Abstract
Background
Alcohol abuse and dependence are major causes of morbidity and mortality worldwide, and have a strong familial component. Several linkage and association studies have identified chromosomal regions and/or genes that affect alcohol consumption, notably in genes involved in the two-stage pathway of alcohol metabolism.
Methods
Here, we use multiple regression models to test for associations and interactions between two alcohol related phenotypes and SNPs in 17 genes involved in alcohol metabolism in the U.S. Caucasian subset of the Collaborative Genetic Study of Nicotine Dependence (COGEND) participants.
Results
Several SNPs across six genes showed evidence for association with either maximum number of drinks consumed in a 24-hour period or DSM-IV symptom count. The strongest evidence for association was between rs1229984, a non-synonymous coding SNP in ADH1B, and DSM-IV symptom count (P = 0.0003). This SNP was also associated with maximum drinks (P = 0.0004). Each minor allele at this SNP predicts 45% fewer DSM-IV symptoms and 18% fewer max drinks. Another SNP in a splice site in ALDH1A1 (rs8187974) showed evidence for association with both phenotypes as well. Minor alleles at this SNP predict greater alcohol consumption. In addition, pairwise interactions were observed between SNPs in several genes (P = 0.00002).
Conclusions
We replicated the large effect of rs1229984 on alcohol behavior, and although not common (MAF = 4%), this polymorphism may be highly relevant from a public health perspective in European Americans. Another SNP, rs8187974, may also affect alcohol behavior but requires replication. Also, interactions between polymorphisms in genes involved in alcohol metabolism are likely determinants of the parameters that ultimately affect alcohol consumption.
Keywords: Alcoholism, Alcohol Metabolism, Genetic Association
Introduction
Alcoholism is a chronic, disabling disorder with estimated lifetime prevalence of 19% in men and 8% in women in the U.S. (Grant, 1997). The combination of heavy drinking and smoking represents the single greatest cause of preventable morbidity and mortality in Western societies (Edwards, 2004). In addition to numerous social and environmental factors, variation in susceptibility to alcohol dependence is likely influenced by inter-individual differences in pathways involved in alcohol metabolism and neurotransmission (reviewed in (Tyndale, 2003; Dick and Bierut, 2006)). The primary ethanol metabolism pathway is a two-stage process—first, oxidation to acetaldehyde, catalyzed by alcohol dehydrogenases (ADHs), followed by further oxidation to acetate, catalyzed by aldehyde dehydrogenases (ALDHs). The buildup of acetaldehyde in the blood causes the flushing reaction common in several Asian populations and generally results in lower alcohol consumption (Hurley et al., 2002).
There is substantial evidence for genetic linkage to alcoholism phenotypes at the location of the main ADH gene cluster on chromosome 4 (Williams et al., 1999; Long et al., 1998; Prescott et al., 2006; Corbett et al., 2005), including linkage to maximum number of drinks ever consumed in a 24-hour period (max drinks) (Saccone et al., 2000), and alcohol consumption without dependence (Reich et al., 1998). Max drinks has also been linked to chromosomes 15 and 18 (Kuo et al., 2006). There is also evidence for linkage to maximum daily alcohol consumption on chromosome 9 at the location of ALDH1A1 (Bergen et al., 2003). In addition, variants in five alcohol-metabolizing genes, ADH4, ADH7, ADH1B, ADH1C and ALDH2, have been associated with alcohol-related phenotypes (Hurley et al., 2002; Li, 2000; Edenberg et al., 2006; Kuo et al., 2008; Luo et al., 2006; Luo et al., 2007; Birley et al., 2008; Matsuo et al., 2007; Ehlers et al., 2007).
In this analysis, we evaluated 190 single nucleotide polymorphisms (SNPs) in 17 ethanol metabolism genes and two alcohol phenotypes in a sample of unrelated European American nicotine dependent cases and nicotine exposed, non-nicotine dependent controls. This control group was chosen to increase the likelihood of identifying SNPs associated with the progression from smoking initiation to nicotine dependence.
The primary phenotype analyzed was max drinks, a phenotype related to alcohol metabolism capacity. Although max drinks may reflect an acute event unrepresentative of chronic alcohol consumption, studies have demonstrated max drinks to be an indicator of alcoholism that differs significantly between alcohol dependent subjects with and without a physiological component (Schuckit et al., 1998; Schuckit et al., 1995). In addition, adult twin studies have shown max drinks to have heritability on the order of 50% [A. Heath, unpublished data], making it an attractive phenotype for genetic analysis. We also tested the number of DSM-IV alcohol dependence symptoms endorsed as a dimensional linear predictor, a model shown to best describe the association between dependence criteria and validating variables (Hasin et al., 2006).
Materials and Methods
Participants
Study participants were selected in the U.S. from a community-based sample participating in the Collaborative Genetic Study of Nicotine Dependence (COGEND) from sites in St. Louis, Detroit and Minneapolis. Participants who reported a lifetime smoking history of 100 or more cigarettes were included. The age range for eligible participants was 25–44 years. An initial telephone screen was used to determine nicotine dependence status based on the Fagerstrom Test for Nicotine Dependence (FTND) (Heatherton et al., 1991). Nicotine dependent cases were defined to have a current FTND scores ≥ 4, while non-nicotine dependent controls had a lifetime FTND scores of zero despite smoking more than 100 cigarettes lifetime. This control group was chosen in order to identify genetic determinants of nicotine dependence based on the original goal of the study. For further study details, see (Bierut et al., 2007; Saccone et al., 2007). The Institutional Review Boards at Washington University, University of Minnesota, and Henry Ford Health Sciences Center approved the protocols for this study. Blood samples collected for extraction of DNA were submitted along with electronic phenotypic and genetic data to the National Institute on Drug Abuse (NIDA) Center for Genetic Studies, which manages the sharing of research data according to guidelines of the National Institutes of Health.
Measures
Participants were assessed for nicotine and alcohol dependence. The alcohol related questions were derived from the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA-II). The SSAGA-II is a comprehensive, diagnostic interview which was developed for Collaborative Study on Genetics of Alcoholism (COGA) (Bucholz et al., 1994; Hesselbrock et al., 1999). Participants were assessed for alcohol dependence on DSM-IV classification system and the maximum drinks that a person ingested in a 24 period was queried.
Genotyping
Candidate genes for addiction, including the ADH genes on chromosome 4 and the aldehyde dehydrogenase genes ALDH1A1, ALDH1A3 and ALDH2, were selected by a panel of expert members of the NIDA Genetics Consortium for their putative biological significance in addiction (Saccone et al., 2007). The set of SNPs selected to cover candidate genes was derived from Phase II of the HapMap Project. The number of SNPs genotyped was reduced using linkage disequilibrium (LD) binning using the method described in (Carlson et al., 2004). Briefly, SNPs were grouped into “LD bins” in which at least one tag SNP satisfied r2 ≥ 0.8 with every other SNP in the bin. The candidate gene SNP set included tag SNPs for any LD bin that intersected the physical footprint of a candidate gene. Coverage of additional alcohol metabolizing genes (ALDH1L1, ALDH1L2, ALDH1A2, ALDH3B1, ALDH5A1, ALDH6A1, and ALDH18A1) was attained through the genome-wide component of the study (Bierut et al., 2007). Since these genes were not initially targeted in the candidate gene component of the study, SNP coverage is generally sparser. Because this study is evaluating alcohol-related phenotypes, we have chosen to analyze all available ADH and ALDH genes. Initial SNP genotyping was performed by Perlegen Sciences for the Genome Wide Association and Candidate Gene Studies of Nicotine Dependence and details are available in (Bierut et al., 2007 and Saccone et al., 2007). Additional genotyping was then performed at the Center for Inherited Disease Research (CIDR) to improve coverage of the ADH and ALDH genes. Data cleaning to insure high quality genetic data included a missing genotype rate < 2%. Out of 190 SNPs successfully typed, 15 were not analyzed; two SNPs were out of Hardy-Weinberg equilibrium (P < 0.001), and thirteen had a minor allele frequency less than 1%, leaving 175 SNPs across the seven alcohol dehydrogenase(ADH) and ten aldehyde dehydrogenase (ALDH) genes for association tests.
