Abstract
Chemical stabilizers are widely used to enhance protein stability, both in nature and in the laboratory. Here, the molecular mechanism of chemical stabilizers is studied using a disulfide trapping assay to measure the effects of stabilizers on thermal backbone dynamics in the Escherichia coli galactose/glucose binding protein. Two types of backbone fluctuations are examined: (a) relative movements of adjacent surface α-helices within the same domain and (b) interdomain twisting motions. Both types of fluctuations are significantly reduced by all six stabilizers tested (glycerol, sucrose, trehalose, l-glucose, d-glucose, and d-galactose), and in each case larger amplitude motions are inhibited more than smaller ones. Motional inhibition does not require a high-affinity stabilizer binding site, indicating that the effects of stabilizers are nonspecific. Overall, the results support the theory that effective stabilizing agents act by favoring the most compact structure of a protein, thereby reducing local backbone fluctuations away from the fully folded state. Such inhibition of protein backbone dynamics may be a general mechanism of protein stabilization in extreme thermal or chemical environments.
Structural dynamics play an integral role in the functions of proteins (Hayward & Go, 1995; Holzwarth, 1995; Peng & Wagner, 1994; Wade et al., 1994; Dagget & Levitt, 1993), yet the need for structural dynamics must be carefully balanced against the need for protein stability (Jaenicke, 1996; Matthews, 1996; Shortle, 1996; Larzaridis et al., 1995; Bryson et al., 1995; Chakrabartty & Baldwin, 1995; Dill et al., 1995). Sugars and polyhydric alcohols have been known to stabilize proteins in solution for several decades. Among the most effective stabilizers are glycerol, sucrose, glucose, and trehalose (Back et al., 1979; Gerlsma, 1968). Such compounds are used as natural protective agents by many organisms exposed to extreme environments (Nwaka et al., 1994; Attfield, 1987; Neves & Francois, 1992; Hottiger et al., 1994) and have found wide applications in research and biotechnology. Sufficiently high concentrations of these compounds preserve both biological activity and native protein structure, generally increasing the thermal denaturation temperature of a given protein by as much as 20 °C (Gekko & Timasheff, 1981b). Hydrogen exchange studies have shown that stabilizers inhibit the global or partial unfolding reactions of several proteins, as detected by decreases in slow exchange rates for buried residues in the protein interior (Wang et al., 1995; Gregory, 1988; Calhoun & Englander, 1985; Knox & Rosenberg, 1980). In contrast, no effects on intermediate exchange rates were observed, implying that thermal fluctuations within the folded state might be unaffected by stabilizers. In principle, however, the observed effects of sugars and polyols on protein stability could arise from the inhibition of backbone dynamics within the folded state itself, since such dynamics yield random thermal fluctuations away from the most compact conformation, thereby initiating the more dramatic unfolding processes that eventually expose the protein interior. Although stabilizers are known to decrease the volume and compressibility of proteins (Priev et al., 1996; Cioni & Strambini, 1994), their effects on specific, local backbone motions within the folded state have not yet been investigated.
The present study utilizes the disulfide trapping method (Careaga et al., 1995; Careaga & Falke, 1992a; Falke & Koshland, 1987) to probe the effects of protein stabilizers on local backbone dynamics within a representative folded protein. The Escherichia coli galactose/glucose binding protein (GBP) is a 32 kDa soluble periplasmic receptor which mediates both chemotaxis toward and uptake of d-galactose and d-glucose (Hottiger et al., 1994; Zukin et al., 1977; Boos et al., 1972). As illustrated in Figure 1, the structure of GBP consists of two distinct αβ globular domains connected by a three-strand hinge (Vyas et al., 1988,1987). The C-terminal domain contains an EF-hand-like Ca2+ binding site, while sugar binding occurs in the cleft formed by the interface of the N- and C-terminal domains. Upon sugar binding, the interdomain cleft closes at least 18°, thereby trapping the ligand inside (Luck & Falke, 1991); thus, protein backbone movements play an essential role in the function of this soluble receptor. The folded protein exhibits intermediate thermal and chemical stability, yielding a simple, two-state cooperative unfolding transition at a temperature of 61 °C or at a urea concentration of 3.1 M.
