Abstract
Distal myopathies are a heterogeneous group of disorders characterized by progressive weakness and muscular atrophy, beginning in distal limb muscles and affecting proximal limb muscles at a later stage. We studied a large German kindred with 10 affected members. Weakness and atrophy of the anterior tibial muscles started between the ages of 8 and 16 years, followed by atrophy of intrinsic hand muscles. Progression was slow, and patients retained the ability to walk until the seventh decade. Serum creatinine kinase levels were increased in the range of 150–1400 U/l. Muscle biopsies showed myopathic changes, whereas immunohistochemistry showed normal expression of marker proteins for muscular dystrophies. Patients had reduced sensation with stocking-glove distribution in the distal limbs in later life. Nerve conduction studies revealed no evidence of neuropathy. Genome-wide linkage analysis in this family revealed a new locus for distal myopathy at 9p21.2-p22.3 (multipoint logarithm of the odds ratio = 4.21). By positional cloning we found a heterozygous mutation L95F in the Kelch-like homologue 9 gene, encoding a bric-a-brac Kelch protein. Molecular modelling indicated that the mutation may interfere with the interaction of the bric-a-brac domain with Cullin 3. Coimmunoprecipitation experiments confirmed that the mutation reduces association with Cullin 3 in the Kelch-like homologue 9-Cullin 3–E3 ubiquitin ligase complex, which is involved in ubiquitin-dependent protein degradation. We identified a unique form of early onset autosomal dominant distal myopathy which is associated with a Kelch-like homologue 9 mutation and interferes with normal skeletal muscle through a novel pathogenetic mechanism.
Keywords: early onset, distal myopathy, bric-a-brac-BACK-kelch protein, Cullin 3, ubiquitin ligase
Introduction
Distal myopathies form a heterogeneous group of disorders characterized by progressive muscular atrophy and weakness, beginning in distal and proceeding to proximal limb muscles. Age at onset, inheritance pattern and clinical as well as histopathological features have been used to distinguish different forms with linkage to different genetic loci or genes. The autosomal dominant inherited distal myopathies are: (i) Welander myopathy, a late adult onset myopathy (MIM 604454) with clinical onset in the hands and linkage to chromosome 2p13 (Ahlberg et al., 1999); (ii) Udd distal myopathy (MIM 600334), which starts with weakness in the anterior compartment of the lower leg and is caused by mutations in the Titin (TTN) (MIM 188840) gene (Haravuori et al., 2001; Hackman et al., 2002); (iii) the late adult onset zaspopathy (Markesbery–Griggs) caused by mutations in the LIM domain binding 3 ZASP (MIM 605906) gene (Griggs et al., 2007), which begins after 40 years of age with ankle dorsiflexion weakness, progressing to finger and wrist extensors; (iv) a new adult onset form caused by mutations in Matrin3 (MATR3) has recently been described (Senderek et al., 2009). A distinguishing feature of MATR3 mutations is vocal cord paralysis and pharnygeal weakness but these were not present in all affected family members; (v) early adult/infantile onset Laing myopathy (MIM 160500), which starts in the anterior compartment of the lower leg, shows autosomal dominant inheritance and is caused by mutations in the tail of myosin heavy chain 7 (MYH7) (MIM 160760) gene located at chromosome 14q11 (Laing et al., 1995; Voit et al., 2001; Meredith et al., 2004); and (vi) Mahjneh et al. (2003) reported a distal myopathy from Finland (MIM 610099) with adult onset and rimmed vacuoles, which showed significant linkage to two separate loci, 8p22-q11 and 12q13-q22 (Mahjneh et al, 2003; Haravuori et al., 2004). The autosomal recessively inherited types include: (i) early onset Nonaka myopathy (MIM 605820), which starts with onset in the distal legs. Mutations in the UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) (MIM 603824) gene on chromosome 9p12-p13 are associated with the hereditary inclusion body myopathy (Eisenberg et al., 2001) (MIM 600737) and are also described in Nonaka myopathy (Kayashima et al., 2002); (ii) early adult onset Miyoshi myopathy (MIM 254130), which affects calf muscles, is caused by mutations in the Dysferlin gene (MIM 603009) on chromosome 2p13 (Liu et al., 1998; Aoki et al., 1999) and is allelic to limb girdle muscular dystrophy 2B (MIM 253601); and (iii) homozygous missense mutations in the Nebulin (NEB) gene which cause childhood to early adulthood onset distal myopathy with ankle dorsiflexor, finger extensor and neck flexor weakness (MIM 161650). The progression is mild, and the patients remain ambulant (Wallgren-Pettersson et al., 2007).
Furthermore, families with uncomplicated distal myopathy but without a known genetic cause have been reported (Udd, 2007, 2009), and several reports describe families with distal myopathy associated with features such as joint contractures and kyphoscoliosis (Bautista et al., 1978), external ophthalmoplegia, weakness of pharynx muscles and respiratory failure (MIM 607569) (Uyama et al., 1998). Interestingly, a family with adult onset dominant distal vacuolar myopathy with additional features including dysphonia, dysphagia, pes cavus and areflexia was mapped to chromosome 19p13.3 (Servidei et al., 1999). This locus has been further confirmed in a family with autosomal dominant distal myopathy, but the additional clinical features were less striking (Di Blasi et al., 2004).
The myofibrillar myopathies are an important differential diagnosis, e.g. the above mentioned zaspopathy, and other gene mutations such as in the Desmin and Myotilin genes, which mainly present as a distal myopathy (Udd, 2007, 2009).
Here, we describe a novel form of early onset distal myopathy with autosomal dominant inheritance in a large German family. Affected family members developed mild sensory disturbances in stocking-glove distribution later in life, a distinguishing feature for distal myopathy. We mapped the disease to a new locus on chromosome 9p21.2-p22.3. Screening of candidate genes resulted in the identification of a heterozygous missense mutation c.796 T>C (NM_018847.1) leading to a p.L95F mutation in the kelch-like homologue-9 (KLHL9) gene, which segregated with the disease phenotype. Molecular modelling and immunoprecipitation studies showed that the mutation reduces interaction with Cullin 3 (Cul3) in the KLHL9–Cul3–Roc1 E3 ubiquitin ligase complex.
Materials and methods
Patients and clinical evaluation
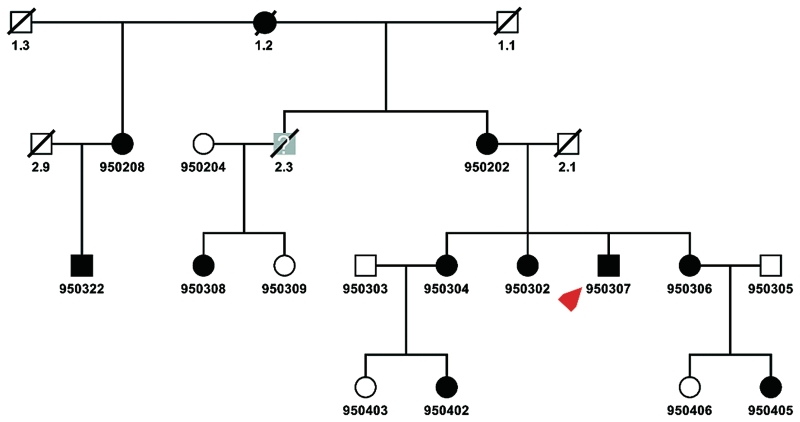
We studied four generations of a German family with distal myopathy (Fig. 1). Ten affected members, three unaffected members and two with unclear status from three generations were examined physically, while the remaining family members were contacted by phone. The muscle biopsies were obtained from the index patient (950307) at the age of 17 years from the gastrocnemius muscle during an Achilles tendon release surgery, and at the age of 32 years from the vastus lateralis muscle via Bergström needle. The biopsy specimens were processed in the neuropathology laboratory using standard protocols for haematoxylin-eosin, nicotinamide adenine dinucleotide and trichrome staining. Immunhistochemistry of muscle sections were performed as described elsewhere (Herrmann et al., 2000). The study was approved by the Ethical Review Board of the University of Essen. Informed consent was obtained from all family members.
Figure 1.