Statistical Analysis
We tested max drinks and DSM-IV symptom count phenotypes for genetic association. These data were analyzed using linear regression models implemented in SAS v.9.1 and PLINK (Purcell et al., 2007). In all models, genotypic effects were modeled additively as the number of minor alleles increased. Due to its skewed distribution, max drinks were log transformed to better approximate a normal distribution and analyzed in a linear regression model. The number of DSM-IV alcohol dependence criteria endorsed was also analyzed using a linear regression model. To better approximate a normal distribution, the maximum symptom count was set at five, and 60 participants who endorsed six or seven symptoms had their counts truncated at five. In all models, we adjusted for the effects of gender and nicotine dependence, and also for age in the max drinks analysis. Covariates were included in the models if the associated P-values were ≤ 0.05. Eight participants who reported never having consumed an alcoholic beverage were excluded from analysis.
The tests for each SNP by SNP interaction were also done performed using PLINK’s linear regression option and were adjusted for the same covariates as the corresponding main effects models. LD and correlation (r2) between SNPs was measured using Haploview v.3.2 (Barrett et al., 2005).
Haplotype blocks were created if 95% of informative marker comparisons were in “strong LD,” which is defined by the pairwise 95% confidence bounds by the algorithm described in (Gabriel et al., 2002). Haplotypes were tested for association using adjusted haplotype regression models in PLINK, which assigns a probabilistic set of haplotypes for each individual. (Purcell et al., 2007). PLINK was also used to test for population stratification by grouping the sample according to genome-wide average identity by state sharing at more than 32,000 markers spaced across the genome. Stratification is assumed to be present if multiple groups are identified.
We corrected for multiple testing based on the number of independent association tests. To calculate this number, we used the method described in (Li and Ji, 2005). This method adjusts the number of independent tests based on the Eigenvalues of the N x N SNP correlation matrix. We then used a Bonferroni correction based on the number of independent association tests, also factoring in the correlation between the two phenotypes.
Results
A total of 1588 individuals had genotype and phenotype data. The sample was 62.5% female and had a mean age of 36 years. Approximately 12% of the sample was classified as alcohol dependent according to DSM-IV. Figure 1 shows the sample distribution of max drinks, and figure 2 shows the distribution of DSM-IV symptom count. Due to ascertainment, approximately half of the sample was nicotine dependent. Clustering the sample by average genome wide IBD sharing yielded a single cluster, indicating no evidence for population stratification.
Figure 1.
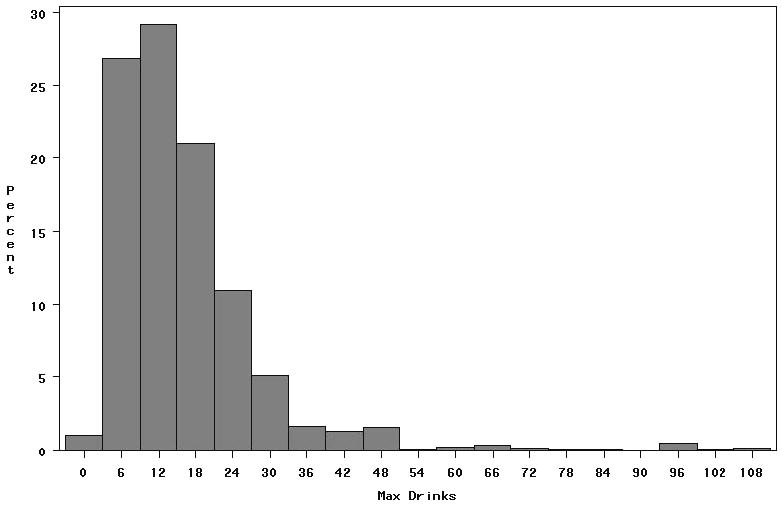
Distribution of maximum lifetime 24-hour drink consumption
Overall mean (SD) = 16.0 (13.7)
Mean (SD) = 22.9 (16.8) in men
Mean (SD) = 11.9 (9.3) in women
The eight participants who reported never having consumed an alcoholic beverage were excluded from analysis.
Figure 2.
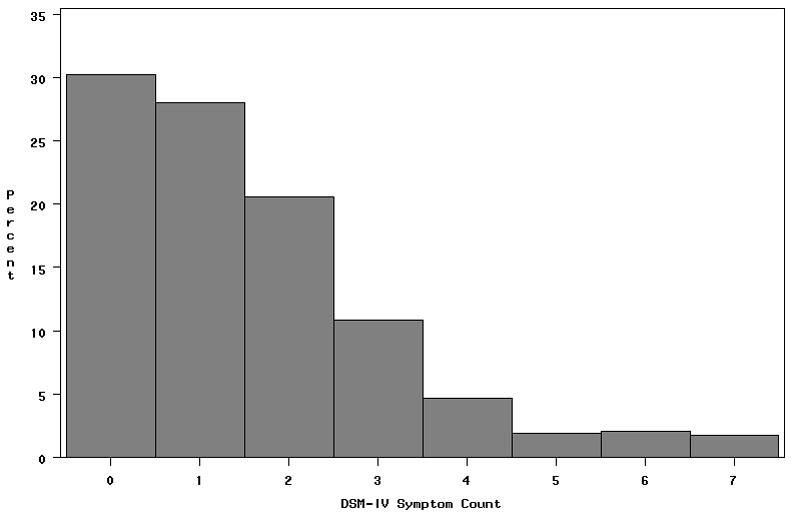
Distribution of DSM-IV alcohol dependence symptom count
Overall mean (SD) = 1.5 (1.6)
Mean (SD) = 1.9 (1.7) in men
Mean (SD) = 1.5 (1.6) in women
Four or more symptoms indicatedependence.
Men reported 10.5 more max drinks than women and endorsed, on average, one half an extra DSM-IV dependence criterion (βmale = 0.51). . Nicotine dependent participants had 3.2 more max drinks than controls endorsed more DSM-IV criteria (βcase = 0.39) greater odds of increased alcohol dependence severity. Older participants reported fewer max drinks. There is substantial correlation between the phenotypes; the Spearman rank order correlation is 0.50 between max drinks and the number of DSM-IV symptoms endorsed (P < .0001).
In total, 175 SNPs in 17 genes on nine chromosomes were tested for association. Several SNPs across six genes showed evidence for association (P < 0.05) with one or both of the phenotypes analyzed. The strongest association was between rs1229984, a non-synonymous coding SNP in ADH1B, and DSM-IV symptom count (P = 0.0003). This SNP was also associated with max drinks (P = 0.0004). For both phenotypes, minor alleles were associated with less severe drinking behavior. In addition, SNP rs8187974 in ALDH1A1 was also nominally associated with both phenotypes. Finally, there was a haplotype of six contiguous and correlated SNPs in ALDH1A2 that were all nominally associated with max drinks (P = 0.01). Table 1 shows the complete single SNP association results for both phenotypes analyzed.