Figure 1.
Crystallographic structure of the galactose/glucose binding protein (Vyas et al., 1988, 1987). (A) Ribbon diagram of the full backbone structure with bound Ca2+ and d-glucose indicated. The locations of the six engineered cysteine residues are indicated by sulfhydryl collision spheres (2.3 Å radius about Cβ); disulfide formation can occur only if two collision spheres overlap during a structural fluctuation. (B) Expanded stereoview of helices α1 and α10 with the 26/260 and 26/274 dicysteine pairs indicated as collision spheres. (C) Expanded stereoview of the sugar binding cleft with the 182/43 and 182/15 dicysteine pairs shown as collision spheres.
Previous disulfide trapping studies of GBP have characterized relative motions of parallel helices α1 and α10, located on the surface of the N-terminal domain, as well as relative rotational motions of the N- and C-terminal domains (Careaga et al., 1995; Careaga & Falke, 1992a). These studies utilized pairs of engineered cysteine residues located at nonadjacent positions in the known crystal structure, such that, for a given cysteine pair, a collision between their sulfhydryl moeities could occur only during a significant thermal fluctuation away from the crystallographic structure of the fully folded protein. Upon the addition of a redox catalyst, such sulfhydryl–sulfhydryl collisions were covalently trapped by disulfide bond formation. The resulting time course of disulfide formation placed limits on the collision frequency, while analysis of the crystallographic coordinates provided the minimum spatial amplitudes of the motions. These studies detected local relative helix movements ranging up to 15.2 Å (Careaga & Falke, 1992a) and relative twisting motions of the interdomain hinge as great as 36° (Careaga et al., 1995), where both motions occurred at frequencies of approximately 10 s−1. Such motions are significantly faster than global unfolding, which occurs over a period of 1–2 days when GBP is diluted into chemical denaturants (Careaga & Falke, 1992a).
Here, a set of four dicysteine GBP mutants are used to measure the effects of protein stabilizers on both intradomain helix–helix motions and interdomain rotational motions. Altogether, six representative stabilizers are examined, including those most commonly used in pharmaceutical and research applications. Each of the stabilizers is observed to enhance the thermal stability of GBP. In addition, each stabilizer significantly reduces the probabilities of relative helix–helix motions and interdomain rotations, and larger amplitude motions are uniformly inhibited to a greater extent than smaller motions. These results provide important insights into the molecular mechanisms underlying the stabilization of protein structure and function in extreme environments.
MATERIALS AND METHODS
Materials
Stabilizers used for this study were purchased from Fluka (d-galactose) or Sigma (all others, Ultra grade). 35S-Labeled cysteine was purchased from Amersham.
Purification of GBP
Wild-type and dicysteine mutants of GBP were generated by expression of engineered pSF5 plasmids and grown in the E. coli strain NM303, which is deleted for endogenous GBP production, as described previously (Careaga & Falke, 1992a). Protein was isolated by osmotic shock of the periplasm and dialyzed into buffers appropriate for each experiment, with or without sugar, as needed. Concentration was adjusted by ultrafiltration (Amicon), and protein was quick frozen in liquid N2 and stored at −70 °C until use (Careaga & Falke, 1992a).
Thermal Melting Measurements
The Tm of wild-type GBP in each stabilizer was determined using intrinsic tryptophan fluorescence to indicate the folding state of the protein. The protein was brought to 5 μM in 2 mL of 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.0, with NaOH, 50 mM KCl, 50 mM NaCl, 1.0 mM d-glucose, 0.2 mM CaCl2, and the indicated stabilizer. Measurements were performed on an SLM 48000 spectrofluorometer (λex = 285 nm, λem = 337 nm, bandwidths = 4 nm). Temperature was increased stepwise in increments of 2 °C in the transition region, and the samples were allowed to equilibrate up to 45 min before measurements were recorded. Melting of GBP was evident as a distinct, reversible decrease in the fluorescence signal. Sigmoidal curves generated were processed by vant Hoff analysis (Brandts, 1964a,b). Detailed procedures are published (Gekko & Morikawa, 1981).
