Abstract
The confidence in peptide (and protein) identifications with ion mobility spectrometry time-of-flight mass spectrometry (IMS-TOFMS) is expected to drastically improve with the addition of information from an efficient ion dissociation step prior to MS detection. High throughput IMS-TOFMS analysis imposes a strong need for multiplexed ion dissociation approaches where multiple precursor ions yield complex sets of fragment ions that are often intermingled with each other in both the drift time and m/z domains. We have developed and evaluated an approach for collision-induced dissociation (CID) using IMS-TOFMS instrument. It has been shown that precursor ions activated inside an rf-device with an axial dc-electric field produce abundant fragment ions which are radially confined with the rf-field and collisionally cooled at an elevated pressure, resulting in high CID efficiencies comparable or higher than those measured in triple-quadrupole instruments. We have also developed an algorithm for deconvoluting these complex multiplexed tandem MS spectra by clustering both the precursor and fragment ions into matching drift time profiles and by utilizing the high mass measurement accuracy achievable with TOFMS. In a single IMS separation from direct infusion of tryptic digest of bovine serum albumin (BSA), we have reliably identified 20 unique peptides using a multiplexed CID approach downstream of the IMS separation. Peptides were identified based upon the correlation between the precursor and fragment drift time profiles and by matching the profile representative masses to those of in silico BSA tryptic peptides and their fragments. The false discovery rate (FDR) of peptide identifications from multiplexed MS/MS spectra was less than 1%.
Keywords: ion mobility spectrometry, multiplexed CID, time-of-flight, peptide identifications, false discovery rate
Introduction
In a typical “bottom-up” proteomics approach, proteins are enzymatically digested to a mixture of peptides, and subjected to a condensed phase separation, such as reverse-phase liquid chromatography (RPLC) [1]. Species eluting off the chromatographic column are, typically, ionized by electrospray ionization (ESI) and analyzed with tandem mass spectrometry. The latter involves isolation of a single precursor ion species followed by its fragmentation. These fragmentation products are then used to obtain sequence related information for identification of the peptide precursor ion. Different techniques have been used to induce ion fragmentation. Notably, collision induced dissociation (CID) [2], electron capture dissociation (ECD) [3, 4] and electron transfer dissociation (ETD) [5, 6] have been widely employed in proteomics. However, as the number of precursor candidates increases, the “one-at-a-time” precursor ion activation in the course of a fast RPLC separation results in the “under sampling”, i.e. only a limited set of candidate precursor ions can be targeted for sequence elucidation in tandem MS experiments. Two possible solutions to the under sampling issue include i) increasing the RPLC gradient time to provide slower separation and ii) increasing the rate of ion activation for a given RPLC separation. Increasing the RPLC gradient time increases sample size requirement and decreases the analysis throughput and is most useful only for a limited applications, including generation of an accurate time and mass (AMT) tag database [7]. Increasing the rate of ion activation ultimately requires a brighter ion source and higher fragmentation efficiency. Alternatively, multiplexed MS/MS approach has been proposed to address these challenges. In the multiplexed MS/MS approach, all candidate precursor ion species are subjected to concurrent activation and the corresponding products and their precursor ions are detected and identified in a single spectrum.
One of the main challenges of the multiplexed MS/MS approach is interpreting the complex fragmentation spectra due to the convolution of fragment species from different precursor ions. Experimental approaches to correlate fragment species to their precursors in the multiplexed MS/MS studies have included using high resolution and high mass measurement accuracy (MMA) of FTICR MS [8-10], LC retention time [11-13], the inherent mass bias during variable accumulation times in ion trap [14], or using ion mobility [15-19]. Utilizing the high MMA and resolution of FT-ICR for both precursors and fragments, Masselon et al. [8, 9] and Li et al. [10] were able to reduce the number of false identifications by matching the multiplexed MS/MS spectra to a large database. It was emphasized that the high mass accuracy and resolving powers were of importance and that additional constraints might be needed for lower resolution instruments, such as TOFMS, to reduce false identifications to acceptable levels. It is noteworthy that accurate mass information was sufficient for assignment of the multiplexed MS/MS spectra in FTICR experiments [8-10]. Alternatively, alignment of RPLC retention time profiles for both the precursors and their fragments was invoked to establish the correlation between precursors and their fragments [12, 13, 20]. This approach is based upon the observation that retention time profiles of the fragments resemble those of their precursors, and if more than one precursor co-elute a correlation can be drawn between each precursor and its fragments, provided the retention profiles are distinguishable [20]. Geromanos et al. demonstrated the advantage of the multiplexed MS/MS over conventional data-dependent MS/MS for identifying peptides in the presence of a complex matrix [13, 21].
The correlation between precursors and their fragments can be also established using ion mobility spectrometry (IMS). In IMS, ions are separated according to differences in their gas phase mobilities [22] while traveling in an inert buffer gas (e.g. helium or nitrogen) under the influence of a weak electric field. When IMS is coupled to TOF MS, ions spend most of their drift-time inside an IMS drift tube, while the time needed to traverse a mass spectrometer is considerably shorter. Therefore, when mobility-separated ions are subjected to fragmentation at the interface between the drift tube and the mass spectrometer, the fragments would have arrival time distributions similar to that of their precursor. This fragmentation approach will be referred to as IMS-CID-MS for the remainder of this manuscript [23]. Alignment of the drift time profiles of fragments ions with their precursor establishes an important correlation that could be used in interpretation of the multiplexed MS/MS data. Alignment of the drift time profiles of fragments ions with their precursor establishes an important correlation that could be used in interpretation of the multiplexed MS/MS data. Though grouping of precursor and their fragment ions was mentioned [24] as a potential effective label in IMS-CID-MS, actual peptide identifications from a single protein digest were performed with the use of m/z values and some correlation in the drift time domain corresponding to the maximum of precursor ion profile. Neither mass accuracy nor false discovery rates (FDR) were reported, and annotation of IMS-CID-MS spectra were performed only for precursor species resolved in the drift time domain [24]. In complex sample analysis, correlation of drift time profiles would be superior over single-point correlation for minimizing FDR. Importantly, drift time profiles of fragment ions could only be recorded at increased sensitivity, which inextricably couples instrument's limit of detection in MS/MS mode with confidence in peptide identifications. Other constraints impeding the practical use of IMS-CID-MS can include low sensitivity due to inefficient ion trapping prior to IMS separation and poor collection efficiency for both precursor and fragment ions in the IMS-MS interface region. Whether CID is performed downstream of IMS separation in an orifice-skimmer cone region [25] or in a split-field modulated region coupled to a conical lens [24], confinement of spatially dispersed ion packets represents a major challenge. Additional fragment ion scattering upon collisional activation can further degrade sensitivity.
Recently released commercial hybrid quadrupole-IMS-TOFMS instrument (Synapt and Synapt G2, Waters Corp., Manchester, UK) employs a travelling wave ion guide (TWIG) for IMS separation and is capable of performing CID before and after IMS [26]. Detailed description of ion transport principles through the TWIG has been reported elsewhere [27]. Synapt has recently been employed for targeted CID of species mobility-separated in the TWIG [28, 29] and high-quality CID spectra were reported after summation over 1 min experiments [29]. However, to the best of our knowledge, no identifications from IMS-multiplexed CID experiments (e.g., MS/MS-based peptide identifications from overlapping ion mobility peaks) and no CID efficiencies were reported. In general, little or no information on the efficiency of IMS-CID-MS platforms is available in the literature compared to other instruments, such as triple quadrupole mass spectrometers.
In this work, we have addressed the aforementioned shortcomings of the IMS-CID-MS approach. Specifically, we have i) developed and evaluated a novel CID approach inside an rf-quadrupole operating at an elevated pressure of ∼ 200 mTorr and employing an axial dc electric field; ii) developed an algorithm for deconvolution of intermingled multiplexed MS/MS spectra, and iii) rigorously evaluated CID efficiencies in two different configurations and determined a false discovery rate for identification of peptides from complex proteolytic digests.