Pedigree. Circles denote females, squares denote males, slashed symbols indicate death, filled symbols indicate the diagnosis of distal myopathy. The index patient is indicated with a red arrow. The symbols filled with grey colour and a question mark indicate that the affection status of these family members could not be identified because of either death or that, at the age of examination, they were considerably younger than the age of onset. All family members with a six-digit identification number were genotyped. This shows the pedigree that was used for the exclusion of distal myopathy loci and the genome scan.
Analysis of linkage with known distal myopathy loci
DNA was isolated from peripheral lymphocytes using standard protocols. For exclusion of previously described loci for distal myopathy we used the following polymorphic microsatellite markers: locus 2p12-p14 (Miyoshi myopathy and Welander myopathy): D2S303, D2S292, D2S291, D2S169 and D2S329; locus 2q31-q33 (Udd myopathy): D2S138, D2S148, D2S300, D2S385, D2S324, D2S2310, D2S1391, D2S152, D2S389 and D2S315; Locus 9p12-p13 (Nonaka myopathy): D9S43, D9S248, D9S165 and D9S50; and locus 14q11 (Laing myopathy): D14S72, D14S283, D14S50, MYH7, D14S64, D14S54 and D14S49. Polymerase chain reactions (PCRs) were performed with fluorescent end-labelled microsatellite markers and semi-automated genotyping was performed on an ABI 377 (Applied Biosystems) Sequencer using the Genotyper V 3.7 (Applied Biosystems) software.
Genome scan
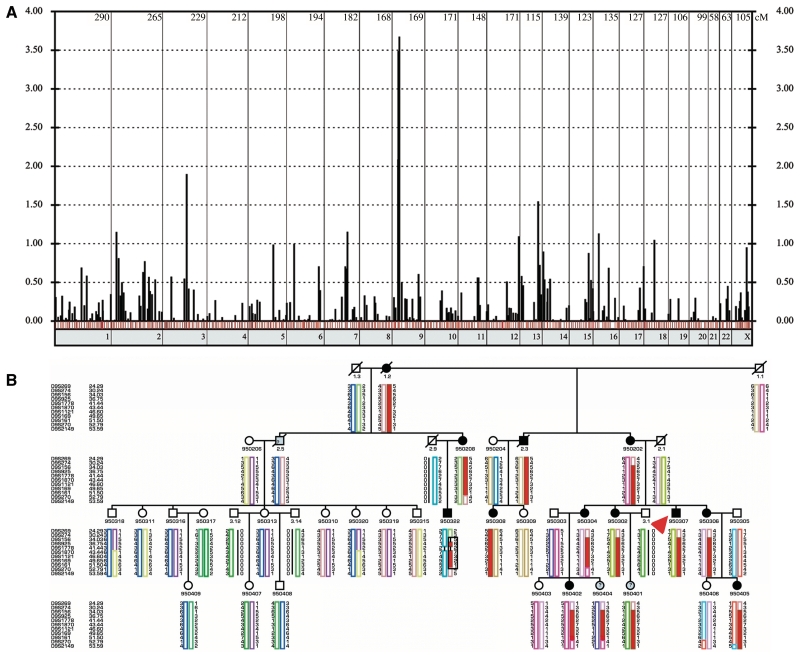
The genome scan was performed using the core pedigree shown in Fig. 1. We performed a genome-wide scan with 439 microsatellite markers that had an average marker density of 7.76 ± 3.80 cm (Marshfield) (Hoffmann et al., 2007). PCRs of fluorescent labelled microsatellite markers were performed as described elsewhere (Thiele et al., 2004). The genotyping was done by a MegaBACE-1000 (Amersham) analysis System using the Genetic Profiler Software 1.5 (Amersham). PedCheck (O'Connell and Weeks, 1998) was used for detection of Mendelian errors. The correct relationships in families were confirmed by the program Graphical Relationship Representation (Abecasis et al., 2001). Two-point linkage analyses were performed using the software FASTLINK v4.1 (Cottingham et al., 1993), multipoint linkage analyses using Genehunter v2.1 (Kruglyak et al., 1996) and Simwalk v2.86 (Sobel and Lange, 1996). We assumed an autosomal-dominant model with 100% penetrance, equally distributed alleles and a disease allele frequency of 0.0001. Haplotype reconstruction was done by Simwalk2 v2.86 and Genehunter v2.1. The fine mapping was performed using the extended pedigree shown in Fig. 3B. Subsequent fine mapping included markers D9S274, D9S156, D9S1778, D9S161 and D9S270.
Figure 3.
Whole genome scan and haplotypes of the disease locus at chromosome 9p21.2-p23.3. (A) Genome-wide two point LOD scores are shown, one bar for every genotyped marker. The red bars indicated a maximum LOD score at recombination fraction θ = 0. The blue bars show markers with a positive LOD score with θ > 0, the black bars stand for markers with negative LOD scores. The numbers of the chromosomes are shown under the x-axis, the marker distances are drawn according to the DeCODE Map. (B) This figure shows the haplotypes of the fine mapping using the extended pedigree. Circles denote females, squares denote males, slashed symbols indicate death, and filled symbols indicate the diagnosis of distal myopathy. The symbols filled with grey colour and a question mark indicate that the affection status of these family members could not be identified because of either death or that at the age of examination, they were considerably younger than the age of onset. All family members with a six-digit identification number were genotyped. The haplotypes were reconstructed using the Simwalk2 program. Uninformative or non-reconstructable markers are indicated with a black bar. The order of the microsatellite makers are from p-telomer to centromer according to the DeCODE Map. The disease haplotype is marked with a black square. Note that all affected family members carry the red haplotype. The index case is indicated with a red arrow.
Sequencing of candidate genes and mutational analysis of KLHL9
The following genes were chosen as candidate genes according to NCBI Human Genome draft number 35: ADAMTSL (a disintegrin and metalloproteinase with thrombospondin motif-like), KIAA1354 (KLHL9), FLJ39267, TEK (endothelial tyrosine kinase) and CCDC2 (coiled coil domain containing 2). Based on the predicted and annotated sequences, primers pairs were designed using the Primer 3 program (Rozen and Skaletsky, 2000) (primer sequences used available on request). All coding exons with exon-intron boundaries were amplified from genomic DNA and both strands of the PCR products were sequenced using Big-Dye Terminator 1.1 (Applied Biosystems) on an ABI 3730xl sequencer (Applied Biosystems). We performed sequence comparisons using the DNASTAR package (Lasergene). Mutations indentified in the KLHL9 gene were confirmed by restriction enzyme digestion with Hind III (Fermentas) with the following primer pair: primer A (forward) CCCTTGATAAGGAAACGAGC, primer B (reverse) TGTTAGCAATTCGTCCAACC, which yielded a 602 bp product. PCR fragments containing the wild-type sequence had two restriction sites resulting in three digestion fragments 106, 207 and 289 bp. The mutation KLHL9 abolishes the last restriction site and results in four digestion fragments of 106, 207, 289 and 496 bp in the heterozygous patients.
Reverse transcriptase polymerase chain reactions
The tissue expression patterns of KLHL9 (NM_018847) and β-actin (NM_001101) as a control (NM_001101) were examined by reverse transcriptase polymerase chain reaction (RT-PCR) using a human 24 tissue rapid-scan cDNA panel (HSCA101; OriGene) with the following PCR primers:
KLHL9—FOR 2146 5′-GACCCCAATTGCCGCCATGTTA-3′;
KLHL9—REV—2340 5′-CAGGCTCGAATGCCACCAAGTG-3′;
β-actin—FOR 736 5′-GGACTTCGAGCAAGAGATGG-3′;
β-actin—REV 969 5′-AGCACTGTGTTGGCGTACAG-3′.
Each 25 µl PCR reaction consisted of: 2.5 µl 10× PCR buffer, 1 pmol forward primer, 1 pmol reverse primer, 1.25 µl 1 mM deoxynucleoside triphosphates, 1.5 mM MgCl2 and five units of Taq polymerase (Invitrogen, USA). The PCR conditions followed the manufacturer’s recommendations: one cycle at 94°C for 3 min; 35 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 2 min; one cycle at 72°C for 5 min; 4°C hold. RT-PCR primers were designed using Primer3 (Rozen and Skaletsky, 2000). The products were resolved by agarose gel electrophoresis, visualized on an ultraviolet transilluminator and the images captured using a charge-coupled device camera system (Fotodyne Inc., USA). The expression levels of KLHL9 and β-Actin in each tissue type were quantified with MultiGauge v. 2.3 (Fujifilm Medical Systems, USA) and the ratios of KLHL9:β-actin expression were calculated and normalized to the expression levels of KLHL9 and β-actin in muscle tissue, which was assigned a value of ‘1’, as shown in Supplementary Table 1.
