Abstract
Heparan sulfate proteoglycans (HSPGs) play vital roles in every step of tumor progression allowing cancer cells to proliferate, escape from immune response, invade neighboring tissues, and metastasize to distal sites away from the primary site. Several cancers including breast, lung, brain, pancreatic, skin, and colorectal cancers show aberrant modulation of several key HS biosynthetic enzymes such as 3-O Sulfotransferase and 6-O Sulfotransferase, and also catabolic enzymes such as HSulf-1, HSulf-2 and heparanase. The resulting tumor specific HS fine structures assist cancer cells to breakdown ECM to spread, misregulate signaling pathways to facilitate their proliferation, promote angiogenesis to receive nutrients, and protect themselves against natural killer cells. This review focuses on the changes in the expression of HS biosynthetic and catabolic enzymes in several cancers, the resulting changes in HS fine structures, and the effects of these tumor specific HS signatures on promoting invasion, proliferation, and metastasis. It is possible to retard tumor progression by modulating the deregulated biosynthetic and catabolic pathways of HS chains through novel chemical biology approaches.
Keywords: Proteoglycan, Cancer, Heparanase, Sulfotransferase, Sulfatase, Heparan Sulfate
Introduction
It has been known for decades that heparan sulfate proteoglycans (HSPGs) and chondroitin sulfate proteoglycans (CSPGs) are involved in the progression of cancer at various stages [1–3]. Many excellent reviews have described cancer growth, development, and metastasis [4–6]. However, knowledge on the detailed molecular interactions between tumor cells and ECM components including HS chains has only recently been devolved. In lieu of this new knowledge, this review article focuses on the role of HSPGs in several types of cancer and provides insights into how future cancer therapies can be developed based on changes in HS biosynthesis and catabolism.
Tumor Transformation, Growth, Invasion and Metastasis
The development of cancer typically involves four distinct stages: transformation into a cancerous phenotype, sequestration of nutrition through angiogenesis, invasion into nearby tissue, and metastasis to distal sites away from the primary location. While the transformation of normal cells into a cancerous phenotype hasn’t yet been linked to HSPGs, there is an abundance of evidence relating HSPG fine structures to cancer growth, invasion, and metastasis.
Tumor cells upregulate the production of several angiogenic factors such as fibroblast growth factor (FGF) and vascular endothelial cell growth factor (VEGF) [7]. In order to support altered growth patterns and metabolism, these molecules trigger angiogenesis, the growth of blood vessels, which provides nearby cells with increased nutrition and oxygen-supply [8].
Malignant cancers are characterized by their invasiveness into nearby tissues and metastasis to distal locations away from the primary tumor site. In order for these processes to take place, tumor cells breakdown the surrounding ECM by activating or releasing various proteases such as matrix metalloproteases (MMPs) and serine proteases [9–12]. These proteases make the ECM more permeable for invading cells to pass through established ECM boundaries [13]. Malignant cancers can even invade into the blood stream and metastasize to distal locations in the body by cleaving off their adhesions to the ECM, entering the bloodstream, and then binding to ECM at a distal location. Since HSPGs are implicated in the above-mentioned critical processes, controlling their fine structures can significantly impact the growth, invasion and metastatic properties of tumor cells.
Common HSPG- Growth factor and Cytokine interactions
One of the primary methods by which HSPGs control cancer progression is by regulating the interactions between cells and signaling molecules such as growth factors and cytokines. Several insightful reviews discuss the roles that HSPGs play in binding to signaling molecules and localizing them to their cognate receptors on cell surfaces, storing them in tissues for later use, and/or helping to form molecular gradients for directional cellular activities during the development [14–16].
FGF2-HS interactions are well-studied example of GAG-growth factor interactions. The binding of FGF2 to its receptor causes autophosphorylation of the receptor’s tyrosine kinase domains and leads to increased growth, migration and differentiation in several cancers [17,18]. HSPGs play an integral role in the FGF2-FGFR interactions by localizing FGF2 near the receptor and forming a bridge that stabalizes the ligand-receptor complex and allows for signal transduction [19,20].
However, only specific HS fine structures allow FGF2-FGFR-mediated signaling. To bind FGF2, HS chains require N-Sulfated glucosamine units and 2-O-Sulfonated iduronic acid units [21–23]. Concurrently, for HS chains to bind FGFR, they require 6-O-Sulfonated glucosamine residues in addition to 2-O-Sulfonated iduronic acid and N-Sulfonated glucosamine residues [21,23,24].
Similar to HS-FGF2 interactions, HSPGs are essential to the signaling pathways triggered by other growth factors and cytokines as well. The following are some particularly important signaling molecules affected by HSPG interactions: VEGF, Heparin binding epidermal growth factor (HB-EGF), Transforming growth factor (TGF), Bone morphogenic protein (BMP), and FGF. Manipulating HS biosynthesis and catabolism can drastically affect several of these signaling pathways.
Proteoglycans: Biosynthesis and Catabolism
Cancer cells have inherently altered proteoglycan fine structures that allow them to invade, metastasize and grow uncontrollably [25,26]. These fine structural changes are due to errors in the regulation of biosynthetic and catabolic enzymes that control proteoglycan fine structures. The following section details several of the enzymes that are responsible for PG synthesis and catabolism.
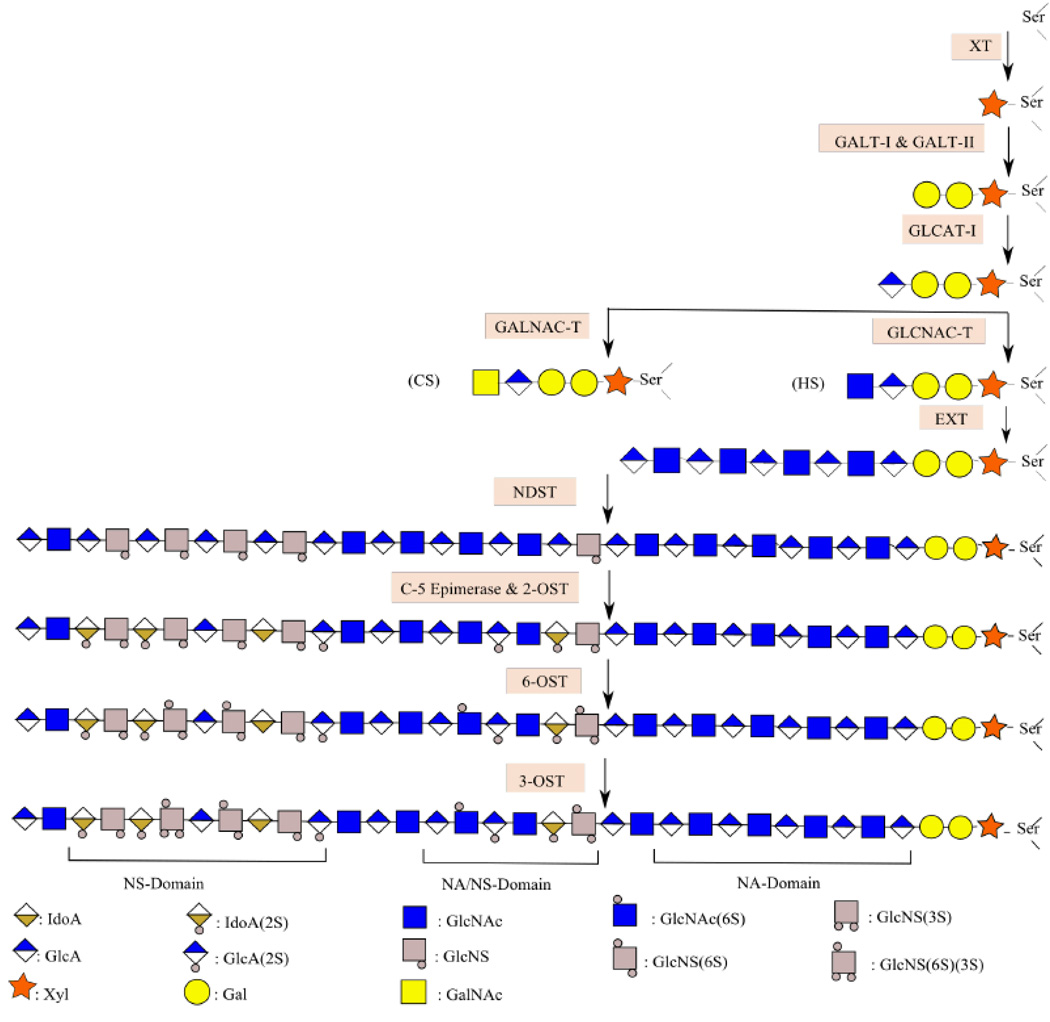
Proteoglycans are composed of a core protein substituted with one or more glycosaminoglycan (GAG) polysaccharide side chains [27,28]. PG assembly involves the following steps: (1) assembly of the linkage tetrasaccharide (2) concurrent elongation and modification of the GAG backbone such as epimerization, deacetylation and N-/O- sulfonation (Fig. 1).
Fig. 1. Proteoglycan Biosynthesis.
Biosynthesis begins with the addition of xylose to serine residues of the core protein. Subsequently, glycosyl transferases add two Gal residues and one GlcA residue. EXT-1 and EXT-2 then extend the GAG chain by alternatively adding GlcNAc and GlcA. While elongation takes place, NDST-1 and NDST-2 convert certain GlcNAc to GlcNS residues. Epimerization and 2-O Sulfonation also occur in parallel; certain GlcA are converted to IdoA while 2-OST acts on certain GlcA and IdoA residues. Subsequent action of 6-OST and 3-OST yields the final PG chain that contains regions that have several N-Sulfated residues (NS Domain), others that have several N-Acetylated residues (NA Domain), and short regions that have a mixture of both (NA/NS Domain).
Synthesis begins with the transfer of xylose residues to certain serine amino acids in the core protein. Next, transfer of galactose and glucuronic acid residues by glycosyl transferases such as GalT-I, GalT-II, and GlcAT-I results in the formation of a tetrasaccharide linkage region [29,30]. Subsequently, the GAG chain is elongated via the alternate addition of D-glucuronic acid (GlcA) and N-acetyl-D-hexosamine (N-acetyl-D-glucosamine, GlcNAc or N-acetyl-D-galactosamine, GalNAc) to the tetrasaccharide linker [31–34]. HS contains GlcNAc and GlcA/Iduronic acid (IdoA), CS contains GalNAc and GlcA, and dermatan sulfate (DS) contains GalNAc and GlcA/IdoA disaccharide units.