Experimental Arrangement
Experiments were performed with a home built ESI-IMS instrument (Figure 1a) coupled to an Agilent 6210 TOFMS instrument (Agilent Technologies, Santa Clara, CA, USA). Briefly, ions were generated by an ESI source and transferred through 64 mm long 500 μm i.d. stainless steel inlet capillary heated to 150°C. Ions exiting the heated capillary entered an ion funnel trap [30, 31] where they were initially focused in the ion funnel section and then transferred into the trap section. Inside the trap, ions were accumulated for a predetermined interval (∼4 ms) by applying a stopping voltage to a dual grid [31] and then released from the trap into the drift tube as ion packets by lowering the voltage on the dual grid for 200 μs. After being ejected from the trap, ion packets were focused radially by the converging section of the ion funnel trap before being released into the drift tube. The voltages applied to the ESI source, inlet capillary and the ion funnel trap were biased relative to the drift tube entrance voltage.
Figure 1.

a) Schematic diagram of the ESI-IMS-TOFMS instrument. b) Schematic diagram of the segmented quadrupole. Only two quadrupole rods are shown for clarity.
The drift tube was 88 cm long and was filled with nitrogen gas at a pressure of 4 Torr at a temperature of 23°C. A small counter pressure gradient (∼40 mTorr) was established between the drift tube and the ion funnel trap regions to minimize contamination of the drift tube by neutrals from the ESI source. Ion packets traveled along the drift tube under the influence of a weak electric field of 17 V/cm. Ions of different mobilities were separated according to their collision cross sections, which in turn depend on their masses, charges and tertiary structures. Dispersed ion packets were collimated at the IMS drift tube exit with an electrodynamic ion funnel [32]. The rear ion funnel is 86-mm long with a 51 mm acceptance orifice i.d. and linearly decreasing ring electrodes i.d. to an exit orifice electrode i.d. of 2.5 mm. The ion funnel electrodes were 0.5 mm thick and separated by 0.5 mm Teflon washers [33]. An rf amplitude of 80 V (peak-to-peak) at a frequency of 500 kHz was applied to the rear funnel using a home-built rf power supply. The dc electric field for the rear funnel was adjusted to match the electric field within the IMS drift cell.
Ions exiting the rear ion funnel were transmitted through a differentially-pumped segmented quadrupole evacuated to 200 mTorr, which was separated from the rear ion funnel with a dc-only conductance limiting orifice of 2.5 mm i.d. The segmented quadrupole consisted of four rods (6.4 mm diameter, 2.8 mm inscribed radius) sectioned into six segments each (Figure 1b). Each segment was 11.7 mm long and separated by 0.5 mm PEEK washers nested in between the segments so not to be exposed to the ion beam. A voltage drop of ∼ 5 V/segment was applied to the segmented quadrupole to reduce ions residence time and thus minimize any effect on ion mobility resolving power. As shown in Figure 1b, two independent resistor chains were used to control the dc gradient of the first four segments (section 1) and the last two segments (section 2). An rf amplitude of 220 V (peak-to-peak) and a frequency of 800 kHz was applied to the segmented quadrupole using a home-built rf power supply. A dc-only conductance limiting orifice (2.2 mm i.d., 1.4 mm from segmented quadrupole) separated the segmented quadrupole from a subsequent differentially-pumped octopole which constituted the first component of a commercial time-of-flight mass spectrometer (Agilent Technologies, Santa Clara, CA). The dc bias voltages of the segmented quadrupole and the conductance limiting orifice were kept close (∼3-5 V) to the octopole dc-bias for optimum sensitivity. Signal from the Agilent TOFMS detector was fed into a 1 GS/s 8-bit AP240 analog-to-digital converter (ADC) board (Acqiris, Geneva, Switzerland). A custom C# code was developed to incorporate the ADC board into the IMS–TOFMS data acquisition system. A typical IMS-TOF experiment involves accumulating ions in the ion funnel trap for a few ms (below trap saturation) and releasing them into the drift tube in 200 μs pulses followed by an IMS separation. In the IMS experiment, 600 TOF spectra were collected sequentially to form a 60-ms long IMS separation, thus forming a nested IMS-TOF 2D map. In the current setup 300 nested IMS-TOF maps were added to form one IMS-TOF frame.
Ion fragmentation was performed by collision-induced-dissociation (CID) whose efficiency was evaluated in two regions. The first region was inside the segmented quadrupole, and ion activation was accomplished by raising the bias voltage of the first four segments of the quadrupole (section 1) relative to the bias of the last two segments (section 2). The second region was in between the segmented quadrupole exit (exit of section 2 in Figure 1b) and the following conductance limiting orifice (CL). The region between an rf-only quadrupole operating at a pressure of ∼170 mTorr and the following conductance limit was previously examined for ion fragmentation in IMS-CID-TOFMS experiments [17] and we have compared this approach to the collisional activation inside the segmented quadrupole. Attempts were also made to fragment ions at the entrance of the first octopole of the Agilent TOFMS (i.e. between the conductance limit orifice and the octopole entrance) and at the entrance to the segmented quadrupole. In the former case, a pronounced loss of ions transmission was observed at increased voltage gradients, while in the latter case the higher-density collision gas emanating from the IMS drift tube at elevated pressure (∼ 4 torr) caused reduced fragmentation efficiency.
Note that collision energies (C.E., eV) for IMS-CID-TOF experiments are reported in terms of collision voltage gradient (Va, V/mm) where
| (1) |
z is the charge state of the ions and d is the distance upon which the voltage gradient was applied.
The CID efficiencies of the IMS-CID-TOF platform were also compared to those obtained from a triple-quadrupole instrument (TSQ Quantum Ultra, Thermo Scientific, San Jose, CA). In order to calculate the CID efficiency, the detector gain was kept constant between the precursor and MS/MS spectra. The precursor ion intensity was obtained by operating Q1 in the MS scan mode, no gas in the collision gas and Q3 in the transmission mode. MS/MS spectra were obtained by selecting the precursor of interest in Q1; inducing CID in the collision cell at an Ar pressure of 1.5 mTorr, and then scanning Q3 to identify the fragmentation products. The collision energy was varied to determine the optimum fragmentation and CID efficiencies for each peptide studied. Q1 and Q3 were operated with a peak width of 0.7 amu.
Samples
Neurotensin, fibrinopeptide A, leucine enkephalin and methionine enkephalin were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used without further purification. Samples were diluted in 49.5:49.5:1 methannol/water/acetic acid solution to a concentration of 1 μM each. Proteolytic digestions of bovine serum albumin (BSA; Pierce Biotechnology, Rockford, IL, USA) were conducted using sequencing grade trypsin (Promega, Madison, WI, USA) and previously described procedures [34]. The resulting solution was then diluted to 0.05 μg/μl in 49.5:49.5:1 methannol/water/acetic acid buffer. All samples were ionized in positive ESI mode and infused at a flow rate of 500 nl/min.
Data Processing
A flow-chart diagram for data processing pipeline is shown in the Supplementary Materials section. In brief, candidate precursors were identified from a low energy (no-CID) spectrum followed by searching for their fragments in the CID spectra using precursor sequences, drift times and mass accuracy information. The raw data were acquired in a 3D format which contained ions' time-of-flight (in nanoseconds), ion mobility scan number (IMS drift time = IMS scan number × TOF scan length) and intensity. The TOF axis was converted from the raw data to m/z using a calibration procedure. A list of peaks was then generated for each mass spectrum using Decon2LS software developed in-house [35, 36]. Deconl2LS converts isotopic envelops to monoisotopic masses using THRASH algorithm [37]. The processed data represent a list of peaks along with their monoisotopic masses, charge states, intensities, TOF scan numbers, signal-to-noise ratios (S/N) and isotopic envelope matching fit parameters.
