Abstract
Due to the risk of insertional mutagenesis, viral transduction has been increasingly replaced by non-viral methods to generate induced pluripotent stem (iPS) cells. One technique that has not yet been explored is the use of “minicircle” DNA, a novel compact vector that is free of bacterial DNA and capable of persistent high level expression in cells. Here, we report the use of a single minicircle vector to generate transgene-free iPS cells from adult human cells.
Keywords: minicircle DNA, reprogramming, iPS cells, viral-free, human adipose stem cells
Non-viral methods for generating iPS cells using adenovirus1, plasmids2, or excision of reprogramming factors using Cre/LoxP3,4 or piggyBAC transposition5 have been reported, but in general are restricted to mouse, suffer from low reprogramming efficiencies (<0.003%), and may leave behind residual vector sequences. Recently, successful reprogramming of human neonatal foreskin fibroblasts was reported using episomal vectors derived from the Epstein-Barr virus6. However, this technique required three individual plasmids carrying a total of seven factors, including the oncogene SV40, and has not been shown to successfully reprogram cells from adult donors, a more clinically-relevant target population. Further, expression of the EBNA1 protein, as was required for this technique, may increase immune cell recognition of transfected cells7, thus potentially limiting clinical application if the transgene is not completely removed. Protein-based iPS cell generation in mouse8 and human9 fetal and neonatal cells has also been published, but required either chemical treatment (valproic acid)8 or greater than four rounds of treatment9. Lastly, protein-based methods necessitate expertise in protein chemistry and handling-skills that many laboratories do not have. Most DNA-based methods require only minimal molecular biology background, and so remain a more attractive option for a wider population of researchers interested in cellular reprogramming.
Compared to plasmids, minicircle DNA benefits from higher transfection efficiencies and longer ectopic expression due to its lower activation of exogenous silencing mechanisms10,11, and thus may represent an ideal mechanism for generating iPS cells. We constructed a plasmid (P2PhiC31-LGNSO) that contained a single cassette of four reprogramming factors (Oct4, Sox2, Lin28, Nanog) plus a green fluorescent protein (GFP) reporter gene, each separated by selfcleavage peptide 2A sequences12,13 (Supplementary Fig. 1a,b). We next took advantage of the PhiC31-based intramolecular recombination system that allows the plasmid backbone to be excluded and degraded in bacteria, and the minicircle to be purified and isolated as described10,11 (Supplementary Fig. 1c). Expression of individual protein factors was validated in 293FT cells (Supplementary Fig. 2). To determine the reprogramming ability of the minicircle vector, we chose to induce pluripotency in human adipose stem cells (hASCs). hASCs have a number of advantages over other somatic cell types such as fibroblasts since they can be isolated in large quantities (100 ml of human adipose tissue yields about 1 × 106 cells) with minimal morbidity14. Furthermore, quantitative PCR (qPCR) of hASCs revealed 3–4 times higher relative expression of Klf4 compared to human ES cells, and ~1.3 higher relative expression of c-Myc (Supplementary Fig. 3).
Using hASCs derived from three adult patients, we introduced the minicircle vector into cells using nucleofection and achieved 10.8±1.7% GFP-positive cells, compared to only 2.7±0.8% using a standard plasmid carrying the same expression cassette (Supplementary Fig. 4). The GFP-positive cell population was enriched 72h post-transfection by flow cytometetry, and then seeded on inactivated mouse embryonic fibroblast (MEF) feeder layers. Due to the dilution of minicircle vector with proliferation, we observed gradual loss of GFP expression in cells that was concurrent with activation of endogenous Oct4 expression (Supplementary Fig. 5). Additional transfections at days 4 and 6 with the minicircle vector were performed to supplement this loss of transgene expression. Importantly, compared to standard plasmids that carry a GFP/firefly luciferase reporter gene, minicircle DNA with the same reporter gene maintains higher expression for a longer period of time in hASCs, as confirmed by photon counts and qPCR (Supplementary Fig. 6). Thus, we believe minicircle DNA is a more robust gene expression platform compared to regular plasmid.