Subsequently EXT enzymes and CS synthases elongate HS and CS chains, respectively, by adding the appropriate sugar building blocks [35–40]. While the elongation is in progress, parts of the GAG chain are modified via sulfonation and epimerization to yield the final complex PG structure [41–43]. A host of HS biosynthetic and catabolic enzymes that have been implicated in cancer progression are listed below:
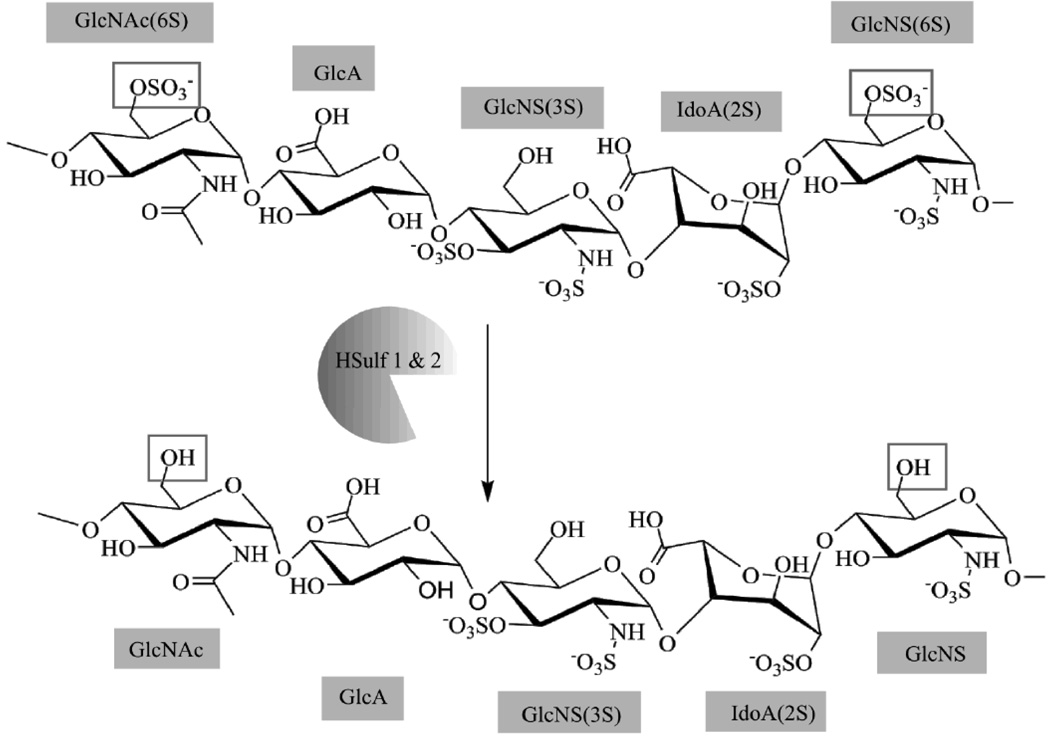
HSulf-1 and HSulf-2
A set of HS 6-O-endosulfatases that are secreted from the Golgi and localized in the ECM. HSulfs selectively remove 6-O sulfate groups on GlcN residues of HS; preferentially catalyzing the desulfonation on trisulfated disaccharides (Fig. 2) [44–48].
Fig. 2. HSulf 1 & 2 Enzymatic Activity.
HSulfs provide arylsulfatase activity selectively removing 6-O-Sulfate groups from GlcNAc and GlcNS residues. Deviant H-Sulf mRNA expression is present in several cancers.
3-O Sulfotransferase (3-OST)
A group of 7 related enzyme isoforms that catalyze the 3-O sulfonation of GlcN residues, a rare HS modification [49–52].
6-O Sulfotransferase (6-OST)
A group of three enzyme isoforms that catalyze the 6-O sulfonation of GlcNAc as well as GlcNS residues [53–56].
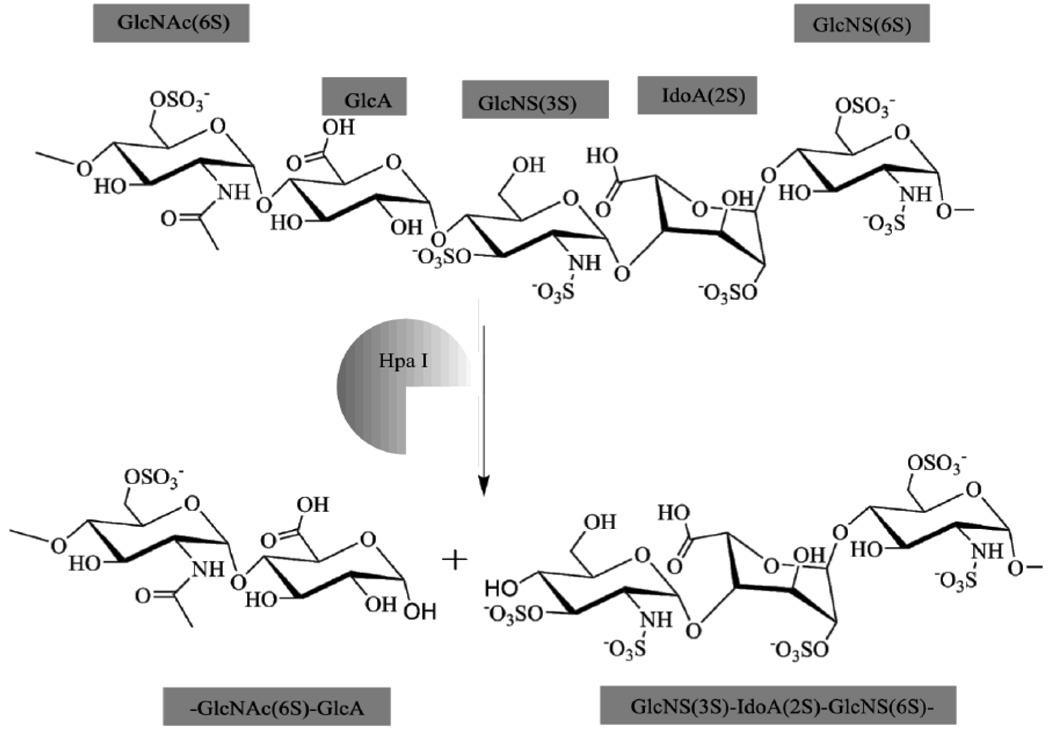
Heparanase (Hpa)
The Heparanase (Hpa) family consists of endo-β-D-glucuronidases that cleave HS into several smaller chains ~5–7 kDa in size (Fig. 3) [57–59].
Fig. 3. Hpa I Enzymatic Activity.
The Heparanase family is the only mammalian enzyme family that can cleave HS to form oligosaccharride units. Due to its unique activity, Hpa modulates growth factor signaling, ECM permeability and remodeling, cell clustering and adhesion, and several other cancer-related functions. Increased extracellular Hpa is an early indicator of malignant carcinomas.
In several cancer cells, these enzymes and their isoforms are either up- or down- regulated. Bret et al. found that RT-PCR analysis of several melanoma cells revealed a significant difference in the expression of genes encoding for EXT2, HS3ST2 (3-OST-2), HS2ST1(2-OST-1), HPSE (Heparanase), and SULF2 (HSulf-2) between normal and malignant plasma cells [60]. Determining enzymes and their isoforms that are misregulated, decoding their respective PG fine structural modifications, and understanding how these changes affect cancer progression, are essential for directing future research endeavors that could lead to the development of anti-cancer drugs.
Roles of HS and Important HS-Protein Interactions in Cancer
Growth
Cell-surface HSPGs, especially glypican-1 (Gpc1) and syndecan-1 (Sdc1), are greatly upregulated in late-stage (malignant) breast cancer tissue [61]. It is possible that these changes allow increased growth factor signaling since certain sulfated residues of HS are required for FGF receptor (FGFR) activation [62,63]. These HSPGs could also be responsible for protecting the cancer cells from natural killer cell (NK) recognition [64]. Since NK cells recognize particular HS fine structural patterns, cancer cells modify their HS patterns to evade NK cells and the immune system. Emerging evidence suggests that interactions between HS and HSulf-1, HSulf-2, and 3-OST, may also play an important role in breast tumor growth. The gene encoding HSulf-1 is down regulated in breast cancer cells [46]. Lower HSulf-1 expression increases autocrine activation of the EGFR-ERK (epidermal growth factor receptor-extracellular signal regulated kinase) pathway and stimulates cell growth [46]. Breast carcinoma cells also express 8-fold more HSulf-2 mRNA than normal tissues [65] Increased HSulf-2 mRNA correlates with increased angiogenesis and FGF binding capacity [65]. Several cancers also demonstrate 3-OST-2-gene silencing via methylation [66]. While it is unknown how 3-OST-2 specifically modulates growth factor binding to HS, it is possible that 3-OST-2 has anti-tumor properties that are stifled in cancer cells.
Invasion and Metastasis
Over expression of Hpa, which cleaves HS chains and facilitates migration of tumor cells through the ECM, is a key indicator of malignant breast cancers. In a clinical study, Hpa-1 expression was significantly upregulated in microinvasive lesions in ductal carcinoma in situ (DCIS) [67]. Hpa over-expressing breast tumors are 7 times larger and significantly more vascularized [68]. Increased Hpa also induces shedding of cell surface Sdc-1, which forms a paracrine signaling complex and in-turn signals distal breast cancer cells to proliferate [69,70]. Surprisingly, breast cancer can increase Hpa expression throughout the body. Lymphocytes in peripheral blood mononuclear cell fractions (PBMCs) from breast cancer patients express Hpa [71]. When serum containing these leukocytes is introduced to fresh lymphocytes, Hpa expression is stimulated in the normal lymphocytes [71]. Such tumor-system interactions lead to increased systemic heparanase expression in breast cancer [71]. Given the multitude of functions of Hpa, systemic Hpa amplification can perpetuate the tumor-promoting autocrine, paracrine, and growth factor signaling. Heparanase inhibitors can be effective against invasive cancers. However, it is also possible to control Hpa activity by controlling HS sulfonation since Hpa substrate recognition requires certain sulfonation and acetylation patterns [72].
Pancreatic Cancer
Growth
HSPGs such as Gpc-1 and Sdc-1 play key roles in the interactions between stromal elements and pancreatic cancer cells. Pancreatic ductal adenocarcinoma (PDAC) cells and adjacent fibroblasts over express Gpc-1, which is involved in FGF2, HB-EGF, VEGF, BMP, activin, and TGF-β signaling [73–76]. Down-regulation of Gpc-1 in PANC-1 cells attenuates tumor growth, angiogenesis, and metastasis in vivo in athymic mice [75]. Both HSulf-1 and HSulf-2 are also significantly upregulated in PDAC cell lines and stimulate autocrine Wnt signaling, which augments cancer growth [77]. HSulf-2 silencing blocks Wnt signaling and significantly reduces PDAC cell growth in an immuno-compromised mouse model. As with breast cancer, pancreatic cancer cells also protect themselves from NK activity. Since NK cells can only recognize cancer cells exhibiting certain 6-O-Sulfonation and N-Acetylation patterns, pancreatic cancer cells that express increased extracellular Hpa and aberrant HSulf activity exhibit much lower NK cell recognition [78].
Invasion and Metastasis
Sdc-1 is overexpressed in the majority of the pancreatic cancer tissue, surrounding metastatic lesions, and a significant amount is shed into serum [79]. Sdc-1 shedding correlates with increased mitogenic activity and cell invasive potential [79]. Hpa plays an important role in pancreatic cancer invasion and metastasis as well. Hpa1 mRNA is upregulated 7.9- and 30.2-fold in chronic pancreatitis and pancreatic cancer cells, respectively [80,81]. Hpa mRNA over expression is preferentially higher in primary tumor sites and correlates with decreased postoperative survival of patients [80]. Overexpression of Hpa leads to Sdc-1 and Gpc-1 cleavage, which in-turn leads to cellular growth via FGF-2 signaling and invasion of surrounding tissue [3].
Skin Cancer
Growth
Smetsers et al. found that melanoma cells only displayed certain GAG epitopes with particular sulfonation patterns on their cell-surface [82]. While the identity of these GAGs wasn’t devolved in their findings, it is possible that the GAGs represented a combination of fragments from Glypican, Syndecan, and Perlecan (Plc) HSPGs. Gpc, Sdc, and Plc are involved in growth factor binding. Plc, overexpressed (upto 150 fold) in human melanoma samples, also acts as a co-receptor for growth factors such as FGF-2 and FGF-7 [83,84]. Plc knockdown greatly inhibits murine melanoma tumor growth and neovascularization [85].