The list of peptides from in silico digestion of bovine serum albumin (BSA) was generated using Protein Digestion Simulator software [38]. The FASTA file of BSA (P02769|ALBU_BOVIN) was obtained from Universal Protein Resource (UniProt) database (http://www.uniprot.org). Protein Digestion Simulator performs in silico protein digestion using different enzymes (e.g., trypsin) and digestion rules (e.g., full), and generates a list of peptides sequences along with their monoisotopic masses. Fully tryptic rules (R or K except before P) with a maximum of three missed cleavages were chosen to generate 302 peptides for BSA. To evaluate the false discovery rate, glycogen phosphorylase (P00489|PYGM_RABIT) was in silico digested to provide 389 entries for a decoy database. Identifications of peptides observed in IMS-multiplexed CID-TOF MS experiments were accomplished using a custom C++ code. Candidate precursors were determined by matching the masses of peptides from the BSA in silico digest to those of the experimental IMS-TOFMS features. Candidate precursor features were clustered to precursor drift time profiles using observations from consecutive TOFMS spectra. Only the precursors observed in at least three consecutive TOF spectra (forming an IMS drift time profile) and exhibiting a mass measurement accuracy of better than 15 ppm were considered. Each precursor drift time profile is characterized by a profile representative (or unique precursor) that is the precursor species at the maximum intensity of the drift time profile. The masses of profile representatives were matched to those of in silico digested peptides at an MMA of ± 15 ppm to generate a list of unique candidate precursors. A list of theoretical candidate fragments (b, y, a, x) was then generated from the list of the unique candidate precursors. For the experimental fragment species, IMS-multiplexed CID-TOFMS features were grouped into fragment drift time profiles, a procedure identical to that employed for the clustering of precursor IMS-TOFMS features. Three consecutive TOF spectra and a mass measurement accuracy of 15 ppm were typically used for combining IMS-CID-TOFMS features into fragment drift time profiles. Each fragment drift time profile was represented by a single deisotoped feature (a profile representative), using the same routine as that employed for treating precursors. The masses of fragment profile representatives were matched against the masses of theoretical fragments at a MMA of ± 15 ppm to yield a list of candidate fragment identifications. A matched fragment profile was considered “unique” if satisfied the following constraints: (a) MMA of ±15 ppm; (b) the fragment species is observed in, at least, three consecutive TOF spectra; (c) the charge state of the fragment, zf, is less than that of the precursor, zp (zf < zp) and (d) drift time profile of the fragment aligns with and is enclosed in the drift time profile of the precursor as follows:
where scan_start and scan_end indices denote the beginning and the end of the IMS drift profile. A combination of the unique candidate precursor and a minimum of three unique candidate fragments was set as a criterion to confidently identify a unique peptide. If identified with more than one charge state, a unique peptide was counted only once.
Results
Collection, fragmentation and CID efficiencies
The IMS-CID-TOF MS approach was assessed using the figures of merits for the CID efficiency and the contributing factors. A CID experiment is characterized by the collection, fragmentation and CID efficiencies as follows [39, 40]:
| (2) |
| (3) |
| (4) |
where P0 is the intensity of the precursor ion (obtained from the precursor mass spectrum under conditions optimum for the transmission of the precursor), P is the surviving precursor ion intensity in the CID spectrum, Σfi is the sum of all fragment intensities in the CID spectrum. Ec accounts for the losses due to ion scattering/defocusing during the collision process, Ef reflects the efficiency of producing fragment ions and ECID is the overall CID efficiency, which incorporate both the fragmentation and collection efficiency. Note that in the triple-quadrupole platform Ec accounts for the ion losses at the entrance of the collision cell (Q2) and due to transmission through Q3 in the mass resolving mode.
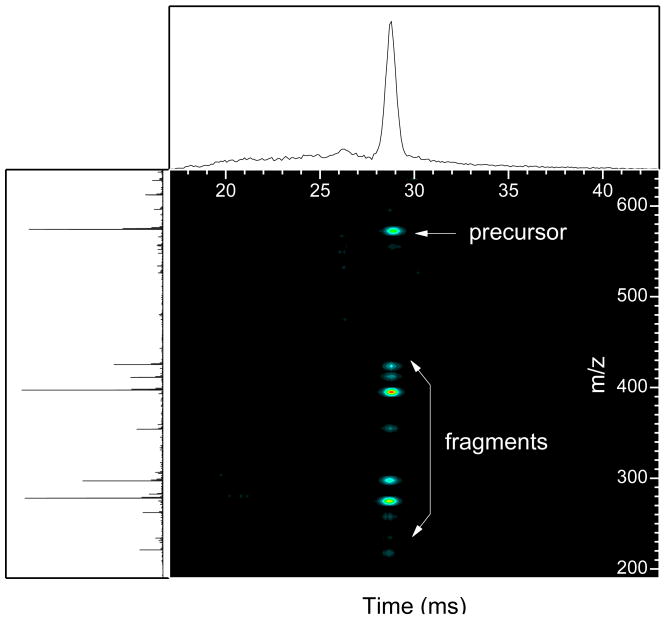
A characteristic feature of the IMS-CID-TOFMS approach is that the fragments drift time profiles match that of their precursor. Though a precursor and its fragments are separated in the m/z domain, their drift times are similar, enabling a correlation between the precursor and fragment ions. This is illustrated in Figure 2 for IMS-CID-TOF 2D display of methionine enkephalin where the fragments are evident as vertically-aligned horizontal bands. Each band represents a drift time profile projected onto the IMS-TOF 2D map. A slight shift (∼200 μs) in drift time profiles between the fragment of the lowest m/z and their precursor was noticed. The observed shift is due to the fact that fragments continue to drift in the last two segments of the quadrupole. This slight shift is negligible compared to ion drift time in our drift tube and minimally, if not at all, affecting our fragments assignments and our developed software allow for controlled shift in drift time profiles.
Figure 2.
2D display of the IMS-CID-TOF spectra of methionine enkephalin.
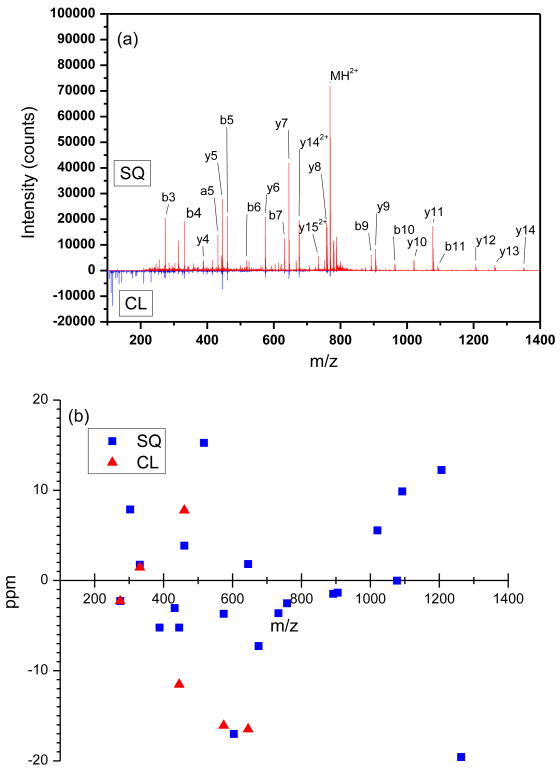
To evaluate the CID efficiency, we have examined CID spectra of a number of peptides. Figure 3a shows a comparison of the summed mass spectra for the doubly-charged fibrinopeptide A ions (768.8498 m/z) collisionally activated inside the segmented quadrupole (SQ) and at the conductance limiting orifice (CL), as was previously reported [17]. The spectra in Figure 3a were generated by summing 20 TOF mass spectra in the IMS drift time range of 24.0 – 26.0 ms. While only 6 lower intensity fragments were observed in experiment with the CL approach, inducing dissociation inside the SQ resulted in detection of 21 higher intensity fragments. Not only was the number of fragments markedly different, but also m/z distribution was found to be broader for the collisional activation inside the SQ. This drastic difference in the number and m/z distribution of observed fragments as well as their intensities reflect, mainly, the difference in the fragment collection efficiencies.