Table 1.
| Chr | SNP | Positon | Gene | Min/Maj | MAF | MaxD (β) | MaxD (P) | DSM4 (β) | DSM4 (P) |
|---|---|---|---|---|---|---|---|---|---|
| 3 | rs2069033 | 127335336 | ALDH1L1 | A/C | 0.47 | −0.02 | 0.42 | 0.01 | 0.89 |
| 4 | rs1230154 | 100207682 | ADH5 | C/T | 0.29 | 0.02 | 0.51 | −0.02 | 0.77 |
| 4 | rs1061187 | 100211876 | ADH5 | A/G | 0.10 | −0.01 | 0.75 | 0.04 | 0.59 |
| 4 | rs6827292 | 100212017 | ADH5 | C/T | 0.04 | −0.03 | 0.56 | 0.10 | 0.40 |
| 4 | rs1803037 | 100212174 | ADH5 | T/C | 0.10 | −0.01 | 0.72 | 0.04 | 0.61 |
| 4 | rs17595424 | 100212892 | ADH5 | T/G | 0.10 | −0.002 | 0.94 | 0.05 | 0.54 |
| 4 | rs7683802 | 100214161 | ADH5 | C/A | 0.10 | −0.01 | 0.85 | 0.05 | 0.56 |
| 4 | rs4699699 | 100216202 | ADH5 | G/C | 0.10 | −0.01 | 0.85 | 0.05 | 0.56 |
| 4 | rs4699700 | 100217358 | ADH5 | C/T | 0.14 | −0.01 | 0.64 | 0.07 | 0.33 |
| 4 | rs1154415 | 100218056 | ADH5 | C/T | 0.42 | 0.004 | 0.85 | −0.04 | 0.44 |
| 4 | rs1154412 | 100220270 | ADH5 | T/C | 0.19 | 0.02 | 0.47 | −0.06 | 0.36 |
| 4 | rs7683704 | 100223249 | ADH5 | A/G | 0.10 | −0.01 | 0.87 | 0.06 | 0.43 |
| 4 | rs1154405 | 100227773 | ADH5 | G/A | 0.28 | 0.01 | 0.54 | −0.01 | 0.92 |
| 4 | rs17216887 | 100229296 | ADH5 | G/C | 0.04 | 0.03 | 0.52 | 0.01 | 0.91 |
| 4 | rs7667261 | 100230323 | ADH5 | G/C | 0.04 | −0.02 | 0.65 | 0.08 | 0.50 |
| 4 | rs2851275 | 100243906 | G/A | 0.43 | 0.01 | 0.79 | −0.04 | 0.44 | |
| 4 | rs1042364 | 100264597 | ADH4 | A/G | 0.29 | 0.02 | 0.41 | −0.01 | 0.86 |
| 4 | rs1126672 | 100266835 | ADH4 | A/G | 0.29 | 0.02 | 0.40 | −0.01 | 0.85 |
| 4 | rs10009145 | 100266952 | ADH4 | A/G | 0.43 | 0.0002 | 0.99 | −0.04 | 0.46 |
| 4 | rs1126671 | 100267437 | ADH4 | A/G | 0.31 | 0.02 | 0.42 | −0.04 | 0.47 |
| 4 | rs1126670 | 100271756 | ADH4 | C/A | 0.31 | 0.02 | 0.44 | −0.04 | 0.45 |
| 4 | rs4699714 | 100279561 | ADH4 | C/T | 0.28 | 0.02 | 0.38 | 0.001 | 0.98 |
| 4 | rs17028609 | 100289908 | ADH4 | A/G | 0.17 | −0.01 | 0.60 | 0.05 | 0.47 |
| 4 | rs1893883 | 100343739 | ADH6 | C/G | 0.45 | −0.001 | 0.95 | −0.07 | 0.16 |
| 4 | rs3857224 | 100348708 | ADH6 | A/G | 0.34 | −0.003 | 0.89 | 0.09 | 0.08 |
| 4 | rs6833176 | 100350186 | ADH6 | C/G | 0.45 | −0.003 | 0.90 | −0.07 | 0.14 |
| 4 | rs4147544 | 100353537 | ADH6 | C/A | 0.47 | −0.003 | 0.88 | −0.05 | 0.28 |
| 4 | rs4699733 | 100356557 | ADH6 | G/C | 0.29 | 0.01 | 0.52 | −0.01 | 0.79 |
| 4 | rs13104485 | 100359846 | ADH6 | A/T | 0.46 | −0.01 | 0.67 | −0.03 | 0.49 |
| 4 | rs3819197 | 100419532 | ADH1A | T/C | 0.25 | −0.01 | 0.70 | 0.04 | 0.45 |
| 4 | rs1229967 | 100426601 | ADH1A | C/G | 0.22 | 0.02 | 0.37 | 0.003 | 0.96 |
| 4 | rs4147532 | 100430809 | ADH1A | C/T | 0.47 | −0.003 | 0.89 | −0.02 | 0.63 |
| 4 | rs4147531 | 100431220 | ADH1A | A/G | 0.47 | −0.004 | 0.84 | −0.02 | 0.61 |
| 4 | rs1229966 | 100432456 | ADH1A | G/A | 0.36 | −0.001 | 0.98 | 0.04 | 0.47 |
| 4 | rs1042026 | 100447489 | ADH1B | G/A | 0.29 | 0.01 | 0.73 | 0.05 | 0.39 |
| 4 | rs17033 | 100447968 | ADH1B | C/T | 0.09 | −0.03 | 0.35 | 0.10 | 0.27 |
| 4 | rs10033960 | 100450365 | ADH1B | A/G | 0.29 | 0.01 | 0.73 | 0.04 | 0.40 |
| 4 | rs1789883 | 100455398 | ADH1B | A/G | 0.03 | −0.03 | 0.66 | −0.21 | 0.13 |
| 4 | rs2066701 | 100457436 | ADH1B | A/G | 0.29 | 0.01 | 0.73 | 0.05 | 0.39 |
| 4 | rs2075633 | 100458021 | ADH1B | G/A | 0.29 | 0.01 | 0.76 | 0.05 | 0.38 |
| 4 | rs1229984 | 100458342 | ADH1B | A/G | 0.04 | −0.19 | 0.0004 | −0.45 | 0.0003 |
| 4 | rs1229983 | 100459025 | ADH1B | C/T | 0.04 | −0.04 | 0.52 | −0.27 | 0.04 |
| 4 | rs1235416 | 100459944 | ADH1B | T/G | 0.03 | −0.03 | 0.58 | −0.22 | 0.11 |
| 4 | rs1353621 | 100460598 | ADH1B | C/T | 0.37 | 0.01 | 0.70 | −0.04 | 0.43 |
| 4 | rs1159918 | 100462032 | ADH1B | T/G | 0.36 | −0.01 | 0.76 | 0.01 | 0.83 |
| 4 | rs6810842 | 100462468 | ADH1B | T/G | 0.37 | −0.02 | 0.30 | −0.001 | 0.99 |
| 4 | rs1229982 | 100462955 | ADH1B | A/C | 0.19 | −0.03 | 0.22 | −0.002 | 0.98 |
| 4 | rs1789896 | 100476007 | ADH1C | G/A | 0.47 | −0.004 | 0.86 | −0.04 | 0.46 |
| 4 | rs2298753 | 100476930 | ADH1C | C/T | 0.10 | −0.005 | 0.90 | −0.01 | 0.86 |
| 4 | rs1614972 | 100477178 | ADH1C | A/G | 0.30 | −0.01 | 0.73 | 0.06 | 0.23 |
| 4 | rs1789898 | 100477359 | ADH1C | C/A | 0.41 | 0.01 | 0.61 | −0.03 | 0.55 |
| 4 | rs1662060 | 100478864 | ADH1C | G/A | 0.41 | 0.01 | 0.51 | −0.03 | 0.56 |
| 4 | rs698 | 100479812 | ADH1C | G/A | 0.41 | 0.01 | 0.49 | −0.03 | 0.57 |
| 4 | rs904096 | 100482607 | ADH1C | G/T | 0.41 | 0.01 | 0.51 | −0.03 | 0.57 |
| 4 | rs2241894 | 100485156 | ADH1C | G/A | 0.23 | −0.01 | 0.57 | 0.06 | 0.28 |
| 4 | rs1789921 | 100488225 | ADH1C | A/G | 0.30 | 0.02 | 0.45 | −0.03 | 0.58 |
| 4 | rs3133158 | 100489635 | ADH1C | G/C | 0.30 | 0.02 | 0.40 | −0.04 | 0.51 |
| 4 | rs17586023 | 100491973 | ADH1C | G/A | 0.10 | −0.004 | 0.91 | −0.01 | 0.90 |
| 4 | rs4147541 | 100493180 | ADH1C | G/C | 0.26 | −0.01 | 0.82 | 0.06 | 0.28 |
| 4 | rs1789924 | 100493309 | ADH1C | T/C | 0.41 | 0.01 | 0.56 | −0.03 | 0.60 |
| 4 | rs11499823 | 100493772 | ADH1C | G/A | 0.10 | −0.002 | 0.95 | −0.003 | 0.97 |