Invasion and Metastasis
Exogenous Hpa and cell-surface Gpc expression moderate the ability of skin cancer to metastasize. Lung metastatic melanoma cells overexpress Hpa1 mRNA (upto 29-fold) compared to normal lung tissue [86]. In melanoma patients with lung metastases, Hpa1 is found around vascularized regions and in blood vessels near the invasion front, whereas tumor nodules contain very little Hpa [86,87]. Hpa1-expressing melanoma cells demonstrate increased invasion in vitro and metastasis to lung and liver in vivo [88,89]. Gpc expression also affects melanoma invasion and metastasis characteristics since antisense-mediated-Gpc1-knockdown reduces B16-F10 melanoma cell pulmonary metastasis by attenuating Gpc1-HBGF (Heparin binding growth factor) signaling [75]. In combination with Hpa overexpression, melanoma cells exhibit 3-OST gene hypermethylation and subsequent gene silencing [90]. While the implications of this modification are not yet known, it is possible that certain patterns of 3-O sulfonation impart cancerous phenotypic changes. Ma et al. have found that P-Selectin, a cell adhesion molecule, bound to HS-like molecules on melanoma cells even in the absence of its recognition motif [91]. Interplay between 3-OST, 6-OST, and HSulf-1/2 might play a role in modifying HS to confer P-Selectin binding ability and hence promote metastasis by allowing cells to migrate to secondary sites.
Colorectal Cancer
Growth
In human colorectal cancers, Sdc-1 and Sdc-4 are downregulated while Sdc-2 is upregulated [92,93]. In conjunction with these HSPGs, 6-OST-2 is also upregulated. Sdc-2 knockdown leads to G0/G1 cell cycle arrest, increased expression of tumor suppressor proteins, and consequentially reduced tumorigenic activity [93]. In contrast, Sdc-2 expression in colorectal cancer cells contributes to cell evolution into a migratory mesenchymal-like phenotype (flatter shape, more membranal projections, and loss of intercellular contacts) [94]. 6-OST-2 mRNA is also significantly upregulated in colonic mucinous adenocarcinoma whereas 6-OST-3 is the predominant isoform present in normal mucosa [95]. Evidence from ovarian cancer suggests that 6-OST overexpression correlates with FGF-2 signaling and cell proliferation [96].
Invasion and Metastasis
Hpa over- expression correlates with increased colon cancer proliferation, invasion, and metastatic potential. Friedmann et al. have found high levels of Hpa in lung, liver, and lymph tumor metastases, while the highest amounts were found in deeply invading colon carcinoma cells [97]. Hpa activity is critical in colon cancer progression because stored FGFs attached to cell-surface HS are released by Hpa and MMP-7 [98]. Thus, in addition to making the ECM more permeable and susceptible to invasion, Hpa expression promotes cell proliferation and other growth factor related signals.
Lung and Brain Cancer
Growth
The roles of HS in lung and brain cancer are largely unknown, perhaps due to the delicacy of lung and brain tissue. Furthermore, since both these tissues are highly vascularized, cancers here are usually secondary metastases [99]. For lung cancer patients, serum Sdc-1 (S-Sdc-1) levels decline following tumor resection; whereas recurrent lung cancers demonstrate elevated S-Sdc-1 levels [100]. High S-Sdc-1 levels probably indicate high Hpa activity and extensive paracrine growth and proliferation signals.
Similarly, malignant glioma cells express high levels of Sdc-1 whereas nonneoplastic tissues do not [101]. Cell-surface Sdc-1 expression may correspond with FGF based growth factor signaling. It is also possible that overexpression of Sdc-1 and similar HSPGs is a distinguishing mark of cancer. Steck et al. generated antibodies that bound favorably to HSPGs from higher-grade gliomas [102]. From this they hypothesized that alternate HSPG expression patterns, and possibly sulfonation patterns, pertained to malignant transformation and growth potential of glial-cells. However, it is uncertain whether these alterations in HSPG expression are a cause or an effect of the transformation process.
Invasion and Metastasis
Hpa activity and mRNA are upregulated in lung and brain cancers as well. In a clinical study involving 114 lung cancer patients, cancer cells from 75% of the patients overexpressed Hpa [103]. Hpa over expression indicates increased invasive potential since Hpa knockdown in A549 lung cancer cells decreases their invasive potential in vivo [104]. Similarly in brain tumors, melanoma cells that are highly metastatic to the brain overexpress Hpa1 [105]. Hpa1 pretreated cells invade brain tissue in greater numbers and to greater tissue depths [105].
Other Cancers
While the focus of this review is on cancers of the brain, breast, lung, skin, pancreas, and colon, aberrant HS biosynthesis and catabolism play a role in several other cancers as well. EXT-1 and EXT-2, involved in heparan sulfate biosynthesis, are known tumor suppressor genes involved in preventing progression of osteochondromas to chondrosarcomas [106]. Several cancers, including ovarian cancer and head and neck squamous carcinoma, demonstrate extensive HSulf-1 silencing [107,108]. Re-expression of HSulf-1 in these cancers suppresses proliferation, enhances apoptosis in vitro and reduces angiogenesis [109]. Chen et al. found that the HSulf-1 promoter was more frequently methylated in DNA samples from cell-free serum samples of gastric cancer patients (55%) compared to healthy patients (19%) [109]. They proposed that this methylation-induced silencing of HSulf-1 could be used as an early diagnostic tool for cancer. Hpa is overexpressed in the invading front of endometrial cancers in humans where Hpa expression correlates with tumor-associated angiogenesis and invasion into lymph vascular space and myometrium [110,111]. It is also over expressed in renal cell carcinomas and head and neck carcinomas [112,113].
Underlying patterns of HSPG involvement in all cancers
Based on the previously stated roles of HS biosynthetic and catabolic enzymes in cancer, it is now possible to see several overarching patterns (Table 1, Fig. 4). Hpa, the sole mammalian enzyme to degrade HS, is overexpressed in most cancers. Hpa activity breaks down proteoglycans, increases endothelium permeability, releases stored cell-surface FGFs, and permits serum-HS to interact with FGFs. 3-OST, an enzyme that moderates rare HS fine structural changes, is silenced by hypermethylation in several cancers [66]. Although demethylation of 3-OST in breast cancer cells didn’t produce any visible changes, there must be a reason behind 3-OST silencing in such an array of cancers [66]. HSulf-1 also seems to be downregulated (via methylation) in several cancers. Since re-expressing HSulf-1 decreases cancer mitogenesis, cancer cells probably utilize hyper-sulfated residues that are normally controlled by HSulf-1 for growth signaling via FGF-2 signaling [114]. Interplay between Hpa and Sdc-1 in several cancers is also noteworthy. Both entities seem to act together to induce proliferation and autocrine growth signaling. Changes in HS biosynthetic and catabolic enzymes are essential to cancer growth. HS is a profound target for developing novel cancer therapeutics because modifying HS chains would affect Hpa, HSulf-1, HSulf-2, and 3-OST activity in tumor cells, which in turn would affect angiogenesis, growth-factor signal over-amplification, and tumor growth, invasion, and metastasis.
Table 1.
Summary of HS and HS-related-enzyme expression aberrations.
| HSPGs | ||
|---|---|---|
| Sdc-1 | Gpc-1 | |
|
Breast Cancer |
↑ | ↑ |
|
Pancreatic Cancer |
↑ | ↑ |
|
Skin Cancer |
- | - |
|
Colorectal Cancer |
↓ | - |
|
Lung Cancer |
↑ | - |
|
Brain Cancer |
↑ | - |
| HS Catabolic Enzymes | |||
|---|---|---|---|
| Hpa-1 | HSulf-1 | HSulf-2 | |
|
Breast Cancer |
↑ | ↓ | ↑ |
|
Pancreatic Cancer |
↑ | ↑ | ↑ |
|
Skin Cancer |
↑ | - | - |
|
Colorectal Cancer |
↑ | - | - |
|
Lung Cancer |
↑ | - | - |
|
Brain Cancer |
↑ | - | - |
| HS Biosynthetic Enzymes | ||
|---|---|---|
| 3-OST-2 | 6-OST-2 | |
|
Breast Cancer |
X, ↑ | - |
|
Pancreatic Cancer |
X | - |
|
Skin Cancer |
X | - |
|
Colorectal Cancer |
X, ↑ | ↑ |
|
Lung Cancer |
X | - |
|
Brain Cancer |
- | - |
↑: Upregulated ↓: Downregulated x: Gene silenced --: Not known
Fig. 4. The roles of HS in cancer.
From gathered evidence presented in this article, HS is involved in several key functions regarding cancer progression. Cell metastasis depends on HS to provide P-Selectin binding so cells can adhere. As an antenna molecule, HS also plays a role in transducing signals from external growth factors by forming complexes with growth factor receptors. Additionally it is necessary for efficient activation of Heparanase, which is necessary for invasion, angiogenesis, and inflammation. By binding P-Selectins on platelets, HS also protects cancer cells from natural killer (NK) cell activity. Systemically, shed cell-surface HS can form paracrine signaling complexes FGFRs. Additionally, though not completely understood, cancer cells instigate circulating lymphocytes to upregulate heparanase activity; as an antenna molecule, HS is probably involved in this pathway as well.
Current HS-based treatments
It has been known for decades that HS and other GAGs modulate tumor biology and are essential to cell survival. However, HS-specific treatments have only recently begun preliminary clinical trials. Several different treatment strategies have been the focus of recent research efforts. These strategies include: inhibition of Hpa with HS analogs such as PI-88, inhibition of tumor invasion and metastasis using non-anti-coagulant LMWH analogs, and inhibition of tumor growth using carbodiimide modified GAGs or ‘neoglycans’.
Difficulties associated with cancer treatment
Treating cancer poses a challenge because cancer cells have several inherent defense mechanisms. Not only do cancer cells originate from the host system, but they also use natural cellular metabolic pathways to grow. Additionally, due to the genetic errors that manifest cancer, tumors are composed of heterogeneous populations of cells that respond differently to treatments and impart multi-drug resistance to tumors [115]. Moreover, cancer patients exhibit a hyper-coagulable state because cancer cells employ platelets and modified HS to evade immune cells. Several insightful studies implicate platelets in forming barriers that prevent NK cells from recognizing and destroying tumors by coagulating around cancer cells [116–118]. Depletion of platelet P-Selectins, which allow platelets to adhere to tumor cells, leads to reduced tumor dissemination, and increased tumor cell death [119]. To further complicate cancer diagnosis, there is an epigenetic aspect to the development of cancer. The probability of being diagnosed with cancer increases with age, as published by the American Cancer Society [120]. Since the disease manifests at varying ages, it is not yet possible to predict the onset of cancer. To combat such a variety of defense mechanisms employed by cancer cells, effective cancer therapies should be based on multiple anti-tumor mechanisms while utilizing pathways that are essential to survival of all cells.