Figure 3.
(a) Comparison between the CID spectra of fibrinopeptide A inside the segmented quadrupole (SQ, 90 V/mm) and at the conducting limiting orifice (CL, 60 V/mm). The spectra shown are the sum of TOF spectra for fibrinopeptide A (24.0 - 26.0 ms). (b) Mass measurement accuracy for the CID products performed at CL and in SQ.
To find the optimum CID efficiency for both the CL and SQ approaches, we varied the collision energy by adjusting the electric field strength in the corresponding regions of ion activation (to be referred to as collision voltage gradient). Table 1 shows comparison of the Ec, Ef and ECID efficiencies between the CL and SQ methods for fibrinopeptide A 2+ ions at different voltage settings. Although the fragmentation efficiencies (Ef) were found to be comparable between the CL and SQ approaches, the collection efficiencies (Ec) were not. Higher collection efficiency of the SQ technique is ascribed to better ion confinement in the segmented quadrupole following the collision process. At low collision voltage gradient (and low collision energy) the losses due to ion defocusing and scattering are low, leading to high collection efficiency of the SQ approach (0.75). The SQ collection efficiency decreases to 0.60 at a higher acceleration voltage gradient of 110 V/mm, while the collection efficiency of the CL approach remained limited to ∼ 0.1. As a result, the CID efficiency (ECID) of the SQ approach (0.61) was higher than that of the CL approach by a factor of ∼ 6. Another observation is that CID products in the CL activation (Figure 3a) are shifted toward the low m/z end of a mass spectrum while those observed with the SQ approach are distributed across a broader m/z range, producing rich informational content. The MMAs for the fragment signals acquired under the optimum conditions for both the CL and SQ methods are shown in Figure 3b. These results indicate an MMA of ±10 ppm, limited mainly by the ion statistics. As shown later in the manuscript, higher MMA of both precursor and fragment species is key for high confidence peptide identifications.
Table 1.
CID efficiencies of fibrinopeptide A (2+) and neurotensin 3+ inside the segmented quadrupole (SQ)
| Segmented quadrupole | Conductance limit orifice | ||||||
|---|---|---|---|---|---|---|---|
| V | Ec | Ef | ECID | V | Ec | Ef | ECID |
| fibrinopeptide A 2+ | |||||||
| 74 | 0.75 | 0.37 | 0.27 | 44 | 0.10 | 0.37 | 0.04 |
| 78 | 0.74 | 0.47 | 0.35 | 50 | 0.08 | 0.67 | 0.05 |
| 80 | 0.73 | 0.55 | 0.40 | 54 | 0.07 | 0.84 | 0.06 |
| 84 | 0.67 | 0.64 | 0.43 | 60 | 0.10 | 0.96 | 0.09 |
| 90 | 0.62 | 0.84 | 0.52 | 64 | 0.08 | 0.98 | 0.08 |
| 94 | 0.64 | 0.92 | 0.59 | 70 | 0.09 | 1.00 | 0.09 |
| 100 | 0.63 | 0.97 | 0.61 | ||||
| 110 | 0.60 | 1.00 | 0.60 | ||||
| neurotensin 3+ | |||||||
| 80 | 0.64 | 0.94 | 0.60 | 60 | 0.02 | 0.94 | 0.02 |
V: collision voltage gradient, V/mm; Ec: collection efficiency; Ef: fragmentation efficiency; ECID: CID efficiency.
Experiments with triply-protonated neurotensin have also demonstrated SQ superior performance, as reflected in the CID spectra included in the Supporting Materials section. Using an MMA of ±10 ppm, a total of 22 fragments were confidently identified with the SQ approach, while only 6 fragments were matched in the CL method. Table 1 reports ECID of 0.60 and 0.02 for the SQ and CL approaches under the optimum conditions. Similar CID efficiency trends have been observed for number of other peptides, including leucine enkephalin, methionine enkephalin, bradykinin and angiotensin I. Experiments with singly-charged methionine enkephalin and leucine enkephalin in SQ revealed a CID efficiency of 39% and 36 %, respectively. Since ions are confined radially in the segmented quadrupole, ion losses can likely be attributed to ion defocusing in the acceleration field region that could not be mitigated by the quadrupole effective potential. This argument is supported by the observation that triply-charged neurotensin ions exhibit higher collection efficiency than singly-charged leucine enkephalin ions at similar collision energies. The effective potential is proportional to the charge state squared which leads to better confinement of multiply-charged species. Higher effective potential is expected to improve the collection efficiency inside the segmented quadrupole. The difference in ECID between multiply-charged and singly-charged species can be also attributed to the ion multiplication [40]. Since charges can be located on various sites along the peptide backbone each multiply-charged precursor ion can fragment into two or more ions while singly-charged ions fragment into an ion and a neutral. Accordingly, ion multiplication results in higher collection efficiency for multiply-charged species.
A plausible explanation for ion losses at the conductance limit is that the fragments retain part of their kinetic energy in the TOF interface (due to low number of collisions at reduced pressure between quadrupole exit and CL) that increases their velocity component perpendicular to the TOF axis in the TOF extraction region. An increase in the initial velocity of ions entering the TOFMS leads to an increase in the incident angle in the TOF flight tube and is accompanied by the loss of sensitivity, as reflected in the lower collection efficiency of the CL approach.
The CID efficiencies of IMS-CID-TOF for singly-charged species were comparable to those obtained from the triple-quadrupole instrument. The CID efficiency for singly-charged leucine enkephalin (m/z 556) in the triple-quadrupole instrument was measured as 36% which is similar to 36% obtained from IMS-CID-TOF and is comparable to the 34% efficiency for ions at m/z 500, as reported by Thomson et al. [40]. A notable difference between MS/MS spectra from the triple-quadrupole and that from IMS-CID-TOF is the reduction in the number of internal fragments in the spectrum obtained with the latter (see Figure S2 in the supplementary materials).
The difference in ECID between the triple-quadrupole and IMS-CID-TOF approaches becomes more pronounced when comparing multiply charged species such as doubly-charged fibrinopeptide A and triple-charged neurotensin. An optimum ECID of 17% and 10% were obtained for the double-charged fibrinopeptide A and triply-charged neurotensin ions, respectively, with the triple-quadrupole instrument. These ECID values are lower than those obtained in the IMS-CID-TOF instrument by factors of 3.6 and 6, respectively, (see Table 1). These data demonstrate the advantages of inducing ion fragmentation at the higher pressures (∼200 mTorr) inside the segmented quadrupole which is characterized by more effective radial confinement of both the precursor and fragment ions.
