To the Editor
From the discovery of DNA to the sequencing of the human genome, the template-dependent formation of biological molecules from gene to RNA and protein has been the central tenet of biology. Yet the origins of many diseases, including allergy, Alzheimer’s disease, asthma, autism, diabetes, inflammatory bowel disease, Lou Gehrig’s disease, multiple sclerosis, Parkinson’s disease and rheumatoid arthritis, continue to evade our understanding. Expectations that defined variation in the DNA blueprint would serve to pinpoint even multigenic causes of these diseases remain unfulfilled. Studies of distinct populations have implicated different genes, and those genes that are identified contribute to disease in a small fraction of the individuals diagnosed1–3. The genetic parts list seems insufficient to account for the origin of many grievous illnesses. Environmental factors including diet and microorganisms are also origins of disease. For example, type 2 diabetes, which affects hundreds of millions of people, is linked to a high-fat diet4, and this mechanism of disease onset is common to diverse species. When disease arises from a cellular response to a pathogen or environmental stimulus, genomics alone is unlikely to provide all the answers. This view is underscored by the observation that surprisingly similar numbers of genes exist in even the most divergent of life forms. Moreover, while the genome provides the framework and basic instruction upon which the cell develops and operates, the full complexity of cellular life cannot be directly encoded by it.
As indivisible units of life, the cells of all organisms consist of four fundamental macromolecular components: nucleic acids (including DNA and RNA), proteins, lipids and glycans. From the construction, modification and interaction of these components, the cell develops and functions. The struggle to comprehend this interplay is the preoccupation of biologists, and more recently those engaged in systems biology. But do we readily take into account all of the components of biological systems to model health and disease accurately? To do this, the basic composition of all cells must be evident.
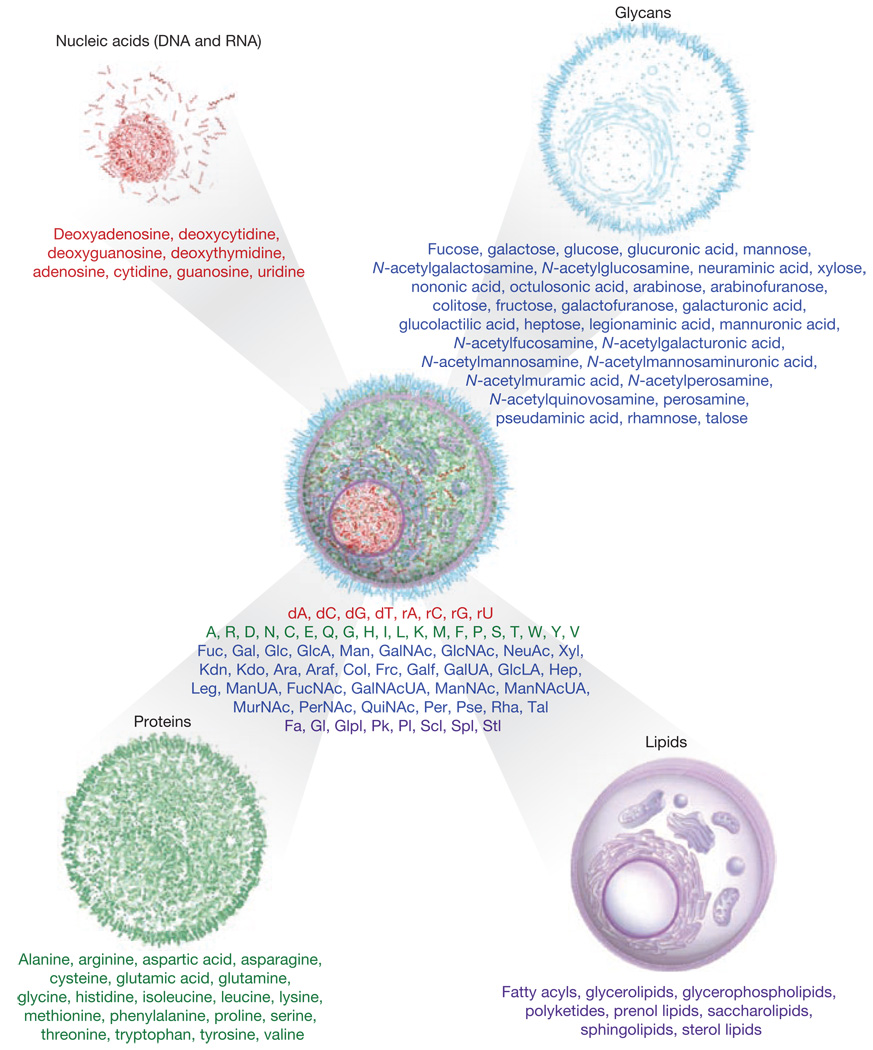
The physical sciences developed the periodic table of the elements to convey the composition and relatedness of matter. A related construct for biology may provide a more balanced view of the cell and its biochemistry. The four fundamental components of cellular life are derived from 68 molecular building blocks (Fig. 1). Unlike the genome and proteome, the glycome and lipidome are not directly encoded by DNA. Nevertheless, the glycome and the lipidome contribute to the pathogenesis and severity of an increasing number of diseases, and are usurped by pathogens as receptors for infection5–9. Scientific discussions that encompass these components remain relatively infrequent in the protein centric world of cell biology. Some scientists lament the ‘complexity of the molecules’. Yet our alphabet of 26 characters, let alone Chinese characters, is rather easily assimilated. Imagine a world in which each of us knew only a fraction of the alphabet.