Molecular modelling
The structural model of the bric-a-brac (BTB)–Cul3 complex was generated through superposition of the Skp1–Cul1 domain (Protein Data Bank id: 1ldj) (Zheng et al., 2002) and the promyelocytic leukemia zinc finger protein BTB domain (Protein Data Bank id: 1buo) (Ahmad et al., 1998) crystal structures (Stogios et al., 2005). Molecular modelling and illustrations were generated with PyMOL (DeLano Scientific) (DeLano, 2003). The location of the KLHL9 p.L95F mutation within the complex was identified by a sequence alignment of the BTB domains from promyelocytic leukemia zinc finger protein and KLHL9.
Expression vectors
The expression vector for human influenza hemagglutinin (HA) Tag-cullin 3 (HA-Cul3) has previously been described (Zhang et al., 2004). The cDNA for human KLHL9 (KIAA1354) was obtained from the Kazusa DNA Research Institute (Chiba, Japan) and sequenced to confirm its identity. The p.L95F amino-acid substitution was introduced with overlapping primers via a PCR-based approach with Pfu Turbo polymerase (Stratagene). For cloning purposes, flanking BamHI and XhoI restriction sites were introduced by PCR with Pfu Turbo polymerase (Stratagene). To express wild-type or L95F mutant KLHL9 as N-terminal FLAG epitope-tagged proteins in mammalian cells, the cDNAs encoding wild-type KLHL9 and mutant L95F KLHL9 were cloned in frame into the pCMV-Tag2A expression vector (Stratagene). The wild-type and mutant p.L95F KLHL9 cDNAs were cloned into the BamHI and XhoI sites of pCMV-Tag2A (Stratagene). The integrity of the entire coding region of both wild-type and p.L95F KLHL9 were verified by sequence analysis.
Human embryonic kidney 293T cell culture and transfections
Human embryonic kidney 293T cells (ATCC CRL-11268) were cultured in a humidified 37°C, 5% CO2 incubator on 0.1% gelatine-coated tissue culture dishes in high glucose (4.5 g/l) Dulbecco’s modified Eagle’s medium supplemented with 2 mM l-glutamine, 10% heat-inactivated foetal bovine serum, amphotericin and gentamycin. For transfections, 9 × 105 human embryonic kidney 293 T cells were seeded onto 0.1% gelatine-coated 60 mm tissue culture dishes on the day prior to transfection. Each 60 mm dish of human embryonic kidney cells was re-fed with 2 ml of Dulbecco’s modified Eagle’s medium lacking any additives and transfected with 3 µg of total plasmid DNA comprised of 750 ng of wild-type or p.L95F KLHL9 and either 0, 83.3, 250, 750 or 2250 ng of HA-Cul3 and empty pcDNA3.1 (Invitrogen) vector up to 3 µg total with 8 µl PLUS reagent (Invitrogen) and 12 µl Lipofectamine (Invitrogen) in the 37°C, 5% CO2 incubator. After a 4 h incubation period, the cells were re-fed with complete Dulbecco’s modified Eagle’s medium and returned to the 37°C, 5% CO2 incubator. Expression of the HA-Cul3, wild-type and p.L95F FLAG-KLHL9 constructs was confirmed by immunoblot analysis.
Immunoprecipitations and immunoblots
At 40 h post-transfection, the transfected cells were washed with cold 1× phosphate buffered saline (pH 7.4) and cell lysates were collected in erythrocyte lysis buffer (50 mM HEPES, pH7.4, 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100) supplemented with 1 mM dithiothreitol, 1 mM NaF, 0.4 mM NaVO3, 1 mM phenylmethanesulphonyl fluoride and Protease Inhibitor Cocktail (Sigma) and clarified by centrifugation at 14 000g for 10 min at 4°C. Total protein concentration was determined by Bradford assay (Bio-Rad). Equivalent amounts of total protein were pre-cleared with protein A agarose (Sigma) for 1 h on the rotator at 4°C and subjected to overnight anti-FLAG M2 (F3165; Sigma) immunoprecipitations at 4°C, on the rotator. The immunoprecipitates were collected with protein A agarose (Sigma), extensively washed with erythrocyte lysis buffer, resolved by sodium dodecyl sulphate polyacrylamide gel electrophoresis, transferred to nitrocellulose and subjected to rabbit anti-HA (Covance) immunoblot analysis using the SuperSignal West Dura Extended Duration Substrate system (Thermo Fisher Scientific) enhanced chemiluminescence system. Images were captured using a charge-coupled device camera system (Fujifilm Medical Systems) and the expression levels of wild-type FLAG-KLHL9, mutant FLAG-L95F-KLHL9 and HA-Cul3 were determined using MultiGauge v. 2.3 (Fujifilm Medical Systems). The results are shown in Supplementary Table 2.
Results
Family and index patient
The pedigree encompassed 33 family members from four generations and indicated a dominant inheritance (Figs 1 and 3B). The family originated from the north of Germany. In total, 15 family members underwent physical examination, 10 of whom were affected. Two unaffected children were younger than the mean age at onset at the time of investigation. Hence their affection status was defined as ‘unknown’ for linkage analysis.
First symptoms of weakness appeared between the ages of 8 and 16 years, resulting in difficulties in walking on heels, followed by selective weakness and slow wasting of ankle extensors leading to a high stepping gait. Progression of weakness was very slow. The proximal muscle groups were generally spared. Common features of the affected family members were ankle contractures, reduced sensation in stocking distribution in lower limbs and weakness in intrinsic hand muscles. The ankle joint contractures might be a secondary phenomenon to drop foot. Some affected family members also had sensory symptoms in the hands. Nerve conduction studies available in some patients did not confirm a definite peripheral neuropathy. The serum creatine kinase levels were elevated and ranged between 200 and 1400 U/L. A summary of the clinical findings in other affected family members is provided in Table 1. The index patient is the most severe case in this family; all other affected family members have milder difficulties.
Table 1.
Clinical details of the affected individuals
| Patient ID | Age at examination (years) | Age of onset (years) | Atrophy of anterior tibial muscles | Contractures of ankles | Reduced sensation | Weakness of intrinsic hand muscles | CK (U/l) | Other features |
|---|---|---|---|---|---|---|---|---|
| 950202 | 63 | ? | + | + | Lower leg, hand | + | 170 | Reduced motor nerve conduction velocity of peroneal nerve (39 m/s) |
| 950206 | 67 | ? | + | + | Lower and upper leg, hand | + | 200 | Deep tendon reflexes in legs not preserved |
| 950302 | 40 | 16 | + | + | Lower leg, hand | − | 110 | |
| 950304 | 39 | 10 | + | + | Lower leg | + | 190 | |
| 950306 | 34 | 13 | − | (+) | Lower leg | (+) | 260 | Reduced amplitude 5.7 mV and increased distal motor latency with 6.9 ms of peroneal nerve with 51 m/s |
| 950308 | 25 | 13 | + | + | Lower leg, hand | + | 140 | |
| 950307 | 25 | 8 | + | + | Lower leg, hand | + | 700–1400 | Deep tendon reflexes in legs reduced, surgical lengthening of the right tendon Achilles at age 17 years Peripheral axonal neuropathy |
| 950402 | 18 | 16 | − | (+) | − | − | 170 | |
| 950405 | 12 | 10 | + | (+) | − | − | 230 |
Patient ID according to pedigree in Fig. 1.
The index patient (950307, Figs 1, 2 and 3) experienced first symptoms at the age of 12 years with foot drop, heel walk difficulties and unsteady gait due to atrophy of anterior tibial muscles. Clinical examination at the age of 16 years showed a ‘naked tibia’ with ankle contractures and distinct atrophy of ankle extensors. Mild intrinsic hand muscle atrophy with formation of a tabatière was noticed at the age of 16 years. At 17 years of age, surgical lengthening of the right Achilles tendon was performed.