Analysis of a bovine serum albumin digest
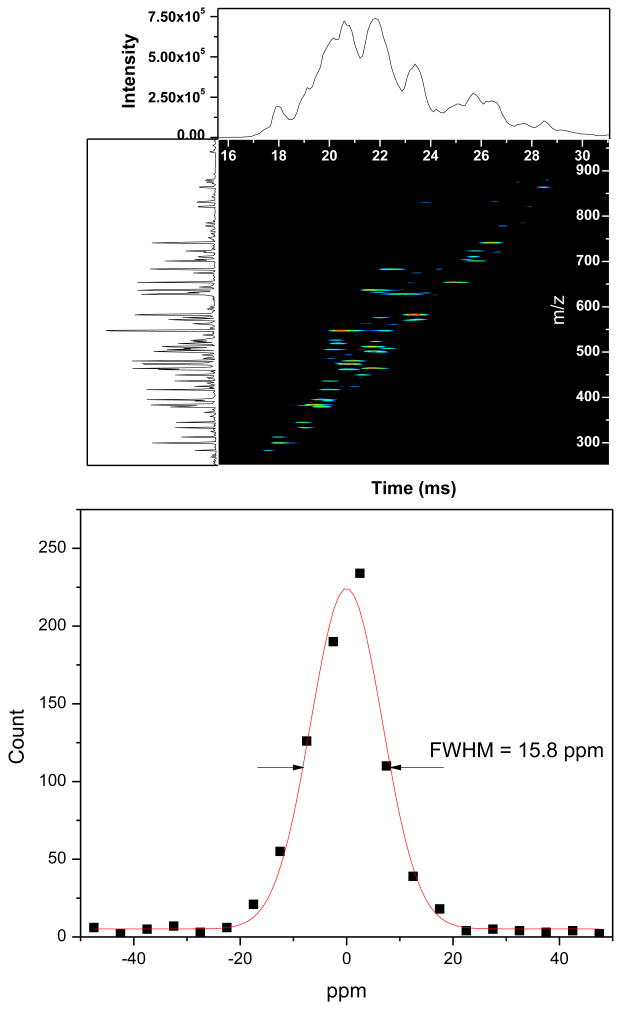
To further validate the SQ approach, a BSA tryptic digest has been examined in direct infusion experiments with the IMS-TOF MS instrument. BSA tryptic digest was used to mimic the complexity of a single IMS-TOFMS frame typically acquired in the course of LC-IMS-TOFMS studies with biological fluids. BSA was also commonly used for method development, evaluation of platform performance and false discovery rate in comparative studies.[41-46] Figure 4a shows a 2D display of the IMS-TOFMS map for the BSA tryptic digest acquired at low collision energy condition (no CID). Matching of the deisotoped IMS-TOFMS features against the in silico BSA tryptic peptides at an MMA of ±50 ppm resulted in a histogram with a full width at half magnitude (fwhm) of 15.8 ppm, as shown in Figure 4b. Searching against the in silico digest within ± 15 ppm gave rise to 37 putative matches in the precursor IMS-TOFMS frame. It is noteworthy that all unique precursor species and their fragments were matched against in silico BSA peptides and their fragments in a fully automated fashion using the algorithm developed for the deconvolution of complex multiplexed IMS-TOFMS and IMS-multiplexed CID-TOFMS signals. The same algorithm was then used to evaluate the false discovery rate (FDR) of peptide identifications. Figure 5 shows an example of IMS-CID-TOFMS frame at collision voltage gradient of 56 V/mm inside the segmented quadrupole. The vertical streaks in the 2D display represent concurrent fragmentation of multiple peptides at IMS drift times of 19-23 ms. The collision voltage gradient was varied from 28 to 140 V/mm to determine the optimum conditions for all ions in IMS-TOFMS separation. As shown later in the manuscript, a collision energy ramp may not be required to efficiently fragment peptide ions, since many of these species are present as multiple charged ions and efficiently fragment at the same collision energy. Instead, dissociation of all ions exiting the IMS stage can be attained at two or three different collision energies.
Figure 4.
(a) 2D display of the IMS-TOF spectra for BSA tryptic digest under conditions of no CID induced. (b) Mass measurement accuracy histogram for matched candidates of BSA peptides (±50 ppm) from the precursor ion spectrum across all IMS scans.
Figure 5.
2D display of the IMS-MS/MS-TOF spectra for a BSA tryptic digest sample under collision voltage gradient of 56 V/mm inside SQ.
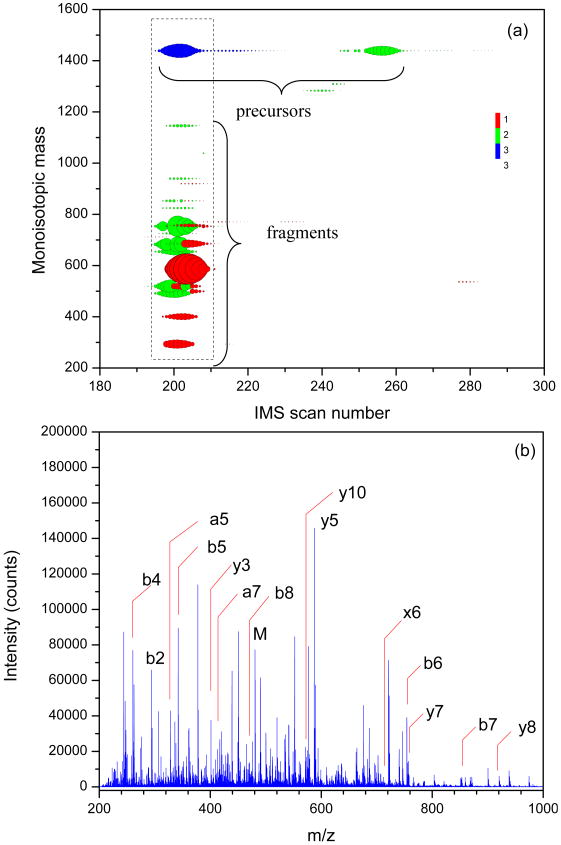
To illustrate identification of peptides with IMS-multiplexed CID-TOFMS approach, we now focus on one of the BSA peptides. Figure 6a shows the monoisotopic masses of RHPEYAVSVLLR peptide and its fragments as a function of the TOF spectrum number (also referred to as IMS scan number) at a collision voltage gradient of 56 V/mm. Although RHPEYAVSVLLR peptide has 2+ and 3+ charge states (labeled precursors in Figure 6a), only 3+ ions (IMS scan number ∼200) fragmented at this voltage gradient, while the 2+ ions (IMS scan number ∼260) needed higher voltage gradient to induce dissociation. Figure 6a illustrates that fragments are aligned with their precursor in the IMS domain (see vertical streaks), so that ions located outside of the dashed rectangle in Figure 6a should be disregarded. Accordingly, the deisotoped clusters at IMS scan numbers of ∼240 and ∼ 232, and monoisotopic masses of 1287 and 773, respectively, were excluded from consideration. Fragments with MMA worse than ±15 ppm were also discarded. The fragment at an IMS scan number of 208 and a monoisotopic mass of 1038.5 was also disregarded because it was observed in less than three consecutive scans which does not constitute a drift time profile. The summed mass spectrum for the drift time range within the rectangle in Figure 6a is shown in Figure 6b. The mass spectrum also displays fragments from other multiple precursors along with labeled fragments of RHPEYAVSVLLR precursor. A total of 25 fragments (a2, a4-8, b2-9, y2-11, x6, x11) were identified for the RHPEYAVSVLLR precursor. Another illustration for the utility of the developed algorithm in deconvoluting data for peptides which overlap in the IMS dimension is shown in Figure 7. The doubly-charged ions of the two identified peptides, HLVDEPQNLIK and FKDLGEEHFK, are partially overlapped in the IMS dimension as shown by the extracted IMS profiles of each peptide. As mentioned in the data processing section, the first step involve the identification of each peptide sequence based on the matching of monoisotopic mass of each profile representative to a list of candidates masses within a mass accuracy tolerance. Since the two peptides profiles shown in Figure 7 have different masses, the profiles representatives are readily assigned despite the overlap in the IMS dimension. In cases of complete overlap of IMS profiles, the algorithm will still be able to assign the profile representative as long as the monoisotopic masses of the overlapped ions differ by greater than the mass accuracy tolerance. The profile representatives for the IMS-aligned fragments are also determined similar to the precursors. Finally, only the fragments of monoisotopic masses that match the theoretical list of candidate fragment generated based upon the precursor sequences and within the mass tolerance are considered.
Figure 6.
(a) Plot of monoisotopic mass vs. IMS scan number for the identified fragments of RHPEYAVSVLLR. The circles size is scaled to reflect the ions' monoisotopic peak intensity. The colors denote the observed charge state (1+, red; 2+ green; 3+, blue). (b) MS/MS spectra (sum of IMS scans 185-205) for BSA at acceleration voltage gradient of 56 V/mm. For clarity, annotations for some of the fragments are not shown.
