Abstract
Background
The Human Microbiome Project has ushered in a new era for human metagenomics and high-throughput next generation sequencing strategies.
Content
This review will describe evolving strategies in metagenomics with a special emphasis on the core technology of DNA pyrosequencing. The challenges of microbial identification in the context of microbial populations are described.
Summary
Both 16S rDNA amplicon and whole genome sequencing approaches may be useful for human metagenomics, and numerous bio-informatics tools are being deployed to tackle such vast amounts of microbiological sequence diversity. Metagenomics or studies of microbial communities may ultimately contribute to a more comprehensive understanding of human health, disease susceptibilities, and the pathophysiology of infectious and immune-mediated diseases.
Keywords: 454, DNA Sequencing, Informatics, Metagenomics, Microbial Ecology, Microbes, Microbiome, Molecular Microbiology, Pathogens, Pyrosequencing
Metagenomics and The Human Microbiome
Metagenomics refers to culture-independent studies of the collective set of genomes of mixed microbial communities, and may be applied to the exploration of all microbial genomes in consortia that reside in environmental niches, in plants or in animal hosts. Examples in mammalian biology include studies of microbial communities on various mucosal surfaces and the human skin. This review article will focus on analytical strategies for identifying pathogens in mixed microbial communities via metagenomics.
Metagenomics and associated meta-strategies have arrived at the forefront of biology primarily due to two major developments. The deployment of next generation DNA sequencing technologies in many centers has provided greatly enhanced capabilities for sequencing large meta-datasets. Technology has created new opportunities for the pursuit of large scale sequencing projects that were difficult to imagine just several years ago. The second key development is an emerging appreciation for the importance of complex microbial communities in mammalian biology and human health and disease. The human microbiome project (HMP) was approved in May 2007 as one of two major components (in addition to the human epigenomics program) of RoadMap version 1.5 of the U.S. National Institutes of Health (1). The demands of this project have resulted in the intense interest and focus in genome centers to apply parallel DNA sequencing technologies to human biology in a scale not previously witnessed.
The human microbiome refers to the entire population of microbes that colonize the human body including the gastrointestinal tract, genitourinary tract, oral cavity, nasopharynx, respiratory tract, and human skin. Different microorganisms compose the microbiome including bacteria, fungi (mostly yeasts), and viruses. Depending on the context, parasites may also be considered to compose part of the indigenous microbiota. The ‘metagenome’ of microbial communities that occupy various sites in the body is estimated to be approximately 100-fold greater in terms of gene content than the human genome. These diverse and complex collections of genes encode a wide array of biochemical and physiologic functions that may benefit the host as well as neighboring microbes (1). This review will focus on bacterial populations because bacteria form a predominant group of the microbiome with the most comprehensively documented phylogenetic datasets and classification systems. Most of the data gathered to date have been compiled with Sanger (dideoxy) sequencing platforms, but this review will focus on emerging parallel DNA sequencing technologies based on pyrosequencing. Such next generation sequencing systems introduce the possibilities for deeply sequenced data collections and strategies aimed at microbial identification via single genetic targets or whole genome methodologies.
Several important issues have emerged in the past several years with respect to metagenomics and microbes. One issue is that the science of metagenomics, in contrast to single microbial or animal genomes, is ultracomplex and challenged with vast unknown or knowledge “deserts.” Of the immense microbial taxonomic “space” in nature, only a restricted set of bacterial populations have been identified in the human body. As an example, the colonic microbiota is a vast ecosystem with approximately 800–1000 species per individual, but these estimates are in flux due to the new science of metagenomics and microbial pan-arrays. Approximately 62% of bacteria from the human intestine were previously unknown, and 80% of bacteria identified by metagenomic sequencing were considered noncultivable (2). Only nine of 70 bacterial phyla (divisions that vary in number depending on taxonomic scheme) have been found in the human intestine, and two phyla of bacteria, the Firmicutes and Bacteroidetes, predominate in number (1, 3). As an example within the Firmicutes, the genus Lactobacillus includes >100 different species (www.bacterio.cict.fr/l/lactobacillus.htm). To date, fewer than 20 Lactobacillus species have been found consistently in the mammalian gastrointestinal (GI) tract. These findings indicate that indigenous community membership is restricted to a limited subset of all bacteria, and bacterial populations are not randomly distributed in and on the human body. Preliminary studies suggest that the predominant species in the genitourinary tract and skin sites are fundamentally different from the predominant populations in the GI tract (4, 5).
In contrast to the human genome, the human metagenome may differ depending on location (site) within the human body, age, and environmental factors such as diet. A key remaining question is whether a core human microbiome is definable (1). Different regions of the body, even in related and contiguous sites, may differ with respect to bacterial quantities and species composition. Bacterial species may not be randomly distributed in space or time. The large intestine contains highly complex microbial populations, and the relative proportions of different bacteria may vary in different regions of the large intestine. Culture-independent studies of cecal bacteria indicate that facultative anaerobes compose a greater proportion of luminal bacteria, in contrast to the distal colon where obligate anaerobes predominate. Metagenomics studies highlighted differences between colonic mucosa-associated and fecal bacterial populations in humans and nonhuman primates (6–12). In addition to differences with respect to specimen type or body location, differences have also been noted with respect to gender in nonhuman primates, implying sexual dimorphism with respect to the human microbiome (12). In order to address the central challenge of microbial identification in the context of mixed species communities, primary strategies for DNA sequencing-based bacterial identification must be refined.