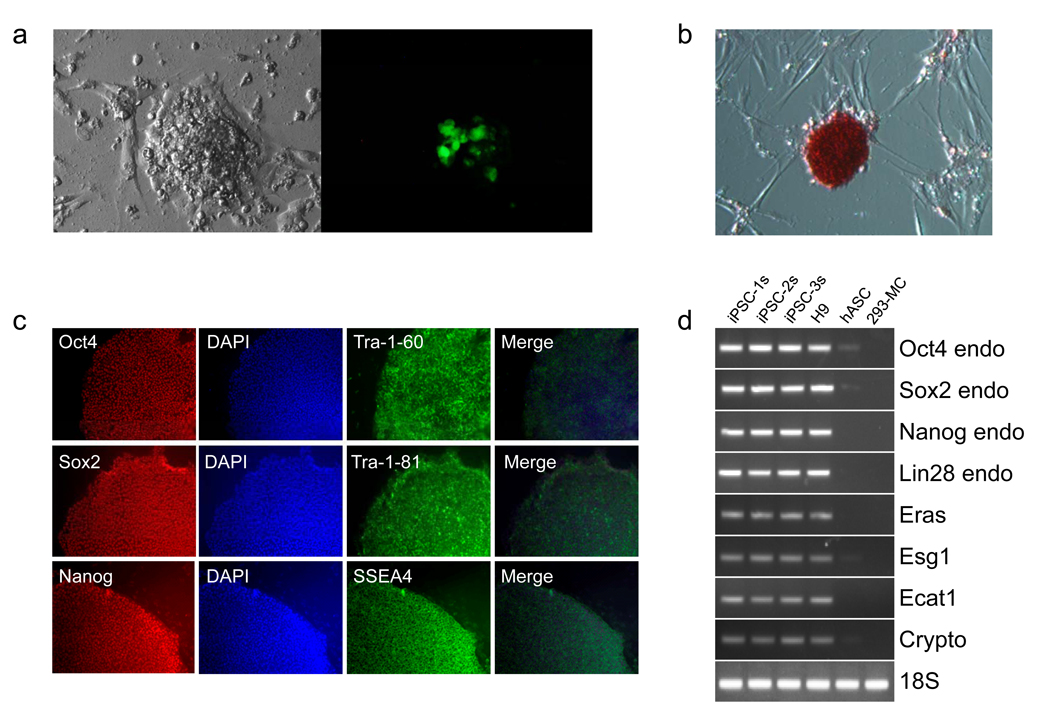
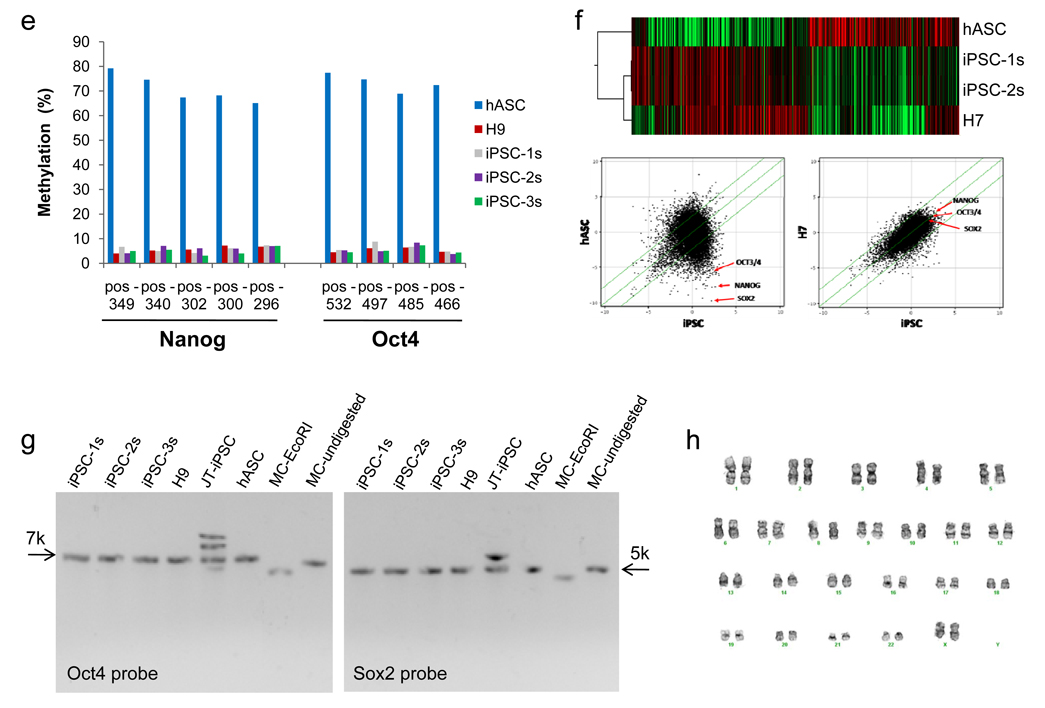
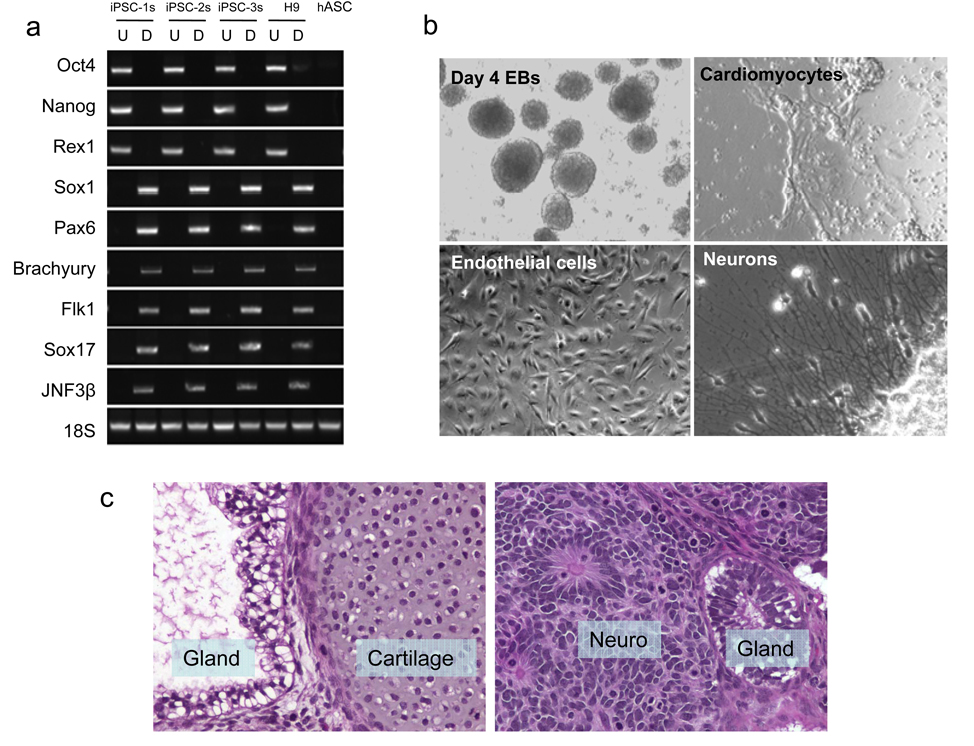
On days 14–16, we observed GFP-positive clusters that had morphologic similarities with human ES cell colonies (Fig. 1a). Many clusters showed no GFP expression at all, suggesting loss of transgene expression, and these were isolated for further analysis. Minicircle-derived iPS cell (mc-iPS cell) colonies stained positive for embryonic markers (Fig. 1b,c), and exhibited reactivation of endogenous Oct4, Sox2, and Nanog genes (Fig. 1d,e). Global gene expression profiling of mc-iPS cell subclones demonstrated a high degree of similarity with human ES cells (Fig. 1f), as well as with iPS cells derived using both lentiviral and non-viral vectors (Supplementary Fig. 7). Importantly, Southern blot analysis did not detect genomic integration of the minicircle transgene in the subclones (Fig. 1g, see Supplementary Fig. 8 for probe sensitivities). Finally, mc-iPS cells had normal diploid karyotype (Fig. 1h). The pluripotency of mc-iPS cells was examined both in vitro through the formation of embryoid bodies (EBs), and in vivo by teratoma formation. EBs expressed genes of all three embryonic germ layers (Fig. 2a) and formed multi-lineage cell types (Fig. 2b, Supplementary Fig. 9, Supplementary Video 1). Lastly, subcutaneous injection of each of the three mc-iPS subclones into immunodeficient mice resulted in a teratoma (Fig. 2c).