| 4 | rs1908965 | 100495016 | ADH1C | A/G | 0.41 | 0.01 | 0.53 | −0.03 | 0.58 |
| 4 | rs1662037 | 100497842 | ADH1C | A/G | 0.30 | 0.02 | 0.46 | −0.03 | 0.59 |
| 4 | rs284793 | 100549317 | ADH7 | A/G | 0.30 | −0.04 | 0.09 | −0.06 | 0.26 |
| 4 | rs729147 | 100552290 | ADH7 | C/T | 0.21 | −0.005 | 0.85 | −0.07 | 0.22 |
| 4 | rs284787 | 100552579 | ADH7 | A/G | 0.25 | −0.05 | 0.03 | −0.10 | 0.07 |
| 4 | rs894369 | 100552869 | ADH7 | C/G | 0.21 | −0.01 | 0.81 | −0.08 | 0.18 |
| 4 | rs3805329 | 100552635 | ADH7 | C/T | 0.06 | 0.02 | 0.60 | 0.09 | 0.38 |
| 4 | rs3805331 | 100552955 | ADH7 | G/A | 0.06 | 0.03 | 0.55 | 0.10 | 0.33 |
| 4 | rs284786 | 100553000 | ADH7 | A/T | 0.31 | −0.04 | 0.09 | −0.06 | 0.24 |
| 4 | rs17588403 | 100553472 | ADH7 | T/A | 0.18 | 0.004 | 0.88 | 0.03 | 0.62 |
| 4 | rs284784 | 100554897 | ADH7 | A/C | 0.25 | −0.05 | 0.04 | −0.10 | 0.09 |
| 4 | rs2851011 | 100555006 | ADH7 | T/C | 0.21 | −0.01 | 0.84 | −0.08 | 0.18 |
| 4 | rs1827567 | 100555125 | ADH7 | A/G | 0.25 | −0.05 | 0.03 | −0.10 | 0.08 |
| 4 | rs284779 | 100557284 | ADH7 | C/G | 0.47 | 0.04 | 0.05 | −0.03 | 0.56 |
| 4 | rs1154454 | 100557365 | ADH7 | C/T | 0.17 | −0.01 | 0.84 | −0.06 | 0.34 |
| 4 | rs2584464 | 100558072 | ADH7 | A/G | 0.49 | −0.03 | 0.14 | −0.05 | 0.27 |
| 4 | rs1154458 | 100559545 | ADH7 | C/G | 0.42 | 0.02 | 0.27 | 0.06 | 0.28 |
| 4 | rs1154459 | 100560126 | ADH7 | A/G | 0.33 | −0.01 | 0.74 | −0.08 | 0.15 |
| 4 | rs1154460 | 100560666 | ADH7 | T/C | 0.45 | −0.01 | 0.61 | −0.09 | 0.07 |
| 4 | rs971074 | 100560884 | ADH7 | T/C | 0.12 | −0.01 | 0.75 | −0.05 | 0.51 |
| 4 | rs1573496 | 100568692 | ADH7 | G/C | 0.11 | −0.02 | 0.48 | −0.03 | 0.70 |
| 4 | rs1154468 | 100573280 | ADH7 | T/A | 0.33 | −0.01 | 0.78 | −0.07 | 0.16 |
| 4 | rs17529530 | 100574513 | ADH7 | A/T | 0.04 | −0.07 | 0.21 | −0.06 | 0.64 |
| 4 | rs4147549 | 100575035 | ADH7 | A/G | 0.01 | 0.05 | 0.57 | −0.13 | 0.56 |
| 4 | rs1154470 | 100575360 | ADH7 | T/C | 0.33 | −0.01 | 0.74 | −0.07 | 0.16 |
| 4 | rs1530320 | 100585664 | C/T | 0.12 | −0.03 | 0.35 | 0.04 | 0.64 | |
| 4 | rs749407 | 100586814 | T/C | 0.38 | −0.01 | 0.77 | −0.07 | 0.17 | |
| 4 | rs2119888 | 100599171 | G/A | 0.49 | −0.02 | 0.46 | −0.08 | 0.10 | |
| 6 | rs2744605 | 24652311 | ALDH5A1 | A/G | 0.04 | 0.12 | 0.03 | −0.03 | 0.81 |
| 9 | rs8188000 | 72745223 | ALDH1A1 | C/T | 0.01 | 0.09 | 0.31 | 0.21 | 0.32 |
| 9 | rs8187999 | 72745363 | ALDH1A1 | G/C | 0.03 | −0.05 | 0.49 | −0.08 | 0.60 |
| 9 | rs8187981 | 72751492 | ALDH1A1 | C/T | 0.50 | −0.01 | 0.48 | −0.01 | 0.84 |
| 9 | rs8187974 | 72756420 | ALDH1A1 | A/C | 0.01 | 0.32 | 0.004 | 0.56 | 0.02 |
| 9 | rs7043217 | 72772449 | ALDH1A1 | G/A | 0.46 | 0.04 | 0.05 | 0.04 | 0.39 |
| 9 | rs8187915 | 72775436 | ALDH1A1 | G/A | 0.49 | 0.03 | 0.15 | 0.04 | 0.45 |
| 9 | rs348462 | 72776723 | ALDH1A1 | C/G | 0.30 | 0.01 | 0.69 | 0.001 | 0.98 |
| 9 | rs3909559 | 72789123 | ALDH1A1 | T/G | 0.41 | 0.03 | 0.21 | 0.01 | 0.78 |
| 9 | rs7858367 | 72824110 | ALDH1A1 | G/T | 0.43 | 0.02 | 0.28 | 0.01 | 0.86 |
| 9 | rs348483 | 74704934 | ALDH1A1 | C/T | 0.15 | 0.02 | 0.57 | 0.03 | 0.61 |
| 9 | rs348484 | 74705145 | ALDH1A1 | G/A | 0.15 | 0.02 | 0.55 | 0.04 | 0.56 |
| 9 | rs3764435 | 74706696 | ALDH1A1 | C/A | 0.49 | −0.02 | 0.37 | −0.01 | 0.82 |
| 9 | rs1888202 | 74709071 | ALDH1A1 | G/C | 0.49 | 0.01 | 0.48 | 0.01 | 0.78 |
| 9 | rs348472 | 74710880 | ALDH1A1 | A/G | 0.05 | −0.004 | 0.93 | 0.05 | 0.66 |
| 9 | rs17058224 | 74712655 | ALDH1A1 | G/T | 0.05 | 0.01 | 0.84 | 0.08 | 0.47 |
| 9 | rs10781106 | 74712742 | ALDH1A1 | G/A | 0.50 | −0.01 | 0.56 | −0.005 | 0.92 |
| 9 | rs63319 | 74714604 | ALDH1A1 | C/A | 0.50 | −0.01 | 0.56 | −0.01 | 0.89 |
| 9 | rs348475 | 74715699 | ALDH1A1 | G/A | 0.50 | −0.01 | 0.57 | −0.01 | 0.87 |
| 9 | rs348458 | 74719708 | ALDH1A1 | T/C | 0.44 | 0.03 | 0.16 | 0.01 | 0.77 |
| 9 | rs348457 | 74720374 | ALDH1A1 | C/G | 0.44 | 0.03 | 0.14 | 0.02 | 0.69 |
| 9 | rs610529 | 74723144 | ALDH1A1 | G/A | 0.44 | 0.03 | 0.14 | 0.02 | 0.72 |
| 9 | rs348460 | 74724580 | ALDH1A1 | A/C | 0.06 | 0.02 | 0.62 | 0.10 | 0.38 |
| 9 | rs348459 | 74727410 | ALDH1A1 | G/A | 0.06 | 0.02 | 0.72 | 0.08 | 0.46 |
| 9 | rs2161811 | 74730007 | ALDH1A1 | T/C | 0.49 | 0.03 | 0.11 | 0.04 | 0.46 |
| 9 | rs2303317 | 74731762 | ALDH1A1 | C/A | 0.49 | 0.03 | 0.11 | 0.03 | 0.49 |
| 9 | rs2288087 | 74733858 | ALDH1A1 | A/T | 0.49 | 0.03 | 0.11 | 0.03 | 0.49 |
| 9 | rs722921 | 74734119 | ALDH1A1 | A/T | 0.49 | 0.03 | 0.13 | 0.03 | 0.52 |
| 9 | rs2017362 | 74734211 | ALDH1A1 | A/G | 0.36 | 0.02 | 0.40 | −0.01 | 0.90 |
| 9 | rs8187919 | 74734617 | ALDH1A1 | G/A | 0.03 | −0.06 | 0.38 | 0.004 | 0.98 |
| 9 | rs348461 | 74734890 | ALDH1A1 | A/T | 0.36 | 0.02 | 0.40 | −0.01 | 0.90 |
| 9 | rs3815836 | 74735458 | ALDH1A1 | T/G | 0.49 | 0.03 | 0.10 | 0.04 | 0.46 |
| 9 | rs348463 | 74737432 | ALDH1A1 | G/A | 0.27 | 0.01 | 0.69 | −0.04 | 0.52 |
| 9 | rs7851899 | 74740495 | ALDH1A1 | C/T | 0.04 | −0.05 | 0.40 | 0.10 | 0.46 |
| 9 | rs1330286 | 74742773 | ALDH1A1 | C/G | 0.33 | −0.01 | 0.79 | −0.03 | 0.55 |