DNA Sequencing and Bacterial Identification
Pathogen identification in infectious diseases relies mostly on routine cultures and biochemical testing using semi-automated platforms in the clinical laboratory. The shift towards widespread adoption of nucleic acid sequencing for identification of microbial pathogens has been slowed by the user-intensive, highly technical nature of Sanger DNA sequencing. Nevertheless, several studies published in the 1990s indicated that 16S rRNA gene sequencing could be useful for pathogen discovery and identification (13, 14). Studies of bacterial evolution and phyogenetics in the 1980s provided the foundation for subsequent applications of 16S rRNA gene (or 16S rDNA) based sequencing for microbial identification (15). Initial studies were based on Sanger sequencing strategies that included targeted sequencing of 16S rRNA genes (approximately 1.5 kB target sequence). Such “long read” approaches enabled investigators and medical laboratory scientists to identify many individual genera and species that could not be identified by biochemical methods. Sequence-based identification could be established with a reasonable amount of confidence based on relatively long reads and sequence classifier algorithms that included most of the 16S rDNA coding sequence. However, less than half of the coding sequence (approximately 500 bp) including several hypervariable regions may be sufficient for genus- and species-level pathogen identification by Sanger sequencing (14, 16). As sequence targets for microbial identification were more precisely defined, the introduction of pyrosequencing provided a user-friendly approach for the clinical laboratory that enabled more extensive sampling of microbial diversity with improved labor efficiencies (17).
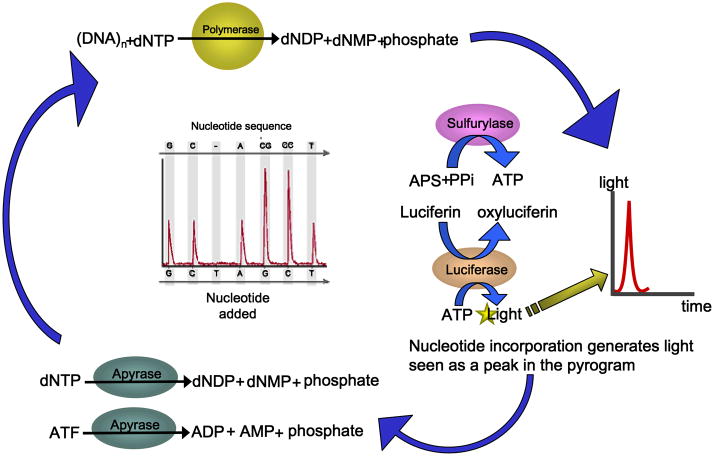
Although a large body of phylogenetic data for microbial identification was gathered using Sanger sequencing, new sequencing technologies have emerged that offer particular attractions to research and diagnostic laboratories. Specific genetic targets such as hypervariable regions within bacterial 16S rRNA genes may be amplified by PCR and subjected to DNA pyrosequencing. DNA pyrosequencing or sequencing-by-synthesis was developed in the mid-1990s as a fundamentally different approach to DNA sequencing (18). Sequencing-by-synthesis occurs by DNA polymerase-driven generation of inorganic pyrophosphate, resulting in the formation of adenosine triphosphate (ATP) and ATP-dependent conversion of luciferin to oxyluciferin (Fig. 1). The generation of oxyluciferin results in the emission of pulses of light, and the amplitude of each signal is directly related to the presence of one or more nucleosides. One important limitation of pyrosequencing is its relative inability to sequence longer stretches of DNA, rarely exceeding 100–200 bases with first- and second-generation high throughput pyrosequencing chemistries. For the purposes of this review, pyrosequencing will refer to the core chemistry, core technology, and low-throughput sequencing platforms (e.g. PSQ 96, Biotage Inc.) currently implemented in clinical laboratories. The term “454 sequencing” will refer to high-throughput sequencing platforms (Roche/454 Life Sciences) for metagenomics that are based on pyrosequencing chemistry.
Figure 1. Pyrosequencing chemistry.
This figure shows the biochemical reactions and enzymes involved in the generation of light signals by DNA pyrosequencing (18). Each peak in the pyrograms represents a pulse of light detected in the instrument. ATP, adenosine triphosphate; ADP, adenosine diphosphate; dNDP, deoxy-nucleotidyl diphosphate; dNMP, deoxy-nucleotidyl monophosphate; PPi, pyrophosphate. Adapted from www.biotagebio.com
DNA pyrosequencing has been successfully applied in a variety of applications including genotyping, single nucleotide polymorphism (SNP) detection, and microorganism identification (19). Pyrosequencing has been used to detect point mutations in antimicrobial or antiviral resistance genes in order to explore the presence of drug-resistant microbes (20–22). The relatively short read lengths of DNA pyrosequencing have placed a premium on careful target selection and oligonucleotide primer placement. Pyrosequencing has been successfully applied to microbial identification by combining informative target selection (hypervariable regions within 16S rRNA gene) and signature sequence matching (23, 24). Despite the fact that DNA pyrosequencing yields relatively short read lengths and limited amounts of sequence data per pathogen or microbe, this strategy has been useful for microbial identification in different settings. As one example, careful selection of highly informative hypervariable regions within the 16S rRNA genes facilitated the implementation of routine pyrosequencing strategies for pathogen identification in a hospital setting (17).