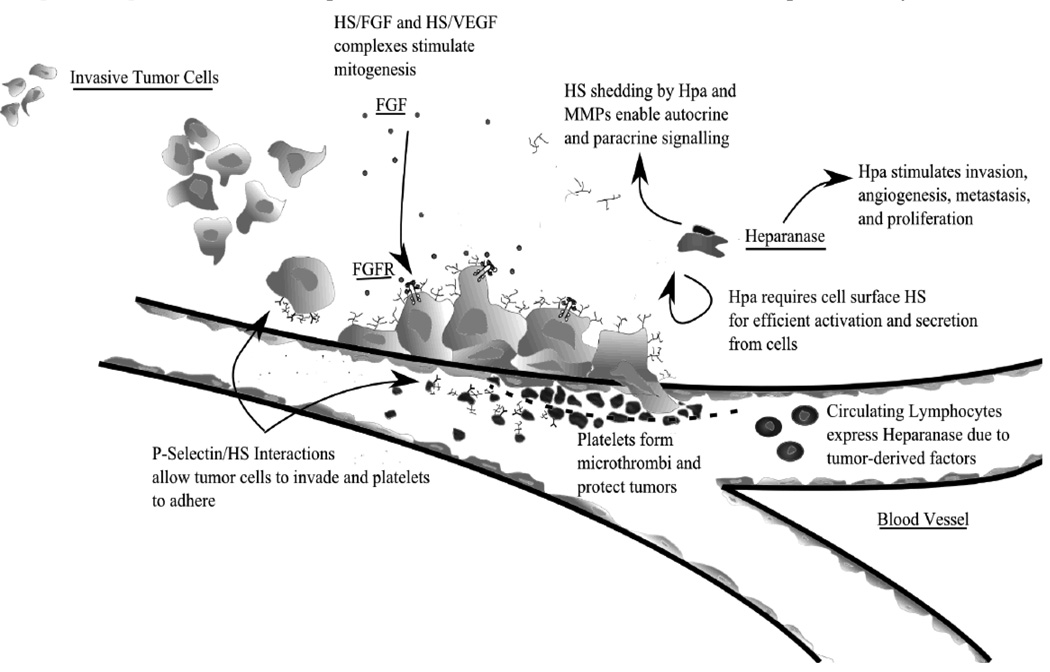
Heparanase, a potent anti-cancer target
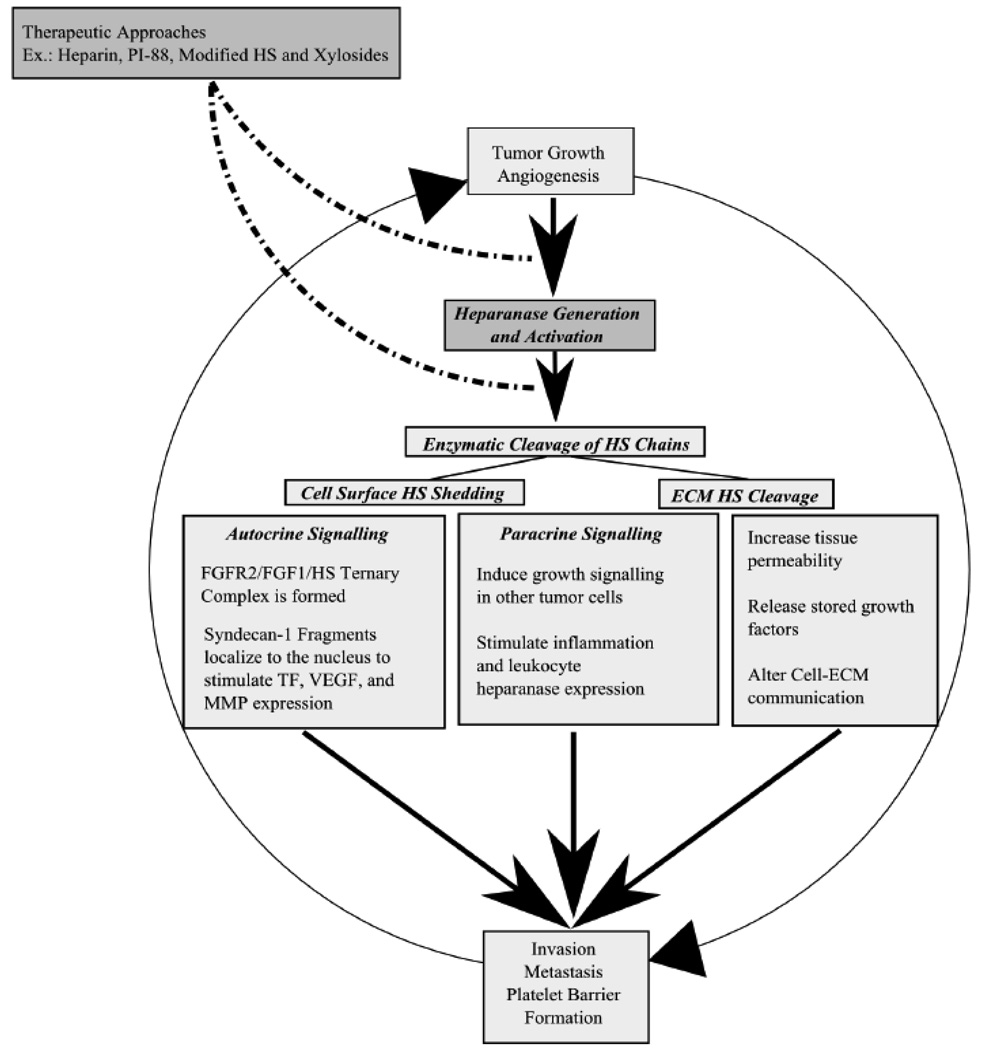
Hpa is a potent anti-cancer target because it is upregulated in a majority of cancers and plays a major role in tumor-induced angiogenesis, invasion and metastasis (Fig. 5). Based on available information, it is possible to propose one comprehensive pathway by which Hpa aids cancer by providing tumor cells with a self-induced proliferation loop. During hypoxia, cancer cells overexpress Hpa well above their normal Hpa levels [121]. Overexpressed Hpa is secreted in a pro-enzyme form. After reuptake, the proenzyme is cleaved, activated, and secreted once again [57,122]. The active enzyme then cleaves cell surface HS (i.e. Sdc-1 shedding) to form paracrine and autocrine signaling complexes [69].
Fig. 5. Current HS-Based drug development strategies and their target signaling pathways in the Heparanase-induced self-proliferation loop.
PI-88 and related oligosaccharides are potent anti-cancer agents because they interrupt Heparanase activity and also affect several growth signaling pathways such as the FGF2 pathway. Heparin and its low molecular weight anti-coagulant derivatives are natural Hep inhibitors that competitively inhibit Heparnase enzymatic activity by acting as the enzyme substrate. However, it is possible to affect both Heparanase as well as HS binding growth factors by utilizing modified HS and xylosides to tailor endogenous HS on cells to resist Hpa enzymatic activity and growth factor binding.
Autocrine signaling complexes enhance FGF-HS-FGFR interactions and upregulate cellular signaling pathways. On one hand shed Sdc-1 are taken up by cancer cells and localized to the nucleus where they affect expression of MMP-9, VEGF, and tissue factors [123,124]. On the other hand, shed Sdc-1 form trimeric complexes to stabilize FGFR-1 and FGF-1 interactions [114] Paracrine signaling complexes induce other tumor cells to proliferate as in breast cancer. The MMP activity then potentiates tumor invasion whereas VEGF and FGF activity increase tumor lymph node metastasis, angiogenesis and growth. Moreover, tissue factors promote clotting, angiogenesis, and platelet barrier formation around the tumor [125–129]. Newly formed cancer cells and improved nutrient supply then perpetuate tumor growth and heparanase production so that the self-proliferation loop continues (Fig. 5).
PI-88 and related Heparan Sulfate analogs
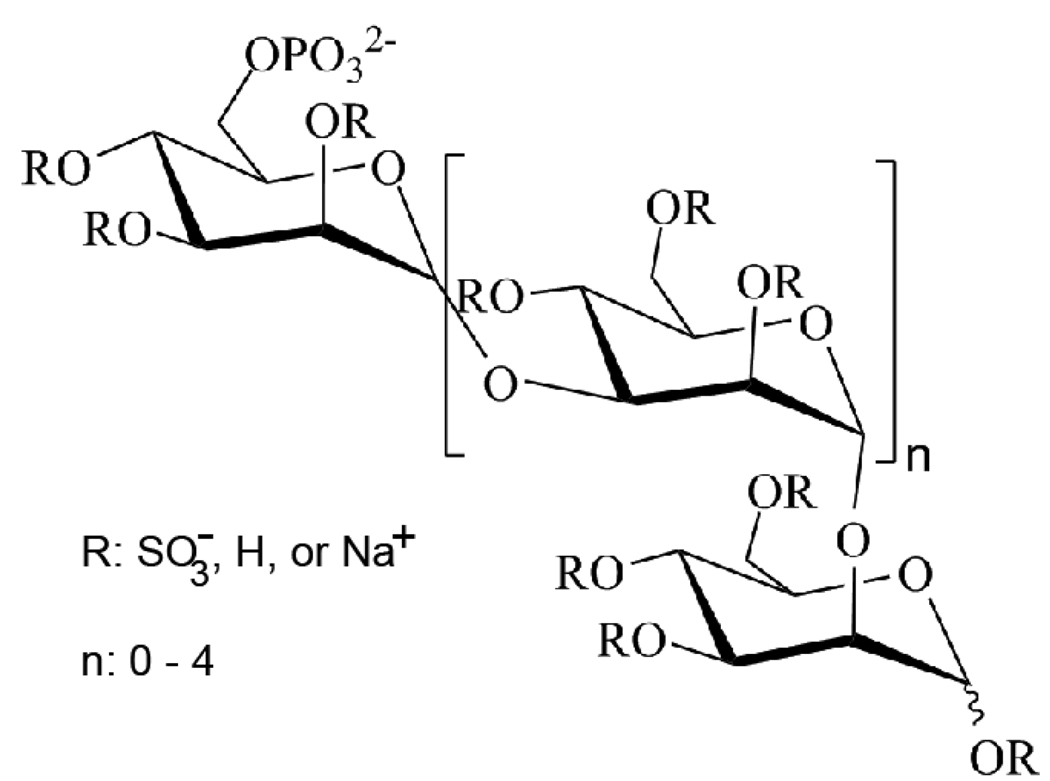
The structures, advantages, and disadvantages of several Hpa inhibitors have been discussed in detail by Ferro et al.[130] One particularly interesting Hpa inhibitor is PI-88 (Fig. 6), a highly sulfated oligosaccharide mixture, which is currently undergoing phase II clinical trials for treating patients with metastatic melanoma [131,132]. It prevents angiogenesis and tumor growth by blocking FGF-1, FGF-2, and VEGF interactions with their receptors and HS [131] PI-88 reduces the growth of invasive rat mammary adenocarcinoma cells by 50%. It also reduces lymph node and blood metastases [133]. Among 400 patients already tested, PI-88 has shown minimal side-effects and excellent efficacy whereby one patient with melanoma had a partial response for >50 months [131]. Based on improvements to PI-88 chemistry, several new analogs of PI-88, known as the PG500 series, have been developed [134]. Some advantages of these new molecules include improved inhibition of FGF- and VEGF- induced endothelial cell tube formation and lower plasma clearance levels compared to PI-88 [131]
Fig. 6. Chemical Structure of PI-88.
A potent Heparanase inhibitor, PI-88 is a hyper-sulfated oligosaccharide that blocks Hpa and several HS/Growth factor interactions.
Low molecular weight non-anticoagulant heparins
Another current HS-based treatment strategy is based on heparin’s natural anti-cancer properties. While the molecular details of heparin’s effects on cancer have not been completely resolved, it is believed that heparin inhibits tumor metastasis by inhibiting Hpa activity, preventing P-Selectin mediated cell-platelet interactions, and releasing tissue factor pathway inhibitor, a potent endogenous anti-angiogenic agent [135,136]. However, heparin is also a potent anti-coagulant and cannot be used to treat cancer patients for a prolonged period of time because it may lead to bleeding complications. Several groups have prepared low molecular weight anti-coagulant heparin (LMWH) derivates to mimic heparin’s anti-mitogenic activity without its anti-coagulant properties. Non-anticoagulant LMWH are produced by separating the anti-thrombin binding sequence from unfractionated heparin, chemically desulfating heparin at the 2-O and 3-O sulfonation sites or by using periodate oxidation (also known as the ‘glycol split’) to decrease the number of glucuronic acid residues which are critical for binding to antithrombin III [136] Dalteparin, a 5kD non-anticoagulant LMWH, has been shown to improve 1- year survival of cancer patients with small cell lung cancer by 11% and 2-year survival by 17% in clinical studies without presenting bleeding complications [137]. Anti-coagulant LMWHs have shown promise in mouse models but have not yet entered clinical trials [136].