| 9 | rs8187895 | 74746729 | ALDH1A1 | G/A | 0.08 | 0.06 | 0.11 | 0.08 | 0.36 |
| 9 | rs8187890 | 74749669 | ALDH1A1 | C/T | 0.08 | 0.06 | 0.11 | 0.08 | 0.38 |
| 9 | rs1424482 | 74753377 | ALDH1A1 | G/A | 0.35 | 0.00 | 1.00 | −0.03 | 0.57 |
| 9 | rs8187876 | 74754774 | ALDH1A1 | A/G | 0.05 | 0.07 | 0.12 | 0.06 | 0.58 |
| 10 | rs12359272 | 97355153 | ALDH18A1 | A/G | 0.35 | 0.00 | 0.87 | 0.04 | 0.41 |
| 10 | rs11188394 | 97355448 | ALDH18A1 | C/T | 0.36 | 0.004 | 0.84 | 0.04 | 0.43 |
| 10 | rs10882640 | 97359929 | ALDH18A1 | T/C | 0.49 | 0.01 | 0.81 | 0.03 | 0.52 |
| 10 | rs943343 | 97360233 | ALDH18A1 | A/G | 0.48 | 0.005 | 0.83 | 0.04 | 0.45 |
| 10 | rs10882644 | 97378569 | ALDH18A1 | T/C | 0.35 | 0.01 | 0.73 | 0.04 | 0.42 |
| 10 | rs11188411 | 97387334 | ALDH18A1 | C/T | 0.35 | 0.01 | 0.73 | 0.04 | 0.39 |
| 10 | rs10882649 | 97416076 | ALDH18A1 | G/A | 0.34 | 0.003 | 0.90 | 0.02 | 0.72 |
| 11 | rs2075626 | 67556502 | ALDH3B1 | C/T | 0.24 | 0.002 | 0.92 | 0.03 | 0.61 |
| 12 | rs4281549 | 103930831 | ALDH1L2 | G/T | 0.13 | −0.01 | 0.74 | 0.06 | 0.39 |
| 12 | rs4964316 | 103935544 | ALDH1L2 | C/T | 0.38 | 0.01 | 0.50 | 0.08 | 0.13 |
| 12 | rs10861340 | 103956564 | ALDH1L2 | G/A | 0.18 | −0.03 | 0.25 | −0.07 | 0.26 |
| 12 | rs886205 | 110667147 | ALDH2 | G/A | 0.17 | −0.03 | 0.34 | 0.04 | 0.57 |
| 12 | rs16941667 | 110707133 | ALDH2 | T/C | 0.07 | −0.06 | 0.11 | −0.07 | 0.43 |
| 14 | rs9652369 | 73599413 | ALDH6A1 | G/T | 0.46 | −0.001 | 0.96 | −0.03 | 0.56 |
| 14 | rs12587903 | 73611071 | ALDH6A1 | C/T | 0.15 | −0.01 | 0.66 | −0.03 | 0.61 |
| 15 | rs1837853 | 56050473 | ALDH1A2 | G/C | 0.03 | −0.15 | 0.01 | −0.10 | 0.49 |
| 15 | rs1964429 | 56058994 | ALDH1A2 | T/G | 0.04 | −0.14 | 0.01 | −0.12 | 0.33 |
| 15 | rs2012147 | 56059036 | ALDH1A2 | A/G | 0.04 | −0.14 | 0.01 | −0.13 | 0.32 |
| 15 | afd3555077 | 56062530 | ALDH1A2 | G/C | 0.03 | −0.16 | 0.01 | −0.13 | 0.38 |
| 15 | rs8041922 | 56063468 | ALDH1A2 | C/A | 0.03 | −0.15 | 0.01 | −0.05 | 0.71 |
| 15 | rs2218261 | 56081314 | ALDH1A2 | T/C | 0.03 | −0.15 | 0.01 | −0.10 | 0.49 |
| 15 | rs4646561 | 56140802 | ALDH1A2 | C/T | 0.07 | 0.03 | 0.55 | 0.001 | 0.99 |
| 15 | rs3809523 | 99236205 | ALDH1A3 | T/C | 0.14 | 0.05 | 0.09 | 0.10 | 0.15 |
| 15 | rs4646649 | 99240250 | ALDH1A3 | T/C | 0.17 | 0.04 | 0.12 | 0.05 | 0.48 |
| 15 | rs4646651 | 99240989 | ALDH1A3 | G/C | 0.17 | 0.04 | 0.17 | 0.04 | 0.55 |
| 15 | rs9944290 | 99242258 | ALDH1A3 | T/C | 0.18 | 0.04 | 0.17 | 0.05 | 0.42 |
| 15 | rs4646660 | 99245521 | ALDH1A3 | C/A | 0.01 | −0.09 | 0.39 | 0.28 | 0.23 |
| 15 | rs4646662 | 99249217 | ALDH1A3 | G/A | 0.16 | 0.05 | 0.06 | 0.03 | 0.68 |
| 15 | rs4646669 | 99253830 | ALDH1A3 | T/C | 0.16 | 0.05 | 0.07 | 0.005 | 0.94 |
| 15 | rs7182884 | 99256385 | ALDH1A3 | C/A | 0.41 | −0.002 | 0.91 | 0.10 | 0.04 |
| 15 | rs4646672 | 99257737 | ALDH1A3 | T/C | 0.03 | −0.02 | 0.77 | −0.005 | 0.97 |
| 15 | rs4646678 | 99259959 | ALDH1A3 | A/G | 0.18 | 0.05 | 0.09 | −0.01 | 0.89 |
| 15 | rs4246323 | 99260525 | ALDH1A3 | T/C | 0.03 | −0.02 | 0.74 | 0.01 | 0.92 |
| 15 | rs4246326 | 99260953 | ALDH1A3 | A/G | 0.03 | −0.01 | 0.82 | 0.001 | 1.00 |
| 15 | rs4646681 | 99262441 | ALDH1A3 | G/C | 0.03 | −0.02 | 0.70 | 0.01 | 0.94 |
| 15 | rs3803430 | 99263338 | ALDH1A3 | G/A | 0.03 | −0.02 | 0.68 | 0.07 | 0.62 |
| 15 | rs4246328 | 99263793 | ALDH1A3 | G/A | 0.48 | −0.03 | 0.16 | −0.10 | 0.03 |
| 15 | rs11854028 | 99265497 | ALDH1A3 | G/T | 0.44 | −0.03 | 0.17 | −0.11 | 0.03 |
| 15 | rs4646688 | 99267747 | ALDH1A3 | G/T | 0.44 | −0.03 | 0.19 | −0.10 | 0.04 |
| 15 | rs12914598 | 99268002 | ALDH1A3 | T/C | 0.18 | 0.04 | 0.17 | −0.02 | 0.73 |
| 15 | rs3803428 | 99268433 | ALDH1A3 | C/T | 0.44 | −0.03 | 0.15 | −0.10 | 0.05 |
| 15 | rs4646690 | 99272532 | ALDH1A3 | C/G | 0.04 | −0.03 | 0.53 | 0.003 | 0.98 |
| 15 | rs3803426 | 99272709 | ALDH1A3 | T/C | 0.03 | −0.01 | 0.83 | −0.05 | 0.72 |
| 15 | rs1130738 | 99273363 | ALDH1A3 | G/A | 0.44 | −0.02 | 0.25 | −0.11 | 0.02 |
| 15 | rs14226 | 99273840 | ALDH1A3 | A/G | 0.18 | 0.04 | 0.10 | −0.001 | 0.98 |
| 15 | rs7045 | 99274306 | ALDH1A3 | A/G | 0.18 | 0.04 | 0.12 | −0.005 | 0.94 |
Min/Maj = Minor/Major allele
MAF = minor allele frequency
MaxD = max drinks
DSM4 = number of DSM-IV alcohol dependence symptoms endorsed
MaxD β = percent change in max drinks per copy of the minor allele
DSM4 β = absolute change in symptom count per copy of the minor allele
OR = odds ratio
P = P-value
SNPs in bold were associated with at least one phenotype at P ≤ 0.05
SNPs underlined were associated with both phenotypes at P ≤ 0.05
We also tested the nominally significant SNPs for dominant and recessive allelic effects. Minor alleles at SNP rs1229984 showed evidence for dominant effects on both phenotypes, although the dominant model did not increase the significance of the associations over additive allelic coding. One SNP that was nominally associated with max drinks in the additive model, rs284787 in ADH7 (P = 0.04), showed evidence for recessive allelic effects on max drinks (P = 0.01) and DSM-IV symptom count (P = 0.0009).