Due to the relatively short read lengths, DNA pyrosequencing applications for microbial identification have focused attention on hypervariable regions within small ribosomal subunit RNA genes, especially 16S rRNA genes. Specific hypervariable region have been preferentially used to identify different classes of bacteria by pyrosequencing (24, 25). Once DNA sequence data are generated, sequences must be analyzed with special considerations in mind to facilitate accurate bacterial identification. First, different taxonomic classifications are used for identification, and different species identifications may be generated depending on the taxonomic scheme. The oldest and most traditional bacterial classification system is based on Bergey’s taxonomy which has attempted to merge phenotypic (e.g. biochemical) and molecular data to create a higher-order taxonomy in recent years (26). More recently developed taxonomic schemes include systems proposed by Pace (27), Ludwig (28), Hugenholtz (29), and the NCBI. Multiple on-line databases have been developed on the basis of these different taxonomic schemes and provide convenient access to large ribosomal RNA sequence databases for clinical laboratories and research teams. The most prominent databases include the Ribosomal Database Project II (RDP II) (http://rdp.cme.msu.edu/) (30), Greengenes (greengenes.lbl.gov) (31), and ARB-Silva (32). RDP II is based on Bergey’s taxonomy which contains a relatively small number of phyla (divisions). Greengenes includes multiple taxonomic schemes so that query results with this database can be compared using different classification systems. The ARB-Silva database also offers a choice of microbial taxonomies, although it is more limited in its flexibility than Greengenes. Microbial identification depends on the taxonomic curation. As a case in point, the Pace and Hugenholtz lineages separately named 12 phylum-level lineages, and RDP II had not named any of these lineages (31). The taxonomic schemes varied with respect to numbers of phyla, for example, with a maximum of 88 phyla for the Pace and Hugenholtz curations and 31 phyla for the RDP (based on Bergey’s) classification system (31). So, in addition to the routine issues of “splitting” and “lumping” taxa in different schemes, one is confronted with different phyla (divisions) and corresponding sub-groupings (e.g. class, order, family).
On-line ribosomal RNA databases include a variety of software tools for sequence classification and multiple sequence alignments in order to facilitate microbial identification. The ARB software package is a widely used program suite that includes open source, directly interacting software tools that are linked to an integrated microbial sequence database (ARB-Silva) (28). These software environments (Greengenes, RDP, ARB-Silva) contain sequence query tools, sequence alignment programs and sequence editors. Greengenes provides a 16S rRNA workbench for sequence-based microbial identification with different query and sequence alignment tools (31). Greengenes uses the NAST aligner tool (33) and generates output that is compatible with ARB software tools so that different open source environments may be linked via the internet for comprehensive microbial population studies. Different supervised sequence classifier tools are available for matching test with query sequences. Compared to BLAST, supervised classifiers like RDP Seqmatch demonstrated greater accuracy in finding most similar rDNA sequences (34). The RDP-based SeqMatch k-NN classifier is effective at determining probable sequence identities on the basis of pairwise aligned distances. Alternatively, the RDP II group has developed its own naive Bayesian classifier which can be easily retrained as new sequences are incorporated into rapidly expanding microbial sequence databases (35). The Bayesian classifier uses information averaged within the entire genus and is less influenced by individual misplaced sequences. Sequence query tools such as SeqMatch in RDP II enable relatively short query sequences that are 50 or more bases in length to yield accurate microbial identifications. Despite using two supervised classifiers with the same database, different results were generated for particular sequences, particularly with phylogenetically broad genera such as Clostridium (35).
Next Generation DNA Sequencing Technologies – Pyrosequencing and 454
Until recently, most microbial genome and metagenomics sequencing projects have been generated primarily by Sanger sequencing methods. The rapid development of parallel, high throughput sequencing technologies during the current decade has resulted in the commercialization and widespread adoption of next generation (NexGen) sequencing technologies. In contrast to a relatively homogeneous DNA sequencing enterprise in the 1990s, current large scale genome and metagenome sequencing projects are deploying multiple platforms and different sequencing chemistries in parallel. As of June 2008, three NexGen platform vendors commercially distribute machines for high throughput sequencing and include Roche/454 Life Sciences (GS20, FLX, LXR); Illumina/Solexa (Illumina G2) and Applied Biosystems (SOLiD). Different generations of the machines have been created, with different levels of performance (36, 37). The company 454 Life Sciences (now a subsidiary of Roche Diagnostics) was the one company that commercially developed pyrosequencing for metagenomics, and thus “454 sequencing” is the term used in this article for high throughput pyrosequencing.
With respect to next generation DNA pyrosequencing (termed “454 sequencing” in this article), third-generation platforms are now emerging and providing longer read lengths. The first generation instrument or genome sequencer 20 (GS20) yielded 100 bp reads and 30–60 Mb per run. The second- and third-generation instruments include the 454-FLX and 454-LXR platforms, respectively. The 454-FLX was released in 2006 and yielded 250 bp reads and ~ 150 Mb/run. The 454-XLR released in 2008 yielded demonstrably higher read lengths exceeding 350 bp and ~ 400 Mb/run. The 454 instruments are the most widely deployed Nexgen sequencing systems currently in the scientific community, and these pyrosequencing-based platforms preceded other high throughput platforms such as Solexa (Illumina) and Solid (Applied Biosystems) technologies. Each 454 platform uses a modern adaptation of DNA pyrosequencing chemistry (36, 37). More than 100 publications are cited on the vendor’s website, including 15 bacterial and 13 metagenomics papers (www.454.com/news-events/publications.asp). The first human genome sequencing project based on 454 sequencing was recently published (38). Generally, the sequencing community regards the 454 technology as advantageous because of the technical robustness of the chemistry. The relatively “long” reads generated by 454 sequencing allow more frequent unambiguous mapping to complex targets, than do the products of the other ‘short read’ NexGen technologies. During the past decade, sequencing read lengths have improved due to refinements in pyrosequencing biochemistry such as addition of recombinant enzymes including single-stranded binding protein (39, 40). Advances in microfluidics technologies within instruments have increased the speed of sequencing reaction cycles such that more cycles can be performed per unit time in second- and third-generation sequencers. Additionally, the large number of reads possible per run with 454 technology delivers much greater depth of coverage for metagenomic sequencing when compared to Sanger sequencing.