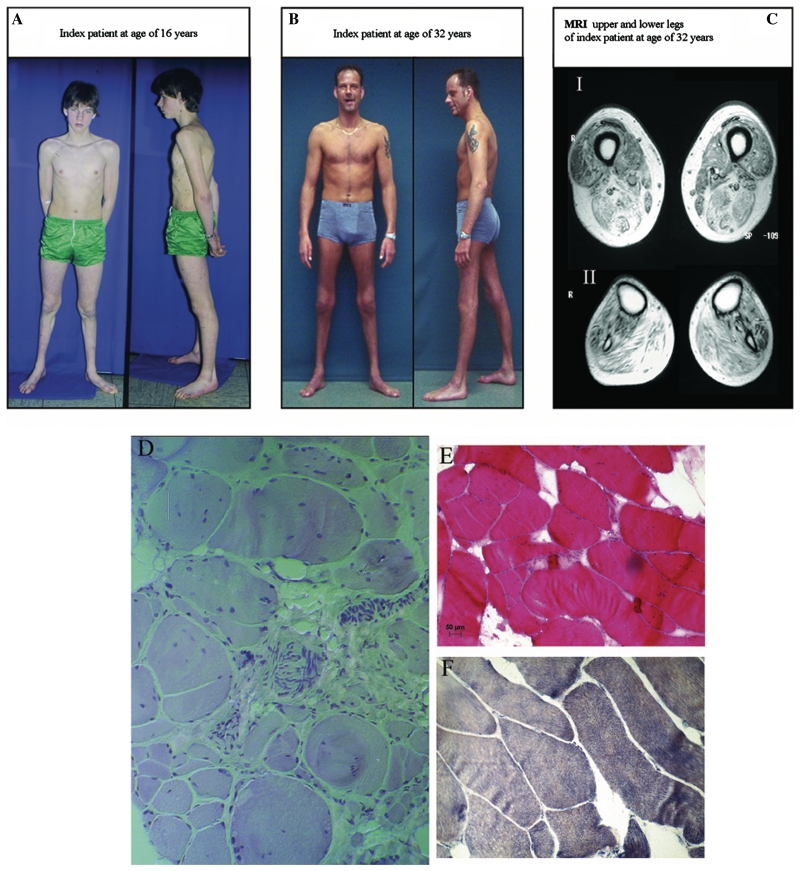
Figure 2.
Clinical information from index Patient 950307. (A) Index patient at the age of 16 years, clearly with atrophy of anterior tibial muscles. (B) Index patient at the age of 32 years with the sign of the ‘naked tibia’ due to distal atrophy. (C) Axial T1-weighted spin echo images (Magnetom Symphony, Siemens, FGR: 1.5 Tesla system, repetition time of 377 ms, echo time 20 ms, two acquisitions, 10 mm slice thickness, field of view of 300 × 300 mm) showing the muscle MRI of distal thigh (I) and proximal lower limb (II). In the lower thigh, striking changes are evident in the semimembranosus, biceps femoris and vastus intermedius muscles. Fatty atrophy in the lower leg was pronounced in the tibialis anterior, gastrocnemius and soleus muscles with relative preservation of tibialis posterior and peroneus longus muscles. (D) Trichrom stain of the muscle biopsy from the gastrocnemius muscle at the age of 15 years showing a myopathic pattern with variation fibre size, atrophic fibres, replacement by fat and connective tissue and internal nuclei. Furthermore some peripheral nerves are visualized, which show normal myelinization. (E) Haematoxylin-eosin stain of the vastus lateralis muscle at the age of 32 years confirming the myopathic features and (F) the nicotinamide adenine dinucleotide stain of the same biopsy showing a loss of fibre typing. All images were captured with 20× objective lens.
Follow-up of the index patient at the age of 32 years showed muscle wasting of the lower limb muscles in particular the tibialis anterior muscles. Dorsal flexion and inversion of the ankles were not possible; flexion and eversion were very weak. There was moderate weakness of limb girdle muscles, particularly of the hip extensors. Hopping on one leg was impossible. The patient was still ambulant, with the use of callipers. Strength of proximal muscle groups in the arms was normal. Impairment of sensation, in particular touch and vibration, in the lower legs (up to the knees) and in the hands was observed. There was a slight postural tremor, and the intrinsic hand muscles showed weakness and atrophy. The Achilles and patellar muscle stretch reflexes could not be elicited. Touch and vibration senses were reduced. Cranial nerve examination was normal. Echocardiography, electrocardiogram and lung function testing were normal.
Nerve conduction studies and electromyography
Table 2 summarizes the results of nerve conduction studies performed in two patients.
Table 2.
Nerve conduction studies
|
Motor conduction studies |
Sensory conduction studies |
|||||||
|---|---|---|---|---|---|---|---|---|
| Age (years) | Distal latency (ms) | Conduction velocity (m/s) | Amplitude (mV) | Age (years) | Conduction velocity (m/s) | Amplitude (µV) | ||
| Patient 950307 | ||||||||
| Median R | 14 | 4.8 | 41 | n. a. | Sural L | 14 | 38 | n.a. |
| Normal valuesa | [3.2±0.7] | [49.6±3.4] | [7.2±1.6] | Normal valuesa | [40.6±4.8] | [18.7±4.4] | ||
| Peroneal L | 14 | 5.8b | 44 | n. a. | Sural L | 31 | 60 | 6.9 |
| Normal valuesa | [3.2±0.7] | [49.6±3.4] | [7.2±1.6] | Normal valuesc | [47.0±4.6] (35.8–62.0) | [17.2±10.1] (1.7–67.3) | ||
| Peroneal L and R | 31 | n.o. | n.o. | n.o. | ||||
| Tibial L | 31 | 3.7 | 55 | 11.8 | ||||
| Tibial R | 31 | 4.5 | 55 | 9.7 | ||||
| Normal valuesb | [4.5 ± 0.8] (3.2–6.9) | [47.0 ± 5.4] (32.7–61.0) | [11.0 ± 5.2] (0.3–25.6) | |||||
| Patient 950306 | ||||||||
| Peroneal L | 34 | 6.9b | 51 | 5.7 | Sural L | 34 | 53 | 13.8 |
| 43 | 5.6 | 59 | 5.5 | 43 | 55 | 18.6 | ||
| Normal valuesc | [4.6 ± 0.7] (3.5–6.4) | [48.6 ± 5.1] (32.4–59.6) | [5.6 ± 2.6] (0.1–12.3) | Normal valuesc | [47.0 ± 4.6] (35.8–62.0) | [17.2 ± 10.1] (1.7–67.3) | ||
| Tibial L | 43 | 4.6 | 60 | 35.1 | Median L | 43 | 62 | 34.6 |
| Normal valuesc | [4.5 ± 0.8] (3.2–6.9) | [47.0 ± 5.4] (32.7–61.0) | [11.0 ± 5.2] (0.3–25.6) | Normal valuesc | [52.0 ± 5.3] (35.4–65.0) | [32.9 ± 17.6] (3.1–86.0) | ||
| Median L | 43 | 3.8 | 52 | 18.4 | Ulnar L | 43 | 53 | 47.6 |
| Normal valuesc | [3.6 ± 0.6] (2.4–5.9) | [56.0 ± 3.9] (43.8–63.5) | [9.1 ± 3.3] (1.7–19.5) | Normal valuesc | [52.4 ± 4.1] (40.3–62.9) | [29.8 ± 17.6] (4.4–90.7) | ||
L = left; R = right; Amplitude = amplitude of compound muscle action potential; n.a.= not available; n.o =not obtained. Normal values are presented as mean ± SD (range). All the reference values and the range are in italics.
a Normal values from Cai and Zhang (1997).
b ≥2 SD from control (increase for distal latencies).
c Normal values from Benatar et al. (2009).
Few distal motor latencies were found to be increased. In one study, the amplitude of the sural sensory nerve action potential was reduced. All other nerve conduction velocities were within normal limits.