Figure 7.
2D display for part of the IMS-TOF spectra of the BSA tryptic digest at low collision energy. The mass spectra as well as the IMS profiles for the two peptides overlapping in the IMS dimension (highlighted in rectangles) are also shown.
Mass errors of all candidate fragments identified within ±50 ppm from all IMS scans, at all collision energies and using ±15 ppm for precursors ions were composed to a histogram whose fwhm was found to be 18.5 ppm (Figure S3 in the Supplementary Information section). This indicates that collisional activation in the segmented quadrupole at relatively high pressure (∼200 mTorr) has negligible effect on the mass error distribution of the fragments as compared to the precursors (see Figure 4), implying that the fragments are thermalized and have narrow energy distribution before exiting the quadrupole into the TOF MS. Searching for fragment matches within ±15 ppm and considering only the unique precursors with at least three unique fragments, we have identified 283 unique fragments that belonged to 20 unique peptides of the initial 37 candidate precursors. Masselon et al. reported that a minimum of three fragments per precursor was required to reduce the false positives [9]. As described in the Experimental section, the term “unique” refers to the representatives of the fragment/precursor drift time profiles that passed all the constraints. In this initial evaluation, we have employed stringent matching criteria and considered only a, x, b and y fragments types. In the future, the multiplexed IMS approach will be extended to account for post-translational modifications and neutral losses, such as H2O and NH3. Figure 8 shows the number of fragments matched per unique peptide. The number of matches ranged from 3 to 31 fragments with an average of 14 fragments per unique peptide which indicates a high level of confidence in identifications of peptides in IMS-multiplexed CID-TOFMS experiment.
Figure 8.
Plot of number of unique fragments for each uniquely identified precursor ion.
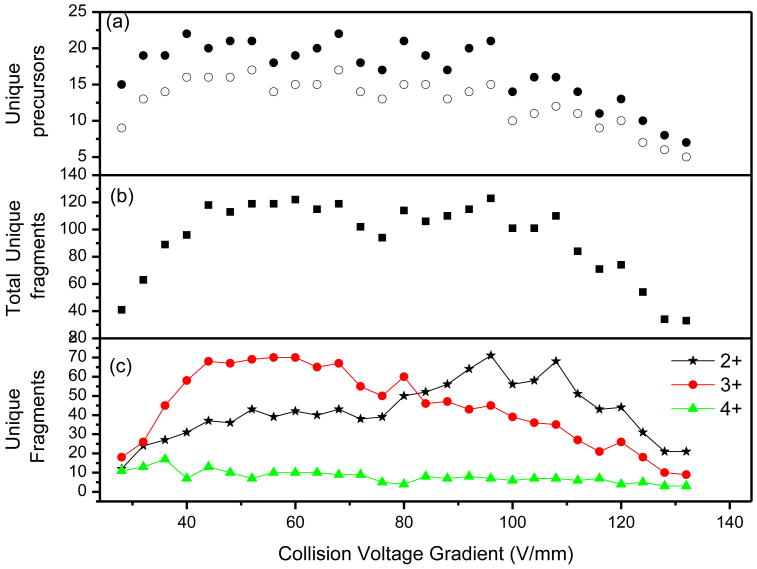
Figure 9a shows the trend in peptide identifications as a function of the collision energies. With an increase in the collision energy, the number of unique precursors increases, levels off and then decreases at high collision energies. The maximum number of unique peptides identified at the optimum collision energy was 22. As indicated in Figure 9a, many of the precursors were identified from different charge states which fragmented at different collision energies. Shown in Figure 9b, the number of unique fragments followed the similar trend observed in Figure 9a for the unique precursors. The number of matched unique fragments initially increases, mainly due to fragmentation of the higher charge state ions. The number of unique fragments was then found to be nearly constant (∼110 unique fragments) over wide range of collision energies followed by a decrease at high collision energies due to over fragmentation (internal fragments). The maximum number of BSA unique fragments identified at any acceleration voltage was 123. Figure 9b is further analyzed according to the different precursors charge states and is shown in Figure 9c. Figure 9c indicates that, generally, doubly-charged ions have optimum collision voltages higher than triply-charged species. There were not enough data points for the quadruply-charged ions to visualize a trend. It should be noted that a peptide having multiple charge states could be identified at different collision voltage gradients. Since the collision energy in the center-of-mass reference frame is proportional to the analyte charge state and inversely proportional to the analyte mass, higher charge state ions readily fragment at lower collision voltage gradients, while higher collision voltage gradients are needed to decompose their singly-charged counterparts. Although a range of collision voltages were used in this study we found that two or three voltage was needed to cover 100 % of those BSA peptides identified at all voltages. Table S1 (supplementary Materials) list all BSA peptides identified with different charge states at all collision voltages ranges. Also reported in Table S1 those BSA peptides identifies at low and moderately higher collision voltage (40 and 96 V/mm). As indicated in Table S1 all peptides can be identified at those two points. Given the limited complexity of BSA digest it seems that three rather than two collision voltages are sufficient for more complex mixture.
Figure 9.
Number of unique (a) precursors and (b) fragments identified at each collision voltage gradient. Redundancy due to precursors multiple charge states was either removed (open circles) or not (filled circles). Panel c) represent the data on b) partitioned according to the charge state of the precursors.
Finally, the dynamic range of the identified peptide from the direct infusion of BSA digest (as obtained from the mass spectra peak intensities) spanned two orders of magnitude (see Figure S4 in supplementary materials) and extends to a factor of 25 when considering fragments. The dynamic range of this experiment is limited by ADC acquisition system, the mode of analysis (i.e., direct infusion versus LC) and the sample complexity. Approaches to extend the ADC-limited dynamic range involve data-dependent accumulation in the trap [47] in addition to using multiplexed ion mobility-TOF approach [41, 48].
Estimate of false discovery rate
To evaluate the confidence in peptides identifications with IMS-multiplexed CID-TOFMS, we have determined the false discovery rate (FDR) using a decoy database. Glycogen phosphorylase (P00489|PYGM_RABIT, 843 amino acids) was chosen for this study. Using the same digestion rules (R and K but not before P) and three missed cleavages, in silico PYGM digest yielded 388 peptides while that of BSA had 302. No overlap in sequences for the PYGM and BSA tryptic peptides with masses above 400 Da exists. Similarly, within ±15 ppm there is no overlap in monoisotopic masses between BSA and PYGM tryptic peptides. Therefore, IMS-TOFMS features obtained in the analysis of the BSA sample would represent only the random matches against PYGM tryptic peptides. Using both the precursor and fragment ion information, 20 unique features were confidently identified as BSA tryptic peptides and only one feature was matched against PYGM sequence, resulting in an FDR of 0.6 %. The FDR was calculated as a ratio of the number of unique features matching PYGM to that of the identified BSA tryptic peptides at all collision energies studied. Importantly, this high confidence in peptide identifications results from a combination of both the precursor and fragment ion mobility information derived in the IMS-multiplexed CID-TOFMS analysis as well as MMA. Alternatively, relying only on the MMA of ±15 ppm and allowing all the IMS-TOFMS features to be counted as a valid match irrespective of the IMS drift time profiles resulted in an FDR of 35.3 %. Using only the MMA of ±15 ppm and the constraint of three fragments per peptide while discarding the IMS information, brought about an FDR of 8.8 %. Therefore, a combination of physical constraints, such as drift time information for the precursors and fragments, high MMA, and the requirement to detect, at least, three unique fragments per peptide reduced the FDR by a factor of > 35 in IMS-multiplexed CID–TOF MS experiments with complex proteolytic digests. We have also evaluated the confidence in identification of BSA peptide using much larger decoy database (> 156,000 entries) which incorporated all possible peptides (no cleavage rules) of six proteins (horse myoglobin, haptoglobin, carbonic anhydrase, glycogen phosphorylase, transferrin, phosphorylase b). Interestingly, the FDR remained <1% when the requirement of four unique fragments per peptide was employed for peptide identifications.