Metagenomics: 16S rDNA Amplicon Sequencing
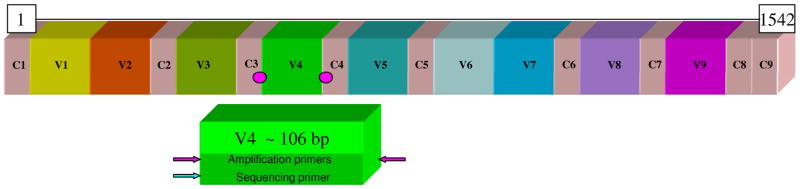
Metagenomics strategies may be directed at examining microbial composition or the broader issue of tackling phylogenomic diversity of highly complex microbial populations. One basic approach is to identify microbes in a complex community by exploiting universal and conserved targets such as rRNA genes. By amplifying selected target regions within 16S rRNA genes (Fig. 2), microbes and specifically, bacteria and Archaea, can be identified by the effective combination of conserved primer binding sites and intervening variable sequences that facilitate genus and species identification (Fig. 3). The 16S rRNA gene in bacteria is comprised of interspersed conserved and variable sequences including a total of 9 hypervariable regions (V1–V9) (Fig. 2). These hypervariable regions range in size from approximately 50–100 bases in length, and sequences differ with respect to variation and corresponding utility for universal microbial identification. Reads obtained by 454 sequencing will encompass multiple hypervariable regions with second-generation platforms such as FLX. Third-generation 454 sequencing such as LXR will generate reads exceeding 350 bp and further facilitate multi-hypervariable region sequencing. A recent study documented that the longest stretch of totally conserved bases in 16S rDNA numbered only 11 bases in length, but the longest strings of absolutely conserved bases numbered only 1–4 bases in most areas of this gene (41). This stark reality in a highly conserved gene highlights the enormous challenge with any metagenomics strategy. Different hypervariable regions demonstrated varying efficacies with respect to species calls in different genera, and the V2-V3 regions were most effective for universal genus identification (42). In a separate study, parallel analysis of three different hypervariable regions including the V2–V3, V4–V5 and V6–V8 regions of 16S rDNA sequence was effective for determining the composition of bacterial consortia in maize rhizospheres (43). As a universal approach to bacterial pathogen identification, a two-region approach yielded bacterial genus identifications in approximately 90% of isolates not amenable to biochemical identification (17). These studies highlighted the degree of variability regarding OTU representation, depending on the hypervariable region used for the analysis.
Figure 2. Conserved and hypervariable regions in the 16S rRNA gene.
The interspersed conserved regions (C1–C9) are shown in gray, and the hypervariable regions (V1–V9) are depicted in different colors. An example of primer selection for DNA amplification and sequencing-based microbial identification is provided in the figure (V4 subregion with pink circles and arrows representing primer binding sites).
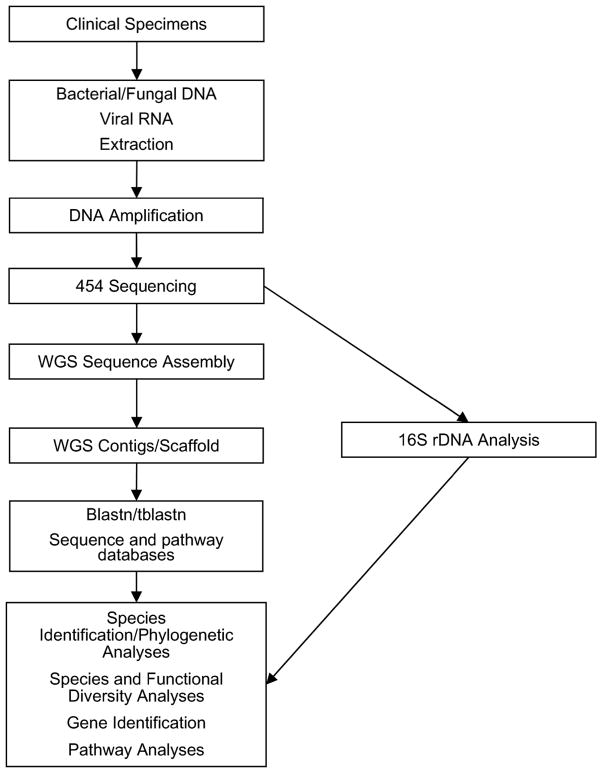
Figure 3. Deployment of 454 sequencing technology for metagenomics.
A proposed pipeline is shown for high throughput 454 sequencing and associated bio-informatics strategies in metagenomics. Each box represents a discrete step in the process using either whole genome sequencing (WGS) or 16S rDNA amplicon sequencing. Note that WGS may be performed with prior whole genome amplification (WGA) or without prior amplification.