An electromyography of anterior tibial, gastrocnemius and interosseus I dorsalis muscles in Patient 950307, performed at 14 years of age, revealed signs compatible with active denervation with fibrillation potentials and positive waves, as well as chronic neurogenic alteration most pronounced in lower limbs. Motor unit potentials were strikingly polyphasic, and recruitment of motor unit potentials was mildly reduced in the interosseus I dorsalis muscle and markedly reduced in tibial and gastrocnemius muscles. These changes might be partly secondary to myopathy and do not prove a peripheral neuropathy.
Imaging (Patient 950307 at 32 years of age)
The muscle MRI (Fig. 2C) with T1 images of the lower limbs showed symmetric fatty atrophy of muscles. In the lower thigh, the most significant changes were seen in the semimembranosus muscle, in the biceps femoris muscle as well as in the vastus intermedius muscle, whereas the lateral and medial vastus muscles and parts of the adductor muscles gracilis and sartorius were preserved. Fatty atrophy of the lower leg was pronounced in the tibialis anterior, gastrocnemius and soleus muscles with relative preservation of the tibialis posterior and peroneus longus muscles (Fig. 2C).
Muscle biopsy (Patient 950307)
A muscle biopsy of the gastrocnemius muscle, taken at age 17 years, showed an increased variation in fibre size with atrophy and hypertrophy, disseminated atrophic fibres, replacement of muscle by fat and connective tissue, and an increase of internalized nuclei. No autophagic vacuoles or inclusions were seen (Fig. 2D). A sural nerve biopsy, taken at 16 years of age, did not show any signs of axonal or demyelinating neuropathy. A second muscle biopsy of the vastus lateralis muscle, at 32 years of age, showed myopathic changes with fibre angulation and size variation, internal nuclei and increased connective tissue (Fig. 2E). The NADH staining showed loss of fibre typing and angulated fibres (Fig. 2F). As the vastus lateralis is less affected then distal muscle groups, it may not show the full range of histopathological changes. Immunocytochemistry showed normal expression of the cell-surface proteins dystrophin, caveolin, laminin α2 and of the dystroclycan-sarcoglycan complex (data not shown).
Linkage analysis
Previously described loci for autosomal dominant distal myopathy on chromosomes 2p12-p14 (Miyoshi- and Welander-myopathy), 2q31-q33 (Udd-myopathy), 9p1-q1 (Nonaka-myopathy) and 14q11 (Laing-myopathy) were excluded by haplotype reconstruction and did not show evidence for linkage [logarithm of the odds ratio (LOD) < −2]. Therefore, we performed a genome wide linkage scan and subsequently identified a single disease region with a maximum two-point parametric LOD = 3.69 (D9S1870) at θ = 0 (Fig. 3A, Table 3). Further fine mapping with an extended pedigree, with additional 10 markers, confirmed the locus at 9p21.2-p22.3 with a multipoint LOD = 4.21 within the interval between D9S1870 and D9S161 (Supplementary Fig. 1). Haplotype analysis revealed critical recombination events in the affected individuals 950 322 (proximal border) and 950304 (distal border), defined D9S274 and D9S270 as the outer border markers of the disease haplotype (Fig. 3B) and comprised 22.59 cM according to the DeCODE Map.
Table 3.
Two point LOD scores generated at various recombination fractions (θ = 0–0.4) for the marker in the region 9p22.3-p21.2
| Marker | cMa | 0 | 0.001 | 0.01 | 0.05 | 0.1 | 0.15 | 0.2 | 0.3 | 0.4 |
|---|---|---|---|---|---|---|---|---|---|---|
| D9S269 | 24.29 | −∞ | −4.842 | −1.889 | −0.006 | 0.62 | 0.861 | 0.936 | 0.828 | 0.504 |
| D9S274 | 30.24 | −4.292 | −3.183 | −1.85 | −0.699 | −0.258 | −0.057 | 0.042 | 0.094 | 0.055 |
| D9S156 | 34.03 | 2.873 | 2.87 | 2.844 | 2.707 | 2.501 | 2.263 | 1.996 | 1.388 | 0.704 |
| D9S925 | 36.75 | 3.458 | 3.452 | 3.397 | 3.148 | 2.824 | 2.485 | 2.13 | 1.372 | 0.589 |
| D9S1778 | 41.44 | 0.32 | 0.319 | 0.311 | 0.273 | 0.227 | 0.184 | 0.142 | 0.069 | 0.017 |
| D9S1870 | 43.44 | 3.694 | 3.688 | 3.629 | 3.361 | 3.012 | 2.645 | 2.259 | 1.425 | 0.554 |
| D9S1121 | 46.6 | 2.185 | 2.179 | 2.128 | 1.897 | 1.598 | 1.289 | 0.973 | 0.37 | −0.003 |
| D9S169 | 49.65 | 2.216 | 2.212 | 2.179 | 2.026 | 1.826 | 1.615 | 1.391 | 0.906 | 0.395 |
| D9S161 | 51.5 | 2.197 | 2.194 | 2.161 | 2.012 | 1.817 | 1.611 | 1.395 | 0.93 | 0.444 |
| D9S270 | 52.79 | −∞ | −0.859 | 0.104 | 0.634 | 0.72 | 0.677 | 0.583 | 0.341 | 0.128 |
| D9S2149 | 53.59 | −∞ | −1.745 | −0.719 | 0.038 | 0.338 | 0.467 | 0.515 | 0.469 | 0.292 |
a Marker position in cM refers to the DeCODE Map.
Candidate gene selection and mutation analysis
For the positional cloning we used the NCBI Human genome Build 35 Version 1. At the time of analysis (August 2004), 81 genes (including pseudogenes and predicted genes) were located between the Marker D9S274 and D9S270. Of these 81 genes, 19 were genes or pseudogenes for interferons, which we did not consider as candidate genes, since the clinical and pathological examinations did not suggest an inflammatory aetiology. The locus contained 19 in silico predicted genes, and 9 transcripts were pseudogenes. We prioritized the remaining 35 genes using the following criteria: (i) expression in skeletal muscle; (ii) sharing of homology and/or functional domains of the known genes for distal myopathies: MYH7, GNE, TTN and dysferlin; (iii) structural proteins of skeletal muscle; and (iv) evidence of a functional role in skeletal muscle. A number of possible candidate genes were identified. ADAMTSL1 is expressed in the extracellular matrix, which is essential for muscle integrity, as exemplified by congenital muscular dystrophy resulting from mutations in collagen VI alpha chains. FLJ39267 contains a myosin tail, and coiled coil domain containing two has 24% homology to the myosin heavy chain. Mutations in the myosin heavy chain 7 tail cause Laing type distal myopathy (Meredith et al., 2004). Endothelial tyrosine kinase contains fibronectin type III domains similar to titin (Hackman et al., 2002), which if mutated causes distal myopathy type Udd. Finally, KLHL9 was selected as candidate gene because it contains six kelch domains that function as oligomeric ring canal actin organizers in Drosophila (Robinson and Cooley, 1997). KLHL9 is also expressed in skeletal muscle according to genetic databases such as Genecards (http://www.genecards.org).
Sequencing coding exons and intron-exon boundaries of the candidate genes ADAMTSL1, FLJ39267, TEK and CCDC2 did not reveal any mutation. In contrast, a heterozygous missense mutation was detected in KLHL9, c.796 T>C (NM_018847.1), leading to a leucine to phenylalanine substitution at amino-acid position 95, situated in the conserved BTB-domain of the KLHL9 protein. This mutation co-segregated with the disease phenotype as shown in Supplementary Fig. 2 and was not observed in 300 healthy unrelated German control individuals.
Alignment of Kelch proteins
KLHL9 is widely distributed across the vertebrates, and genes encoding proteins with >90% sequence identities with human KLHL9 are present in mammalian, fish and bird genomes (Fig. 4D). All of the putative vertebrate KLHL9 orthologues have a leucine at the equivalent position of Leu-85 in KLHL9. Within the human genome, there are 49 BTB-BACK-Kelch (BBK) proteins and KLHL9 is a typical representative of this group (Stogios and Prive, 2004; Stogios et al., 2005). The residue position equivalent to Leu-95 in KLHL9 is conserved in the human BBK proteins, and is typically a leucine, isoleucine or methionine. The homologous position in other family members (Fig. 4C, red box) also shows a high propensity for large, aliphatic amino acids.