To further illustrate the importance of ion mobility we show the case where two fragments having masses of 711.4209 and 711.4267 amu, which are separated only by 8 ppm, and their profile representatives were observed at IMS scans of 193 and 237, respectively. However, based upon drift-times information the fragment of 711.4209 amu was matched to a7 of KVPQVSTPTLVEVSR peptide whose IMS profile spans IMS scans 190-230 while the fragment of 711.4267 amu was matched to ×6 of HPEYAVSVLLR peptide whose IMS profile spans IMS scans 236-246. These results indicate that the developed IMS-multiplexed CID-TOF MS approach can be successfully utilized in the course of capillary LC separation of protein digests from biological fluids. Importantly, this peptide sequencing technology can be now extended to the analysis of LC-IMS-MS features which are separated only in the IMS domain, while having the same retention time and mass as the species identified in the conventional LC-MS/MS experiments. Reliable identification of these LC-IMS-TOFMS features uniquely position IMS-TOF MS technology to augment the proteomics research.
Conclusions
We have evaluated multiplexed collision induced dissociation (CID) in the interface between an ion mobility spectrometer (IMS) and a time-of-flight mass spectrometer (TOF MS). To deconvolute the IMS-multiplexed CID-TOF MS raw data, informatics approaches effectively using information on the precursor and fragment drift profiles and mass measurement accuracy (MMA) have been developed. It was shown that radial confinement of ion packets inside an rf-only segmented quadrupole operating at a pressure of ∼ 200 mTorr and having an axial dc-electric field minimizes ion losses due to defocusing and scattering, resulting in high abundance fragment ions which span a broad m/z range. Efficient dissociation at high pressure (∼ 200 mTorr) and high ion collection efficiency inside the segmented quadrupole resulted in CID efficiencies of singly-charged ions that are comparable to those reported with triple quadrupole mass spectrometers. The modulation of an axial dc-electric field strength inside the segmented quadrupole can be used either to induce or to prevent multiplexed ion fragmentation. In addition, the axial electric field ensures ion transmission through the quadrupole at velocities which do not affect the quality of IMS separation. Importantly, both the precursor and fragment ions were acquired at good MMA (<20 ppm). The IMS-multiplexed CID TOFMS approach was validated using a mixture of peptides and a tryptic digest of BSA. Combining alignment of the precursor and fragment ion drift time profiles, an MMA of ±15 ppm for the precursors and fragments, and the requirement of having greater than 3 unique fragments per unique precursor, we have confidently identified 20 unique BSA tryptic peptides in a single IMS separation. On average, each peptide sequence was corroborated with an average of 14 unique fragments. The peptide level false discovery rate of < 1% was determined when matching IMS-multiplexed CID-TOFMS features against a decoy database composed of the tryptic peptides of glycogen phosphorylase (PYGM) without use of liquid phase separation (e.g., LC). It was also found that incorporating IMS information for the precursors and fragments as well as high MMA for fragments decreased the FDR by a factor of > 35 as compared to that obtained using the MMA only. We have shown that the developed algorithm was able to identify peptides whose IMS profiles overlapped. The algorithm utilized a combination of constraints provided by the ion mobility as well as the time-of-flight mass spectrometer mass accuracy. A modest mass accuracy (<15 ppm) was crucial to the success of the approach so improving mass accuracy is expected to help reduce the false discovery rate specially with more complex mixtures than BSA. This approach relies also on the knowledge of candidate peptides (in silico digestion) as well as candidate fragments (generated using common cleavage rules) and was not evaluated for de novo sequencing which may require better mass accuracy in addition to ion mobility to reduce false identifications. Finally, the developed IMS-multiplexed CID-TOFMS approach has been shown to provide high throughput, high confidence identifications of peptides from complex mixtures and, in the following work, will be applied to identification of the LC-IMS-TOFMS features which can only be detected due to separation in the IMS drift time domain.
Supplementary Material
Acknowledgments
The authors thank William Danielson III for the helpful discussions. Portions of this research were supported by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research, the Environmental Molecular Science Laboratory (EMSL) and the NIH National Center for Research Resources (Grant RR018522). This work was performed in the Environmental Molecular Science Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under contract DE-AC05-76RL0 1830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Wells JM, McLuckey SA. Biol Mass Spectrom. Vol. 402. Elsevier Academic Press Inc; San Diego: 2005. pp. 148–185. [Google Scholar]
- 3.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120(13):3265–3266. [Google Scholar]
- 4.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 5.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. Tandem mass spectrometry for peptide and protein sequence analysis. BioTechniques. 2005;38(4):519–523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- 7.Smith RD. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2(5):513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 8.Masselon C, Anderson GA, Harkewicz R, Bruce JE, Paša-Tolić L, Smith RD. Accurate Mass Multiplexed Tandem Mass Spectrometry for High-Throughput Polypeptide Identification from Mixtures. Anal Chem. 2000;72(8):1918–1924. doi: 10.1021/ac991133+. [DOI] [PubMed] [Google Scholar]
- 9.Masselon C, Paša-Tolić L, Lee SW, Li L, Anderson GA, Harkewicz R, Smith RD. Identification of tryptic peptides from large databases using multiplexed tandem mass spectrometry: simulations and experimental results. Proteomics. 2003;3(7):1279–1286. doi: 10.1002/pmic.200300448. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Masselon CD, Anderson GA, Paša-Tolić L, Lee SW, Shen Y, Zhao R, Lipton MS, Conrads TP, Tolic N, Smith RD. High-Throughput Peptide Identification from Protein Digests Using Data-Dependent Multiplexed Tandem FTICR Mass Spectrometry Coupled with Capillary Liquid Chromatography. Anal Chem. 2001;73(14):3312–3322. doi: 10.1021/ac010192w. [DOI] [PubMed] [Google Scholar]
- 11.Purvine S, Eppel JT, Yi EC, Goodlett DR. Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics. 2003;3:847–850. doi: 10.1002/pmic.200300362. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty AB, Berger SJ, John C. Gebler Use of an integrated MS - multiplexed MS/MS data acquisition strategy for high-coverage peptide mapping studies. Rapid Commun Mass Spectrom. 2007;21(5):730–744. doi: 10.1002/rcm.2888. [DOI] [PubMed] [Google Scholar]
- 13.Geromanos SJ, Vissers JPC, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9(6):1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 14.Wilson J, Vachet RW. Multiplexed MS/MS in a Quadrupole Ion Trap Mass Spectrometer. Anal Chem. 2004;76(24):7346–7353. doi: 10.1021/ac048955d. [DOI] [PubMed] [Google Scholar]
- 15.Lee YJ, Hoaglund-Hyzer CS, Taraszka JA, Zientara GA, Counterman AE, Clemmer DE. Collision-Induced Dissociation of Mobility-Separated Ions Using an Orifice-Skimmer Cone at the Back of a Drift Tube. Anal Chem. 2001;73(15):3549–3555. doi: 10.1021/ac010295e. [DOI] [PubMed] [Google Scholar]