There is significant LD among SNPs within each gene, as well as significant LD across genes in the ADH gene cluster on chromosome 4. Supplementary figures 1–6 show the r2 plots for the SNPs analyzed by chromosome. There were 21 haplotype blocks present, 9 of which are on chromosome 4. No individual haplotypes were associated with either trait at P ≤ 0.01. Two haplotypes in block number three on chromosome 15 (see supplementary figure 1F) in the ALDH1A3 gene, showed the greatest evidence for association to DSM-IV symptom count (P = 0.02). In general, the haplotype association results were less convincing than the individual SNP results within that gene (see table 1). The haplotype association results provide no evidence for untyped, causal loci tagged by our typed markers.
Given the two-stage process of alcohol metabolism and the potential for differences in metabolic rates in one stage to be masked or heightened by rate differences in the other, we also tested for pairwise interactions between SNPs. The strongest interaction effect was between two SNPs in ADH7 that jointly predict more DSM-IV dependence criteria endorsed for each additional minor allele when at least one minor allele is present at each locus (βinteraction = 0.7, P = 0.00002). Two SNPs in high LD in ADH1B show epistatic effects with a SNP in ALDH6A1 as well (βinteraction = −0.4, P = 0.00004). Table 2 shows the epistatic effects between SNP pairs with interaction term P-values < 0.001. Although we show results for each SNP pair with interaction term P-values below this threshold, not all of these findings are independent due to high correlation between the SNPs involved (see supplementary figure 1).
To obtain a more meaningful measure of the significance of our smallest P-values, we used the method of (Li and Ji, 2005) to determine that there were 75 independent tests of association after factoring in the correlations among SNPs. In addition, there was a 50% correlation between the phenotypes tested. Thus, we adjust our significance threshold based on 113 independent tests (75 SNPs times 1.5 phenotypes), yielding an experiment-wide P-value cutoff of 0.0004. Only two P-values, for the associations between rs1229984 and DSM-IV symptom count (P = 0.0003), and the same SNP with max drinks (P = 0.0004), exceed this threshold. The smallest P-values for SNP x SNP interactions are on the order of 2.0 × 10−5, which are unimpressive P-values given the number of pairwise interaction tests performed.
Discussion
To date, this is the first analysis including genotype data on every gene known to play a major role in alcohol metabolism in European Americans recruited from the community. We report several SNPs suggesting association with one of two alcohol phenotypes, max drinks, a measure of an individual’s capacity to metabolize alcohol, and a more clinically relevant phenotypic measure of alcohol dependence, the number of DSM-IV symptoms endorsed.
The strongest evidence for association was between DSM-IV symptom count, max drinks (the P-values differed by only 1.0 × 10−4), and a widely studied non-synonymous coding SNP in ADH1B, rs1229984. This SNP results in a histidine to arginine substitution and has been shown to affect the kinetic properties of the enzyme, with V(max) of ethanol oxidation differing by 100-fold between minor and major allele homozygotes (Matsuo et al., 1989), and has also been shown to affect alcohol-related phenotypes in numerous populations (Shea et al., 2001; Carr et al., 2002; Chai et al., 2005). Our results replicate the previous findings in a community-based sample. The 4% minor allele frequency observed in this population lies within the range observed in previous samples of European Americans (MAF = 0.00 – 0.12), although it is higher than the MAF observed in similar sized alcohol association studies (Edenberg et al., 2006; Kuo et al., 2008; Goedde et al., 1992; Hashibe et al., 2008).
The second SNP of interest due to its association with both phenotypes tested, rs8187974, is located in an intron at a splice site in ALDH1A1. ALDH1A1 is the primary cytosolic aldehyde dehydrogenase, as compared to the mitochondrial form ALDH2. Although common polymorphisms in ALDH1A1 with large effects on alcohol behavior have been described in several non-European populations, these polymorphisms are not present in Caucasians. There are alcohol flushing Caucasians, however, who have much lower ALDH1 enzymatic activity (Yoshida et al., 1989). It is unlikely that rs8187974 is responsible for this observation, however, since minor allele carriers show increased max drinks and more severe alcohol dependence. If this SNP has a real biological effect it is likely to increase the rate of ALDH1 activity or expression. The SNP’s location raises the possibility that it results in an alternately spliced form of the enzyme in the 1% of the population carrying the minor allele, or that it results in a more efficiently translated mRNA.