Neoglycan and carbodiimide modified GAGs
Pumphrey et al. have developed a new class of anti-cancer agents, termed ‘neoglycans’, based on carbodiimide-modified CS and heparin chains. While the actual structure of these chains is unclear, carbodiimide modification of GAG chains could produce crosslinked chains that form GAG complexes via reaction of the free amine groups. These crosslinked meshes may resemble multimeric arrays of GAGs found on syndecans/glypicans. However, it is essential to note that GAG chains have abundant carboxyl groups and limited amine functionality. Therefore, the carbodiimide reaction may not go to completion and thus may produce highly sulfated neoglycans functionalized with several O-acylisourea groups along the chains.
These molecules significantly reduce myeloma and breast cancer cell viability in vitro in a dose dependent manner [138]. NeoCS treatment at 32 µg/ml reduced ARP-1 cell viability by 96%. As reported in this study, the IC50 values for neoHeparin and neoCS on ARP-1 myeloma cells are 21.94 µg/ml and 14.79 µg/ml, respectively. These molecules trigger apoptosis in ARP-1 and MDA-MB-231 (breast cancer) cells, however the mechanism of their action and uptake are still not known. neoCS was a more potent inhibitor for a majority of cell lines tested in vitro and a single dose of neoCS completely eliminated MDA-MB-231 tumors implanted into BALB/c nu/nu mice. After treatment with the neoCS, only 1 tumor reemerged out of 15 separate in vivo tests. These modified GAGs show promise for future anti-cancer drug development. However, one should exercise caution because these modified GAG chains are unlikely to be metabolized in vivo and therefore may possess toxicity.
There are also several other therapeutic strategies based on HS-like molecules. Other areas of research include: developing FGF-2 inhibitors that prevent FGF-HS-induced cellular growth, developing drug delivery vehicles that utilize HS-mediated internalization into cells, and modulating HS-GAG production in cancer cells using novel drugs [139–142]. Suramin, a non-heparin Hpa inhibitors, has also shown promise in vitro and has undergone several phase I clinical trials [143]. Preliminary results from these studies are promising and reinforce our hypothesis that modulating HS fine structures, their biosynthesis and interactions with the tumor microenvironment will lead to potential drug therapies for cancer in the near future.
Future Directions
HS-based treatments can be more effective against cancer
Cancer cells have been suggested to mimic misbehaving stem cells where erroneous cellular machinery causes cells to trigger abnormal signals, misinterpret incoming signals, and differentiate into several families of cancerous cells [144]. HS chains, ubiquitously expressed on cell-surfaces and in ECM throughout the body, act as molecular antennae that send and receive signals through binding to a wide variety of molecules such as FGF-2, VEGF, HB-EGF, P-Selectin, MMP-7 and MMP-9 [15,123,135,145,146] or releasing these sequestered molecules from the cell surface/ECM. Thus, therapeutics that modify HS will be able to attack cancer cells on multiple fronts because they can target P-Selectin interactions, growth factor binding, the coagulation cascade, protease activation and inhibition, Heparanase activation and activity, and possibly tumor evolution/differentiation [147].
Novel chemical biology approaches to treating cancer
Future research should utilize knowledge of the downstream effects of fine structural changes in HS to invent novel therapeutics that affect a wide range of tumor-specific pathological processes. Unraveling the mysteries of ‘GAGOSOMES’, which are responsible for forming and controlling the HS biosynthesis enzyme complex, may also yield a valuable target [148,149]. Modulating HS fine structures will affect HS-ECM, HS-FGF, and HS-protein interactions and prevent cancer growth without causing any adverse downstream effects.
It is possible to inhibit HS/CS biosynthesis by utilizing 4-deoxy-4-fluoro-xylosides [150]. Decreasing overall levels of HS may reduce FGF and VEGF signaling and affect tumor proliferation, invasion and metastasis. Inhibiting HS production may also prevent Hpa activation and hence restrain Hpa activity [122]. It is also possible to stimulate HS and CS GAG production by utilizing xylosides to prime GAG chains in vitro or in vivo [151]. Increasing HS can render cancer cells more adherent and less metastatic due to enhanced integrin binding [152]. However, these methods produce a wide array of GAGs with no specific properties. Further research into the chemistry and biology of these xylosides is necessary to make treatments from xyloside-induced GAGs with fine structural elements that can compete with endogenous GAGs made by cancer cells and disrupt cancer growth factor signaling and Hpa activity.
In summary, this article proposes that controlling GAG biosynthesis and catabolism in general, and HS biosynthesis and catabolism in particular, may be a powerful tool to design effective treatments against cancer and perhaps even other diseases. Key enzymes involved in `HS biosynthesis and catabolism include HSulf-1, HSulf-2, Hpa, 3-OST, and 6-OST; each of these enzymes is differentially expressed in various cancers. By changing HS structure, these enzymes affect several downstream cellular processes that are integral to cancer progression. However, there are still several unanswered questions relating to the use of HS in treating cancer. Which Xylosides prime GAGs with fine structural elements that prevent FGF binding and inhibit cancerous phenotype changes? What is the role of 3-O sulfation, a rare HS modification, in cancer progression? In a novel experiment, Liu et al. injected Heparinase I and III into mice with B16BL6 melanoma and found that the specificity of the enzymes dictated whether tumors regressed (Heparinase III) or advanced (Heparinase I) due to enzymatic cleavage of cellular HS [153]. They found that the tumor cell GAG fragments produced by Heparinase III digestion caused upto 75% tumor growth inhibition whereas fragments produced by Heparinase I digestion significantly enhanced growth. This experiment reinforces the concept that there is an incredible specificity to the structural/biological relationships between HS fine structures and the biological effects of these molecules on cancers. However, the metaphorical HS elephant has many facets that need to be extensively studied before we can develop any effective anti-cancer drugs [154]. Thus, chemical biologists, who are at the interface of understanding both the biological as well as the chemical roles of HS, have ample opportunity to research and collaborate in order to find an effective HS-based scaffold for treating cancer without any side effects.
Acknowledgements
B. K. is supported by grants from the National Institutes of Health (GM075168), Mizutani Foundation for Glycoscience, American Heart Association National Scientist development award, and the Human Frontier Science Program
Abbreviations
- HS
Heparan Sulfate
- CS
Chondroitin Sulfate
- DS
Dermatan Sulfate
- HSPG
Heparan Sulfate Proteoglycan
- CSPG
Chondroitin Sulfate Proteoglycan
- Sdc
Syndecan
- S-Sdc
Shed Syndecan
- Gpc
Glypican
- Hpa
Heparanase
- HSulf
HS 6-O Endosulfatase
- 3-OST
3-O Sulfotransferase
- 6-OST
6-O Sulfotransferase
- FGF
Fibroblast Growth Factor
- VEGF
Vascular Endothelial Cell Growth Factor
- HBGF
Heparin Binding Growth Factor
- HB-EGF
Heparin Binding Epidermal Growth Factor
- FGFR
Fibroblast Growth Factor receptor
- VEGFR
Vascular Endothelial Cell Growth Factor Receptor
- BMP
Bone Morphogenic Protein
- TGF-β
Transforming Growth Factor Beta
- MMP
Matrix Metalloprotease
- GAG
Glycosaminoglycan
- ECM
Extraceullar Matrix
- PG
Proteoglycan
- DCIS
Ductal Carcinoma in situ
- PBMC
Peripheral blood mononuclear cell fraction
- NK
Natural Killer Cell
- LMWH
low molecular weight heparin
References
- 1.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 2.Sanderson RD. Heparan sulfate proteoglycans in invasion and metastasis. Semin Cell Dev Biol. 2001;12:89–98. doi: 10.1006/scdb.2000.0241. [DOI] [PubMed] [Google Scholar]
- 3.Sanderson RD, Yang Y, Kelly T, MacLeod V, Dai Y, Theus A. Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: growth regulation and the prospect of new cancer therapies. J Cell Biochem. 2005;96:897–905. doi: 10.1002/jcb.20602. [DOI] [PubMed] [Google Scholar]
- 4.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 6.Liu D, Shriver Z, Qi Y, Venkataraman G, Sasisekharan R. Dynamic regulation of tumor growth and metastasis by heparan sulfate glycosaminoglycans. Semin Thromb Hemost. 2002;28:67–78. doi: 10.1055/s-2002-20565. [DOI] [PubMed] [Google Scholar]
- 7.Ribatti D, Marimpietri D, Pastorino F, Brignole C, Nico B, Vacca A, Ponzoni M. Angiogenesis in neuroblastoma. Ann N Y Acad Sci. 2004;1028:133–142. doi: 10.1196/annals.1322.014. [DOI] [PubMed] [Google Scholar]
- 8.Folkman J. Tumor angiogensis: role in regulation of tumor growth. Symp Soc Dev Biol. 1974;30:43–52. [PubMed] [Google Scholar]
- 9.Turk V, Kos J, Turk B. Cysteine cathepsins (proteases)--on the main stage of cancer? Cancer Cell. 2004;5:409–410. doi: 10.1016/s1535-6108(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 10.Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 11.Ravanko K, Jarvinen K, Helin J, Kalkkinen N, Holtta E. Cysteine cathepsins are central contributors of invasion by cultured adenosylmethionine decarboxylase-transformed rodent fibroblasts. Cancer Res. 2004;64:8831–8838. doi: 10.1158/0008-5472.CAN-03-2993. [DOI] [PubMed] [Google Scholar]
- 12.Matrisian LM, Wright J, Newell K, Witty JP. Matrix-degrading metalloproteinases in tumor progression. Princess Takamatsu Symp. 1994;24:152–161. [PubMed] [Google Scholar]
- 13.Liabakk NB, Talbot I, Smith RA, Wilkinson K, Balkwill F. Matrix metalloprotease 2 (MMP-2) and matrix metalloprotease 9 (MMP-9) type IV collagenases in colorectal cancer. Cancer Res. 1996;56:190–196. [PubMed] [Google Scholar]
- 14.Whitelock JM, Melrose J, Iozzo RV. Diverse cell signaling events modulated by perlecan. Biochemistry. 2008;47:11174–11183. doi: 10.1021/bi8013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombe DR. Biological implications of glycosaminoglycan interactions with haemopoietic cytokines. Immunol Cell Biol. 2008;86:598–607. doi: 10.1038/icb.2008.49. [DOI] [PubMed] [Google Scholar]