Figure 1.
The molecular building blocks of life. There are 68 molecules that contribute to the synthesis and primary structures of the 4 fundamental macromolecular components of all cells: nucleic acids, proteins, glycans and lipids. DNA and RNA are produced from the 8 nucleosides. Although deoxyribose (d) and ribose (r) are saccharides, they are an integral part of the energetically charged nucleoside building blocks that are used to synthesize DNA and RNA. There are 20 natural amino acids used in the synthesis of proteins. Glycans derive initially from 32, and possibly more, saccharides used in the enzymatic process of glycosylation and are often attached to proteins and lipids, although some exist as independent macromolecules. Lipids are represented by 8 recently classified categories and contain a large repertoire of hydrophobic and amphipathic molecules. The number of molecular building blocks does not directly infer the relative structural complexity of the repertoire of each component. Not shown are the many different post-synthetic modifications of the molecules within these components.
Interdisciplinary education and research can ensure communication of ideas and advances, and will be essential to tackle complex trait diseases. Nonetheless, it is risky for individual scientists to enter into interdisciplinary research. The mechanisms that fund research continue to impede risk-taking behaviour. Meanwhile, the curriculums of universities and the programmes of major symposia rarely demonstrate an integrative vision of twenty-first-century biology. Public and private institutions that design educational programmes and provide funding are responsible for ensuring that the next generations of scientists receive the training, encouragement and resources necessary to engage in teaching and research that can seamlessly encompass all the major components critical to cells.
Defining the molecular building blocks of life provides a conceptual framework for biology that has the potential to enhance education and research by promoting the integration of knowledge. The insights afforded by bridging the divides that exist between disciplines can further moderate the view that researchers must invariably sacrifice breadth of knowledge to acquire depth of understanding. Cultivating this integration would reflect a more holistic and rigorous endeavour, which will ultimately be required if we are to perceive and most effectively manipulate the biological mechanisms of health and disease.
ACKNOWLEDGEMENTS
I thank Jon Pollock, Karen Chin and Ajit Varki for discussions. J.D.M. is supported by the Howard Hughes Medical Institute and National Institutes of Health grants HL57345, HL78784 and GM62116. Illustration by Katie Ris-Vicari.
References
- 1.Shriner D, Vaughan LK, Padilla MA, Tiwari HK. Problems with genome-wide association studies. Science. 2007;316:1840–1842. doi: 10.1126/science.316.5833.1840c. [DOI] [PubMed] [Google Scholar]
- 2.Wright L. Looking deep, deep into your genes. Onearth. 2007;29:32–35. [Google Scholar]
- 3.Garber K. The elusive ALS genes. Science. 2008;319:20. doi: 10.1126/science.319.5859.20. [DOI] [PubMed] [Google Scholar]
- 4.Smyth S, Heron A. Diabetes and obesity: the twin epidemics. Nature Med. 2005;12:75–80. doi: 10.1038/nm0106-75. [DOI] [PubMed] [Google Scholar]
- 5.Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- 6.Fahy E, et al. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Dube DH, Bertozzi CR. Glycans in cancer and inflammation-potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 8.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Wymann MP, Schneiter R. Lipid signaling in disease. Nature Rev. Mol. Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]