In addition to replicating the association between rs1229984 and alcohol behavior, two other SNP associations we observed have been reported in previous papers. A SNP in ADH7 on chromosome 4, rs284779, was associated with DSM-IV dependence in a paper by Edenberg et al. (Edenberg et al., 2006). We found this SNP predicted a 4% increase in max drinks (P = 0.05), but was not associated with DSM-IV symptom count.
The interactions involving one SNP in an ADH and one in an ALDH gene are of biological interest since it is the combination of the relative rates at which these enzymes act which determines the amount of acetaldehyde present in an individual’s system for a given alcohol intake level. Because a genetically determined difference in the rate of one stage in the pathway could theoretically be masked by a variation at the other stage, modeling an interaction term may be the only way to uncover genetic effects. Although variants in genes acting in the same stage of alcohol metabolism may be expected to have a simple additive effect, the potential still exists for the effect of one variant to mask the effect of another. Alternatively, multiple variants may interact to put an individual’s ethanol metabolizing capacity above or below a certain threshold to make the effects detectable.
Previous evidence also supports a role for epistasis in determining alcohol behavior. A study of Taiwanese Han showed evidence for epistasis between class 1 ADH haplotypes and ADH7 genotypes in predicting alcohol dependence (Osier et al., 2004). Although we found no evidence for interaction between class 1 ADH SNPs and ADH7, there was evidence for interaction between ADH7 and ALDH1A1 SNPs.
Finally, one SNP showing an epistatic effect has known functionality. The smallest interaction P-value involved SNPs rs17588403 and rs1573496, a non-synonymous coding SNP in ADH7 with main effects predicting decreases in max drinks and number of DSM-IV symptoms, although these effects are not statistically significant. This SNP also shows evidence for interaction with two other SNPs in ADH7. One of these, rs284784, predicts a nominally significant decrease in max drinks (P = 0.04), raising the possibility that both minor alleles are required to significantly change ADH7 activity.
Strengths and Limitations
The genotyping in this study provides coverage of all the major genes known to be involved in the metabolism of alcohol in a large sample of European Americans. We have relatively complete, comprehensive phenotypic information covering a wide range of alcohol use, abuse, and dependence parameters available to test for genetic association. Also, several of the SNP associations we report replicate previous findings, and/or have potential, or in the case of rs1229984, proven functionality. The association between rs1229984 and both DSM-IV symptom count and max drinks satisfies multiple test corrected significance thresholds based on a Bonferroni adjustment based on the 113 independent tests. Finally, this study extends the findings to a community-based population as opposed to a sample recruited from treatment centers.
Although several potential confounding factors have been identified and controlled for, others may certainly exist. For example, body mass index data was not collected and may affect in individuals’ lifetime maximum drink consumption. Aside from rs1229984, the P-values for the single marker and interaction tests presented in this paper are modest.
Finally, this sample was ascertained to detect genetic factors contributing to nicotine (rather than alcohol) dependence. Given the co-occurrence of drinking and smoking, we may hesitate to generalize these findings due to the potential for alcohol use in this sample greater than that in the whole population. The prevalence of alcohol dependence in this sample is similar to the general population prevalence, however, which along with the fact that we adjusted for nicotine dependence, increases our confidence that our results are unbiased by ascertainment.
Conclusion
We present results of a candidate gene association study between SNPs in 17 alcohol metabolizing genes and two alcohol phenotypes. In this sample of European Americans, SNPs in eight genes suggest association with one or both of the outcomes. Although the effect of the genetic factors presented here on the overall problem of alcohol abuse and dependence is modest, it is likely that these or proximal polymorphisms contribute to the inter-individual variation in alcohol metabolizing capacity and subsequent risk for alcohol dependence. Combinations of polymorphisms in genes involved in both stages of alcohol metabolism may also interact to affect alcohol behavior.
Supplementary Material
Supplementary figure 1. Haplotype block structure across the ADH cluster on chromosome 4
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Includes, in order, the genes ADH5, ADH4, ADH6, ADH1A, ADH1B, ADH1C, and ADH7
Supplementary figure 2. Haplotype block structure of SNPs in the ALDH1A1 gene on chromosome 9
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Supplementary figure 3. Haplotype block structure of SNPs in the ALDH18A1 gene on chromosome 10
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Supplementary figure 4. Haplotype block structure of SNPs on chromosome 12
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Includes, in order, the genes ALDH1L2 and ALDH2
Supplementary figure 5. Haplotype block structure of SNPs in the ALDH6A1 gene on chromosome 14
Pairwise comparisons of SNPs located more than 500 KB apart were ignored. Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Supplementary figure 6. Haplotype block structure of SNPs on chromosome 15
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Includes, in order, the genes ALDH1A2 and ALDH1A3
Acknowledgments
Research funded in part by NIMH grant # T32MH014677.
In memory of Theodore Reich, founding Principal Investigator of COGEND, we are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge with great admiration his seminal scientific contributions to the field. This work was supported by the NIH grant CA89392 from the National Cancer Institute.
The authors wish to acknowledge the contributions of advisors to this project. The NIDA Genetics Consortium, with Jonathan Pollock, and NICSNP committees were vital to the success of the research. The Data Analysis Committee helped oversee analyses for the genome wide association studies and investigated methodological issues in association analyses. Further, the committee assisted in data management and data sharing functions. In addition to the authors, committee members included Andrew Bergen, Gerald Dunn, Mary Jeanne Kreek, Huijun Ring, Lei Yu, and Hongyu Zhao. At Perlegen Sciences, we would like to acknowledge the work of Laura Stuve, Curtis Kautzer, the genotyping laboratory, Laura Kamigaki, the sample group, and John Blanchard, Geoff Nilsen, and the bioinformatics and data quality groups for excellent technical and infrastructural support for this work performed under NIDA Contract HHSN271200477471C. This work is supported by NIH grants CA89392 from the National Cancer Institute, DA12854 and DA015129 from the National Institute on Drug Abuse, and the contract N01DA-0-7079 from NIDA.