- 16.Laguri C, Arenzana-Seisdedos F, Lortat-Jacob H. Relationships between glycosaminoglycan and receptor binding sites in chemokines-the CXCL12 example. Carbohydr Res. 2008;343:2018–2023. doi: 10.1016/j.carres.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 17.Adatia R, Albini A, Carlone S, Giunciuglio D, Benelli R, Santi L, Noonan DM. Suppression of invasive behavior of melanoma cells by stable expression of anti-sense perlecan cDNA. Ann Oncol. 1997;8:1257–1261. doi: 10.1023/a:1008243115385. [DOI] [PubMed] [Google Scholar]
- 18.Su G, Meyer K, Nandini CD, Qiao D, Salamat S, Friedl A. Glypican-1 is frequently overexpressed in human gliomas and enhances FGF-2 signaling in glioma cells. Am J Pathol. 2006;168:2014–2026. doi: 10.2353/ajpath.2006.050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 20.Stauber DJ, DiGabriele AD, Hendrickson WA. Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci U S A. 2000;97:49–54. doi: 10.1073/pnas.97.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guimond S, Maccarana M, Olwin BB, Lindahl U, Rapraeger AC. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2, and FGF-4. J Biol Chem. 1993;268:23906–23914. [PubMed] [Google Scholar]
- 22.Jia J, Maccarana M, Zhang X, Bespalov M, Lindahl U, Li JP. Lack of L-iduronic acid in heparan sulfate affects interaction with growth factors and cell signaling. J Biol Chem. 2009;284:15942–15950. doi: 10.1074/jbc.M809577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, Claesson-Welsh L. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem. 2000;275:24653–24660. doi: 10.1074/jbc.M908930199. [DOI] [PubMed] [Google Scholar]
- 24.Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem. 2008;283:10366–10376. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- 25.Pauli BU, Schwartz DE, Thonar EJ, Kuettner KE. Tumor invasion and host extracellular matrix. Cancer Metastasis Rev. 1983;2:129–152. doi: 10.1007/BF00048966. [DOI] [PubMed] [Google Scholar]
- 26.Nakato H, Kimata K. Heparan sulfate fine structure and specificity of proteoglycan functions. Biochim Biophys Acta. 2002;1573:312–318. doi: 10.1016/s0304-4165(02)00398-7. [DOI] [PubMed] [Google Scholar]
- 27.Jansson L, Lindahl U. Evidence for the existence of a multichain proteoglycan of heparan sulphate. Biochem J. 1970;117:699–702. doi: 10.1042/bj1170699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl U, Roden L. The chondroitin 4-sulfate-protein linkage. J Biol Chem. 1966;241:2113–2119. [PubMed] [Google Scholar]
- 30.Fransson LA, Silverberg I. Carlstedt I. Structure of the heparan sulfateprotein linkage region. Demonstration of the sequence galactosyl-galactosyl-xylose-2-phosphate. J Biol Chem. 1985;260:14722–14726. [PubMed] [Google Scholar]
- 31.Salmivirta M, Lidholt K, Lindahl U. Heparan sulfate: a piece of information. FASEB J. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 32.Rabenstein DL. Heparin and heparan sulfate: structure and function. Nat Prod Rep. 2002;19:312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 33.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 34.Lindahl U, Kusche M, Lidholt K, Oscarsson LG. Biosynthesis of heparin and heparan sulfate. Ann N Y Acad Sci. 1989;556:36–50. doi: 10.1111/j.1749-6632.1989.tb22488.x. [DOI] [PubMed] [Google Scholar]
- 35.Ahn J, Ludecke HJ, Lindow S, Horton WA, Lee B, Wagner MJ, Horsthemke B, Wells DE. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nat Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- 36.Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- 37.Kitagawa H, Shimakawa H, Sugahara K. The tumor suppressor EXT-like gene EXTL2 encodes an alpha1, 4-Nacetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J Biol Chem. 1999;274:13933–13937. doi: 10.1074/jbc.274.20.13933. [DOI] [PubMed] [Google Scholar]
- 38.Izumikawa T, Koike T, Shiozawa S, Sugahara K, Tamura J, Kitagawa H. Identification of chondroitin sulfate glucuronyltransferase as chondroitin synthase-3 involved in chondroitin polymerization: chondroitin polymerization is achieved by multiple enzyme complexes consisting of chondroitin synthase family members. J Biol Chem. 2008;283:11396–11406. doi: 10.1074/jbc.M707549200. [DOI] [PubMed] [Google Scholar]
- 39.Kitagawa H, Uyama T, Sugahara K. Molecular cloning and expression of a human chondroitin synthase. J Biol Chem. 2001;276:38721–38726. doi: 10.1074/jbc.M106871200. [DOI] [PubMed] [Google Scholar]
- 40.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 41.Prydz K, Dalen KT. Synthesis and sorting of proteoglycans. J Cell Sci. 2000;113(Pt 2):193–205. doi: 10.1242/jcs.113.2.193. [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg RD, Shworak NW, Liu J, Schwartz JJ, Zhang L. Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? J Clin Invest. 1997;100:S67–S75. [PubMed] [Google Scholar]
- 43.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, Venkataraman G, Sasisekharan R, Sanderson RD. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–40073. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- 46.Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, Shridhar V. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–14420. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- 47.Ohto T, Uchida H, Yamazaki H, Keino-Masu K, Matsui A, Masu M. Identification of a novel nonlysosomal sulphatase expressed in the floor plate, choroid plexus and cartilage. Genes Cells. 2002;7:173–185. doi: 10.1046/j.1356-9597.2001.00502.x. [DOI] [PubMed] [Google Scholar]
- 48.Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shworak NW, Liu J, Fritze LM, Schwartz JJ, Zhang L, Logeart D, Rosenberg RD. Molecular cloning and expression of mouse and human cDNAs encoding heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J Biol Chem. 1997;272:28008–28019. doi: 10.1074/jbc.272.44.28008. [DOI] [PubMed] [Google Scholar]
- 50.Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1. J Gen Virol. 2004;85:805–809. doi: 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Shworak NW, Sinay P, Schwartz JJ, Zhang L, Fritze LM, Rosenberg RD. Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem. 1999;274:5185–5192. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 52.Mochizuki H, Yoshida K, Gotoh M, Sugioka S, Kikuchi N, Kwon YD, Tawada A, Maeyama K, Inaba N, Hiruma T, Kimata K, Narimatsu H. Characterization of a heparan sulfate 3-O-sulfotransferase-5, an enzyme synthesizing a tetrasulfated disaccharide. J Biol Chem. 2003;278:26780–26787. doi: 10.1074/jbc.M301861200. [DOI] [PubMed] [Google Scholar]
- 53.Smeds E, Habuchi H, Do AT, Hjertson E, Grundberg H, Kimata K, Lindahl U, Kusche-Gullberg M. Substrate specificities of mouse heparan sulphate glucosaminyl 6-O-sulphotransferases. Biochem J. 2003;372:371–380. doi: 10.1042/BJ20021666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Beeler DL, Lawrence R, Lech M, Liu J, Davis JC, Shriver Z, Sasisekharan R, Rosenberg RD. 6-O-sulfotransferase-1 represents a critical enzyme in the anticoagulant heparan sulfate biosynthetic pathway. J Biol Chem. 2001;276:42311–42321. doi: 10.1074/jbc.M101441200. [DOI] [PubMed] [Google Scholar]
- 55.Kusche M, Backstrom G, Riesenfeld J, Petitou M, Choay J, Lindahl U. Biosynthesis of heparin. O-sulfation of the antithrombin-binding region. J Biol Chem. 1988;263:15474–15484. [PubMed] [Google Scholar]
- 56.Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282:15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- 57.Vlodavsky I, Abboud-Jarrous G, Elkin M, Naggi A, Casu B, Sasisekharan R, Ilan N. The impact of heparanese and heparin on cancer metastasis and angiogenesis. Pathophysiol Haemost Thromb. 2006;35:116–127. doi: 10.1159/000093553. [DOI] [PubMed] [Google Scholar]
- 58.Naparstek Y, Cohen IR, Fuks Z, Vlodavsky I. Activated T lymphocytes produce a matrix-degrading heparan sulphate endoglycosidase. Nature. 1984;310:241–244. doi: 10.1038/310241a0. [DOI] [PubMed] [Google Scholar]
- 59.Oosta GM, Favreau LV, Beeler DL, Rosenberg RD. Purification and properties of human platelet heparitinase. J Biol Chem. 1982;257:11249–11255. [PubMed] [Google Scholar]
- 60.Bret C, Hose D, Reme T, Sprynski AC, Mahtouk K, Schved JF, Quittet P, Rossi JF, Goldschmidt H, Klein B. Expression of genes encoding for proteins involved in heparan sulphate and chondroitin sulphate chain synthesis and modification in normal and malignant plasma cells. Br J Haematol. 2009;145:350–368. doi: 10.1111/j.1365-2141.2009.07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsuda K, Maruyama H, Guo F, Kleeff J, Itakura J, Matsumoto Y, Lander AD, Korc M. Glypican-1 is overexpressed in human breast cancer and modulates the mitogenic effects of multiple heparin-binding growth factors in breast cancer cells. Cancer Res. 2001;61:5562–5569. [PubMed] [Google Scholar]
- 62.Nurcombe V, Smart CE, Chipperfield H, Cool SM, Boilly B, Hondermarck H. The proliferative and migratory activities of breast cancer cells can be differentially regulated by heparan sulfates. J Biol Chem. 2000;275:30009–30018. doi: 10.1074/jbc.M003038200. [DOI] [PubMed] [Google Scholar]
- 63.Delehedde M, Deudon E, Boilly B, Hondermarck H. [Involvement of sulfated proteoglycans in the control of proliferation of MCF-7 breast cancer cells] Bull Cancer. 1996;83:129–134. [PubMed] [Google Scholar]
- 64.Damiens E, El Yazidi I, Mazurier J, Elass-Rochard E, Duthille I, Spik G, Boilly-Marer Y. Role of heparan sulphate proteoglycans in the regulation of human lactoferrin binding and activity in the MDA-MB-231 breast cancer cell line. Eur J Cell Biol. 1998;77:344–351. doi: 10.1016/S0171-9335(98)80093-9. [DOI] [PubMed] [Google Scholar]
- 65.Morimoto-Tomita M, Uchimura K, Bistrup A, Lum DH, Egeblad M, Boudreau N, Werb Z, Rosen SD. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia. 2005;7:1001–1010. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyamoto K, Asada K, Fukutomi T, Okochi E, Yagi Y, Hasegawa T, Asahara T, Sugimura T, Ushijima T. Methylation-associated silencing of heparan sulfate D-glucosaminyl 3-O-sulfotransferase-2 (3-OST-2) in human breast, colon, lung and pancreatic cancers. Oncogene. 2003;22:274–280. doi: 10.1038/sj.onc.1206146. [DOI] [PubMed] [Google Scholar]
- 67.Maxhimer JB, Pesce CE, Stewart RA, Gattuso P, Prinz RA, Xu X. Ductal carcinoma in situ of the breast and heparanase-1 expression: a molecular explanation for more aggressive subtypes. J Am Coll Surg. 2005;200:328–335. doi: 10.1016/j.jamcollsurg.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 68.Cohen I, Pappo O, Elkin M, San T, Bar-Shavit R, Hazan R, Peretz T, Vlodavsky I, Abramovitch R. Heparanase promotes growth, angiogenesis and survival of primary breast tumors. Int J Cancer. 2006;118:1609–1617. doi: 10.1002/ijc.21552. [DOI] [PubMed] [Google Scholar]
