both the expression in presumably important sites related to organogenesis and the distinct changes in localization during development (Nobukuni et al., 1997; Diekwisch and Marches, 1997; Diekwisch et a, 2002) as well as gain- or loss-of-function studies (Diekwisch and Luan, 2002; Luan and Diekwisch, 2002) suggest that CP27 may play important roles during development.
To understand the expression of the cp27 gene and elucidate the mechanisms that govern it, we have cloned the promoter region of the mouse cp27 gene and characterized the cell-specific elements in the 5′ flanking region in embryonic fibroblasts. Using gel-shift and functional studies, we have identified NF-Y as a transactivator of the CP27 promoter that regulates cp27 gene expression via multiple CCAAT boxes. Our results document for the first time the importance of the 5′ 2-kb flanking region in the expression of the mouse cp27 gene and establish NF-Y as a transcriptional regulator of cp27 gene expression.
2. Material and Methods
2.1. Library Screening and DNA Sequencing
A mouse genomic lambda Fix II 129/SVJ library (Stratagene, La Jolla, CA) was screened with a full-length mouse CP27 cDNA, and five clones were identified. Using the EcoRI restriction enzyme, the DNA insert was cut and fragments were subcloned into the pBluescript vector (Stratagene). The resulting DNA sequence was determined with an ABI 373 automatic sequencer. One of the five genomic clones contained 2.1 kb of the 5′ flanking region of the cp27 gene and was used for further analysis. The transcription factor binding sites within the 5′ flanking region were determined using MatInspector (www.genomatrix.de) and Signal Scan (www-bimas.cit.nih.gov/molbio/signal/).
2.2. Primer Extension Analysis
Primer extension was carried out using the Primer Extension System kit (Promega, Madison, WI). An antisense primer CP 82-61 (5′ GCTACCCACACGACTGCGCCAC 3′) was labeled with r-32P using T4 polynucleotide kinase and annealed in AMV primer extension buffer at 58°C for 40 min to 10 μg of total RNA from NIH 3T3 cells, which have been previously shown to express CP27 (Luan and Diekwisch, 2002) or tRNA. The primer was extended with AMV reverse transcriptase at 42°C for 30 min. Resulting products were electrophoresed in an 8% denaturing urea polyacrylamide gel and autoradiographed. The sizes of the products were determined by 32P-labeled φX 174 Hinf I DNA markers.
2.3. Ribonuclease Protection Assay (RPA)
The 5′ flanking region and partial exon 1 of the cp27 gene were amplified via polymerase chain reaction using sense primer CP 261/-242 (5′ TATTAGCTTGTGAGCAAATT 3′) and antisense primer CP 82/61. The 343 bp fragment was then subcloned into the plasmid pCR II-TOPO (Invitrogen, Carlsbad, CA). Transcription was performed with T7 RNA polymerase and yielded α 32P-labeled antisense RNA that was then used as a probe. The probe was annealed to 10 μg of total RNA from NIH 3T3 cells or yeast RNA at 42°C for 16 h. Following digestion with RNase A and RNase T1 (Ambion, Austin, TX) according to manufacturer’s instructions, the RNase-resistant radioactivity was size-fractionated in an 8% denaturing urea polyacrylamide gel and autoradiographed. The sizes of the protected fragments were determined by 32P-labeled φX 174 Hinf I DNA markers.
2.4. 5′-Rapid Amplification of cDNA Ends(5′ RACE)
Rapid amplification of cDNA 5′ ends was performed using a RLM-RACE kit (Ambion). 10 μg of total RNA was treated with Calf intestine alkaline phosphatase to remove free 5′-phosphate. Tobacco acid pyrophosphatase was added to the reaction to remove the cap structure from full-length mRNA. A 45-base RNA adaptor oligonucleotide was ligated to the RNAs using T4 ligase. The first-strand cDNA was synthesized in a random-primed reverse transcription reaction. Amplification of the 5′ ends of CP27 transcripts was accomplished with two pairs of nested primers: a 5′ RACE outer primer 5′ GCTGATGGCGATGAATGAACACTG 3′ and a CP27 antisense outer primer 5′ TCTCTTCAGTCTCCTCGGCT 3′; a 5′ RACE inner primer 5′ CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG 3′ and a CP27 antisense inner primer 5′ GCTCCTCTTCATCTTCTTCACTGC 3′. The RACE products were subcloned into pCR II-TOPO and then sequenced.
2.5. Promoter-reporter gene Constructs
For this promoter study, a total of 15 promoter-reporter gene constructs were generated. The inserts for 14 of the 15 constructs were amplified by PCR with the screened genomic clone as a template using a common 3′ primer and selected 5′ primers (Tables 1, 2). These primers also introduced a Sac I site at the 5′ end and a Hind III site at the 3′ end of the amplified fragments. The PCR fragments were gel-purified using Qiaquick PCR preps (QIAGEN, Valencia, CA), digested with Sac I and Hind III and subcloned into the pGL3-basic vector (Promega). Correct orientation of all inserts with respect to the pGL3 vector was verified by DNA sequencing. Only the pGL −1475/+48 plasmid was generated using a pGL3 vector into which a 1.5kb Bgl II and Hind III fragment from pGL −1993/+48 was inserted. The constructs used for deletion mutation studies were pGL3-1993, pGL3-1475, pGL3-969, pGL3-720, pGL3-207, pGL3-93, and pGL3-17 (Fig. 3). The constructs pGL-93/-56M10-12, pGL3-1255, pGL3-1190, pGL-93/+48CATm, pGL-93-56M10-12, pGL-1255/+48CAT1m, pGL-1255/+48CAT5m, and pGL-1255/+48CAT1,5m were used for CCAAT box function studies. Plasmids carrying an “m” denominator were subjected to a mutation, and pGL-1255/+48CAT1,5m was subjected to a double mutation. All plasmid constructs contained part of the exon 1 noncoding region.
Table 1.
Primers used in the amplification of PCR fragments for promoter-reporter gene constructs
| Primer | Oligonucleotide Sequence | Orientation |
|---|---|---|
| −1993/−1973 | 5′TACCGAGCTCGGCTAACCTGCTCAACTTTGG3′ | sense |
| −1255/− 1234 | 5′TACCGAGCTCTCTTAGGCTGATTCCCATTGC3′ | sense |
| −1190/−1169 | 5′TACCGAGCTCGCATTGGTTGGTCCTCCCGAT3′ | sense |
| −969/−947 | 5′TACCGAGCTCAGGTGATTTCTGAGGGACTAGGG3′ | sense |
| −720/−699 | 5′TACCGAGCTCTCTAGCACTTTGTGTAGTGGC3′ | sense |
| −207/−186 | 5′TACCGAGCTCTATTAGCTTGTGAGCAAATT3′ | sense |
| −93/−73 | 5′TACCGAGCTCTGAGTGTAGACTGACCAATCGC3′ | sense |
| −17/+4 | 5′TACCGAGCTCCCTCTAGGGCGGCCCTAGCT3′ | sense |
| +48/+25 | 5′TCGCAAGCTTGCGAAGCTAGATATAGGGCGAGAC3′ | antisense |
Each primer was labeled by the location of the 5′ and the 3′ first nucleotide in the CP27 5′ flanking region. Each sense oligonucleotide was paired with the only antisense oligonucleotide for PCR amplification. A Sac I restriction enzyme site was added to the 5′ end of each sense oligonucleotide, while a Hind III site was added to the 5′ end of the antisense oligonucleotide. PCR products were then inserted into the vector pGL3-Basic to generate promoter-reporter gene constructs.
Table 2.
Sense oligonucleotides used for EMSA, the competition and mutation analysis in EMSA and for the construction of mutant plasmids.
 |
Mutated oligonucleotides (mut) are listed below wild-type oligonucleotides and mutation sites are highlighted through vertical lines. Mutations were created directly between the nucleotides −93 and −56 at positions −78 and −77 (CP-93/-56CATm) or between the nucleotides −1255 and −1213 at positions −1225 and −1224 (CP-1255/1213CAT5m). The mutation oligonucleotide CP -93/-56 M10-12 was created by replacing the CGGA motif of the wildtype oligonucleotide with a TACA motif in the mutated sequence.
The deletion oligonucleotides (del) CP -93/-56 D9 and CP -93/-56 D13 were generated by removal of the 3′ region from the wild-type oligonucleotide CP -93/-56 D16. Oligonucleotides cp-1255/-1234 and cp-1233/-1213 were used for competition studies (comp).
Lower cases indicate mutated nucleotides. The corresponding wild-type sequences are marked in bold characters. These mutations were used for EMSA competition assays and for the generation of mutant plasmids. The CCAAT box was underlined.
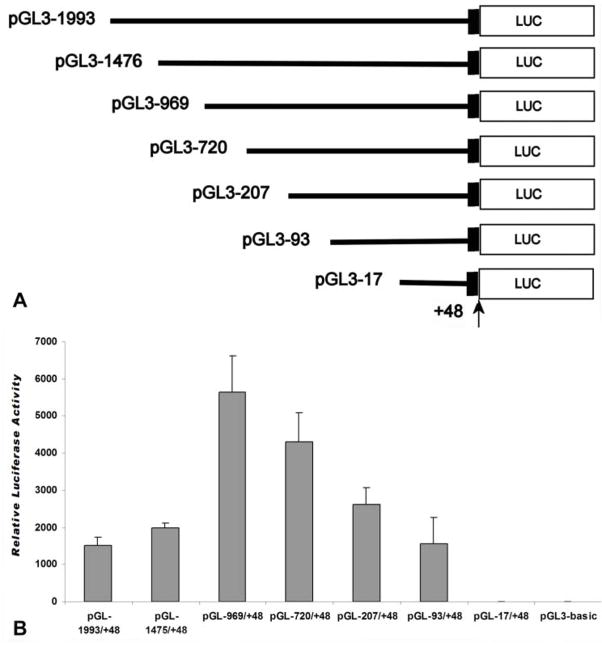
Fig. 3.