Free Cysteine Reactions
In order to measure stabilizer effects on free cysteine reaction rates, [35S]cysteine was spiked into 5 μM cold cysteine in 20 mM NaH2PO4, pH 4.5, with HCl, 50 mM KCl, 50 mM NaCl, 0.1 mM EDTA, and ambient dissolved oxygen (190 μM). Oxidation was initiated at 37 °C by addition of the redox catalyst copper(II) tris(1,10-phenanthroline) to 1.5 mM, and at appropriate time points, aliquots were quenched with 1 M N-ethylmaleimide and 90 mM EDTA (pH 8.0). A 4 μL aliquot of each time point was spotted on a TLC plate (Kieselgel 60 F-254, 0.2 mm) and developed for 2 h in 88% phenol/glacial acetic acid/water (15:1:4, by volume). TLC plates were dried and exposed on phosphorimaging screens for 3–5 days and quantitated using a Molecular Dynamics Phosphorimager and ImageQuant software.
Disulfide Bond Formation and Quantitation
All oxidation reactions utilized 3 μM GBP protein in 20 mM NaH2PO4, pH 7.0, with NaOH, 50 mM NaCl, 50 mM KCl, 1.0 mM d-glucose, 0.2 mM CaCl2, 1.0 mM NaAsO2, ambient dissolved oxygen as the oxidation agent (190 μM), and the specified stabilizer concentration. Oxidation catalyst, 1.5 mM copper(II) tris(1,10-phenanthroline), was added to initiate the reaction at 37 °C, and aliquots were quenched at specific time points with 40 mM N-ethylmaleimide (blocking group for unreacted thiols), 200 mM EDTA [to chelate Cu(II) ions], 130 mM Tris (pH 6.8), 20% (v/v) glycerol, 4% (w/v) SDS, and 0.05% (w/v) bromophenol blue. Since disulfide-trapped molecules have increased electrophoretic mobility, reaction products were separated on 15% SDS–PAGE, stained with Coomassie blue, and quantitated by laser densitometry. Procedures were as previously described (Careaga & Falke, 1992a).
Sugar Binding by Flow Dialysis
Binding data were obtained by flow dialysis, which uses the rate of ligand diffusion across a dialysis membrane to quantitate the free ligand concentration in a binding equilibrium. The apparatus used was based on that described by Colowick and Womack (1969), where signal is proportional to free radiolabeled ligand in the chamber. Starting with equilibrated buffer (20 mM NaH2PO4, pH 7.0, with NaOH, 50 mM NaCl, 50 mM KCl, 0.2 mM CaCl2) at 25 °C, the steps of the experiment were as follows: (1) addition of 10 μM [3H]-d-glucose; (2) addition of 7 μM GBP; (3) first addition of competitor, initiating the titration; (4) final point of titration; (5) addition of 20 mM cold d-glucose to displace all bound radiolabeled sugar. The maximum concentrations of competitors during the titrations were 0.1 mM for d-galactose, 20 mM for sucrose and trehalose, 40 mM for l-glucose, and 80 mM for glycerol. The final concentrations of sucrose, trehalose, l-glucose, and glycerol relative to 10 μM [3H]-d-glucose were chosen to exceed the corresponding concentration ratios in the Tm and disulfide trapping studies by 2-fold or more.