Figure 1. Generation of iPS cells with minicircle vector.
a, Representative day 18 cluster of mc-iPS cells. The cluster shown here exhibited transgene expression, as seen by GFP detection. GFP-negative colonies were isolated for further analysis. b, mc-iPS cells are positive for alkaline phosphatase, c, immunostain for common embryonic markers, and d, express embryonic genes, including reactivation of endogenous Oct4, Sox2, Nanog and Lin28. As negative control, 293FT cells were transfected with the minicircle vector (293-MC) but did not express the same endogenous genes. (Note: H9 is the H9 human ES cell line. iPSC-1s denotes a mc-iPS cell colony subclone derived from Donor 1, iPSC-2s subclone from Donor 2, etc). e, Bisulfate sequencing shows hypomethylation within the promoters of Oct4 and Nanog in the three mc-iPS cell subclones. f, Microarray data. Upper panel, heat map showing two mc-iPS cell subclones are similar to H7 human ES cells and distinct from hASCs. Lower panel, scatter plots highlighting Oct3/4, Sox2, and Nanog expression (red arrows). Green lines indicate 5-fold changes in expression levels between paired samples (iPSC represents the average of both subclones). g, Southern blot analysis of genomic DNA confirms that mc-iPS subclones are transgene free, as determined by the number of copies of Oct4 and Sox2. As positive control, lentivirallyreprogrammed iPS cells from the James Thomson lab (JT-iPSC) exhibited multiple bands for each of the two genes. h, mc-iPS cells (passage 5) had normal diploid karyotype.
Figure 2. Pluripotency of mc-iPS cells.
a, RT-PCR analysis of pluripotent markers (Oct4, Nanog and Rex1) and various differentiation markers for the three germ layers (ectoderm: Sox1, Pax6; mesoderm: Brachyury, Flk1; endoderm: Sox17, JNF3β). Undifferentiated (U) mc-iPS cells; differentiated (D) mc-iPS cells after 8 days suspension culture followed by 8 days adherent culture. b, Multiple cell types were differentiated from mc-iPS cells. See Supplementary Figure 9 and Supplementary Video 1 for further characterization. c, Subcutaneous injection of mc-iPS cells causes teratomas in SCID mice consisting of all three embryonic germ layers, including epithelial cells (ectoderm), cartilage (mesoderm), and glandular structures (endoderm). Tissue sections from subclone mc-iPSC-1s are shown as representative.