Promoter activity in the 5′ flanking region of mouse cp27 gene. A, schematic representation of promoter-reporter plasmids. 1993bp DNA and 5′-truncated fragments of the CP27 promoter upstream of the preferred transcription start site were inserted into the luciferase reporter vector pGL3-Basic in sense orientation. The arrow indicates the preferred transcription start site. The name of each reporter construct was assigned according to the 5′-end nucleotide number and the inserted promoter sequence. All constructs contain a partial exon 1. B, Luciferase activity resulting from the expression of the CP27 promoter-reporter gene constructs. The promoter-reporter gene constructs were transfected into NIH 3T3 cells, and specific luciferase activity was measured 48 hours post transfection. The results were normalized by co-transfection with a pRL-TK reporter plasmid. Error bars represent the standard error for three samples in five independent experiments. The activity of the pGL3-Basic vector transfected in the same experiment is indicated.
2.6. Transient Transfection and Dual Luciferase Assay
The mouse embryonic fibroblast cell line NIH 3T3 was used as the recipient cell line for transient transfection assays. NIH 3T3 cells (3× 105 cells/well) were placed in 6-well plates and cultured for 24 h. For each transfection, cells were incubated with 1 μg of each promoter-reporter plasmid, 0.01 μg of pRL-TK (Promega), which was used as internal control for transfection efficiency, 4 μl of LipofectAMINE PLUS REAGENT (Invitrogen), and 2 μl of LipofectAMINE reagent (Invitrogen) in serum-free medium for 3h. For co-transfection, 1 μg of the pGL-1475/+48 construct was introduced with 0.4 or 0.8 μg of expression vector pIRES-NFYA or with 0.8 μg of pIRES-NFYAm29, a domain negative NF-YA (courtesy of Dr. S. Chen, UTHSCSA). After removal of the DNA-PLUS-LipofectAMINE complex, cells were incubated in 2 ml of complemented medium for 48 h and then subjected to a dual luciferase assay according to the manufacturer’s instructions (Promega). In this dual luciferase system, CP27 promoter fragments were linked to the firefly luciferase gene while the co-transfected renilla luciferase gene (pRL-TK) was driven by the SV40 promoter. The firefly and renilla luciferase activities were measured using TD-20/20 (Promega). Promoter activity measurements were a reflection of the ratio of firefly/renilla luciferase for each construct. For luciferase activity measurements, the means of luciferase activity measurements from five independent sets of experiments using a triplicate set of wells in each experiment were determined for each construct.
2.7. Nuclear Extract Preparation
NIH 3T3 cells (5×107) in 100 mm dishes were washed twice with cold phosphate-buffered saline (pH 7.4) and scraped off in 1 ml of lysis buffer (10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DDT, 0.5 mM phenyl-methylsulphonyl fluoride (PMSF), 0.05% Nonidet P-40 (NP-40). Cell lysates were homogenized with 20 strokes using a tight-fitting Dounce homogenizer and centrifuged at 250 μg for 10 min at 4°C to pellet the nuclei. The pelleted nuclei were resuspended in 1 ml of nuclear extraction buffer (5 mM Hepes (pH 7.9), 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) with a final concentration of 300 mM NaCl, and mixed on a rotator at 4°C for 1 h. Nuclear debris was centrifuged at 24,000 μg for 20 min at 4°C. Aliquots were frozen at −70°C. Protein concentrations were determined using the Bio-Rad Laboratories protein assay reagent (Bio-Rad, Philadelphia, PA).
2.8. Electrophoretic Mobility Shift Assay (EMSA)
Double-stranded oligonucleotides (Table 2) for EMSAs were labeled with (r-32P) ATP using T4 kinase and purified with the Qiaquick nucleotide removal kit (QIAGEN). EMSAs were performed by incubating 5 μg of nuclear extract with labeled double-stranded oligonucleotide in 20 μl reaction buffer [(10 mM Hepes (pH 7.9), 50 mM KCl, 2.5 mM MgCl2, 1 mM DDT, 10% glycerol, 2 μg of poly (dI-dC)] at room temperature for 20 min. For competition analysis, 25- or 50-fold molar excess of unlabeled double-stranded oligonucleotide was added to the nuclear extracts prior to the addition of the labeled probe. For supershift assays, polyclonal antibodies CBFA, CBFB, CBFC and Est 1/2 (Santa Cruz Biotechnology), NF1 and SOX5 (Abcam, Cambridge, MA), were incubated with the nuclear extracts for 15 min followed by the addition of radio-labeled probe. DNA-protein complexes were resolved in a 5% non-denaturing polyacrylamide gel in 1x TBE.
2.9. Immunoblotting
Nuclear extracts separated by EMSA under native conditions were electrophoretically transferred to nitrocellulose in a blotting apparatus filled with transfer buffer (25 mM Tris, 190 mM glycine, 20% methanol) for 1 hour at 75 mA. Nitrocellulose filters were blocked in 2% BSA overnight at room temperature. The blot was incubated with 1:200 diluted anti-mouse CBF-B antibody (Santa Cruz) for 2 h, and a 1:2000 diluted AP-conjugated rabbit anti-goat secondary antibody (Invitrogen) for 1 h, and then with NB/BCIP substrate. For controls, the primary antibody was omitted.
2.10. Mutagenesis
Site-directed mutagenesis was performed using the GeneEditor in vitro Site-Directed Mutagenesis System (Promega). The sense oligonucleotides (Table 2) used in the mutation analysis of competition EMSA served as mutagenesis primers. The templates for the mutagenesis of the CCAAT box and the CGGA site within the proximal promoter region and the CCAAT box in a distant region were constructs pGL −93/+48 and pGL −1255/+48, respectively. The DNA templates, sense oligonucleotide and bottom selection oligonucleotide were annealed at 75°C for 5 min and then cooled slowly to 37°C. Mutant strands were synthesized and ligated in synthesis buffer using T4 DNA polymerase and T4 DNA ligase (Promega). To confirm the fidelity of mutations, plasmids pGL-93/+48CATm, pGL-93-56M10-12, pGL-1255/+48CAT1m, pGL-1255/+48CAT5m, and pGL-1255/+48CAT1,5m were analyzed by DNA sequencing.
2.11. Chromatin immunoprecipitation (ChIP)
ChIP was performed using a Chromatin Immunoprecipitation Assay kit (Invitrogen). For this study, NIH 3T3 cells were fixed in 1% formaldehyde. Nuclei were isolated, sonicated and pre-cleaned with protein A Agarose/Salmon Sperm DNA. For ChIP, the pre-cleaned chromatin solution was set as input or incubated with 5 μg of either anti-CBF-B (Santa Cruz) or anti-Flag antibody (SIGMA, St Luis, MO) on a rotation platform at 4°C overnight. After reversal of the cross-links, the DNA was purified from the immune complex and amplified using PCR primer sets CP−93/−73 and CP+48/+25 or CP−1255/−1234 (Table 1) and CP−1132/−1152 (5′ATCCGTAGGAACAACCAATA3′) specific for the CP27 promoter region.