In order to obtain a medically meaningful microbial identification, genus- or species-level classification is important. Using 16S rRNA gene sequence data, genera and species are typically distinguished at levels of 95% and 97% pairwise sequence identities, respectively (44). Strains may be distinguished at the level of 99% pairwise sequence identity, although alternative molecular methods provide more feasible strain typing approaches than DNA sequencing in today’s clinical laboratory. Ultimately strain-level resolution will depend on whole genome sequencing strategies, and these methods may eventually supplant established molecular typing methods. Sequencing accuracy becomes mission-critical when metagenomics is combined with the need for genus/species level identifications that may affect medical management in the future. In order to maximize specificity of DNA amplification prior to 454 sequencing, accurate, proofreading thermostable DNA polymerases and the application of temperature gradients during PCR amplification represent key considerations. By improving the accuracy of target nucleic acid amplification, subsequent errors by high throughput pyrosequencing can be minimized.
Nexgen pyrosequencing of individual genomes and assembly of many overlapping reads appear to yield comparable sequencing accuracy when compared to the current gold standard of Sanger sequencing, with error rates between 0.03–0.07% depending on the study (45–48). The generation of consensus sequences based on assembly of overlapping reads from a single genome is not an available option for metagenomics studies, and newly developed strategies are required to minimize error rates for such community sequencing endeavors. One recent study with first-generation parallel pyrosequencing (GS20) reported the quantification of per-base error rates and error reduction strategies such as removal of all reads containing any sequence ambiguities, inexact matches to the primer sequences, and read length anomalies (49). The final result of this report is that parallel pyrosequencing can surpass the accuracy of Sanger capillary sequencing in metagenomics applications. High throughput NexGen sequencing greatly increases depth of coverage in sequencing projects so that rich amounts of microbial diversity can be analyzed. Current 454 sequencing runs typically generate 300,000–400,000 reads per run. Our own investigations suggest that rare minority organisms in a microbial community may be detected by 454 sequencing, whereas the same microbes are missed entirely by relatively low depth of coverage approaches such as Sanger sequencing (S.H. and J.F.P, unpublished data).
Several metagenomics studies of the human gastrointestinal tract based on Sanger sequencing of 16S rDNA amplicons have been published in the past several years, indicating differences in composition and relative predominance of a few bacterial phyla. One metagenomic study describes the complex gastrointestinal microbiota as spanning only 9 of 55 bacterial phylogenetic groups, with two predominant phyla including Bacteroidetes and Firmicutes (50). In these studies, it was clear that 7 other bacterial phyla were less well represented (Actinobacteria, Fusobacteria, Proteobacteria, Verrucomicrobia, Cyanobacteria, Spirochaeates and VadinBE97) (2, 51, 52). Thus far, a single Archaea, Methanobrevibacter smithii, has been identified as a member of the gut microbiota. Recent 454 sequencing-based metagenomics studies based on 16S rDNA amplicons provided glimpses into the relative power of such investigations. A survey of the non-human primate macaque gut microbiome by 454 sequencing yielded 141,000 sequences from 100 uncultured samples obtained from 12 macaques and demonstrated clear differences depending on anatomic location, age, and gender of animals (12). Comparative metagenomics of gut microbiomes of humans, mice, and macaques showed clearly defined clusters depending on mammalian species (12). The extension of 16S rDNA amplicon sequencing strategies to whole genome sequencing strategies potentially expands the abilities of high throughput sequencing systems to comprehensively assess microbial diversity and identify pathogens or “pathogenic communities.”
Whole Genome Shotgun Sequencing
Microbial 16S rDNA sequencing is considered the gold standard for characterization of microbial communities, but 16S rDNA sequencing may not be sufficiently sensitive for comprehensive microbiome studies. Ribosomal RNA gene-based sequencing can detect the predominant members of the community, but these approaches may not detect the rare members of a community with divergent target sequences. Primer bias and the low depth of sampling account for some of these limitations, which could be improved using 454 sequencing of whole microbial genomes. In order to circumvent the limitations of single gene-based amplicon sequencing by pyrosequencing, whole genome shotgun sequencing has emerged as an attractive strategy for assessing complex microbial diversity in mixed populations. Because such a strategy is not limited by sequence conservation or primer binding site variability within a specific target, whole genome-based approaches offer the promise of more comprehensive coverage by high-throughput, parallel DNA sequencing platforms (Fig. 3). Whole genome approaches enable scientists to identify and annotate diverse arrays of microbial genes encoding many different biochemical or metabolic functions. Novel genes and functions are being discovered as a result of massive datasets obtained by whole genome shotgun sequencing of marine samples (53). Ultimately the assessment of aggregate biological functions or community phenotypes based on functional metagenomics may depend on whole genome metagenomic sequencing strategies. Arguably, whole genome approaches provide the only bona fide strategies for true metagenomics studies. The challenges and limitations of whole genome strategies include the relatively large amounts of starting material required, potential contamination of metagenomic samples with host genetic material, and high numbers of genes of unknown function or lacking quality annotation.
One key aspect of whole genome sequencing strategies is the requirement for greater amounts of input genomic DNA for comprehensive metagenomics studies. Whole genome amplification (WGA) may be deployed to generate ample amounts of DNA for whole genome shotgun sequencing. Effectively, WGA represents an enabling technology for whole genome shotgun sequencing as precious human samples (e.g. skin) may yield limiting amounts of total DNA following extraction. However, this strategy may introduce amplification bias prior to high throughput sequencing. Roberts et al. used WGA on samples from a deep mine and concluded that sequence bias may be minimized (54). In our estimation, WGA represents a viable approach for studies of metagenomic DNA samples and may be performed with commercially available polymerases such as the Phi29 DNA polymerase (REPLI-G, Qiagen). In addition to the possibility of amplification bias, amplification of human (host) contaminating DNA with WGA poses a significant challenge, possibly overwhelming the bacterial DNA sequence data in the sample. Different human DNA sequence subtraction strategies are being developed to minimize this possible barrier.