- 16.Hoaglund-Hyzer CS, Lee YJ, Counterman AE, Clemmer DE. Coupling Ion Mobility Separations, Collisional Activation Techniques, and Multiple Stages of MS for Analysis of Complex Peptide Mixtures. Anal Chem. 2002;74(5):992–1006. doi: 10.1021/ac010837s. [DOI] [PubMed] [Google Scholar]
- 17.Baker ES, Tang K, Danielson WF, III, Prior DC, Smith RD. Simultaneous Fragmentation of Multiple Ions Using IMS Drift Time Dependent Collision Energies. J Am Soc Mass Spectrom. 2008;19(3):411–419. doi: 10.1016/j.jasms.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker C, Fernandez-Lima FA, Gillig KJ, Russell WK, Cologna SM, Russell DH. A Novel Approach to Collision-Induced Dissociation (CID) for Ion Mobility-Mass Spectrometry Experiments. J Am Soc Mass Spectrom. 2009;20(6):907–914. doi: 10.1016/j.jasms.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 19.Valentine SJ, Koeniger SL, Clemmer DE. A Split-Field Drift Tube for Separation and Efficient Fragmentation of Biomolecular Ions. Anal Chem. 2003;75(22):6202–6208. doi: 10.1021/ac030111r. [DOI] [PubMed] [Google Scholar]
- 20.Purvine S, Eppel JT, Yi EC, Goodlett DR. Wiley-V C H Verlag Gmbh. 2003:847–850. [Google Scholar]
- 21.Li GZ, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Structural Analysis of Synthetic Polymer Mixtures Using Ion Mobility and Tandem Mass Spectrometry. Proteomics. 2009;9(6):1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 22.McDaniel EW, Mason EA. The Mobility and Diffusion of Ions in Gases. John Wiley & Sons; New York: 1973. [Google Scholar]
- 23.Hoaglund-Hyzer CS, Li J, Clemmer DE. Mobility labeling for parallel CID of ion mixtures. Anal Chem. 2000;72(13):2737–2740. doi: 10.1021/ac0000170. [DOI] [PubMed] [Google Scholar]
- 24.Koeniger SL, Valentine SJ, Myung S, Plasencia M, Lee YJ, Clemmer DE. Development of field modulation in a split-field drift tube for high-throughput multidimensional separations. J Proteome Res. 2005;4(1):25–35. doi: 10.1021/pr049877d. [DOI] [PubMed] [Google Scholar]
- 25.Lee JL, Hoaglund-Hyzer CS, Taraszka JA, Zientara GA, Counterman AE, Clemmer DE. Collision-induced dissociation of mobility-separated ions using an orifice-skimmer cone at the back of a drift tube. Anal Chem. 2001;73(15) doi: 10.1021/ac010295e. [DOI] [PubMed] [Google Scholar]
- 26.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int J Mass Spectrom. 2007;261(1):1–12. [Google Scholar]
- 27.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequencyonly stacked ring ion guide. Rapid Comm Mass Spectrom. 2004;18(20):2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 28.Vakhrushev SY, Langridge J, Campuzano I, Hughes C, Peter-Katalinic J. Ion mobility mass spectrometry analysis of human glycourinome. Anal Chem. 2008;80(7):2506–2513. doi: 10.1021/ac7023443. [DOI] [PubMed] [Google Scholar]
- 29.Thalassinos K, Grabenauer M, Slade SE, Hilton GR, Bowers MT, Scrivens JH. Characterization of Phosphorylated Peptides Using Traveling Wave-Based and Drift Cell Ion Mobility Mass Spectrometry. Anal Chem. 2009;81(1):248–254. doi: 10.1021/ac801916h. [DOI] [PubMed] [Google Scholar]
- 30.Ibrahim YM, Belov ME, Tolmachev AV, Prior DC, Smith RD. Ion Funnel Trap Interface for Orthogonal Time-of-Flight Mass Spectrometry. Anal Chem. 2007;79(20):7845–7852. doi: 10.1021/ac071091m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clowers BH, Ibrahim YM, Prior DC, Danielson WF, Belov ME, Smith RD. Enhanced ion utilization efficiency using an electrodynamic ion funnel trap as an injection mechanism for ion mobility spectrometry. Anal Chem. 2008;80(3):612–623. doi: 10.1021/ac701648p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaffer SA, Tang K, Anderson GA, Prior DC, Udseth HR, Smith RD. A Novel Ion Funnel for Focusing Ions at Elevated Pressure Using Electrospray Ionization Mass Spectrometry. Rapid Commun Mass Spectrom. 1997;11:1813–1817. [Google Scholar]
- 33.Belov ME, Gorshkov MV, Udseth HR, Anderson GA, Tolmachev AV, Prior DC, Harkewicz R, Smith RD. Initial implementation of an electrodynamic ion funnel with Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2000;11(1):19–23. doi: 10.1016/S1044-0305(99)00121-X. [DOI] [PubMed] [Google Scholar]
- 34.Kinter MM, Sherman NE. Protein Sequencing and Identification Using Tandem Mass Spectrometry. Wiley-Interscience; New York: 2000. [Google Scholar]
- 35.Jaitly N, Mayampurath A, Littlefield K, Adkins J, Anderson G, Smith R. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10(1):87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decon2ls, http://omics.pnl.gov/software/
- 37.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectrom. 2000;11(4):320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 38.Protein Digestion Simulator, http://omics.pnl.gov/software/
- 39.Yost RA, Enke CG, McGilvery DC, Smith D, Morrison JD. High efficiency collision-induced dissociation in an RF-only quadrupole. Int J Mass Spectrom Ion Phys. 1979;30(2):127–136. [Google Scholar]
- 40.Thomson BA, Douglas DJ, Corr JJ, Hager JW, Jolliffe CL. Improved Collisionally Activated Dissociation Efficiency and Mass Resolution on a Triple Quadrupole Mass Spectrometer System. Anal Chem. 1995;67(10):1696–1704. [Google Scholar]
- 41.Belov ME, Clowers BH, Prior DC, Danielson WF, Iii, Liyu AV, Petritis BO, Smith RD. Dynamically Multiplexed Ion Mobility Time-of-Flight Mass Spectrometry. Anal Chem. 2008;80(15):5873–5883. doi: 10.1021/ac8003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong YZ, Zhang LH, Tao DY, Liang Y, Zhang WB, Zhang YK. Miniaturized two-dimensional capillary electrophoresis on a microchip for analysis of the tryptic digest of proteins. J Sep Sci. 2008;31(3):588–594. doi: 10.1002/jssc.200700444. [DOI] [PubMed] [Google Scholar]
- 43.Creese AJ, Cooper HJ. Liquid chromatography electron capture dissociation tandem mass spectrometry (LC-ECD-MS/MS) versus liquid chromatography collision-induced dissociation tandem mass spectrometry (LC-CID-MS/MS) for the identification of proteins. J Am Soc Mass Spectrom. 2007;18(5):891–897. doi: 10.1016/j.jasms.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francois I, Cabooter D, Sandra K, Lynen F, Desmet G, Sandra P. Tryptic digest analysis by comprehensive reversed phase x two reversed phase liquid chromatography (RP-LC × 2RP-LC) at different pH's. J Sep Sci. 2009;32(8):1137–1144. doi: 10.1002/jssc.200800578. [DOI] [PubMed] [Google Scholar]
- 45.Kaur-Atwal G, Weston DJ, Green PS, Crosland S, Bonner PLR, Creaser CS. Analysis of tryptic peptides using desorption electrospray ionisation combined with ion mobility spectrometry/mass spectrometry. Rapid Comm Mass Spectrom. 2007;21(7):1131–1138. doi: 10.1002/rcm.2941. [DOI] [PubMed] [Google Scholar]
- 46.Satoh T, Sato T, Tamura J. Development of a high-performance MALDI-TOF mass spectrometer utilizing a spiral ion trajectory. J Am Soc Mass Spectrom. 2007;18(7):1318–1323. doi: 10.1016/j.jasms.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Ibrahim YM, Belov ME, Liyu AV, Smith RD. Automated Gain Control Ion Funnel Trap for Orthogonal Time-of-Flight Mass Spectrometry. Anal Chem. 2008;80(14):5367–5376. doi: 10.1021/ac8003488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belov ME, Buschbach MA, Prior DC, Tang K, Smith RD. Multiplexed Ion Mobility Spectrometry-Orthogonal Time-of-Flight Mass Spectrometry. Anal Chem. 2007;79(6):2451–2462. doi: 10.1021/ac0617316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.