3. Results
3.1. Sequence analysis of the mouse CP27 promoter region
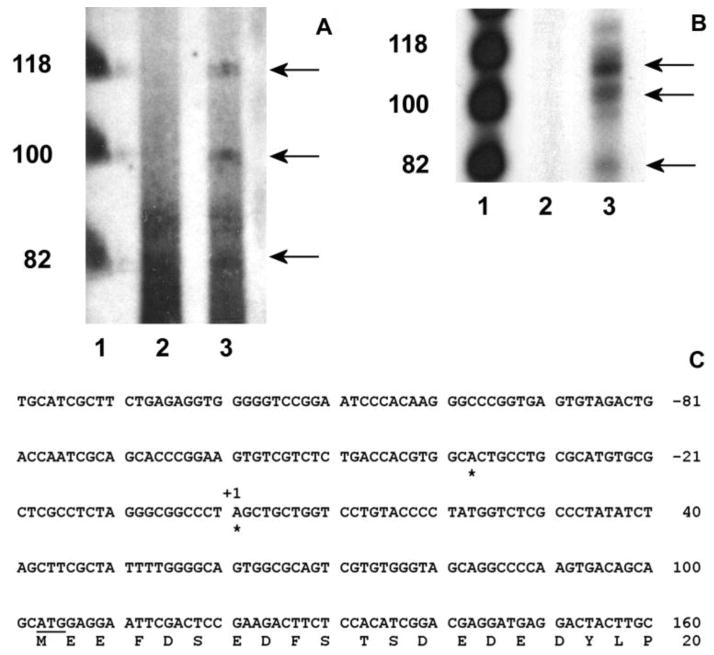
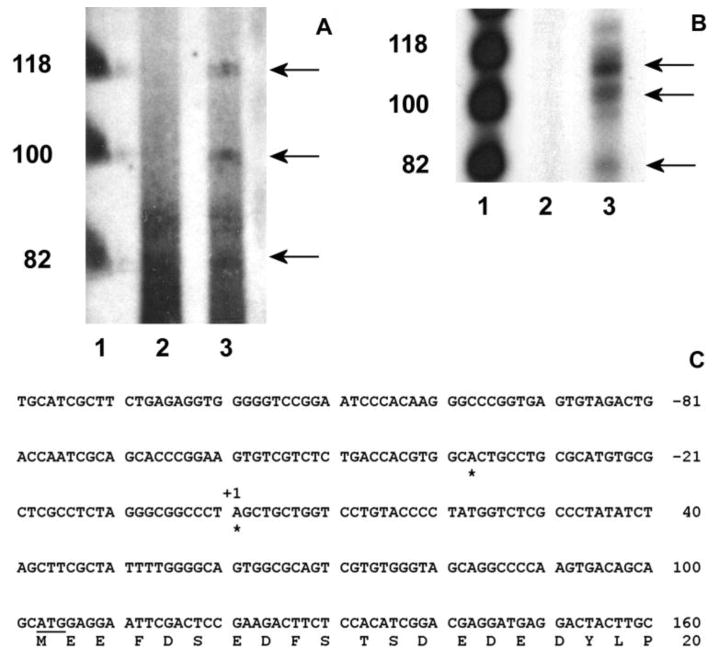
Our sequence analysis of the mouse CP27 promoter region was based on a clone that contained 2.1 kb of the 5′ CP27 flanking sequence. The clone was isolated from a murine 129/SVJ genomic library using the CP27 cDNA as a probe. The sequence of the 5′ flanking region and partial exon 1 is shown in Fig. 1. Computer searches using MatInspector from www.genomatrix.de and Signal Scan from www.bimas.cit.nih.gov/molbio/signal/revealed a CCAAT box at position −79 and an unknown motif (CGGA) in the CCAAT 3′ flanking region. In addition, multiple CCAAT elements were also identified between −1227 and −785. The CCAAT box is a DNA element present in the promoter region of many constitutive, inducible and cell-cycle regulating eukaryotic genes (Caretti et al., 2003; Hu et al., 2000). CCAAT box binding by the transcription factor NF-Y is thought to be a major mechanism required for transcriptional activation (Ceribelli et al., 2008; Donati et al., 2008; Gatta and Mantovani, 2008). Our sequence analysis identified several potential transcription factor binding sites in the proximal promoter region, including an E box core consensus (ccaCGTGg) site at position −44, and a c-myb core consensus (ttCAACggt) site at position −160. Interestingly, there were seven Octamer-binding factor 1 (Oct 1) binding sites in the 5′ CP27 flanking sequence, suggesting a potential regulation of the cp27 gene by the POU homeodomain transcription factor. Two binding sites for the homeobox factor Csx/Nkx 2.5 were located in the promoter sequence as well. In addition, our CP27 promoter sequence analysis revealed the presence of consensus binding sequences for common transcription factors such as AP1 and GATA (Fig. 1).
Fig. 1.
Nucleotide sequence and putative regulatory elements of the 5′ flanking region of the mouse cp27 gene. The numbering of the nucleotides starts at the first nucleotide of the cDNA (+1). The first nucleotide 5′ of the preferred transcription start site is labeled –1. Selected potential transcription factor binding sequences have been labeled. Abbreviations: consensus binding site for the transcription factor oct-1 (OCT-1), consensus binding site for the gata transcription factor (GATA), consensus binding site for the transcription factor nxk-25 (NXK25), consensus binding site for the transcription factor c-myb (C-MYB), consensus binding site for transcription factor ap1 (AP1), CCAAT box (CAT BOX), E-box (E-BOX), unknown binding site (UNKOWN).
3.2. Mapping of CP27 transcription start sites
A primer extension study was performed to map the mouse cp27 gene transcription start sites. For this study, CP27 antisense oligonucleotides CP 82-61 corresponding to nucleotides in exon 1 were synthesized and annealed to total RNA extracted from NIH 3T3 cells. The primer extension reaction yielded three bands of about 80, 100 and 120 nucleotides, respectively (Fig. 2A, lane 3). tRNA was used as a source of control RNA. No primer extension product was detected in the control RNA from these cells (Fig. 2A, lane 2).
Fig. 2.
Determination of the transcription start sites of the mouse cp27 gene. A, primer extension. The oligonucleotide CP 82/61 was end-labeled with (r-32P) ATP and hybridized with 10 μg of total RNA from NIH 3T3 cells or tRNA. The products of the primer extension reaction were size fractionated and autoradiographed. Lane 1, φX174 Hinf I DNA markers; lane 2, primer extension with tRNA; lane 3, primer extension reaction with RNA from NIH 3T3 cells. B, ribonuclease protection assay. The (r-32P) ATP-labeled RNA probe was generated using the TOPO-207/+82 plasmid containing a PCR fragment amplified by the sense oligonucleotide CP –207/-186 and the antisense oligonucleotide CP82/61. The labeled probe was hybridized with 10 μg of total RNA from NIH 3T3 cells or yeast RNA. The RNase-resistant products were sized-fractioned and autoradiographed. Lane 1, φX174 Hinf I DNA markers; lane 2, no RNase-resistant product from yeast RNA; lane 3, RNase-resistant products from NIH 3T3 cells. C, mapping transcription start sites by 5′ RACE assay. Rapid amplification of cDNA 5′ ends using total RNAs extracted from NIH 3T3 cells mapped two transcription start sites of the cp27 gene indicated with stars. The first start site corresponded to an A purine residue 140 nucleotides upstream of the ATG initiation codon, and the second one to an A purine residue located 102 nucleotides upstream of the ATG codon.
To confirm the identity of the transcription start sites determined by primer extension, ribonuclease protection assays were performed. An RNA probe was generated from a TOPO-5′ CP27 plasmid containing a 289 bp PCR product from −207 to +82 bp of the cp27 gene, hybridized to total RNA extracted from NIH 3T3 cells or yeast RNA, and digested with RNase A and T1. The RNA protection assay yielded protected fragments of about 80, 110 and 120 bp in length (Fig. 2B, lane 3). No protected bands were observed in the control RNA (Fig. 2B, lane 2). The size of two protected fragments was similar to the length of two primer extension products, since the 5′-end of the antisense probe used in the RPA was defined by the same antisense oligonucleotide CP 82-61 used in the primer extension assay. However, the size of the second-largest protected fragment ran at a higher molecular weight in the RNA protection assay compared to the primer extension study (Figs. 2A, B, lane 3).
For precise determination of transcription start sites, 5′-RACE assays were performed on NIH 3T3 RNA using two nested CP27 antisense primers. Sequencing 10 RACE subclones yielded two different transcription start sites, as indicated in Fig. 2C. The 5′ end of the longer product (five of ten) corresponded to an adenine purine residue 140 nucleotides upstream from the ATG initiation codon, and the shorter product (four of ten) mapped a second start site to an adenine purine residue located 102 nucleotides upstream of the ATG codon. The two products resulting from the 5′ RACE were identical to the sizes of the first and third bands of the primer extension and ribonuclease protection assays.
3.3. Promoter activity of the 5′ flanking region of the mouse cp27 gene
The function of the 5′ flanking region in the regulation of the mouse CP27 gene was determined by luciferase reporter gene expression. We inserted a 2.0 kb fragment including the 5′ CP27 flanking region and +48 bp of the CP27 exon 1 into the promoterless expression vector pGL3-basic upstream of the luciferase reporter gene (Fig. 3A). Using this assay, the expression of luciferase via the pGL −1993/+48 vector was 500-fold greater than the background measured with pGL-basic.
To localize putative cis-acting elements regulating transcription of the cp27 gene, progressive 5′ deletion mutations of the 2.0 kb promoter (−1993 to +48) were performed using reporter gene constructs. Expression of the reporter gene driven by the mutants was examined by transient transfection assays. The initial deletion removed approximately 500bp from the upstream end of the pGL −1993/+48 construct. The resulting construct (pGL −1475/+48) produced a slight increase in luciferase activity in NIH 3T3 cells. Deletion of another 506 bp from the 5′ end generated the construct pGL−969/+48 and resulted in a 2.5-fold increase compared with the construct pGL −1475/+48. Further deletions from −969 to −93 led to a progressive decrease in promoter activity. Removal of approximate 250 bp from the upstream end of the pGL −969/+48 plasmid generated the construct pGL −720/+48 which resulted in a 1.4-fold decrease in luciferase expression acitivity compared to the construct pGL −969/+48. Deletion of the next 520 nucleotides also resulted in a 1.4 fold decrease in expression activity. A further deletion of 104 bp generated the construct pGL −93/+48 and led to a further 1.6-fold decrease in luciferase activity. The activity of the pGL −17/+48 construct was on the same level with the pGL3-basic vector (Fig. 3B).
Together, the results of the deletion analysis revealed the following three regulatory elements: (i) a positive element in the small interval between −17 to −93 bp, (ii) several enhancer elements located within the first −969 bp, and (iii) a general repression region between nucleotides −969 to −1993.