- 69.Yang Y, Macleod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD, Jr, Sawyer J, Li JP, Zcharia E, Vlodavsky I, Sanderson RD. Heparanase enhances syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282:13326–13333. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 70.Su G, Blaine SA, Qiao D, Friedl A. Shedding of syndecan-1 by stromal fibroblasts stimulates human breast cancer cell proliferation via FGF2 activation. J Biol Chem. 2007;282:14906–14915. doi: 10.1074/jbc.M611739200. [DOI] [PubMed] [Google Scholar]
- 71.Theodoro TR, de Matos LL, Sant Anna AV, Fonseca FL, Semedo P, Martins LC, Nader HB, Del Giglio A, da Silva Pinhal MA. Heparanase expression in circulating lymphocytes of breast cancer patients depends on the presence of the primary tumor and/or systemic metastasis. Neoplasia. 2007;9:504–510. doi: 10.1593/neo.07241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pikas DS, Li JP, Vlodavsky I, Lindahl U. Substrate specificity of heparanases from human hepatoma and platelets. J Biol Chem. 1998;273:18770–18777. doi: 10.1074/jbc.273.30.18770. [DOI] [PubMed] [Google Scholar]
- 73.Kleeff J, Ishiwata T, Kumbasar A, Friess H, Buchler MW, Lander AD, Korc M. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J Clin Invest. 1998;102:1662–1673. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kayed H, Kleeff J, Keleg S, Jiang X, Penzel R, Giese T, Zentgraf H, Buchler MW, Korc M, Friess H. Correlation of glypican-1 expression with TGF-beta, BMP, and activin receptors in pancreatic ductal adenocarcinoma. Int J Oncol. 2006;29:1139–1148. [PubMed] [Google Scholar]
- 75.Aikawa T, Whipple CA, Lopez ME, Gunn J, Young A, Lander AD, Korc M. Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells. J Clin Invest. 2008;118:89–99. doi: 10.1172/JCI32412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Kleeff J, Kayed H, Felix K, Penzel R, Buchler MW, Korc M, Friess H. Glypican-1 antisense transfection modulates TGF-beta-dependent signaling in Colo-357 pancreatic cancer cells. Biochem Biophys Res Commun. 2004;320:1148–1155. doi: 10.1016/j.bbrc.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 77.Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS ONE. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bloushtain N, Qimron U, Bar-Ilan A, Hershkovitz O, Gazit R, Fima E, Korc M, Vlodavsky I, Bovin NV, Porgador A. Membrane-associated heparan sulfate proteoglycans are involved in the recognition of cellular targets by NKp30 and NKp46. J Immunol. 2004;173:2392–2401. doi: 10.4049/jimmunol.173.4.2392. [DOI] [PubMed] [Google Scholar]
- 79.Conejo JR, Kleeff J, Koliopanos A, Matsuda K, Zhu ZW, Goecke H, Bicheng N, Zimmermann A, Korc M, Friess H, Buchler MW. Syndecan-1 expression is up-regulated in pancreatic but not in other gastrointestinal cancers. Int J Cancer. 2000;88:12–20. doi: 10.1002/1097-0215(20001001)88:1<12::aid-ijc3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 80.Koliopanos A, Friess H, Kleeff J, Shi X, Liao Q, Pecker I, Vlodavsky I, Zimmermann A, Buchler MW. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 2001;61:4655–4659. [PubMed] [Google Scholar]
- 81.Kim AW, Xu X, Hollinger EF, Gattuso P, Godellas CV, Prinz RA. Human heparanase-1 gene expression in pancreatic adenocarcinoma. J Gastrointest Surg. 2002;6:167–172. doi: 10.1016/s1091-255x(01)00087-7. [DOI] [PubMed] [Google Scholar]
- 82.Smetsers TF, van de Westerlo EM, ten Dam GB, Clarijs R, Versteeg EM, van Geloof WL, Veerkamp JH, van Muijen GN, van Kuppevelt TH. Localization and characterization of melanoma-associated glycosaminoglycans: differential expression of chondroitin and heparan sulfate epitopes in melanoma. Cancer Res. 2003;63:2965–2970. [PubMed] [Google Scholar]
- 83.Iozzo RV, Cohen IR, Grassel S, Murdoch AD. The biology of perlecan: the multifaceted heparan sulphate proteoglycan of basement membranes and pericellular matrices. Biochem J. 1994;302(Pt 3):625–639. doi: 10.1042/bj3020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen IR, Murdoch AD, Naso MF, Marchetti D, Berd D, Iozzo RV. Abnormal expression of perlecan proteoglycan in metastatic melanomas. Cancer Res. 1994;54:5771–5774. [PubMed] [Google Scholar]
- 85.Sharma B, Handler M, Eichstetter I, Whitelock JM, Nugent MA, Iozzo RV. Antisense targeting of perlecan blocks tumor growth and angiogenesis in vivo. J Clin Invest. 1998;102:1599–1608. doi: 10.1172/JCI3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murry BP, Greiter-Wilke A, Paulsen DP, Hiatt KM, Beltrami CA, Marchetti D. Selective heparanase localization in malignant melanoma. Int J Oncol. 2005;26:345–352. [PubMed] [Google Scholar]
- 87.Komatsu N, Waki M, Sue M, Tokuda C, Kasaoka T, Nakajima M, Higashi N, Irimura T. Heparanase expression in B16 melanoma cells and peripheral blood neutrophils before and after extravasation detected by novel anti-mouse heparanase monoclonal antibodies. J Immunol Methods. 2008;331:82–93. doi: 10.1016/j.jim.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 88.Roy M, Reiland J, Murry BP, Chouljenko V, Kousoulas KG, Marchetti D. Antisense-mediated suppression of Heparanase gene inhibits melanoma cell invasion. Neoplasia. 2005;7:253–262. doi: 10.1593/neo.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, Spector L, Pecker I. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 90.Furuta J, Umebayashi Y, Miyamoto K, Kikuchi K, Otsuka F, Sugimura T, Ushijima T. Promoter methylation profiling of 30 genes in human malignant melanoma. Cancer Sci. 2004;95:962–968. doi: 10.1111/j.1349-7006.2004.tb03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma YQ, Geng JG. Heparan sulfate-like proteoglycans mediate adhesion of human malignant melanoma A375 cells to P-selectin under flow. J Immunol. 2000;165:558–565. doi: 10.4049/jimmunol.165.1.558. [DOI] [PubMed] [Google Scholar]
- 92.Giordano RJ. Heparanase-2 and syndecan-1 in colon cancer: the ugly ducklings or the beautiful swans? Eur J Gastroenterol Hepatol. 2008;20:716–718. doi: 10.1097/MEG.0b013e3282fc2660. [DOI] [PubMed] [Google Scholar]
- 93.Park H, Kim Y, Lim Y, Han I, Oh ES. Syndecan-2 mediates adhesion and proliferation of colon carcinoma cells. J Biol Chem. 2002;277:29730–29736. doi: 10.1074/jbc.M202435200. [DOI] [PubMed] [Google Scholar]
- 94.Contreras HR, Fabre M, Granes F, Casaroli-Marano R, Rocamora N, Herreros AG, Reina M, Vilaro S. Syndecan-2 expression in colorectal cancer-derived HT-29 M6 epithelial cells induces a migratory phenotype. Biochem Biophys Res Commun. 2001;286:742–751. doi: 10.1006/bbrc.2001.5459. [DOI] [PubMed] [Google Scholar]
- 95.Seko A, Nagata K, Yonezawa S, Yamashita K. Ectopic expression of a GlcNAc 6-O-sulfotransferase, GlcNAc6ST-2, in colonic mucinous adenocarcinoma. Glycobiology. 2002;12:379–388. doi: 10.1093/glycob/12.6.379. [DOI] [PubMed] [Google Scholar]
- 96.Backen AC, Cole CL, Lau SC, Clamp AR, McVey R, Gallagher JT, Jayson GC. Heparan sulphate synthetic and editing enzymes in ovarian cancer. Br J Cancer. 2007;96:1544–1548. doi: 10.1038/sj.bjc.6603747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Friedmann Y, Vlodavsky I, Aingorn H, Aviv A, Peretz T, Pecker I, Pappo O. Expression of heparanase in normal, dysplastic, and neoplastic human colonic mucosa and stroma. Evidence for its role in colonic tumorigenesis. Am J Pathol. 2000;157:1167–1175. doi: 10.1016/S0002-9440(10)64632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vlodavsky I, Elkin M, Pappo O, Aingorn H, Atzmon R, Ishai-Michaeli R, Aviv A, Pecker I, Friedmann Y. Mammalian heparanase as mediator of tumor metastasis and angiogenesis. Isr Med Assoc J. 2000;(2 Suppl):37–45. [PubMed] [Google Scholar]
- 99.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 100.Anttonen A, Leppa S, Heikkila P, Grenman R, Joensuu H. Effect of treatment of larynx and hypopharynx carcinomas on serum syndecan-1 concentrations. J Cancer Res Clin Oncol. 2006;132:451–457. doi: 10.1007/s00432-006-0090-z. [DOI] [PubMed] [Google Scholar]
- 101.Watanabe A, Mabuchi T, Satoh E, Furuya K, Zhang L, Maeda S, Naganuma H. Expression of syndecans, a heparan sulfate proteoglycan, in malignant gliomas: participation of nuclear factor-kappaB in upregulation of syndecan-1 expression. J Neurooncol. 2006;77:25–32. doi: 10.1007/s11060-005-9010-3. [DOI] [PubMed] [Google Scholar]
- 102.Steck PA, Moser RP, Bruner JM, Liang L, Freidman AN, Hwang TL, Yung WK. Altered expression and distribution of heparan sulfate proteoglycans in human gliomas. Cancer Res. 1989;49:2096–2103. [PubMed] [Google Scholar]
- 103.Cohen E, Doweck I, Naroditsky I, Ben-Izhak O, Kremer R, Best LA, Vlodavsky I, Ilan N. Heparanase is overexpressed in lung cancer and correlates inversely with patient survival. Cancer. 2008;113:1004–1011. doi: 10.1002/cncr.23680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Uno F, Fujiwara T, Takata Y, Ohtani S, Katsuda K, Takaoka M, Ohkawa T, Naomoto Y, Nakajima M, Tanaka N. Antisense-mediated suppression of human heparanase gene expression inhibits pleural dissemination of human cancer cells. Cancer Res. 2001;61:7855–7860. [PubMed] [Google Scholar]
- 105.Murry BP, Blust BE, Singh A, Foster TP, Marchetti D. Heparanase mechanisms of melanoma metastasis to the brain: Development and use of a brain slice model. J Cell Biochem. 2006;97:217–225. doi: 10.1002/jcb.20714. [DOI] [PubMed] [Google Scholar]
- 106.Schrage YM, Hameetman L, Szuhai K, Cleton-Jansen AM, Taminiau AH, Hogendoorn PC, Bovee JV. Aberrant heparan sulfate proteoglycan localization, despite normal exostosin, in central chondrosarcoma. Am J Pathol. 2009;174:979–988. doi: 10.2353/ajpath.2009.080623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR, Shridhar V. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 108.Lai JP, Chien J, Strome SE, Staub J, Montoya DP, Greene EL, Smith DI, Roberts LR, Shridhar V. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 109.Chen Z, Fan JQ, Li J, Li QS, Yan Z, Jia XK, Liu WD, Wei LJ, Zhang FZ, Gao H, Xu JP, Dong XM, Dai J, Zhou HM. Promoter hypermethylation correlates with the Hsulf-1 silencing in human breast and gastric cancer. Int J Cancer. 2009;124:739–744. doi: 10.1002/ijc.23960. [DOI] [PubMed] [Google Scholar]
- 110.Inamine M, Nagai Y, Hirakawa M, Mekaru K, Yagi C, Masamoto H, Aoki Y. Heparanase expression in endometrial cancer: analysis of immunohistochemistry. J Obstet Gynaecol. 2008;28:634–637. doi: 10.1080/01443610802323542. [DOI] [PubMed] [Google Scholar]