References
- 1.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 2.Bergen AW, Yang XR, Bai Y, Beerman MB, Goldstein AM, Goldin LR Framingham Heart Study. Genomic regions linked to alcohol consumption in the Framingham Heart Study. BMC Genet. 2003;4(Suppl 1):S101. doi: 10.1186/1471-2156-4-S1-S101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, Fugman D, Goate AM, Hinrichs AL, Konvicka K, Martin NG, Montgomery GW, Saccone NL, Saccone SF, Wang JC, Chase GA, Rice JP, Ballinger DG. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Whitfield JB, Martin NG. Association of the gastric alcohol dehydrogenase gene ADH7 with variation in alcohol metabolism. Hum Mol Genet. 2008;17:179–189. doi: 10.1093/hmg/ddm295. [DOI] [PubMed] [Google Scholar]
- 5.Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 6.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr LG, Foroud T, Stewart T, Castelluccio P, Edenberg HJ, Li TK. Influence of ADH1B polymorphism on alcohol use and its subjective effects in a Jewish population. Am J Med Genet. 2002;112:138–143. doi: 10.1002/ajmg.10674. [DOI] [PubMed] [Google Scholar]
- 8.Chai YG, Oh DY, Chung EK, Kim GS, Kim L, Lee YS, Choi IG. Alcohol and aldehyde dehydrogenase polymorphisms in men with type I and Type II alcoholism. Am J Psychiatry. 2005;162:1003–1005. doi: 10.1176/appi.ajp.162.5.1003. [DOI] [PubMed] [Google Scholar]
- 9.Corbett J, Saccone NL, Foroud T, Goate A, Edenberg H, Nurnberger J, Porjesz B, Begleiter H, Reich T, Rice JP. A sex-adjusted and age-adjusted genome screen for nested alcohol dependence diagnoses. Psychiatr Genet. 2005;15:25–30. doi: 10.1097/00041444-200503000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep. 2006;8:151–157. doi: 10.1007/s11920-006-0015-1. [DOI] [PubMed] [Google Scholar]
- 11.Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 12.Edwards R. The problem of tobacco smoking. BMJ. 2004;328:217–219. doi: 10.1136/bmj.328.7433.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehlers CL, Montane-Jaime K, Moore S, Shafe S, Joseph R, Carr LG. Association of the ADHIB*3 allele with alcohol-related phenotypes in Trinidad. Alcohol Clin Exp Res. 2007;31:216–220. doi: 10.1111/j.1530-0277.2006.00298.x. [DOI] [PubMed] [Google Scholar]
- 14.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 15.Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, Bhatia K, Chen LZ, Fang B, Lisker R. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88:344–346. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- 16.Grant BF. Prevalence and correlates of alcohol use and DSM-IV alcohol dependence in the United States: results of the National Longitudinal Alcohol Epidemiologic Survey. J Stud Alcohol. 1997;58:464–473. doi: 10.15288/jsa.1997.58.464. [DOI] [PubMed] [Google Scholar]
- 17.Hashibe M, McKay JD, Curado MP, Oliveira JC, Koifman S, Koifman R, Zaridze D, Shangina O, Wunsch-Filho V, Eluf-Neto J, Levi JE, Matos E, Lagiou P, Lagiou A, Benhamou S, Bouchardy C, Szeszenia-Dabrowska N, Menezes A, Dall’Agnol MM, Merletti F, Richiardi L, Fernandez L, Lence J, Talamini R, Barzan L, Mates D, Mates IN, Kjaerheim K, Macfarlane GJ, Macfarlane TV, Simonato L, Canova C, Holcatova I, Agudo A, Castellsague X, Lowry R, Janout V, Kollarova H, Conway DI, McKinney PA, Znaor A, Fabianova E, Bencko V, Lissowska J, Chabrier A, Hung RJ, Gaborieau V, Boffetta P, Brennan P. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40:707–709. doi: 10.1038/ng.151. [DOI] [PubMed] [Google Scholar]
- 18.Hasin DS, Liu X, Alderson D, Grant BF. DSM-IV alcohol dependence: a categorical or dimensional phenotype? Psychol Med. 2006;36:1695–1705. doi: 10.1017/S0033291706009068. [DOI] [PubMed] [Google Scholar]
- 19.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 20.Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- 21.Hurley TD, Edenberg HJ, Li T. Pharmacogenomics: The Search for Individualized Therapies. Wiley-VCH; 2002. The Pharmacogenomics of alcoholism; pp. 417–441. (none ed) [Google Scholar]
- 22.Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Association of ADH and ALDH Genes With Alcohol Dependence in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) Sample. Alcohol Clin Exp Res. 2008 doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D, Kendler KS, Prescott CA. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol Clin Exp Res. 2006;30:1807–1816. doi: 10.1111/j.1530-0277.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 25.Li TK. Pharmacogenetics of responses to alcohol and genes that influence alcohol drinking. J Stud Alcohol. 2000;61:5–12. doi: 10.15288/jsa.2000.61.5. [DOI] [PubMed] [Google Scholar]
- 26.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 27.Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31:1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- 28.Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Multiple ADH genes modulate risk for drug dependence in both African- and European-Americans. Hum Mol Genet. 2007;16:380–390. doi: 10.1093/hmg/ddl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo K, Hiraki A, Hirose K, Ito H, Suzuki T, Wakai K, Tajima K. Impact of the alcohol-dehydrogenase (ADH) 1C and ADH1B polymorphisms on drinking behavior in nonalcoholic Japanese. Hum Mutat. 2007;28:506–510. doi: 10.1002/humu.20477. [DOI] [PubMed] [Google Scholar]
- 30.Matsuo Y, Yokoyama R, Yokoyama S. The genes for human alcohol dehydrogenases beta 1 and beta 2 differ by only one nucleotide. Eur J Biochem. 1989;183:317–320. doi: 10.1111/j.1432-1033.1989.tb14931.x. [DOI] [PubMed] [Google Scholar]
- 31.Osier MV, Lu RB, Pakstis AJ, Kidd JR, Huang SY, Kidd KK. Possible epistatic role of ADH7 in the protection against alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2004;126:19–22. doi: 10.1002/ajmg.b.20136. [DOI] [PubMed] [Google Scholar]
- 32.Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, Robinson VP, Neale MC, van den Oord EJ, Walsh D, Riley BP, Kendler KS. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- 35.Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 36.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, Swan GE, Goate AM, Rutter J, Bertelsen S, Fox L, Fugman D, Martin NG, Montgomery GW, Wang JC, Ballinger DG, Rice JP, Bierut LJ. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuckit MA, Smith TL, Daeppen JB, Eng M, Li TK, Hesselbrock VM, Nurnberger JI, Jr, Bucholz KK. Clinical relevance of the distinction between alcohol dependence with and without a physiological component. Am J Psychiatry. 1998;155:733–740. doi: 10.1176/ajp.155.6.733. [DOI] [PubMed] [Google Scholar]
- 38.Schuckit MA, Tipp JE, Reich T, Hesselbrock VM, Bucholz KK. The histories of withdrawal convulsions and delirium tremens in 1648 alcohol dependent subjects. Addiction. 1995;90:1335–1347. doi: 10.1046/j.1360-0443.1995.901013355.x. [DOI] [PubMed] [Google Scholar]
- 39.Shea SH, Wall TL, Carr LG, Li TK. ADH2 and alcohol-related phenotypes in Ashkenazic Jewish American college students. Behav Genet. 2001;31:231–239. doi: 10.1023/a:1010261713092. [DOI] [PubMed] [Google Scholar]
- 40.Tyndale RF. Genetics of alcohol and tobacco use in humans. Ann Med. 2003;35:94–121. doi: 10.1080/07853890310010014. [DOI] [PubMed] [Google Scholar]
- 41.Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida A, Dave V, Ward RJ, Peters TJ. Cytosolic aldehyde dehydrogenase (ALDH1) variants found in alcohol flushers. Ann Hum Genet. 1989;53:1–7. doi: 10.1111/j.1469-1809.1989.tb01116.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Haplotype block structure across the ADH cluster on chromosome 4
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Includes, in order, the genes ADH5, ADH4, ADH6, ADH1A, ADH1B, ADH1C, and ADH7
Supplementary figure 2. Haplotype block structure of SNPs in the ALDH1A1 gene on chromosome 9
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Supplementary figure 3. Haplotype block structure of SNPs in the ALDH18A1 gene on chromosome 10
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Supplementary figure 4. Haplotype block structure of SNPs on chromosome 12
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Includes, in order, the genes ALDH1L2 and ALDH2
Supplementary figure 5. Haplotype block structure of SNPs in the ALDH6A1 gene on chromosome 14
Pairwise comparisons of SNPs located more than 500 KB apart were ignored. Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Supplementary figure 6. Haplotype block structure of SNPs on chromosome 15
Pairwise comparisons of SNPs located more than 500 KB apart were ignored.
Blocks were defined according to the algorithm described by Gabriel et al, 2002.
Includes, in order, the genes ALDH1A2 and ALDH1A3