3.4. Identification of NFY as the nuclear factor binding to the CP27 proximal promoter CCAAT box
The transient transfection experiments described above indicated the presence of positive regulatory elements within 93 bp of the mouse CP27 proximal promoter region. To answer whether the CCAAT box is a basal regulatory element in the mouse CP27 proximal promoter, an oligonucleotide containing the CCAAT box (CP-93/-56) was subjected to an EMSA. The EMSA revealed formation of a protein-DNA complex using the radioactively labeled oligonucleotide CP-93/-56 and nuclear extracts from NIH 3T3 cells (Fig. 4A, lane2). Competition experiments were performed to determine the sequence specificity of the protein-DNA complex. Molar excesses (25- or 50-fold) of unlabeled CP-93/-56 eliminated the formation of the complex (Fig. 4A, lane 3 and 4). However, when a mutation in the CCAAT box was introduced by mutating CCAAT to CACAT (Table 2), the mutated CP-93/-56 did not affect protein/DNA binding (Fig 4A, lanes 5 and 6).
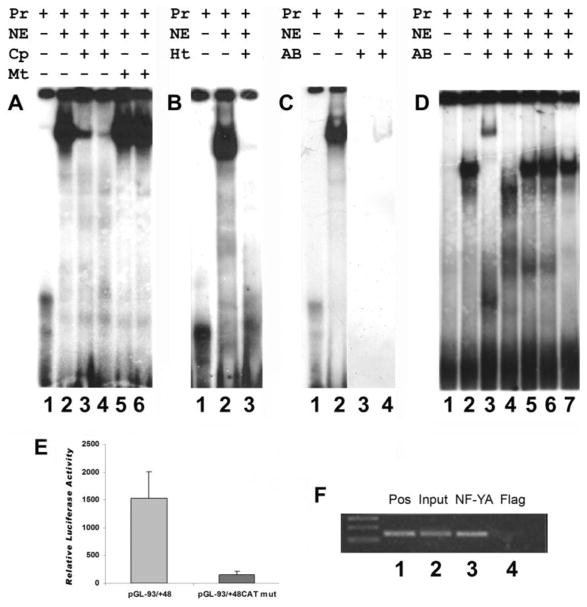
Fig. 4.
Identification of NF-Y as the CCAAT box binding protein of the mouse CP27 proximal promoter. Nuclear extracts were prepared from NIH 3T3 cells. The extracts were incubated with the 32P-labeled double-stranded oligoncleotides CP-93/-56 containing the CCAAT box of the mouse CP27 proximal promoter. A, Electrophoretic mobility shift analysis. Nuclear extracts from NIH 3T3 cells were incubated with 32P-labeled oligonucleotide CP-93/-56 probe in the presence of the various competitors. Lane 1, 32P-labeled probe only; lane 2, probe plus nuclear extracts; lane 3, twentyfive-fold molar excess of unlabeled wild –type(WT) double-stranded oligonucleotides; lane 4, fifty-fold WT; lane 5, twentyfive-fold molar excess of unlabeled mutated (MUT) double-stranded oligonucleotides; lane 6, fifty-fold molar excess of unlabeled mutated (MUT) double-stranded oligonucleotides. The labels above individual lanes read Pr = probe, NE = nuclear extract, Cp = competition, Mt = mutation. B, effect of heat-treated nuclear extract on the DNA-protein complex. Lane 1, probe only; lane 2, nuclear extracts were incubated with the 32P-labeled probe; lane 3, the extracts were heated to 85°C for 5 min, centrifuged and then added to the reaction mixture. C, Immunoblot analysis. Nuclear extracts were analyzed by EMSA in the presence of the double-stranded oligoncleotides CP-93/-56. Lanes 1 and 3, probe only; lanes 2 and 4, probe plus nuclear extracts. The EMSA gels were dried and subjected to autoradiography (lane 1 and 2) or blotted onto a nitrocellulose membrane, which was then immunobloted with anti-NF-YA antibody (lane 3 and 4). D, supershift analysis. Lane 1, probe only; lane 2, nuclear extracts were incubated with a 32P-labeled probe; lane 3–7, the extracts were pre-incubated with anti-NF-YA, NF-YC, NF1, Est1/2 or SOX5 antibodies (respectively) for 15 min and then added to the reaction mixture. The labels above individual lanes read Pr = probe, NE = nuclear extract, Ht = heat treatment, AB = antibody. E, transient transfection experiments. The plasmid pGL-93/-56 CCAATmut is a construct in which the CCAAT box has been mutated. Equal amounts of the mutated or wild-type constructs were transiently transfected into NIH 3T3 cells. pRL-TK was used as internal control for transfection efficiency. The values for relative luciferase activity including standard deviation were derived from at least five independent experiments with triplicate wells. There was a significant difference in luciferase activity between the wild-type and the mutated construct. F, ChIP assay. Chromatin fragments immunoprecipitated with anti-NF-YA antibody were amplified by PCR using primers spanning the CP27 proximal promoter region. Immunoprecipitation with anti-Flag antibody was used as a negative control. Lane 1, positive control; Lane 2, Input DNA fragment amplified by mouse CP27 primers; Lane 3, CP27 target immunoprecipitated by anti-NF-YA antibody; Lane 4, anti-Flag antibody control.
CCAAT box binding proteins may bind to elements containing a CCAAT box. These proteins include the C/EBP family, NF-Y, Y box factor, and CBT/NF1 (Dorn et al., 1987; Didier et al., 1988; Zorbas, 1992; Sylvester et al., 1994). To identify the specific factor(s) that binds to the CCAAT box of the CP27 proximal promoter, nuclear proteins from NIH 3T3 cells were characterized using EMSA. Taking advantage of the heat-stability of the C/EBP family DNA binding proteins (MacDougald and Jump, 1991), nuclear extracts were heated to 85°C prior to incubation with the radioactively-labeled oligonucleotide CP-93/-56. The heat treatment led to an abolishment of the shift and to a loss of the DNA-protein complex band (Fig. 4B, lane 3), indicating that the factor binding to the proximal CP27 promoter is heat-sensitive and thus excluding the heat-stable C/EBP as a candidate factor for CP27 proximal CCAAT box binding. Following exclusion of C/EBP as a candidate CCAAT binding protein, focus was directed toward the heterotrimeric transcription factor NF-Y, which had been characterized as the most ubiquitous and specific factor involved in the regulation of CCAAT box (Mantovani, 1998). To examine whether NF-Y functioned as a binding protein in the DNA-protein complex formed by NIH 3T3 nuclear extracts and oligonucleotide CP-93/-56 in EMSA, anti-NF-YA antibody was applied to EMSA gel blots after Western blotting (Novak and Paradiso, 1995). Immunoblot analysis demonstrated that NF-Y was present in the DNA-protein complex (Fig. 4C, lane 4). Incubation of anti-NF-YA antibody together with the DNA-protein complex led to the formation of a slower migrating supershifted band (Fig. 4D, lane 3), while anti-NF-YC antibody abolished the DNA-protein complex (Fig. 4D, lane 4). However, anti-NF1, anti-EST1/2, and anti-SOX5 antibodies did not affect the formation of the DNA-protein complex (Fig. 4D, lanes 5–7), further confirming that the DNA-protein complex contained the NF-Y transcription factor. These results suggest that NF-Y binds to the CCAAT sequence in the mouse CP27 proximal promoter.
To further confirm that the CCAAT box is involved in the regulation of the CP27 promoter, the CCAAT box of the intact CP27 promoter was mutated by means of a mutated construct (Table 2). Specifically, the activity of the mutated plasmid pGL-93/+48 CAT-mut was compared with that of the wild-type pGL-93/+48. The mutations had significant effects on the activity of the CP27 promoter. Mutation in the CCAAT box reduced the activity by 90% (Fig. 4E). These results demonstrated that the CCAAT box is a significant regulatory element located in the mouse CP27 proximal promoter.
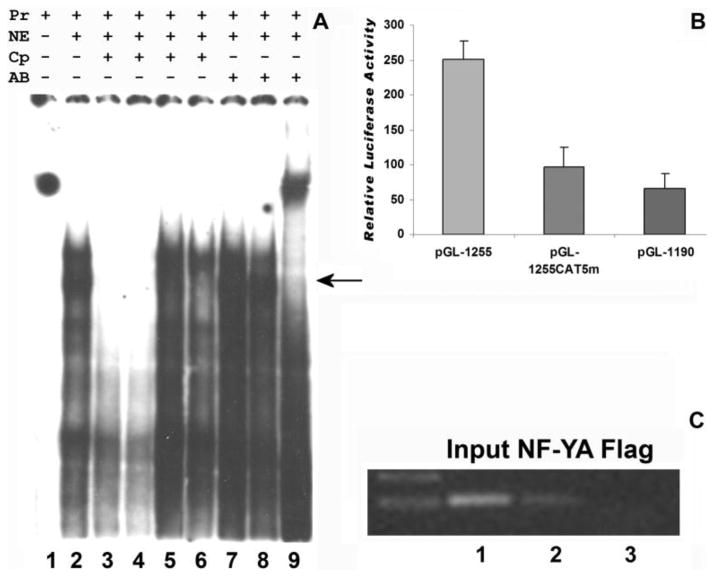
To verify the binding of NFY to the CP27 promoter in vivo in NIH 3T3 cells, Chromatin immuno-precipitation was performed. The cross-linked chromatin was immuno-precipitated with anti-NFYA antibody and subjected to PCR amplification using primers spanning the CCAAT box of the CP27 proximal promoter region. A 140 bp DNA fragment of expected length was amplified (Fig. 4F, lane 3). In contrast, a control anti-flag antibody failed to precipitate chromatin fragments containing the endogenous CP27 promoter (Fig. 4F, lane 4).