RESULTS
Effects of Stabilizers on GBP Denaturation
The six stabilizers chosen for this study, glycerol, sucrose, trehalose, d-glucose, l-glucose, and d-galactose, were found to significantly enhance the stability of fully liganded GBP against thermal denaturation, as summarized in Table 1. The thermal transition temperature, Tm, was obtained by monitoring the intrinsic fluorescence of the five Trp residues which decreased 10-fold upon denaturation. The resulting thermal unfolding curves yielded simple, reversible one-phase transitions from which the Tm values were calculated through vant Hoff analysis (Brandts, 1964a). Individual stabilizers substantially increased the Tm between 5.8 and 17.6 °C (Table 1), yielding the following relative order of thermal stabilization: d-glucose > d-galactose > trehalose > l-glucose > sucrose > glycerol. The highest levels of stabilization were observed for d-galactose and d-glucose (Table 1), the only stabilizers able to bind specifically to the sugar binding cleft under the experimental conditions used (see Figure 2). However, trehalose and l-glucose provided nearly as much thermal stabilization as d-galactose, indicating that the bulk of the stabilization was unrelated to specific ligand binding (Table 1). This result is consistent with analogous findings for other protein systems in which stabilizers also inhibit thermal unfolding even in the absence of specific stabilizer binding sites (Gekko & Timasheff, 1981b).
Table 1.
Summary of Stabilizer Effects
| rel GBP disulfide formation rateb |
|||||||
|---|---|---|---|---|---|---|---|
| stabilizer | concn (mg/mL, M) |
rel control disulfide formation ratea (%) |
thermal stabilization (ΔTm, °C) |
26/260 (8.3 Å) (%) |
26/274 (15.2 Å) (%) |
182/43 (4.3 Å, 0 °C) (%) |
182/15 (7.8 Å, 36 °C) (%) |
| d-glucose | 400, 2.2 | 80 ± 10 | +17.6 | 44 ± 8 | 8 ± 1 | 60 ± 9 | 26 ± 6 |
| d-galactose | 400, 2.2 | 100 ± 10 | +15.9 | 29 ± 6 | 15 ± 3 | 67 ± 13 | 31 ± 5 |
| trehalose | 378, 1.0 | 120 ± 10 | +14.9 | 48 ± 7 | 23 ± 3 | 62 ± 7 | 34 ± 10 |
| l-glucose | 400, 2.2 | 80 ± 20 | +13.8 | 50 ± 12 | 19 ± 3 | 64 ± 7 | 34 ± 5 |
| sucrose | 400, 1.2 | 80 ± 10 | +10.9 | 46 ± 6 | 27 ± 7 | 54 ± 7 | 36 ± 4 |
| glycerol | 378, 4.1 | 130 ± 10 | +5.8 | 38 ± 8 | 10 ± 1 | 66 ± 7 | 28 ± 6 |
Rate of cysteine formation from free cysteine, relative to the same rate in the absence of stabilizer [(2 ± 0.2) × 103 M−1 s−1].
Rates relative to the corresponding disulfide formation rate of each mutant in the absence of stabilizer: (21 ± 3) × 10−3 s−1 molecule−1 for 26/260; (0.24 ± 0.03) × 10−3 s−1 molecule−1 for 26/274; (10 ± 1) × 10−3 s−1 molecule−1 for 182/43; (1.2 ± 0.2) × 10−3 s−1 molecule−1 for 182/15.
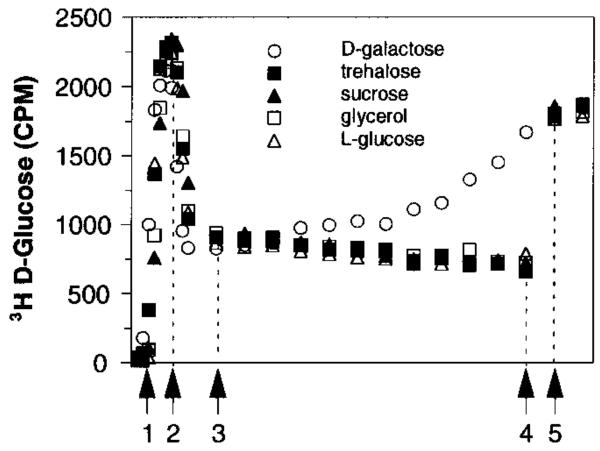
Figure 2.
Comparison of the abilities of stabilizers to displace bound d-glucose from GBP. Shown is a flow dialysis experiment monitoring the binding of a fixed d-glucose population to the protein as a competing ligand is added at 25 °C. The vertical axis is proportional to the concentration of free [3H]-d-glucose in the chamber. The steps of the experiment were as follows: (1) addition of 10 μM [3H]-d-glucose; (2) addition of 7 μM GBP; (3) first addition of competitor, initiating the titration; (4) final point of titration; (5) addition of 20 mM cold d-glucose to displace all bound radiolabeled sugar.