In total, we successfully derived 22 mc-iPS cell colonies from hASCs that had been isolated from three adult donors, yielding an overall reprogramming efficiency of ~0.005% with minicircle DNA. As with other integration-free iPS cell generation approaches, this efficiency is low compared to viral-based methods, which have typically been reported to be ~0.01%15–17. Our efficiency with minicircle DNA was still higher than previous plasmid-based transfection reprogramming methods2,6, though this may be due in part to differences in donor cell types (neonatal fibroblasts vs. hASCs) and the number of reprogramming factors that were used. To address this, we transfected IMR90 neonatal fibroblast with the minicircle vector and were able to create iPS cells, though with 10-fold less efficiency (Supplementary Table 1). Notably, we were unable to generate iPS cells using regular plasmid vector (data not shown). As stated previously, we believe this is due to the higher transfection efficiency, and stronger and more persistent expression, of minicircle DNA in cell cultures.
As the reprogramming field moves from basic science towards clinical translation, efficient derivation of iPS cells that are free of foreign or chemical elements is absolutely critical. Here, we describe a simple method for generating transgene-free iPS cells from adult donor sources, and that requires only a single vector without the need for subsequent drug selection or vector-excision, or inclusion of oncogenes such as SV40. With its basic molecular principles and straightforward protocol, minicircle DNA is ideally suited for facilitating iPS cell research around the world. Finally, the minicircle DNA is already FDA approved, giving our novel method the potential for significant clinical translation.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Andrew J. Connolly for assistance with histological analysis, the Stanford Functional Genomics Facility for assistance with microarrays, and Athena Cherry for assistance with cytogenetics. We thank funding support from Mallinckrodt Foundation, NIH DP2OD004437, RC1HL100490, and AHA 0970394N (JCW); NIH R90 DK 07010301, CIRM T1-00001, CIRM RL1-00662-1, NIH R21 DE018727, NIH R21 DE019274, the Oak Foundation and the Hagey Laboratory for Pediatric Regenerative Medicine (MTL); U01HL099776 (RCR).
REFERENCES
- 1.Stadtfeld Matthias, Nagaya Masaki, Utikal Jochen, et al. Science. 2008;322(5903):945. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okita Keisuke, Nakagawa Masato, Hyenjong Hong, et al. Science. 2008;322(5903):949. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 3.Kaji Keisuke, Norrby Katherine, Paca Agnieszka, et al. Nature. 2009;458(7239):771. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soldner Frank, Hockemeyer Dirk, Beard Caroline, et al. Cell. 2009;136(5):964. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woltjen Knut, Michael Iacovos P, Mohseni Paria, et al. Nature. 2009;458(7239):766. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Junying, Hu Kejin, Smuga-Otto Kim, et al. Science. 2009;324(5928):797. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munz Christian, Bickham Kara L, Subklewe Marion, et al. The Journal of Experimental Medicine. 2000;191(10):1649. doi: 10.1084/jem.191.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Hongyan, Wu Shili, Joo Jin, et al. Cell Stem Cell. 2009;4(5):381. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Dohoon, Kim Chun-Hyung, Moon Jung-Il, et al. Cell Stem Cell. 2009;4(6):472. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Zhi-Ying, He Cheng-Yi, Ehrhardt Anja, et al. Molecular Therapy. 2003;8(3):495. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 11.Chen Zhi-Ying, He Cheng-Yi, Kay Mark. Human Gene Therapy. 2005;16(1):126. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- 12.Ryan Martin D, Drew Jeff. The EMBO Journal. 1994;13(4):928. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan Martin D, King Andrew MQ, Thomas G Paul. The Journal of General Virology. 1991;72(Pt 11):2727. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- 14.Fraser John K, Wulur Isabella, Alfonso Zeni, et al. Trends in Biotechnology. 2006;24(4):150. doi: 10.1016/j.tibtech.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Okita Keisuke, Ichisaka Tomoko, Yamanaka Shinya. Nature. 2007;448(7151):313. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 16.Maherali Nimet, Sridharan Rupa, Xie Wei, et al. Cell Stem Cell. 2007;1(1):55. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Wernig Marius, Meissner Alexander, Foreman Ruth, et al. Nature. 2007;448(7151):318. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.