3.5. Specific sequence determination of the CCAAT box 3′ flanking region required for NF-Y binding
Specific neighboring nucleotides on both 5′ and 3′ sides of the CCAAT box are involved in the efficient binding of NF-Y (Mantovani, 1998). In addition, minimal DNA fragments in both the upstream and downstream distal regions are required for the formation of a stable NF-Y-DNA complex (Sugiura and Takishima, 2003). We therefore investigated what role the CCAAT 3′ flanking region (i.e. the CGGA motif) of the CP27 proximal promoter might play in regulating promoter function and DNA binding by NF-Y.
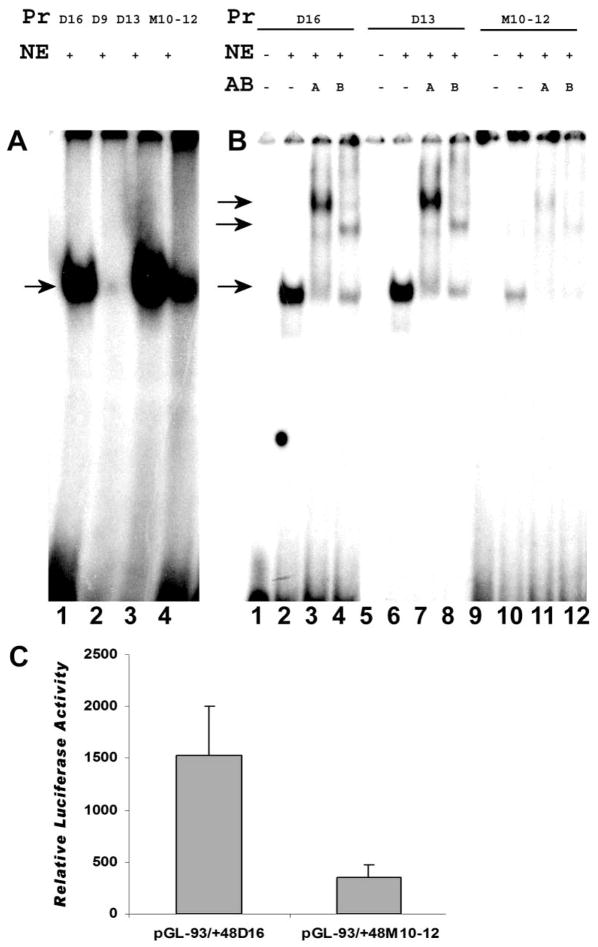
To determine the effect of the CGGA motif on the binding of NF-Y to the CCAAT box of the CP27 proximal promoter, EMSA was performed using oligonucleotides CP-93/-56 (D16) along with its deletion mutants CP-93/-65D9 (D9) and CP-93/-65D13 (D13), and the site-specific mutant CP-93/-65M10-12 (M10-12) (Table 2). In this EMSA, NF-Y efficiently bound to probes D16 and D13 (Fig. 5A, lanes 1 and 3), while binding affinity was substantially reduced when NF-Y bound to probe M10-12 (Fig. 5A, lane 4); and a diminishingly small NF-Y-DNA complex formed between NF-Y and probe D9 (Fig. 5A, lane 2).
Fig. 5.
Effect of site-specific mutation on the NF-Y binding in the 3′ flanking region of the CCAAT box of the mouse CP27 proximal promoter. A, Binding efficiency analysis of the NF-Y on the binding site. Wild-type oligoncleotides or mutated oligonucleotides were labeled with 32P. These probes were incubated with nuclear extracts and the formation of NF-Y-DNA complexes was analyzed using EMSA. Lane 1, wild-type oligonucleotide probe CP-93/-56; lane 2, deletion-mutated probe CP-93/-56D9; lane 3, deletion-mutated probe CP-93/-56D13; lane 4, site-specific mutated probe CP27-93/-56M10-12. B, Antibody supershift assay. Lanes 1–4, wild-type probe CP-93/-56; lanes 5–8, deletion-mutated probe CP-93/-56D13; lanes 9–12, site-specific mutated probe CP-93/-56M10-12; lanes 1,5, and 9, probe only; lanes 2,6, and 10, probe plus nuclear extract; lanes 3,7, and 11, probe plus nuclear extract and anti-NF-YA antibody; lanes 4,8, and 12, probe plus nuclear extract and anti-NF-YB antibody. The labels above individual lanes in Figs. 6A–C read Pr = probe, NE = nuclear extract, Cp = competition, AB = antibody. C. Transient transfection assays. The oligonucleotide containing the mutated CGGA motif in the 3′ flanking region was substituted for the plasmid pGL-93/-56 M10-12. Equal amounts of the wild-type or mutated constructs were transiently transfected into NIH 3T3 cells and luciferase activity was measured. The data represent the means and standard deviation from five separate experiments. There was a significant difference in luciferase activity between the wild-type and the mutated construct.
To determine whether the CGGA motif altered NF-Y binding through interacting with NF-Y heterotrimeric complex, super EMSA was performed using anti-NF-YA and NF-YB antibodies. The reactivity of the NF-Y/M10-12 complex to each antibody was compared with the reactivity of the NF-Y/D16 and NF-Y/D13 complexes. While there were no differences in the supershift pattern between M10-12, D16, and D13, NF-Y binding was greatly reduced in the mutated motif M10-12 compared to D16 and D13 (Fig. 5B). Taken together, these observations suggest that the CGGA motif affects the binding of the NF-Y complexes to the CCAAT box.
To confirm that the CGGA motif indeed had an effect on NF-Y binding to the CCAAT box, a luciferase reporter construct pGL-93/-56M10-12 was generated (Table 2). This construct contained the mutated binding site (tacA vs. CGGA) and the intact CCAAT box. Compared with the wild-type construct (pGL-93/-65), this mutation reduced the activity of the intact promoter by 77%, indicating that the binding site is critical for NF-Y binding to the mouse CP27 proximal promoter (Fig. 5C).
3.6. Involvement of multiple NF-Y/CCAAT complexes in cp27 gene expression
To fully understand CP27 gene expression regulation via CCAAT boxes we explored the function of multiple CCAAT sites harboring in the CP27 promoter subsequent to our characterization of the proximal NF-Y binding CCAAT box. Altogether, a transcriptional cis-regulatory element analysis using MatInspector (www.genomatrix.de) and Signal Scan (www.bimas.dcrt.nih.gov) revealed five CCAAT boxes (CAT 1–5) in the CP27 promoter. The proximal CAT1 (−79 to −83) and CAT2 (−785 to −789) were in forward orientation and located in the enhancing regions (Fig. 1). The CAT boxes 3–5 (−1227 to −1150) were found in ageneral repression region in reverse orientation and located in close proximity to each other, separated only by 33 to 34 bp intervals (Fig. 1). Incubation of nuclear extract from NIH3T3 cells with the 32P -labeled probe CAT5 (CP-1255-1210) produced specific DNA/protein complexes (Fig. 6A, lane2) using EMSA. Twenty-five and fifty-fold molar excess of wild type oligonucleotides CP-1255/-1213 competed the formation of the DNA/protein complex (Fig. 6A lanes 3–4), and the mutated oligonucleotide CP-1255/-1234 did not affect the DNA/protein complexes (Fig. 6A, lane5). However, oligonucleotide CP-1233/-1213 containing a CCAAT box had the competition capability (Fig. 6A, lane 6). Addition of antibodies to NF1 and SOX5 did not recognize these bands (Fig. 6A, lanes 7–8), whereas the anti-NF-YA antibody (Fig. 6A, lane 9) formed a super-shifted band with the DNA/protein complex, indicating that NF-Y binds to the CCAAT-boxes within the CP27 promoter.
Fig. 6.
Multiple NF-Y binding elements in the mouse CP27 promoter. A, EMSA. Nuclear proteins were extracted from NIH 3T3 cells. The extracts were incubated with the 32P-labeled CCAAT box-containing oligonucleotide CP-1255/-1207. lanes 1, probe only; lanes 2, probe plus nuclear extracts; lanes 3, twenty five-fold and lanes 4, fifty-fold molar excess of unlabeled WT double-stranded oligonucleotides CP-1255/-1213; lanes 5, fifty-fold molar excess of unlabeled WT double-stranded oligonucleotides CP-1255/-1234; lanes 6, fifty-fold molar excess of unlabeled WT double-stranded oligonucleotides CP-1233/-1213 (Table 2); lanes 7-9, supershift with anti-NF1, SOX5 or NF-YA antibodies. The labels above individual lanes read Pr = probe, NE = nuclear extract, Cp = competition, AB = antibody. B, Mutation analysis of CAT5 box. The wild type promoter-reporter construct pGL-1255/+48 or its 5′ deletion mutation pGL-1180/+48 or its mutated homologues pGL-1255/+48CAT5-mut was introduced into NIH3T3 cells, and luciferase activity was measured. The values for relative luciferase activity including standard deviation were presented in triplicates in five independent experiments as described before. C, ChIP assay. Chromatin fragments immunoprecipitated with anti-NF-YA antibody were amplified by PCR using primers spanning the CP27 promoter regions from −1254 to −1133bp. Immunoprecipitation with anti-Flag antibody was used as a negative control. Lane1, input DNA fragment amplified by mouse CP27 primers; Lanes 2, CP27 target immunoprecipitated by Anti-NF-YB antibody; Lanes 3, anti-Flag antibody control.