Effects of Stabilizers on a Control Disulfide Trapping Reaction
In order to examine the effects of stabilizers on GBP internal motions, the disulfide trapping method was employed to detect thermal backbone dynamics. First, however, it was necessary to determine whether the stabilizers interfered with the chemistry of a control disulfide trapping reaction. The six stabilizers were found to have no significant effects on the rate of disulfide bond formation between free cysteine sulfhydryls in solution to yield cystine (Table 1). One major implication of this result is that the disulfide trapping reaction was independent of solvent viscosity, which increased 3–6-fold at the highest stabilizer concentrations. It follows that the disulfide trapping reaction was not in the diffusion-controlled limit where the rate would be inversely proportional to viscosity. Instead, the slow reaction limit pertained in which the reactants were in rapid equilibrium with an encounter complex that was slowly converted to products (Steinfeld et al., 1989):
| (1) |
where the equilibrium constant for the formation of the encounter complex is k1/k−1 = K and the overall rate constant of the reaction is Kk2 = k. The diffusible oxidizing agent (OX) is superoxide or hydroxy radical; additional details of the oxidation reaction are published (Oae, 1991).
The failure of stabilizers to change the rate of cystine formation indicated that they did not perturb the chemistry of disulfide trapping. A more complex interpretation, proposing that K and k2 are changed such that the overall rate k = Kk2 remains unchanged, was disfavored by the similar results obtained for six chemically distinct stabilizers. It follows that, even at the highest concentrations of stabilizers utilized, these compounds do not interfere with the chemistry of the disulfide trapping method.
Selection of Engineered Dicysteine GBP Molecules for Disulfide Trapping Studies
For each targeted backbone motion, two engineered cysteine pairs differing in intercysteine distances were used to trap, by disulfide formation, motions of different amplitudes. Relative movements of helices α1 and α10 were detected using the Q26C/N260C and Q26C/D274C engineered cysteine pairs illustrated in Figure 1B. For these cysteine pairs, minimum backbone motions of 8.3 or 15.2 Å were required to bring sulfhydryls into collision range for disulfide formation, respectively. Two other engineered cysteine pairs, M182C/N43C and M182C/N15C, were used to detect relative movements of the N- and C-terminal domains as shown in Figure 1C. The latter pairs required minimum interdomain motions of 4.3 and 7.8 Å for disulfide trapping, corresponding to hinge–twist rotations of 0° and 36°, respectively. All four of the engineered cysteine pairs were located at nonconserved surface positions and were previously shown to yield no significant effects on the structure and dynamics of GBP as judged by four parameters: the equilibrium binding affinity of d-galactose, the dissociation rate of phosphorescent Tb3+ from the Ca2+ binding site, the urea denaturation profile, and the protein conformation as monitored by 19F NMR (Careaga et al., 1995; Careaga & Falke, 1992a).
Effects of Stabilizers on Disulfide Trapping of Thermal Backbone Motions in GBP
Intramolecular disulfide formation between engineered cysteines on the surface of GBP, triggered by thermal backbone motions that allow the two sulfhydryls to collide, displayed two fundamental differences from the free cysteine reaction (Table 1). First, the rate of intramolecular disulfide formation was 2–4 orders of magnitude slower than the free cysteine reaction under identical oxidation conditions. (This slowing factor would be even larger if it included the higher effective cysteine concentrations in the intramolecular GBP reactions.) The slower rates of the intraprotein GBP reactions arose primarily from a decrease in the equilibrium constant K for the formation of the encounter complex (eq 1), since the intrinsic rate k2 of encounter complex conversion to products is approximately constant for cysteine in solution and on the protein surface (Careaga & Falke, 1992a). The lower stability of the encounter complex in the protein suggests that the backbone distortion required to produce a sulfhydryl–sulfhydryl collision exacts a significant energetic cost. As a positive control, disulfide reaction rates were also measured in the presence of urea, which should destabilize the protein structure and thereby decrease the energetic cost of sulfhydryl–sulfhydryl collision. Although urea had no effect on the disulfide formation rate of the free cysteine reaction, it dramatically increased the kinetics of disulfide formation in GBP as predicted. The effect was greatest at urea concentrations sufficient to unfold the protein, as shown in Figure 3. It follows that the structural stability of the folded protein energetically opposes formation of the encounter complex.