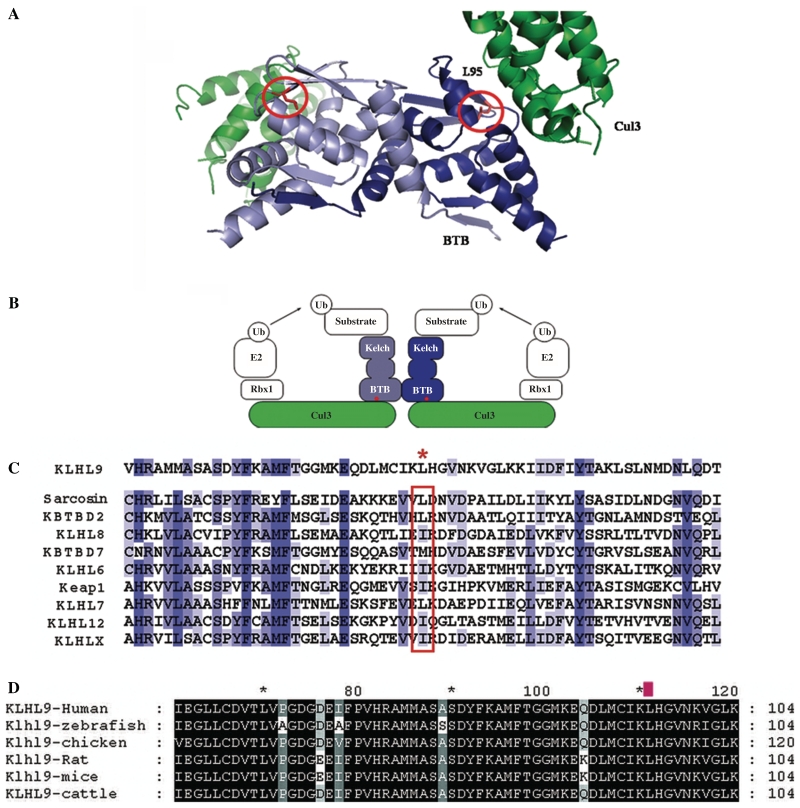
Figure 4.
Structural model of the KLHL9–Cul3 complex and alignment of Kelch proteins. (A) The KLHL9 BTB homodimer is shown in light blue/dark blue ribbons and the N-terminal region of Cul3 is shown as two green ribbons. The complex forms a dimer through self-association of the BTB domain, and each BTB domain interacts with the N-terminus of one Cul3 molecule. The two L95 side chains (circled) in the KLHL9 BTB dimer are in the vicinity of two equivalent and independent Cul3-binding interfaces. (B) Schematic representation of the BTB-BACK-Kelch and Cul3 proteins in the context of a full-length, functional ubiquitin ligase complex. The red dots indicate the approximate position of KLHL9 residue L95. (C) Multiple sequence alignment of the BTB domains from a representative subset of the human BTB-BACK-Kelch proteins, including sequences from Val-65 to Thr-125 in KLHL9. The red asterisk denotes the position of L95 in KLHL9. The homologous position in other family members (red box) shows a high propensity for large, aliphatic amino acids. The blue bars indicate the level of conservation from dark blue (highly conserved) to light blue (less conserved). (D) Alignment of several KLHL9 vertebrate proteins. The L95 residue is labelled with a red mark above and shows 100% conservation in vertebrates.
Molecular modelling
Modelling studies were used to predict the effects of mutation on KLHL9 function. KLHL9 binds to Cul3 via its BTB-domain in the KLHL9–Cul3–Roc1 E3 ubiquitin ligase complex (Furukawa et al., 2003). The homology-based molecular model of the KLHL9/Cul3 complex places residue L95 of KLHL9 near the intermolecular interface with Cul3 (Fig. 4A). Although L95 does not make direct contact with Cul3 and is mostly buried within the BTB domain, this residue is in close proximity of the Cul3 chain, and replacement of an aliphatic residue with a large aromatic residue at this position would probably affect the conformation of the BTB domain, suggesting that the p.L95F mutation could disrupt the KLHL9/Cul3 complex.
Functional analyses
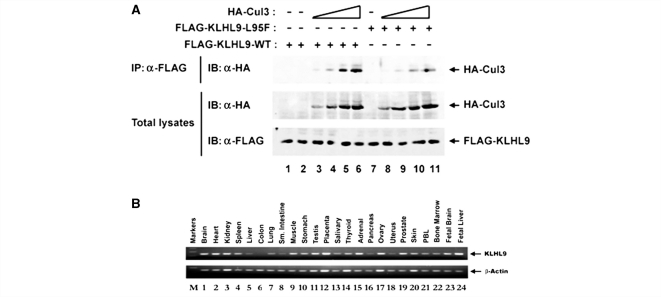
As the p.L95F mutation resides within the BTB domain of KLHL9, we hypothesized that the p.L95F amino-acid substitution reduces association with Cul3. To determine whether the p.L95F mutation within the BTB domain of KLHL9 disrupts association with Cul3, we transfected human embryonic kidney 293T cells with mammalian expression vectors for FLAG epitope-tagged wild-type or p.L95F mutant KLHL9 and increasing amounts of HA-Cul3. The expression levels of HA-Cul3 (Fig. 5A, middle panel) and either wild-type FLAG-KLHL9 (Fig. 5A, lower panel, lanes 1–6) or mutant FLAG-KLHL9-L95F (Fig. 5A, lower panel, lanes 7–11) were confirmed by total α-HA and α-FLAG immunoblots of total lysates, respectively. Our results indicate that while the p.L95F mutation does not alter the total levels of KLHL9, the p.L95F mutation inhibits association with Cul3 (e.g. compare lanes 5–10 and 6–11).
Figure 5.
Functional studies and expression. (A) The L95F mutation in KLHL9 reduces association with Cul3. Human embryonic kidney 293T cells were transfected with wild-type cytomegalovirus FLAG-KLHL9 (lanes 1–6) or mutant cytomegalovirus FLAG-L95F KLHL9 (lanes 7–11) and either 0 ng (lanes 1, 2 and 7), 83.3 ng (lanes 3 and 8), 250 ng (lanes 4 and 9), 750 ng (lanes 5 and 10) or 2250 ng of cytomegalovirus HA-Cul3 (lanes 6 and 11) expression vector. Cell lysates were collected at 40 h post-transfection. Equivalent amounts of total protein were immunoprecipitated with α-FLAG antibody and subjected to an α-HA immunoblot (upper panel). Total α-HA (middle panel) and α-FLAG (lower panel) blots were conducted in parallel to confirm the expression of the respective proteins. Our results clearly demonstrate that the p.L95F mutation reduces association with Cul3 (compare lanes 5 to 10 and 6 to 11, for example). (B) KLHL9 is expressed in multiple tissues. To examine the tissue distribution of KLHL9, a human cDNA panel was screened by RT-PCR. PCR products were analysed on a 2% agarose gel. The results were confirmed to be within the linear range by limited dilution of the cDNA products. Quantification of KLHL9 expression is shown in Supplementary Table 1.
Expression profiling
To examine the tissue distribution of KLHL9, we performed RT-PCR on a human tissue cDNA panel. KLHL9 was expressed in skeletal muscle and multiple tissues at moderate to high levels with the exception of liver, colon, small intestine, the thyroid gland, pancreas and uterus, where KLHL9 was expressed at low levels (Fig. 5B, Supplementary Table 1).
Discussion
In this report, we describe a family with a childhood onset distal myopathy with autosomal dominant inheritance. The phenotype bears similarities to the distal myopathy with childhood onset previously described by Laing and Voit (Laing et al., 1995; Voit et al., 2001; Mastaglia et al., 2002; Meredith et al., 2004; Lamont et al., 2006). Patients with Laing myopathy also have onset of muscle weakness in the anterior lower leg, followed by slowly progressive weakness and involvement of proximal muscles. In both disorders, weakness of finger extensors and hip extensors followed many years later. In contrast to the Laing myopathy phenotype, however, affected members of the family described here did not demonstrate neck flexor weakness or shoulder girdle weakness. The early onset and the slow progression as well as the distribution of affected muscles were also clearly different from the myopathies described by Welander, Markesbery–Griggs, Udd, Nonaka and Miyoshi. Furthermore, the respective loci for these and other relevant conditions were excluded by exclusion mapping. The histopathological investigation of sural nerve was normal at the age of 16 years for the index case, and electrophysiological studies showed only minor abnormalities. Clinical features including areflexia and sensory disturbances in stocking-glove distribution in several family members were distinctive.