Next Generation Microbial Identification Strategies: Metagenomics and Informatics
The primary challenge on the analytical end for metagenomics studies is how to obtain accurate microbial identification of hundreds or thousands of species in a reasonable time and cost framework. Current bio-informatics throughput is too slow and not sufficiently automated for large-scale projects such as the Human Microbiome Project. High throughput methods of metagenomics rDNA analyses are needed by the scientific community and are currently in development. Clearly sufficient computational power is necessary, although distributed computing networks and robust server technology may eventually meet current metagenomics data analysis demands in research settings. The clinical laboratory will need to significantly enhance its computing infrastructure and pipelines in the near future to accommodate this demand, especially in academic centers and universities.
Beginning with sequence collection and verification, algorithms must be in place to trim sequences and vet the quality of individual reads using various strategies (Fig. 3) (49). Sequence trimming removes primer and low quality sequence data prior to sequence assembly using various algorithms. Huse et al. have performed error analyses on V6 sequences generated by 454 sequencing and described methods for filtering low quality sequence data, resulting in robust datasets for 16S rDNA analyses (49). Once the sequence reads are trimmed of primer and low quality sequences, the sequences can be aligned with sequence alignment programs like NAST (33) or MUSCLE (55, 56). Sequence chimeras may be generated by PCR amplification as a result of errors coupling disparate DNA sequences during the amplification process. Chimera checking software has been developed so that amplicons can be vetted for the potential presence of “sequence hybrids” in software environments such as Greengenes (31) and RDP (30) and with tools like Bellerophon (57) or Pintail (58).
Once high quality sequences are obtained from mixed species communities, the next challenge is to generate accurate microbial identification of many microbes in parallel. Sequences can be identified with facile classifiers such as the Bayesian Classifier in the RDP system (35), and compared with robust multiple sequence alignment programs such as NAST (33) or MUSCLE (55, 56). Existing software environments such as RDP, Greengenes, or ARB-Silva include multi-sequence alignment programs that can be effectively coupled with sequence editors in an integrated fashion. For large datasets, 16S rDNA sequences may be binned by programs like FastGroupII (59), and tallies of OTUs may be generated from these bins. Aligned sequences may also be classified against databases such as prokMSA in Greengenes and tallies of phyla may be examined and ultimately displayed as relative abundance histograms so that differences in proportions of different bacterial groups can be compared.
Novel informatics approaches such as CARMA enable protein coding sequences (as short as 27 amino acids) from whole genome sequencing projects to be applied in microbial identification strategies for comparative metagenomics (60). High-throughput informatics approaches must be developed to cope with demands of Nexgen DNA sequencing. One new strategy named Automated Simultaneous Analysis Phylogenetics (ASAP) (61) offers an automated strategy for phylogenomics that may facilitate analyses of high volumes of sequence data especially in future whole genome-based microbiome explorations. In addition to accurate microbial identification, indices and algorithms have been developed to assess microbial diversity in the context of the microbiome. Phylogenetic distance matrices may be constructed in programs such as DNAML (62). Distance matrices may be transferred to DOTUR (63) for construction of collector’s curves, rarefaction curves, calculations of Chao and ACE richness estimates, and computations of Simpson and Shannon indices of diversity. Reductions in microbial diversity have been associated with human disease phenotypes (64), and such diversity indices may be relevant to medical diagnostics in the future.
Special Challenges: Fungal and Viral Metagenomics
Whole genome sequencing of metagenomic samples is likely to reveal many bacteriophage, prophage and eukaryotic viral sequences, but viral metagenomics analyses have shown that 60% of the sequences in a viral preparation are unique, thus representing unknown viral species. As such, viral sequences may be missed by whole genome sequencing. New viral sequences have been isolated from clinical respiratory samples (65–67). Phages have been identified in the oral cavity, urine, sputum and serum (68). Finkbeiner et al. have done a similar study using fecal samples from pediatric patients with diarrhea (69). The Allander and Finkbeiner studies involved using a random PCR technique to amplify nucleic acid for cloning and Sanger sequencing. Preparation of viral nucleic acids may include filtration of samples to remove host and bacterial cells, followed by treatment of the filtrate with nucleases to remove host nucleic acid (65, 67). Such virome sequencing strategies could be easily adapted to high throughput 454 sequencing platforms. In the area of eukaryotic metagenomics, limited studies have been performed on fungal diversity in soil (70–72) and associated with plants (73). The internal transcribed spacer regions downstream of 18S rRNA genes may be useful for fungal identification (74).