Figure 3.
Effects of urea on intramolecular disulfide formation and protein unfolding. (A) Plotted is the fraction of the 26/260 or 26/274 dicysteine protein that forms a disulfide bond within 3 s of oxidation at 25 °C, as a function of urea concentration. Samples were equilibrated in urea for 48 h at 25 °C before oxidation was initiated. (B) Unfolding of WT and 26/274 by urea at 25 °C, as detected by a decrease in the intrinsic tryptophan fluorescence.
The second difference between intramolecular disulfide formation and free cysteine reactions was sensitivity to the presence of chemical stabilizers. All six stabilizers reduced the rate of intraprotein disulfide formation, and this effect increased with stabilizer concentration, as illustrated in Figure 4, although free cysteine reactions were not significantly affected. In some cases, the protein disulfide formation rates were slowed by more than 10-fold (Table 1). For any one cysteine pair, different stabilizers slowed the disulfide formation rate to a similar extent, suggesting that these compounds have analogous effects on backbone dynamics. However, the stabilizers slowed the rates of larger amplitude motions significantly more than those of smaller motions. In the case of relative helix motions, the 15.2 Å motion trapped by the Cys26–Cys274 disulfide was slowed up to 13-fold by stabilizers, while the 8.3 Å motion (Cys26–Cys260) was only slowed 2–3-fold. Similarly, the 7.8 Å interdomain movement (Cys182–Cys15) was slowed up to 4-fold while the 4.3 Å movement (Cys182–Cys43) was slowed less than 2-fold (Table 1). Together, these results indicate that protein stabilizers increase the free energy cost of forming the encounter complex, thereby reducing the equilibrium probability of sulfhydryl–sulfhydryl collisions. Moreover, collisions requiring large amplitude motions are disfavored more than small amplitude collisions.
Figure 4.
Relative rates of intraprotein disulfide formation as a function of stabilizer concentration. (A and B) Disulfide formation rates at 37 °C of the 26/260 and 26/274 dicysteine pairs in varying concentrations of glycerol and sucrose, respectively. (C and D) Disulfide formation rates of the 182/15 and 182/43 dicysteine pairs in glycerol and sucrose, respectively.
DISCUSSION
Overall, the present study reveals inhibition of thermal backbone dynamics in the presence of stabilizing agents. Such inhibition is more pronounced as spatial range increases and is dependent on stabilizer concentration. As the decreased rate of disulfide formation does not stem from a slowing of the chemistry of disulfide formation, it must result from a decrease in the probability of collisions between cysteine residues in the same protein molecule. What is the molecular basis of this collisional slowing? Such slowing is not due to occupancy of the ligand binding site since different stabilizing agents have similar effects of disulfide formation rates, including both sugars that are ligands of GBP (d-glucose, d-galactose) and molecules that do not bind specifically to the sugar binding cleft (glycerol, sucrose, l-glucose, trehalose). Moreover, collisional slowing does not stem from macroscopic viscous damping, since no correlation is observed between inhibition of disulfide formation and the viscosity of the stabilizer solutions. Interestingly, there is a good general correlation between the relative effects of stabilizers on Tm and backbone dynamics (Table 1). The only exception to this correlation is glycerol, which is the least effective thermal stabilizer but the second-best inhibitor of disulfide formation rates. This discrepancy suggests that glycerol may lose efficacy as a stabilizer at the higher temperatures where Tm is measured.