A total genome scan identified the disease locus at 9p21.2-p22.3. A number of possible candidate genes were identified in this interval but only the sequencing of the KLHL9 revealed a mutation c.796 T>C (NM_018847.1) leading to a p.L95F amino-acid substitution in the conserved N-terminal BTB-domain, which we consider to be pathogenic.
KLHL9 binds to Cul3 via its BTB domain to form an E3 ubiquitin ligase complex (Furukawa et al., 2003). Interestingly, Cul3 is expressed in highest levels in skeletal muscle tissue (Du et al., 1998), pointing to a myopathological effect of this mutation. The p.L95F mutation is located in the conserved N-terminal BTB-domain of KLHL9, and it is known that the deletion of the BTB-domain of KLHL9 abolishes Cul3 binding (Furukawa et al., 2003). L95 is located near the intermolecular interface with Cul3 (Fig. 4A). A mutation at this position is likely to affect the conformation of this region of the BTB domain. Indeed, pull-down assays revealed that the p.L95F mutation diminishes association with Cul3 in vitro (Fig. 5A). As mentioned above, KLHL9 forms a part of the KLHL9–Cul3–Roc1 E3 ubiquitin ligase complex and functions as a substrate adaptor that targets substrate proteins via its six Kelch domains for ubiquitination by the KLHL9–Cul3–Roc1 E3 ubiquitin ligase (Furukawa et al., 2003). Hence, the targeted substrates of KLHL9 for ubiquitination and later degradation by the proteasome might accumulate, serving as a possible pathophysiological mechanism of the distal myopathy in our patients. It has recently been published that KLHL9 targets the Aurora B kinase on mitotic chromosomes and regulates mitotic progression and completion of cytokinesis in human cells (Sumara et al., 2007). Aurora B regulates the assembly and disassembly of type III intermediate filaments including vimentin, desmin and peripherin as well as the axonally expressed type IV neurofilaments (Sihag et al., 2007). The clinical pattern of distal myopathy and sensory disturbances could be explained by the hypothetical interference of the KLHL9 mutation with the Aurora B kinase function, which is targeted for degradation by KLHL9 (Sumara et al., 2007).
Our hypothesis of Aurora B kinase missregulation certainly needs to be tested in further experiments. The homologue of KLHL9 in the zebrafish klhl and its human orthologue KLHL display conserved expression patterns in skeletal and cardiac muscles (Wu and Gong, 2004). We did not find clinical evidence of cardiac involvement in this family. The facts and the course of the disease in this family suggest that KLHL9 mutations primarily affect muscle function, but it may be, to a lesser extent, important for the conservation of nerve function.
One possible pathophysiological mechanism may be the interaction between KLHL9 and Smad3 (Grimsby et al., 2004), which is a myogenic transcription factor (Liu et al., 2001). The exact role of KLHL9 in muscle and nerve homeostasis remains to be determined.
The KLHL9 protein belongs to the BTB-Kelch family of proteins that includes gigaxonin, a protein which is mutated in giant-axonal neuropathy (MIM 256550) (Bomont et al., 2000). Gigaxonin controls the degradation of microtubule-associated protein 8 (MAP1B) and tubulin-binding cofactor B, and leads to a neurodegenerative disorder of peripheral neuropathy due to impaired retrograde axonal transport and neuronal death (Ding et al., 2006). The gigaxonin gene is as ubiquitously expressed as the KLHL9 gene (Fig. 5B), and it causes a peculiar neurodegenerative disorder affecting the peripheral nerve and CNS. A mutation in KLHL9 leads to distal myopathy with associated sensory disturbances in several affected patients. Interestingly, several E3 ubiquitin ligases such as atrogin-1 and MuRF1 are markedly up-regulated in muscle atrophy (Jackman and Kandarian, 2004). Trim32, another ubiquitin ligase, is mutated in limb girdle muscular dystrophy 2H (MIM 254110) (Kudryashova et al., 2005). Identification of targeted proteins of the KLHL9–Cul3–Roc1 complex may provide further understanding of the molecular pathogenesis of KLHL9-related distal myopathies and also unravel new candidate genes for distal myopathies. We did not find additional cases of KLHL9 mutations in a cohort of 20 cases representing a broad spectrum of clinical presentations of distal myopathy (data not shown). Further families and cases need to be identified to understand the clinical spectrum of KLHL9 myopathy and the neurogenic component of this disease.
In conclusion, we report on a novel form of early onset autosomal dominant distal myopathy. This novel disorder is caused by the heterozygous missense mutation p.L95F of KLHL9. KLHL9 is a substrate adaptor of the Cul3–E3–ubiquitin ligase. The p.L95F mutation is located in the conserved BTB domain of KLHL9, which mediates interaction with Cul3, while the Kelch repeats mediate association with substrates. We were able to demonstrate that the mutation p.L95F diminishes the binding of KLHL9 to Cul3. The KLHL9–Cul3–E3 ubiquitin ligase represents a novel pathway leading to an early onset distal myopathy in contrast to the proteins encoded by other mutated genes in distal myopathies: UDP-N-acetylglucosamine 2-epimerase is an enzyme (Eisenberg et al., 2001); dysferlin is a membrane protein (Liu et al., 1998); titin (Hackman et al., 2002), nebulin (Wallgren-Pettersson et al., 2007) and myosin heavy chain-7 (Meredith et al., 2004) are all sarcomeric proteins (Udd, 2007). We propose that the distal myopathy observed in this German family should be included in the classification schemes of distal myopathies as an early childhood onset distal myopathy, associated with mutations in the KLHL9 gene.
Funding
National Institute of Health grant AT003389; Muscular Dystrophy Campaign to S.C.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Acknowledgements
We are very grateful to Francesco Muntoni and Ludo van der Pol for their constructive comments and support in finalizing this article. We are grateful to Joerg Schaper for his help with the muscle MRI. We thank Hans-Jürgen Christen and Folker Hanefeld for providing clinical data. We are grateful to Zheng Sun for initiating the work on KLHL9 in the laboratory of Mark Hannink. We thank Shih-Ching Lo for assistance with the KLHL9 expression studies.