To determine whether the CAT5 CCAAT box serves as a stimulatory or a repressive site, we performed a 5′ deletion mutation analysis and also introduced mutations into the box (ATTGG to ATGTG). Three promoter-reporter constructors were generated either as a wild type (pGL-1255/+48) or a 5′ deletion (pGL-1190) or a mutation (pGL-1255CAT5m2) construct, and then transiently transfected into NIH3T3 cells. Our findings indicated that removal of the CAT5 CCAAT box and adjacent regions had a significant effect on the mouse CP27 promoter function, decreasing expression of the reporter gene by 74%. Moreover, mutation of CAT5 CCAAT box resulted in a 62% reduction when compared to the wild type construct(Fig. 6B). The decrease in CP27 promoter activity observed by CAT5 mutation suggests that this site is involved in the control of cp27 gene expression enhancement despite its placement in an overall repressive promoter region.
Chromatin immunoprecipitation was performed to verify the binding of NF-Y to the CAT5 CCAAT box in the CP27 promoter in NIH3T3 cells in vivo. The cross-linked chromatin was immuno-precipitated with anti-NF-YA antibody and subjected to PCR amplification using a pair of primers CP-1255/-1234 and CP-1132/-1152 spanning the CAT5 containing region. A DNA fragment (123 bp) corresponding in size to the CP27 promoter region was amplified (Fig. 6C, lanes 2). In contrast, a control anti-flag antibody failed to precipitate a chromatin fragment containing the endogenous CP27 promoter (Fig. 6C, lanes 3). These results confirmed the presence of the active NF-Y-binding CAT5 CCAAT box in the CP27 promoter.
Oligonucleotides containing the sequence for the CAT2 CCAAT box were used as a probe in EMSA experiments and for the generation of DNA-protein complexes. However, these complexes were not recognized by either anti-NF-YA, NF-YB, or NF-YC antibodies. Oligonucleotides containing the CAT3 or CAT4 CCAAT box sequence did not form substantially shifted bands when incubated with nuclear extracts from NIH3T3 cells (data not shown).
3.7. Requirement for functional cooperation among multiple NF-Y/CCAAT binding sites for cp27 gene expression
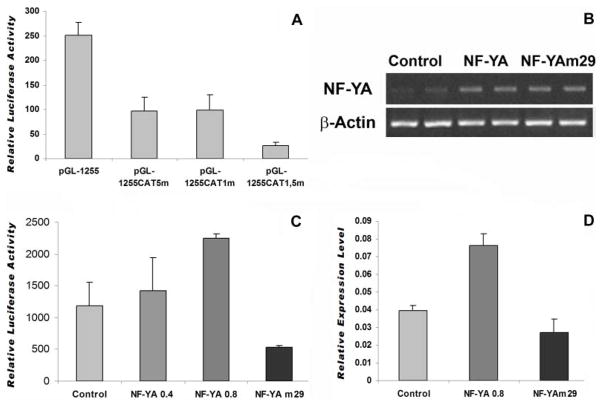
To investigate whether multiple NF-Y/CCAAT binding sites function independently from each other or are required to cooperate in the regulation of the CP27 promoter, four constructs were generated, including one containing the intact CP27 promoter and three constructs in which the CCAAT box was mutated. Specifically, the activity of the three mutated plasmids pGL-1255/+48 CAT1-mut, pGL-1255/+48 CAT5-mut or pGL-1255/+48 CAT 1,5-mut was compared with that of the wild-type pGL-1255/+48. The mutation in the CCAAT-box1 reduced luciferase activity by 61% (Fig. 7A). Mutation of the CAT5 box decreased CP27 promoter activity by 62% (Fig. 7A). Interestingly, simultaneous mutations in the CAT1 and CAT5 boxes reduced CP27 promoter activity by 90% (Fig. 7A). These results suggest that the two CAT boxes cooperate to activate the CP27 promoter.
Fig. 7.
Effect of NF-Y on the cp27 gene expression. A, function study on the cooperation of multiple CCAAT-boxes to regulate the CP27 promoter. The wild type promoter-reporter construct pGL-1255/+48 or its mutated homologues pGL-1255/+48CAT1-mut, pGL-1255/+48CAT5-mut or pGL-1255/+48CAT1, 5-mut was transfected into NIH3T3 cells, and luciferase activity was measured in triplicates in five independent experiments as described before. B, RT-PCR analysis of NF-YA or NF-YAm29 overexpression efficiency. NIH3T3 cells were transfected with expression vectors pIRES-NF-YA or pIRES-NF-YAm29. After twenty-four hours, total RNA was isolated from transfected cells and reverse transcribed. NF-YA expression level was detected by PCR analysis. RT-PCR products of four separate experiments were loaded onto 1% agarose gels and stained with ethidium bromide. β-actin served as an internal control. C, Luciferase activity of CP27 promoter-reporter gene constructs regulated by overexpression of NF- YA or NF-YAm29. The construct pGL-1255/+48 was co-transfected with either the expression vectors pIRES-NF-YA or pIRES-NF-YAm29 into NIH3T3 cells. Transfection with pIRES-GFP was used as control. Forty eight hours after transfection, cells were harvested and luciferase activity was measured as described above. Results are presented as relative luciferase activity. D, qRT-PCR analysis of CP27 mRNA expression modulated by gene manipulation of NF-YA. NIH3T3 cells were transfected with the expression vectors pIRES-NF-YA or pIRES-NF-YAm29. After 48 hours, total RNA was isolated from the transfected cells and reverse transcribed. CP27 expression levels were measured using real time RT-PCR analysis. Changes in expression levels were measured as n-fold increases of stimulation over the control group using the 2−ΔΔCt method. CP27 expression was 1.93-fold higher when cells were transfected with NF-YA.
To determine whether NF-Y can functionally activate the CP27 promoter, an expression vector for NF-YA (pIRES-NF-YA) or NF-Yam29 (pIRES-NF-YAm29) was introduced into NIH 3T3 cells, resulting in significant transfection efficiency as examined by RT-PCR (Fig. 7B). Vectors pIRES-NF-YA or pIRES-NF-Yam29 were then co-transfected with the promoter-reporter construct pGL-CP-1255/+48. Co-transfection resulted in an enhancement of luciferase activity in a dose-dependent manner following NF-YA overexpression. Especially, 0.8 μg of pIRES-NFYA induced a 2.2 fold increase. In contrast, NF-YAm29 reduced the CP27 promoter activity (Fig. 7C).
To confirm our finding that the functional cooperation between NF-Y and CCAAT boxes influences endogenous cp27 gene expression, NIH 3T3 cells were transfected with pIRES-NF-YA or pIRES-NF-Yam29, and CP27 mRNA expression was quantified by real time RT-PCR. Our data indicate that NF-YA overexpression caused a 2-fold increase in endogenous CP27 mRNA expression. There was a slight decrease in CP27 transcription activity following NF-YAm29 treatment when compared to NIH3T3 cells (Fig. 7D). Together, these data suggest that NF-Y interacts with multiple CCAAT boxes to regulate cp27 gene expression.
4. Discussion
The present study is the first analysis of the CP27 promoter structure and function. For our analysis we have cloned the promoter of the mouse CP27 gene, examined its transcriptional activity, and identified a transcription factor in the proximal promoter region. Our studies demonstrated that the similar patterns of CP27 transcriptional activity were displayed in NIH 3T3 cells. Two major transcription start sites were mapped adjacent to exon 1. Functional analysis of the 5′ flanking region by progressive 5′deletion mutations revealed transcription repression elements between −1993 bp to −969 bp as well as several positive elements between −968 bp and the preferred transcription start site. Furthermore, deletion analyses identified an enhancer element within 93 bp of the CP27 proximal promoter. EMSA experiments and function study demonstrated that NF-Y was involved in the activation of the CP27 proximal promoter through two CCAAT boxes. A binding site in the 3′ flanking region of CCAAT box 1 affected the efficiency of NF-Y binding. Together, these studies provide a basis for our understanding of the regulation of the CP27 gene.
NF-Y binding CCAAT boxes have been identified as crucial determinants of proper gene regulation related to cell growth (Bhattacharya et al., 2003; Elkon et al., 2003; Testa et al., 2005). The histone-like NF-Y substitutes H2A-H2B and finely tunes histone methylation and acetylation on CCAAT-containing promoters (Gatta and Mantovani, 2008; Gurtmer et al., 2008). As a bifunctional transcription factor, NF-Y binding is associated with both positive and negative histone marks (Ceribelli et al., 2008; Donati et al., 2008). Most cell-cycle regulated promoters contain CCAAT boxes and promoters of genes with key roles in the G2/M transition have multiple CCAAT motifs, such as cyclin B1 (two), cyclin B2 (three), cdc25c (three), and HSP 70 (two) (Li et al., 1998; Salsi et al., 2003; Muller et al., 2007). These promoters rely on multiple CCAAT boxes activated by NF-Y, whose binding to DNA is temporally regulated during the cell cycle (Alder et al., 1992; Wasner et al., 2003; Zwicker et al., 1995). Inhibition of NF-Y mediated transcription activation arrested cells at G2/M phase and suppressed expression of genes activated at G2/M phase of the cycle (Hu et al., 2006). P53/NF-Y complexes binding to NF-Y target promoters are associated with direct p53 transcriptional repression during the G2/M phase (Di Agostino et al., 2008; Imbriano et al., 2005). The functional implementations of these studies have been confirmed in cell lines as well as in knock-out animal model in vivo. For example, expression of the domain-negative mutant of CBF/NF-Y in mouse fibroblasts resulted in a retardation of cell growth. Furthermore, deletion of both NF-YA alleles caused a specific block of cell proliferation and cell death in embryonic fibroblasts (Bhattacharya et al., 2003; Chae et al., 2004; Hu et al., 2000). Lastly, a conditional knockout animal model in which the inactivation of NF-YA caused early embryonic lethality (Bhattacharya et al., 2003) provided support for a pivotal role of NF-Y-mediated transcription in embryonic development.