Future Directions and Deeper Considerations
The science of metagenomics is currently in its pioneering stages of development as a field and many tools and technologies are undergoing rapid evolution. In addition to paradigmatic shifts towards next generation DNA sequencing technology based on novel chemistries, bio-informatics tools are also being redefined in fundamental ways to accommodate such large data volumes. In addition to massive datasets, new questions are being posited that challenge the abilities of current algorithms to deliver meaningful answers in the context of biology and medicine. The open-source software movement and “wikinomics” or mass collaboration approaches in biology have already established a foundation for the metagenomics arena with software environments such as ARB (28). Complementary strategies for microbial identification that depend on pan-microbial microarrays with known sequences such as the Phylochip (75) or the Virochip (76) may provide practical approaches for metagenomics in the clinical laboratory. Although metagenomics is not ripe yet for routine application in the clinical laboratory setting, rapid progress with the human microbiome in recent years means that the clinical laboratory community must consider how meta- approaches in biology may be relevant to disease risk assessment, diagnosis, and management in the future world of personalized medicine. In addition to the phenotypic dimension of human biology such as gene expression profiling, proteomics, and metabolomics, perhaps we need to extend our concept of the human genome to include the more comprehensive and plastic human metagenome in laboratory medicine. Future diagnostic tests may consider sequence polymorphisms and implied biological functions in our microbial communities as part of the dynamic assessment of health status and disease management.
Acknowledgments
The authors acknowledge Tiffany Morgan for her assistance with manuscript preparation. J.V. currently receives support from the National Institutes of Health (NIH) (NIDDK R01 DK065075; NCCAM R01 AT004326; NCCAM R21 AT003482), the Office of Naval Research, and the Defense Advanced Research Projects Agency (DARPA). R.A.G., J.V., S.H., and J.F.P. have active funding from the NIH (NHGRI) including support for the Jumpstart phase of the Human Microbiome Project. S.H. has current support from the U.S. Department of Agriculture. J.F.P. has current support from the NIH/NIAID sponsored Western Regional Center of Excellence for Biodefense and Emerging Infectious Disease (WRCE, RCE VI).
List abbreviations, in order cited
- rDNA
ribosomal DNA
- HMP
human microbiome project
- GI
gastrointestinal
- rRNA
ribosomal RNA
- SNP
single nucleotide polymorphism
- NCBI
National Center for Biotechnology Information
- RDP II
Ribosomal Database Project II
- BLAST
Basic Local Alignment Search Tool
- k-NN
k-nearest neighbor algorithm
- OTU
operational taxonomic unit
- NexGen
next generation
- GS20
genome sequencer 20
- WGA
Whole genome amplification
- prokMSA
prokaryotic multiple sequence alignment
- ASAP
Automated Simultaneous Analysis Phylogenetics
- DNAML
DNA maximum likelihood
Footnotes
Previous presentation of the manuscript: None
“Human Genes” discussed in the paper: None
References
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson M. Bacteriology of Humans: An Ecological Perspective. Malden, MA: Blackwell Publishing; 2008. pp. 266–326. [Google Scholar]
- 4.Thies FL, Konig W, Konig B. Rapid characterization of the normal and disturbed vaginal microbiota by application of 16S rRNA gene terminal RFLP fingerprinting. J Med Microbiol. 2007;56:755–61. doi: 10.1099/jmm.0.46562-0. [DOI] [PubMed] [Google Scholar]
- 5.Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, et al. A diversity profile of the human skin microbiota. Genome Res. 2008 doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delgado S, Suarez A, Mayo B. Identification of dominant bacteria in feces and colonic mucosa from healthy Spanish adults by culturing and by 16S rDNA sequence analysis. Dig Dis Sci. 2006;51:744–51. doi: 10.1007/s10620-006-3201-4. [DOI] [PubMed] [Google Scholar]
- 7.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55:617–24. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 8.Prindiville T, Cantrell M, Wilson KH. Ribosomal DNA sequence analysis of mucosa-associated bacteria in Crohn’s disease. Inflamm Bowel Dis. 2004;10:824–33. doi: 10.1097/00054725-200411000-00017. [DOI] [PubMed] [Google Scholar]
- 9.Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS microbiology ecology. 2002;39:33–9. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–48. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 11.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Relman DA, Falkow S, LeBoit PE, Perkocha LA, Min KW, Welch DF, Slater LN. The organism causing bacillary angiomatosis, peliosis hepatis, and fever and bacteremia in immunocompromised patients. N Engl J Med. 1991;324:1514. doi: 10.1056/NEJM199105233242117. [DOI] [PubMed] [Google Scholar]
- 14.Kolbert CP, Rys PN, Hopkins M, Lynch DT, Germer JJ, O’Sullivan CE, et al. 16S ribosomal DNA sequence analysis for identification of bacteria in a clinical microbiology laboratory. In: Persing DH, Tenover FD, Versalovic J, Tang Y-W, Unger ER, Relman DA, White TJ, editors. Molecular Microbiology: Diagnostic Principles and Practice. Washington, D.C: ASM Press; 2004. pp. 361–77. [Google Scholar]
- 15.Winker S, Woese CR. A definition of the domains Archaea, Bacteria and Eucarya in terms of small subunit ribosomal RNA characteristics. Syst Appl Microbiol. 1991;14:305–10. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- 16.Kolbert CP, Persing DH. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr Opin Microbiol. 1999;2:299–305. doi: 10.1016/S1369-5274(99)80052-6. [DOI] [PubMed] [Google Scholar]
- 17.Luna RA, Fasciano LR, Jones SC, Boyanton BL, Jr, Ton TT, Versalovic J. DNA pyrosequencing-based bacterial pathogen identification in a pediatric hospital setting. J Clin Microbiol. 2007;45:2985–92. doi: 10.1128/JCM.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–9. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 19.Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007;373:15–24. doi: 10.1385/1-59745-377-3:15. [DOI] [PubMed] [Google Scholar]