Glossary
Abbreviations
- BTB
bric-a-brac
- Cul3
Cullin 3
- HA-Cul3
Human influenza hemagglutinin (HA) Tag-cullin3
- KLHL9
kelch-like homologue 9 gene
- RT-PCR
reverse transcriptase polymerase chain reaction
Reference
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–3. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Ahlberg G, von Tell D, Borg K, Edstrom L, Anvret M. Genetic linkage of Welander distal myopathy to chromosome 2p13. Ann Neurol. 1999;46:399–404. doi: 10.1002/1531-8249(199909)46:3<399::aid-ana16>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Ahmad KF, Engel CK, Prive GG. Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci USA. 1998;95:12123–8. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M, Arahata K, Brown RH., Jr [Positional cloning of the gene for Miyoshi myopathy and limb-girdle muscular dystrophy] Rinsho Shinkeigaku. 1999;39:1272–5. [PubMed] [Google Scholar]
- Bautista J, Rafel E, Castilla JM, Alberca R. Hereditary distal myopathy with onset in early infancy. Observation of a family. J Neurol Sci. 1978;37:149–58. doi: 10.1016/0022-510x(78)90199-5. [DOI] [PubMed] [Google Scholar]
- Benatar M, Wuu J, Peng L. Reference data for commonly used sensory and motor nerve conduction studies. Muscle Nerve. 2009;40:772–94. doi: 10.1002/mus.21490. [DOI] [PubMed] [Google Scholar]
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26:370–4. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- Cai F, Zhang J. Study of nerve conduction and late responses in normal Chinese infants, children, and adults. J Child Neurol. 1997;12:13–8. doi: 10.1177/088307389701200102. [DOI] [PubMed] [Google Scholar]
- Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–63. [PMC free article] [PubMed] [Google Scholar]
- DeLano WL, editor. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, California, USA: 2003. [Google Scholar]
- Di Blasi C, Moghadaszadeh B, Ciano C, Negri T, Giavazzi A, Cornelio F, et al. Abnormal lysosomal and ubiquitin-proteasome pathways in 19p13.3 distal myopathy. Ann Neurol. 2004;56:133–8. doi: 10.1002/ana.20158. [DOI] [PubMed] [Google Scholar]
- Ding J, Allen E, Wang W, Valle A, Wu C, Nardine T, et al. Gene targeting of GAN in mouse causes a toxic accumulation of microtubule-associated protein 8 and impaired retrograde axonal transport. Hum Mol Genet. 2006;15:1451–63. doi: 10.1093/hmg/ddl069. [DOI] [PubMed] [Google Scholar]
- Du M, Sansores-Garcia L, Zu Z, Wu KK. Cloning and expression analysis of a novel salicylate suppressible gene, Hs-CUL-3, a member of cullin/Cdc53 family. J Biol Chem. 1998;273:24289–92. doi: 10.1074/jbc.273.38.24289. [DOI] [PubMed] [Google Scholar]
- Eisenberg I, Avidan N, Potikha T, Hochner H, Chen M, Olender T, et al. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat Genet. 2001;29:83–7. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–7. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Griggs R, Vihola A, Hackman P, Talvinen K, Haravuori H, Faulkner G, et al. Zaspopathy in a large classic late-onset distal myopathy family. Brain. 2007;130:1477–84. doi: 10.1093/brain/awm006. [DOI] [PubMed] [Google Scholar]
- Grimsby S, Jaensson H, Dubrovska A, Lomnytska M, Hellman U, Souchelnytskyi S. Proteomics-based identification of proteins interacting with Smad3: SREBP-2 forms a complex with Smad3 and inhibits its transcriptional activity. FEBS Lett. 2004;577:93–100. doi: 10.1016/j.febslet.2004.09.069. [DOI] [PubMed] [Google Scholar]
- Hackman P, Vihola A, Haravuori H, Marchand S, Sarparanta J, De Seze J, et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haravuori H, Makela-Bengs P, Udd B, Partanen J, Pulkkinen L, Somer H, et al. Assignment of the tibial muscular dystrophy locus to chromosome 2q31. Am J Hum Genet. 1998;62:620–6. doi: 10.1086/301752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haravuori H, Siitonen HA, Mahjneh I, Hackman P, Lahti L, Somer H, et al. Linkage to two separate loci in a family with a novel distal myopathy phenotype (MPD3) Neuromuscul Disord. 2004;14:183–7. doi: 10.1016/j.nmd.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Haravuori H, Vihola A, Straub V, Auranen M, Richard I, Marchand S, et al. Secondary calpain3 deficiency in 2q-linked muscular dystrophy: titin is the candidate gene. Neurology. 2001;56:869–77. doi: 10.1212/wnl.56.7.869. [DOI] [PubMed] [Google Scholar]
- Herrmann R, Straub V, Blank M, Kutzick C, Franke N, Jacob EN, et al. Dissociation of the dystroglycan complex in caveolin-3-deficient limb girdle muscular dystrophy. Hum Mol Genet. 2000;9:2335–40. doi: 10.1093/oxfordjournals.hmg.a018926. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Mattheisen M, Dahm S, Nurnberg P, Roe C, Johnson J, et al. A German genome-wide linkage scan for type 2 diabetes supports the existence of a metabolic syndrome locus on chromosome 1p36.13 and a type 2 diabetes locus on chromosome 16p12.2. Diabetologia. 2007;50:1418–22. doi: 10.1007/s00125-007-0658-4. [DOI] [PubMed] [Google Scholar]
- Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287:C834–43. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- Kayashima T, Matsuo H, Satoh A, Ohta T, Yoshiura K, Matsumoto N, et al. Nonaka myopathy is caused by mutations in the UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase gene (GNE) J Hum Genet. 2002;47:77–9. doi: 10.1007/s100380200004. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–63. [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354:413–24. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Laing NG, Laing BA, Meredith C, Wilton SD, Robbins P, Honeyman K, et al. Autosomal dominant distal myopathy: linkage to chromosome 14. Am J Hum Genet. 1995;56:422–7. [PMC free article] [PubMed] [Google Scholar]
- Lamont PJ, Udd B, Mastaglia FL, de Visser M, Hedera P, Voit T, et al. Laing early onset distal myopathy: slow myosin defect with variable abnormalities on muscle biopsy. J Neurol Neurosurg Psychiatry. 2006;77:208–15. doi: 10.1136/jnnp.2005.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15:2950–66. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet. 1998;20:31–6. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- Mastaglia FL, Phillips BA, Cala LA, Meredith C, Egli S, Akkari PA, et al. Early onset chromosome 14-linked distal myopathy (Laing) Neuromuscul Disord. 2002;12:350–7. doi: 10.1016/s0960-8966(01)00287-5. [DOI] [PubMed] [Google Scholar]
- Meredith C, Herrmann R, Parry C, Liyanage K, Dye DE, Durling HJ, et al. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1) Am J Hum Genet. 2004;75:703–8. doi: 10.1086/424760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Senderek J, Garvey SM, Krieger M, Guergueltcheva V, Urtizberea A, Roos A, et al. Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am J Hum Genet. 2009;84:511–8. doi: 10.1016/j.ajhg.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servidei S, Capon F, Spinazzola A, Mirabella M, Semprini S, de Rosa G, et al. A distinctive autosomal dominant vacuolar neuromyopathy linked to 19p13. Neurology. 1999;53:830–7. doi: 10.1212/wnl.53.4.830. [DOI] [PubMed] [Google Scholar]
- Sihag RK, Inagaki M, Yamaguchi T, Shea TB, Pant HC. Role of phosphorylation on the structural dynamics and function of types III and IV intermediate filaments. Exp Cell Res. 2007;313:2098–109. doi: 10.1016/j.yexcr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–37. [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Prive GG. The BACK domain in BTB-kelch proteins. Trends Biochem Sci. 2004;29:634–7. doi: 10.1016/j.tibs.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, et al. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12:887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Thiele H, Sakano M, Kitagawa H, Sugahara K, Rajab A, Hohne W, et al. Loss of chondroitin 6-O-sulfotransferase-1 function results in severe human chondrodysplasia with progressive spinal involvement. Proc Natl Acad Sci USA. 2004;101:10155–60. doi: 10.1073/pnas.0400334101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udd B. Molecular biology of distal muscular dystrophies–sarcomeric proteins on top. Biochim Biophys Acta. 2007;1772:145–58. doi: 10.1016/j.bbadis.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Udd B. 165th ENMC International Workshop: distal myopathies 6-8th February 2009 Naarden, The Netherlands. Neuromuscul Disord. 2009;19:429–38. doi: 10.1016/j.nmd.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Uyama E, Uchino M, Chateau D, Tome FM. Autosomal recessive oculopharyngodistal myopathy in light of distal myopathy with rimmed vacuoles and oculopharyngeal muscular dystrophy. Neuromuscul Disord. 1998;8:119–25. doi: 10.1016/s0960-8966(98)00002-9. [DOI] [PubMed] [Google Scholar]
- Voit T, Kutz P, Leube B, Neuen-Jacob E, Schroder JM, Cavallotti D, et al. Autosomal dominant distal myopathy: further evidence of a chromosome 14 locus. Neuromuscul Disord. 2001;11:11–9. doi: 10.1016/s0960-8966(00)00158-9. [DOI] [PubMed] [Google Scholar]
- Wallgren-Pettersson C, Lehtokari VL, Kalimo H, Paetau A, Nuutinen E, Hackman P, et al. Distal myopathy caused by homozygous missense mutations in the nebulin gene. Brain. 2007;130:1465–76. doi: 10.1093/brain/awm094. [DOI] [PubMed] [Google Scholar]
- Wu YL, Gong Z. A novel zebrafish kelchlike gene klhl and its human ortholog KLHL display conserved expression patterns in skeletal and cardiac muscles. Gene. 2004;338:75–83. doi: 10.1016/j.gene.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.