The mouse CP27 promoter features multiple CCAAT boxes. NF-Y interacts with these CCAAT boxes and enhances cp27 gene expression. Studies from our laboratory have demonstrated that similar to other NF-Y target genes, CP27 plays significant roles in cell proliferation and mouse organogenesis (Diekwisch and Luan, 2002; Luan and Diekwisch, 2002). Loss of CP27 function inhibited cell growth and induced apoptosis in embryonic fibroblast cells (Luan and Diekwisch, 2002). Deletion of both CP27 alleles in knock-out mice resulted in early embryo lethality and knock-down of the cp27 gene caused cell death (unpublished data). Here we are proposing that NF-Y regulation of CP27 expression provides a meaningful explanation for the powerful effects of CP27 that we have documented in previous studies (Diekwisch and Luan, 2002) and that we are currently confirming in knockout and transgenic models in our laboratory (data not published). Besides CP27, NF-Y affects a number of other genes essential to early embryonic and craniofacial development, including the chondrogenesis gene SOX9 (Pan et al., 2009), the early fibroblast growth factor FGF4 (Bernard et al., 2005), the fibroblast growth factor receptor 2 promoter in osteoblasts (Sun et al., 2009), the tooth enamel gene amelogenin (Xu et al., 2006), and the tooth dentin gene dentin sialophosphoprotein (Chen et al., 2008). Underscoring NF-Y’s essential role in early mouse development and cell proliferation, gene targeting studies have resulted in early embryonic lethality (Bhattacharya et al., 2003).
NF-Y has been demonstrated to interact with the CCAAT box in both forward and reverse orientation in eukaryotic promoters and has been shown to absolutely require all 5 nucleotides of the CCAAT box (Pan et al., 1999; Hu et al., 2000; Xiong et al., 2000; Zhou et al., 2000). Our mutation analysis estalished that a mutation from CCAAT to CACAT almost abolished NF-Y binding activity and CP27 promoter function, which further confirmed the importance of the intact CCAAT pentanucleotides in cp27 gene regulation. In addition to the functional importance of the CCAAT core sequence, database analysis and functional studies revealed that the adjacent flanking nucleotides (C, Pu, Pu, on the 5′-side and C/G, A/G, G A/C, G on the 3′-site) of the CCAAT box are required for efficient binding of NF-Y (Mantovani, 1998). In comparison to the CCAAT consensus sequence including its flanking regions, the mouse CP27 CCAAT motif (TGACCAATCGCAG) has 85% homology with the common consensus sequence (Mantovani, 1998), indicating that the mouse CP27 CCAAT motif may demonstrate similar binding behavior as CCAAT promoter regions of other genes and species.
Interactions between NF-Y and more distal flanking regions of the CCAAT box have been established by previous footprinting experiments (Bi et al., 1997). The sequences protected from hydroxyl radical cleavage are located on both 5′ and 3′ flanking regions. The function of these protected regions is suggested to provide minimal DNA fragments required for proper binding by NF-Y. Partial removal of one of these regions leads to a decrease in binding (Romier et al., 2003). Complete removal of the protected region from the 3′ flanking side not only alters the affinity of NF-Y for its binding site but also the electrophoretic mobility of the NF-Y-DNA complex (Romier et al., 2003; Sugiura and Takishima, 2003). It is not known whether the DNA sequence of the distal region is specific for the NF-Y binding or only necessary for proper distortion of the DNA. In the present study, we have provided evidence that the specific sequence of the protected binding site is critical for NF-Y to interact with the CCAAT box. Sequence analysis of the 3′ protected region in the pro-a1(I) collagen, pro-α2(I) collagen (Bi et al., 1997) and CP27 promoter revealed that the binding sites are located between 10–16bp downstream of the CCAAT box, in an equivalent position to the CP27 CGGA motif. Our supershift assays indicate that a crucial CGGA binding site in the 3′ flanking region of the CCAAT box affected NF-Y binding, resulting in lower binding activity for the mutated CGGA motif (mutated to TACA) interacting with NF-Y. Thus, our studies introduce CGGA as a CP27-related motif for CCAAT-box mediated NF-Y binding.
Using primer extension assay, ribonuclease protection assays and 5′ RACE, we identified two candidate sites as potential CP27 transcription start sites 140 and 98 bp upstream of the ATG initiation codon. The presence of several different transcription start sites is a common feature of mammalian genes. Splicing of exons is one way to shift the location of transcription start sites (Rustighi et al., 1999; Yu et al., 1999). In the present study, we have detected two different transcripts of the cp27 gene in the odontoblast cell line OB1 by Northern blot analysis (data not shown). In other cases, one gene may contain several hitherto unidentified 5′ untranslated regions (Menon et al., 1995; Mu and Burt, 1999). To date, only two full-length mouse CP27 cDNA sequences have been reported (GenBank NM_011801 and BC_005589). These are only a few nucleotides apart from each other. Multiple transcription start sites are often cell line specific (Schroeder and Myers, 2008), and the multiplicity of transcription start sites may play a role in the tissue-specific expression of the cp27 gene at various stages of development. Transcription start sites differ between various sub-species or strains since 5′-RACE assays using different cell lines established from various mouse strains yielded different transcription start sites of the cp27 gene (data not shown). Based on our 5′ RACE study and published information on CP27 cDNA clone regions, we have defined the start site 102 nucleotides upstream of the initiation codon as preferred transcription start site. In addition, this site is the shortest 5′ transcription start site.
Together, our CP27 function studies in tandem with studies on the NF-Y transcriptional network indicate that CP27 may act in synchronicity with other NF-Y regulated genes to modulate cell proliferation and cell survival in craniofacial development. In this context, the present study uncovers the contribution of NF-Y in cp27 gene regulation and provides a further explanation of cp27 gene function in the NF-Y-mediated transcriptional network of genes.
Acknowledgments
Funding for these studies by the National Institute for Dental and Craniofacial Research to TGHD is gratefully acknowledged (DE 13095). Suggestions by three reviewers and by the Editor, Dr. Martin Batzer, greatly improved the quality of this manuscript.