A plausible explanation for the effects of stabilizers on protein dynamics is that they favor the minimization of protein surface area, as proposed by Timasheff (Bhat & Timasheff, 1992; Gekko & Timasheff, 1981a). In molecular terms, stabilizers are thought to be excluded from the solvent layer in contact with the protein, either due to direct repulsion by the protein surface or by an enhanced ordering of the protein hydration shell. These forces provide enthalpic or entropic opposition to protein expansion, respectively, thereby favoring the most compact, folded structure of a protein. Evidence supporting this picture is provided by the demonstrated ability of stabilizers to reduce the average volume and compressibility of proteins (Cioni & Strambini, 1994; Gekko & Timasheff, 1981a) and to significantly decrease the rate of hydrogen exchange from buried, slow exchanging sites in proteins (Wang et al., 1995; Calhoun & Englander, 1985). Since local backbone motions away from the folded state increase protein surface area, stabilizers should decrease the probability of such dynamics, as observed here. The nonspecific nature of stabilizer–protein interactions explains the observed similar effects of different stabilizers on backbone dynamics. More generally, stabilization of the most compact structure, with concomitant reduction of structural fluctuations away from the folded state, may prove to be a fundamental principle underlying other types of thermal stabilization. For example, thermophilic proteins have recently been shown to have smaller surface area-to-volume ratios than their mesophilic homologues (Tanner et al., 1996; Chan et al., 1995), implying that they are also less dynamic. This prediction can be tested by future disulfide trapping studies.
Footnotes
Support provided by NIH Grant GM40731.
REFERENCES
- Attfield PV. FEBS Lett. 1987;225:259–263. doi: 10.1016/0014-5793(87)81170-5. [DOI] [PubMed] [Google Scholar]
- Back JF, Oakenfull D, Smith MB. Biochemistry. 1979;18:5191–5196. doi: 10.1021/bi00590a025. [DOI] [PubMed] [Google Scholar]
- Bhat R, Timasheff SN. Protein Sci. 1992;1:1133–1143. doi: 10.1002/pro.5560010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos W, Gordon AS, Hall RE, Price DH. J. Biol. Chem. 1972;247:917–924. [PubMed] [Google Scholar]
- Brandts JF. J. Am. Chem. Soc. 1964a;86:4291–4301. [Google Scholar]
- Brandts JF. J. Am. Chem. Soc. 1964b;86:4302–4313. [Google Scholar]
- Bryson JW, Betz SF, Lu SF, Suich DJ, Zhou HXX, O'Neil KT, DeGrado WF. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- Calhoun DB, Englander SW. Biochemistry. 1985;24:2095–2100. doi: 10.1021/bi00329a043. [DOI] [PubMed] [Google Scholar]
- Careaga CL, Falke JJ. J. Mol. Biol. 1992a;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga CL, Falke JJ. Biophys. J. 1992b;62:209–219. doi: 10.1016/S0006-3495(92)81806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga CL, Sutherland J, Sabeti J, Falke JJ. Biochemistry. 1995;34:3048–3055. doi: 10.1021/bi00009a036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabartty A, Baldwin RL. Adv. Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- Chan MK, Mukund S, Kletzin A, Adams MWW, Rees DC. Science. 1995;267:1463–1469. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- Cioni P, Strambini GB. J. Mol. Biol. 1994;242:291–301. doi: 10.1006/jmbi.1994.1579. [DOI] [PubMed] [Google Scholar]
- Colowick SP, Womack FC. J. Biol. Chem. 1969;244:774–777. [PubMed] [Google Scholar]