- 111.Watanabe M, Aoki Y, Kase H, Tanaka K. Heparanase expression and angiogenesis in endometrial cancer. Gynecol Obstet Invest. 2003;56:77–82. doi: 10.1159/000072821. [DOI] [PubMed] [Google Scholar]
- 112.Mikami S, Oya M, Shimoda M, Mizuno R, Ishida M, Kosaka T, Mukai M, Nakajima M, Okada Y. Expression of heparanase in renal cell carcinomas: implications for tumor invasion and prognosis. Clin Cancer Res. 2008;14:6055–6061. doi: 10.1158/1078-0432.CCR-08-0750. [DOI] [PubMed] [Google Scholar]
- 113.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments epidermal growth factor receptor phosphorylation: correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–10085. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Escobar Galvis ML, Jia J, Zhang X, Jastrebova N, Spillmann D, Gottfridsson E, van Kuppevelt TH, Zcharia E, Vlodavsky I, Lindahl U, Li JP. Transgenic or tumor-induced expression of heparanase upregulates sulfation of heparan sulfate. Nat Chem Biol. 2007;3:773–778. doi: 10.1038/nchembio.2007.41. [DOI] [PubMed] [Google Scholar]
- 115.Ravna AW, Sager G. Molecular modeling studies of ABC transporters involved in multidrug resistance. Mini Rev Med Chem. 2009;9:186–193. doi: 10.2174/138955709787316065. [DOI] [PubMed] [Google Scholar]
- 116.Palumbo JS, Talmage KE, Massari JV, La Jeunesse CM, Flick MJ, Kombrinck KW, Jirouskova M, Degen JL. Platelets and fibrin(ogen) increase metastatic potential by impeding natural killer cell-mediated elimination of tumor cells. Blood. 2005;105:178–185. doi: 10.1182/blood-2004-06-2272. [DOI] [PubMed] [Google Scholar]
- 117.dos Santos VM, Rodrigues DB, Castro EC, Saldanha JC, Soares S, Teixeira VP, dos Reis MA. Widespread hematogenous metastases and Trousseau's syndrome in gastric adenocarcinoma. Rev Hosp Clin Fac Med Sao Paulo. 2001;56:91–96. doi: 10.1590/s0041-87812001000300005. [DOI] [PubMed] [Google Scholar]
- 118.Kamocka M, Rozalski M, Krajewska U, Wierzbicki R, Mielicki WP. Effect of cancer procoagulant (CP) on the growth and adhesion of MCF-7 cells to vitronectin in vitro. Cancer Lett. 2005;222:89–94. doi: 10.1016/j.canlet.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 119.Ho-Tin-Noe B, Goerge T, Cifuni SM, Duerschmied D, Wagner DD. Platelet granule secretion continuously prevents intratumor hemorrhage. Cancer Res. 2008;68:6851–6858. doi: 10.1158/0008-5472.CAN-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Probability of Developing Invasive Cancers Over Selected Age Intervals by Sex, US, 2001 to 2003. National Cancer Institute. 2007
- 121.He X, Brenchley PE, Jayson GC, Hampson L, Davies J, Hampson IN. Hypoxia increases heparanase-dependent tumor cell invasion, which can be inhibited by antiheparanase antibodies. Cancer Res. 2004;64:3928–3933. doi: 10.1158/0008-5472.CAN-03-2718. [DOI] [PubMed] [Google Scholar]
- 122.Gingis-Velitski S, Zetser A, Kaplan V, Ben-Zaken O, Cohen E, Levy-Adam F, Bashenko Y, Flugelman MY, Vlodavsky I, Ilan N. Heparanase uptake is mediated by cell membrane heparan sulfate proteoglycans. J Biol Chem. 2004;279:44084–44092. doi: 10.1074/jbc.M402131200. [DOI] [PubMed] [Google Scholar]
- 123.Purushothaman A, Chen L, Yang Y, Sanderson RD. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J Biol Chem. 2008;283:32628–32636. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen L, Sanderson RD. Heparanase regulates levels of syndecan-1 in the nucleus. PLoS ONE. 2009;4:e4947. doi: 10.1371/journal.pone.0004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baek SK, Jung KY, Lee SH, Woo JS, Kwon SY, Chung EJ, Kim TH, Chae YS. Prognostic significance of vascular endothelial growth factor-C expression and lymphatic vessel density in supraglottic squamous cell carcinoma. Laryngoscope. 2009 doi: 10.1002/lary.20483. [DOI] [PubMed] [Google Scholar]
- 126.Saito K, Khan K, Sosnowski B, Li D, O'Malley BW., Jr Cytotoxicity and antiangiogenesis by fibroblast growth factor 2-targeted Ad-TK cancer gene therapy. Laryngoscope. 2009;119:665–674. doi: 10.1002/lary.20127. [DOI] [PubMed] [Google Scholar]
- 127.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim S, Choi JH, Lim HI, Lee SK, Kim WW, Cho S, Kim JS, Kim JH, Choe JH, Nam SJ, Lee JE, Yang JH. EGF-induced MMP-9 expression is mediated by the JAK3/ERK pathway, but not by the JAK3/STAT-3 pathway in a SKBR3 breast cancer cell line. Cell Signal. 2009;21:892–898. doi: 10.1016/j.cellsig.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 129.Chen L, Luo G, Tan Y, Wei J, Wu C, Zheng L, Zhang X, Xu N. Immunolocalisation of tissue factor in esophageal cancer is correlated with intratumoral angiogenesis and prognosis of the patient. Acta Histochem. 2009 doi: 10.1016/j.acthis.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 130.Ferro V, Hammond E, Fairweather JK. The development of inhibitors of heparanase, a key enzyme involved in tumour metastasis, angiogenesis and inflammation. Mini Rev Med Chem. 2004;4:693–702. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- 131.Ferro V, Dredge K, Liu L, Hammond E, Bytheway I, Li C, Johnstone K, Karoli T, Davis K, Copeman E, Gautam A. PI-88 and novel heparan sulfate mimetics inhibit angiogenesis. Semin Thromb Hemost. 2007;33:557–568. doi: 10.1055/s-2007-982088. [DOI] [PubMed] [Google Scholar]
- 132.Trial of PI-88 With Dacarbazine in Patients With Metastatic Melanom. Progen Pharmaceuticals. 2005
- 133.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- 134.Dredge K, Hammond E, Davis K, Li CP, Liu L, Johnstone K, Handley P, Wimmer N, Gonda TJ, Gautam A, Ferro V, Bytheway I. The PG500 series: novel heparan sulfate mimetics as potent angiogenesis and heparanase inhibitors for cancer therapy. Invest New Drugs. 2009 doi: 10.1007/s10637-009-9245-5. [DOI] [PubMed] [Google Scholar]
- 135.Hostettler N, Naggi A, Torri G, Ishai-Michaeli R, Casu B, Vlodavsky I, Borsig L. P-selectin- and heparanase-dependent antimetastatic activity of non-anticoagulant heparins. FASEB J. 2007;21:3562–3572. doi: 10.1096/fj.07-8450com. [DOI] [PubMed] [Google Scholar]
- 136.Casu B, Vlodavsky I, Sanderson RD. Non-anticoagulant heparins and inhibition of cancer. Pathophysiol Haemost Thromb. 2008;36:195–203. doi: 10.1159/000175157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gerotziafas GT, Papageorgiou C, Hatmi M, Samama MM, Elalamy I. Clinical studies with anticoagulants to improve survival in cancer patients. Pathophysiol Haemost Thromb. 2008;36:204–211. doi: 10.1159/000175158. [DOI] [PubMed] [Google Scholar]
- 138.Pumphrey CY, Theus AM, Li S, Parrish RS, Sanderson RD. Neoglycans, carbodiimide-modified glycosaminoglycans: a new class of anticancer agents that inhibit cancer cell proliferation and induce apoptosis. Cancer Res. 2002;62:3722–3728. [PubMed] [Google Scholar]
- 139.Hibino S, Shibuya M, Hoffman MP, Engbring JA, Hossain R, Mochizuki M, Kudoh S, Nomizu M, Kleinman HK. Laminin alpha5 chain metastasis- and angiogenesis-inhibiting peptide blocks fibroblast growth factor 2 activity by binding to the heparan sulfate chains of CD44. Cancer Res. 2005;65:10494–10501. doi: 10.1158/0008-5472.CAN-05-0314. [DOI] [PubMed] [Google Scholar]
- 140.Berry D, Lynn DM, Sasisekharan R, Langer R. Poly(beta-amino ester)s promote cellular uptake of heparin and cancer cell death. Chem Biol. 2004;11:487–498. doi: 10.1016/j.chembiol.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 141.Galcheva-Gargova Z, Zhidkova N, Geisler S, Ozug J, Wudyka S, Gunay NS, Qi YW, Shriver Z, Venkataraman G. Overexpression of Merlin in B16F10 mouse melanoma cells reduces their metastatic activity: role of the cell surface heparan sulfate glycosaminoglycans. Int J Oncol. 2008;32:1237–1243. doi: 10.3892/ijo_32_6_1237. [DOI] [PubMed] [Google Scholar]
- 142.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 143.Marchetti D, Reiland J, Erwin B, Roy M. Inhibition of heparanase activity and heparanase-induced angiogenesis by suramin analogues. Int J Cancer. 2003;104:167–174. doi: 10.1002/ijc.10930. [DOI] [PubMed] [Google Scholar]
- 144.Trosko JE. Review paper: cancer stem cells and cancer nonstem cells: from adult stem cells or from reprogramming of differentiated somatic cells. Vet Pathol. 2009;46:176–193. doi: 10.1354/vp.46-2-176. [DOI] [PubMed] [Google Scholar]
- 145.Dai J, Rabie AB. VEGF: an essential mediator of both angiogenesis and endochondral ossification. J Dent Res. 2007;86:937–950. doi: 10.1177/154405910708601006. [DOI] [PubMed] [Google Scholar]
- 146.Tsunezumi J, Higashi S, Miyazaki K. Matrilysin (MMP-7) cleaves C-type lectin domain family 3 member A (CLEC3A) on tumor cell surface and modulates its cell adhesion activity. J Cell Biochem. 2009;106:693–702. doi: 10.1002/jcb.22062. [DOI] [PubMed] [Google Scholar]
- 147.Manduteanu I, Calb M, Lupu C, Simionescu N, Simionescu M. Increased adhesion of human diabetic platelets to cultured valvular endothelial cells. J Submicrosc Cytol Pathol. 1992;24:539–547. [PubMed] [Google Scholar]
- 148.Gorsi B, Stringer SE. Tinkering with heparan sulfate sulfation to steer development. Trends Cell Biol. 2007;17:173–177. doi: 10.1016/j.tcb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 149.Victor XV, Nguyen TK, Ethirajan M, Tran VM, Nguyen KV, Kuberan B. Investigating the elusive mechanism of glycosaminoglycan biosynthesis. J Biol Chem. 2009 doi: 10.1074/jbc.M109.043208. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Garud DR, Tran VM, Victor XV, Koketsu M, Kuberan B. Inhibition of heparan sulfate and chondroitin sulfate proteoglycan biosynthesis. J Biol Chem. 2008;283:28881–28887. doi: 10.1074/jbc.M805939200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kuberan B, Ethirajan M, Victor XV, Tran V, Nguyen K, Do A. "Click" xylosides initiate glycosaminoglycan biosynthesis in a mammalian cell line. Chembiochem. 2008;9:198–200. doi: 10.1002/cbic.200700494. [DOI] [PubMed] [Google Scholar]
- 152.Choi S, Kim Y, Park H, Han IO, Chung E, Lee SY, Kim YB, Lee JW, Oh ES, Yi JY. Syndecan-2 overexpression regulates adhesion and migration through cooperation with integrin alpha2. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.04.093. [DOI] [PubMed] [Google Scholar]
- 153.Liu D, Shriver Z, Venkataraman G, El Shabrawi Y, Sasisekharan R. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc Natl Acad Sci U S A. 2002;99:568–573. doi: 10.1073/pnas.012578299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Varki A. Six blind men and the elephant--the many faces of heparan sulfate. Proc Natl Acad Sci U S A. 2002;99:543–545. doi: 10.1073/pnas.022649499. [DOI] [PMC free article] [PubMed] [Google Scholar]