- AMV
Avian myeloblastosis virus
- AP-1
Activator protein transcription factor
- CBF
Core binding factor
- CDC25c
CDC25 phosphatase
- C/EBP
CCAAT/enhancer binding protein
- CP27
Craniofacial protein with a molecular weight of 27kDa
- Csx/Nkx 2.5
cardiac homeobox transcription factor
- EMSA
Electrophoretic mobility shift assay
- ETS
E-twenty six transcription factor
- FGF
Fibroblast growth factor
- GATA
Transcription factor that binds to the GATA DNA sequence
- HSP 70
Heat shock protein 70
- NF-Y
Nuclear factor Y
- NF-1
Nuclear factor 1
- Oct
Octamer binding factor
- POU
Pit-1, Oct-1, Unc-86 transcription factor family
- RLM-RACE
RNA ligase mediated rapid amplification of cDNA ends
- SOX
Sry-related high mobility group box transcription factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alder H, Yoshinouchi M, Prystowsky MB, Appasamy P, Baserga R. A conserved region in intro 1 negatively regulates the expression of the PCNA gene. Nucleic Acids Res. 1992;11:1769–1775. doi: 10.1093/nar/20.7.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermadt CT, Nowling T, Wiebe MS, Rizzino A. NF-Y behaves as a bifunction transcription factor that can stimulate or repress the FGF-4 promoter in an enhancer-dependent manner. Gene Expr. 2005;12:193–212. doi: 10.3727/000000005783992052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Deng JM, Zhang Z, Behriger R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- Bi W, Wu L, Coustry F, de Crombrugghe B, Maity SN. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J Biol Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J Biol Chem. 2003;278:30435–30440. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]
- Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol. 2008;28:2047–2058. doi: 10.1128/MCB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HD, Yun J, Bang YJ, Shin DY. Cdk2-dependent phosphorylation of the NF-y transcription factor is essential for the expression of the cell cycle-regulatory genes and cell cycle G1/S and G2/M transition. Ongogene. 2004;23:4084–4088. doi: 10.1038/sj.onc.1207482. [DOI] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Martinez M, Li T, Wu Y, Chuang H, Chen L, Dong J, Gay I, MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Later embryogenesis: regulatory circuitry in morphogenetic fields. Development. 1993;118:665–690. doi: 10.1242/dev.118.3.665. [DOI] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekwisch TG, Marches F. cp27novel gene expression during craniofacial development. J Dent Res. 1997;76:27. [Google Scholar]
- Diekwisch TG, Marches F, Williams A, Luan X. Cloning, gene expression, and characterization of CP27, a novel gene in mouse embryogenesis. Gene. 1999;235:19–30. doi: 10.1016/s0378-1119(99)00220-6. [DOI] [PubMed] [Google Scholar]
- Diekwisch TG, Luan X. CP27 function is necessary for cell survival and differentiation during tooth morphogenesis in organ culture. Gene. 2002;287:141–147. doi: 10.1016/s0378-1119(01)00868-x. [DOI] [PubMed] [Google Scholar]
- Diekwisch TG, Luan X, McIntosh JE. CP27 localization in the dental lamina basement membrane and in the stellate reticulum of developing teeth. J Histochem Cytochem. 2002;50:583–586. doi: 10.1177/002215540205000416. [DOI] [PubMed] [Google Scholar]
- Donati G, Gatta R, Dolfini D, Fossati A, Ceribelli M, Mantovani R. An NF-Y-dependent switch of positive and negative histone methyl marks on CCAAT promoters. PLos ONE. 2008;3:e2066. doi: 10.1371/journal.pone.0002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta R, Mantonani R. NF-Y substitutes H2A-H2B on active cell-cycle promoters: recruitment of CoREST-KDM1 and fine-tuning of H3 methylations. Nucleic Acids Res. 2008;36:6592–6607. doi: 10.1093/nar/gkn699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gurtner A, Fuschi P, Magi F, Colussi C, Gaetano C, Dobbelstein M, Sacchi A, Piaggio G. NF-Y dependent epigenetic modifications discriminate between proliferation and postmitotic tissue. Plos One. 2008;23:22047. doi: 10.1371/journal.pone.0002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Lu JF, Luo R, Sen S, Maity SN. Inhibition of CBF/NF-Y mediated transcription activation arrests cells at G2/M phase and suppressed expression of genes activated at G2/M phase of the cell cycle. Nucleic Acid Res. 2006;34:6272–6285. doi: 10.1093/nar/gkl801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Jin S, Scotto KW. Transcriptional activation of the MDR1 gene by UV irradiation. Role of NF-Y and Sp1. J Biol Chem. 2000;275:2979–2985. doi: 10.1074/jbc.275.4.2979. [DOI] [PubMed] [Google Scholar]
- Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Del Sal G, Piaggio G, Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAAT-containing G2/M promoters. Mol Cell Biol. 2006;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko W, Nakatani Y, Wolffe AP. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan X, Diekwisch TG. CP27 affects viability, proliferation, attachment and gene expression in embryonic fibroblasts. Cell Prolif. 2002;35:207–219. doi: 10.1046/j.1365-2184.2002.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougald OA, Jump DB. Identification of functional cis-acting elements within the rat liver S14 promoter. Biochem J. 1991;280 (Pt 3):761–767. doi: 10.1042/bj2800761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Menon RK, Stephan DA, Singh M, Morris SM, Jr, Zou L. Cloning of the promoter-regulatory region of the murine growth hormone receptor gene. Identification of a developmentally regulated enhancer element. J Biol Chem. 1995;270:8851–8859. doi: 10.1074/jbc.270.15.8851. [DOI] [PubMed] [Google Scholar]
- Mu W, Burt DR. The mouse GABA(A) receptor alpha3 subunit gene and promoter. Brain Res Mol Brain Res. 1999;73:172–180. doi: 10.1016/s0169-328x(99)00258-2. [DOI] [PubMed] [Google Scholar]
- Muller GA, Heissig F, Engeland K. Chimpanzee, organgutan, mouse, and human cell cycle promoters exempt CCAAT boxes and CHR elements from interspecies differences. Mol Biol Evol. 2007;24:814–826. doi: 10.1093/molbev/msl210. [DOI] [PubMed] [Google Scholar]
- Nobukuni T, Kobayashi M, Omori A, Ichinose S, Iwanaga T, Takahashi I, Hashimoto K, Hattori S, Kaibuchi K, Miyata Y, Masui T, Iwashita S. An Alu-linked repetitive sequence corresponding to 280 amino acids is expressed in a novel bovine protein, but not in its human homologue. J Biol Chem. 1997;272:2801–28071. doi: 10.1074/jbc.272.5.2801. [DOI] [PubMed] [Google Scholar]
- Novak U, Paradiso L. Identification of proteins in DNA-protein complexes after blotting of EMSA gels. Biotechniques. 1995;19:54–55. [PubMed] [Google Scholar]
- Pan Q, Wu Y, Lin T, Yao H, Yang Z, Gao G, Song E, Shen H. Bone morphogenetic protein-2 induces chromatin remodeling and modification at the proximal promoter of Sox9 gene. BBRC. 2009;379:356–361. doi: 10.1016/j.bbrc.2008.12.062. [DOI] [PubMed] [Google Scholar]
- Pan Z, Hetherington CJ, Zhang DE. CCAAT/enhancer-binding protein activates the CD14 promoter and mediates transforming growth factor beta signaling in monocyte development. J Biol Chem. 1999;274:23242–23248. doi: 10.1074/jbc.274.33.23242. [DOI] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- Rustighi A, Mantovani F, Fusco A, Giancotti V, Manfioletti G. Sp1 and CTF/NF-1 transcription factors are involved in the basal expressionof the Hmgi -c proximal promoter. Biochem Biophys Res Commun. 1999;265:439–447. doi: 10.1006/bbrc.1999.1680. [DOI] [PubMed] [Google Scholar]
- Salsi V, Caretti G, Wasner M, Reinhard W, Engeland K, Mantovani R. Interaction between p300 and multiple NY-Y trimers govern cyclin B2 promogter function. J Biol Chem. 2003;278:6642–6650. doi: 10.1074/jbc.M210065200. [DOI] [PubMed] [Google Scholar]
- Schrodeder DI, Myers RM. Multiple transcription start sites for FOXP2 with varying cellular specificities. Gene. 2008;413:42–48. doi: 10.1016/j.gene.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Slavkin HC, Diekwisch T. Evolution in tooth developmental biology: of morphology and molecules. Anat Rec. 1996;245:131–150. doi: 10.1002/(SICI)1097-0185(199606)245:2<131::AID-AR3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Sugira N, Takishima K. Interaction of NF-Y with the 3′-flanking DNA sequence of the CCAAT box. FEBS Lett. 2003;537:58–62. doi: 10.1016/s0014-5793(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Sun F, Xie Q, Ma J, Yang S, Chen Q, Hong A. Nuclear factor Y is required for basal activation and chromatin accessibility of fibroblast growth factor receptor 2 promoter in osteoblast-like cells. J Biol Chem. 2009;284:3136–3147. doi: 10.1074/jbc.M808992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester SL, ap Rhys CM, Luethy-Martindale JD, Holbrook NJ. Induction of GADD153, a CCAAT/enhancer-binding protein (C/EBP)-related gene, during the acute phase response in rats. Evidence for the involvement of C/EBPs in regulating its expression. J Biol Chem. 1994;269:20119–20125. [PubMed] [Google Scholar]
- Testa A, Donati G, Yan P, Romani F, Huang TH, Vigano MA, Mantovani R. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J Biol Chem. 2005;280:13606–13615. doi: 10.1074/jbc.M414039200. [DOI] [PubMed] [Google Scholar]
- Thesleff I. Tooth morphogenesis. Adv Dent Res. 1995;9:12. doi: 10.1177/0895937495009003S0301. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Wasner M, Haugwitz U, Reinhard W, Tschop K, Lorenz J, Mossner J, Engeland K. Three CCAAT-box and a single cell cycle genes homology region (CHR) are the major regulating sites for transcription from the human cyclin B2 promoter. Gene. 2008;312:225–237. doi: 10.1016/s0378-1119(03)00618-8. [DOI] [PubMed] [Google Scholar]
- Xiong S, Chirala SS, Wakil SJ. Sterol regulation of human fatty acid synthase promoter I requires nuclear factor-Y- and Sp-1-binding sites. Proc Natl Acad Sci U S A. 2000;97:3948–3953. doi: 10.1073/pnas.040574197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhuo Y, Luo W, Zhu Q, Levy D, MacDougald OA, Snead ML. NF-Y and CCAAT/Enhancer-binding protein a synergistically activate the mouse amelogenin gene. J Biol Chem. 2006;281:16090–16098. doi: 10.1074/jbc.M510514200. [DOI] [PubMed] [Google Scholar]
- Yu JH, Schwartzbauer G, Kazlman A, Menon RK. Role of the Sp family of transcription factors in the ontogeny of growth hormone receptor gene expression. J Biol Chem. 1999;274:34327–34336. doi: 10.1074/jbc.274.48.34327. [DOI] [PubMed] [Google Scholar]
- Zhou YL, Snead ML. Identification of CCAAT/enhancer-binding protein alpha as a transactivator of the mouse amelogenin gene. J Biol Chem. 2000;275:12273–12280. doi: 10.1074/jbc.275.16.12273. [DOI] [PubMed] [Google Scholar]
- Zorbas H, Rein T, Krause A, Hoffmann K, Winnacker EL. Nuclear factor I (NF I) binds to an NF I-type site but not to the CCAAT site in the human alpha-globin gene promoter. J Biol Chem. 1992;267:8478–8484. [PubMed] [Google Scholar]
- Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, Muller R. Cell cycle regulation of the cyclin A, cdc25c and cdc2 genes is based on a common mechanism of transcription repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]