- Dagget V, Levitt M. Annu. Rev. Biophys. Biomol. Struct. 1993;22:353–380. doi: 10.1146/annurev.bb.22.060193.002033. [DOI] [PubMed] [Google Scholar]
- Dill KA, Bromberg S, Yue KZ, Fiebig KM, Yee DP, Thomas PD, Chan HS. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Koshland DE., Jr. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- Gekko K, Morikawa T. J. Biochem. 1981;90:51–60. doi: 10.1093/oxfordjournals.jbchem.a133469. [DOI] [PubMed] [Google Scholar]
- Gekko K, Timasheff SN. Biochemistry. 1981a;20:4667–4676. doi: 10.1021/bi00519a023. [DOI] [PubMed] [Google Scholar]
- Gekko K, Timasheff SN. Biochemistry. 1981b;20:4677–4686. doi: 10.1021/bi00519a024. [DOI] [PubMed] [Google Scholar]
- Gerlsma SY. J. Biol. Chem. 1968;243:957–961. [PubMed] [Google Scholar]
- Gregory RB. Biopolymers. 1988;27:1699–1709. doi: 10.1002/bip.360271102. [DOI] [PubMed] [Google Scholar]
- Hayward S, Go N. Annu. Rev. Phys. Chem. 1995;46:223–250. doi: 10.1146/annurev.pc.46.100195.001255. [DOI] [PubMed] [Google Scholar]
- Holzwarth AR. Methods Enzymol. 1995;246:334–362. doi: 10.1016/0076-6879(95)46016-0. [DOI] [PubMed] [Google Scholar]
- Hottiger T, De Vergilio C, Hall MN, Boller T, Wiemken A. FEBS Lett. 1994;219:187–193. doi: 10.1111/j.1432-1033.1994.tb19929.x. [DOI] [PubMed] [Google Scholar]
- Jaenicke R. FASEB J. 1996;10:84–92. doi: 10.1096/fasebj.10.1.8566552. [DOI] [PubMed] [Google Scholar]
- Knox DG, Rosenberg A. Biopolymers. 1980;19:1049–1068. doi: 10.1002/bip.1980.360190509. [DOI] [PubMed] [Google Scholar]
- Larzaridis T, Archontis G, Karplus M. Adv. Protein Chem. 1995;47:231–306. doi: 10.1016/s0065-3233(08)60547-1. [DOI] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. Biochemistry. 1991;30:4248–4256. doi: 10.1021/bi00231a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. FASEB J. 1996;10:35–41. doi: 10.1096/fasebj.10.1.8566545. [DOI] [PubMed] [Google Scholar]
- Neves MJ, Francois J. Biochem. J. 1992;288:859–864. doi: 10.1042/bj2880859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwaka S, Kopp M, Burgert M, Deuchler I, Kienle I, Holzer H. FEBS Lett. 1994;344:225–228. doi: 10.1016/0014-5793(94)00385-8. [DOI] [PubMed] [Google Scholar]
- Oae S. Organic Sulfur Chemistry: Structure and Mechanism. CRC Press; Boca Raton, FL: 1991. [Google Scholar]
- Peng JW, Wagner G. Methods. Enzymol. 1994;239:563–569. doi: 10.1016/s0076-6879(94)39022-3. [DOI] [PubMed] [Google Scholar]
- Priev A, Almagor A, Yedgar S, Gavish B. Biochemistry. 1996;35:2061–2066. doi: 10.1021/bi951842r. [DOI] [PubMed] [Google Scholar]
- Shortle D. FASEB J. 1996;10:27–34. doi: 10.1096/fasebj.10.1.8566543. [DOI] [PubMed] [Google Scholar]
- Steinfeld JI, Francisco JS, Hase WL. Chemical kinetics and dynamics. Prentice-Hall; Englewood Cliffs, NJ: 1989. [Google Scholar]
- Tanner JJ, Hecht RM, Krause KL. Biochemistry. 1996;35:2597–2609. doi: 10.1021/bi951988q. [DOI] [PubMed] [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. Nature. 1987;327:635–638. doi: 10.1038/327635a0. [DOI] [PubMed] [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- Wade RC, Luty BA, Demchuk E, Madura JD, Davis ME, Briggs JM, McCammon JA. Nat. Struct. Biol. 1994;1:65–69. doi: 10.1038/nsb0194-65. [DOI] [PubMed] [Google Scholar]
- Wang A, Robertson AD, Bolen DW. Biochemistry. 1995;34:15096–15104. doi: 10.1021/bi00046a016. [DOI] [PubMed] [Google Scholar]
- Zukin RS, Strange PG, Heavey LR, Koshland DE., Jr. Biochemistry. 1977;16:381–386. doi: 10.1021/bi00622a007. [DOI] [PubMed] [Google Scholar]