- 20.Hopkins KL, Arnold C, Threlfall EJ. Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using Pyrosequencing technology. J Microbiol Methods. 2007;68:163–71. doi: 10.1016/j.mimet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Lindback E, Unemo M, Akhras M, Gharizadeh B, Fredlund H, Pourmand N, Wretlind B. Pyrosequencing of the DNA gyrase gene in Neisseria species: effective indicator of ciprofloxacin resistance in Neisseria gonorrhoeae. Apmis. 2006;114:837–41. doi: 10.1111/j.1600-0463.2006.apm_495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang ZJ, Tu MZ, Liu J, Wang XL, Jin HZ. Comparison of amplicon-sequencing, pyrosequencing and real-time PCR for detection of YMDD mutants in patients with chronic hepatitis B. World J Gastroenterol. 2006;12:7192–6. doi: 10.3748/wjg.v12.i44.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonasson J, Olofsson M, Monstein HJ. Classification, identification and subtyping of bacteria based on pyrosequencing and signature matching of 16S rDNA fragments. Apmis. 2002;110:263–72. doi: 10.1034/j.1600-0463.2002.100309.x. [DOI] [PubMed] [Google Scholar]
- 24.Tarnberg M, Jakobsson T, Jonasson J, Forsum U. Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rDNA variable V1 and V3 regions. Apmis. 2002;110:802–10. doi: 10.1034/j.1600-0463.2002.1101106.x. [DOI] [PubMed] [Google Scholar]
- 25.Monstein H, Nikpour-Badr S, Jonasson J. Rapid molecular identification and subtyping of Helicobacter pylori by pyrosequencing of the 16S rDNA variable V1 and V3 regions. FEMS Microbiol Lett. 2001;199:103–7. doi: 10.1111/j.1574-6968.2001.tb10658.x. [DOI] [PubMed] [Google Scholar]
- 26.Garrity GM, Brenner DJ, Krieg NR, Staley JT. Bergey’s Manual of Systematic Bacteriology. 2. New York: Springer; 2005. [Google Scholar]
- 27.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome biology. 2002;3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–72. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeSantis TZ, Dubosarskiy I, Murray SR, Andersen GL. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics (Oxford, England) 2003;19:1461–8. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 34.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–6. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–52. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 39.Ronaghi M. Improved performance of pyrosequencing using single-stranded DNA-binding protein. Anal Biochem. 2000;286:282–8. doi: 10.1006/abio.2000.4808. [DOI] [PubMed] [Google Scholar]
- 40.Mashayekhi F, Ronaghi M. Analysis of read length limiting factors in pyrosequencing chemistry. Anal Biochem. 2007;363:275–87. doi: 10.1016/j.ab.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–55. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69:330–9. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmalenberger A, Schwieger F, Tebbe CC. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl Environ Microbiol. 2001;67:3557–63. doi: 10.1128/AEM.67.8.3557-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell host & microbe. 2008;3:417–27. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldberg SM, Johnson J, Busam D, Feldblyum T, Ferriera S, Friedman R, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc Natl Acad Sci U S A. 2006;103:11240–5. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, Folta KM, Soltis DE. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gharizadeh B, Herman ZS, Eason RG, Jejelowo O, Pourmand N. Large-scale pyrosequencing of synthetic DNA: a comparison with results from Sanger dideoxy sequencing. Electrophoresis. 2006;27:3042–7. doi: 10.1002/elps.200500834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome biology. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 51.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 52.Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol. 2006;59:1639–50. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
- 53.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS biology. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM, et al. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics (Oxford, England) 2004;20:2317–9. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 58.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–36. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Y, Breitbart M, McNairnie P, Rohwer F. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC bioinformatics. 2006;7:57. doi: 10.1186/1471-2105-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krause L, Diaz NN, Goesmann A, Kelley S, Nattkemper TW, Rohwer F, et al. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36:2230–9. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarkar IN, Egan MG, Coruzzi G, Lee EK, DeSalle R. Automated simultaneous analysis phylogenetics (ASAP): an enabling tool for phlyogenomics. BMC bioinformatics. 2008;9:103. doi: 10.1186/1471-2105-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen GJ, Matsuda H, Hagstrom R, Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–8. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 63.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–6. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, et al. Identification of a third human polyomavirus. J Virol. 2007;81:4130–6. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A. 2001;98:11609–14. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005;102:12891–6. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci. 2005;62:511–9. doi: 10.1007/s00018-004-4403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson IC, Campbell CD, Prosser JI. Diversity of fungi in organic soils under a moorland - Scots pine (Pinus sylvestris L.) gradient Environmental. Microbiology. 2003;5:1121–32. doi: 10.1046/j.1462-2920.2003.00522.x. [DOI] [PubMed] [Google Scholar]
- 71.Anderson IC, Campbell CD, Prosser JI. Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environ Microbiol. 2003;5:36–47. doi: 10.1046/j.1462-2920.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- 72.Hunt J, Boddy L, Randerson PF, Rogers HJ. An evaluation of 18S rDNA approaches for the study of fungal diversity in grassland soils. Microb Ecol. 2004;47:385–95. doi: 10.1007/s00248-003-2018-3. [DOI] [PubMed] [Google Scholar]
- 73.Smit E, Leeflang P, Glandorf B, van Elsas JD, Wernars K. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2614–21. doi: 10.1128/aem.65.6.2614-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White T, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand D, Sninsky J, White T, editors. PCR Protocols: A Guide to Methods and Applications. New York: Academic Press, Inc; 1990. pp. 315–22. [Google Scholar]
- 75.Flanagan JL, Brodie EL, Weng L, Lynch SV, Garcia O, Brown R, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:1954–62. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–92. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]