Abstract
The Escherichia coli CheY protein is activated by phosphorylation, and in turn alters flagellar rotation. To investigate the molecular mechanism of activation, an extensive collection of mutant CheY proteins was analyzed by behavioral assays, in vitro phosphorylation, and 19F NMR chemical shift measurements. Substitution of a positively charged residue (Arg or Lys) in place of Asp13 in the CheY activation site results in activation, even for mutants which cannot be phos-phorylated. Thus phosphorylation plays an indirect role in the activation mechanism. Lys109, a residue proposed to act as a conformational “switch” in the activation site, is required for activation of CheY by either phosphorylation or mutation.
The 19F NMR chemical shift assay described in the preceding article (Drake, S. K., Bourret, R. B., Luck, L. A., Simon, M. I., and Falke, J. J. (1993) J. Biol Chem. 268, 13081–13088) was again used to monitor six phenylalanine positions in CheY, including one position which probed the vicinity of Lys109. Mutations which activate CheY were observed to perturb the Lys109 probe, providing further evidence that Lys109 is directly involved in the activating conformational change. Two striking contrasts were observed between activation by mutation and phosphorylation, (i) Each activating mutation generates a relatively localized perturbation in the activation site region, whereas phosphorylation triggers a global structural change. (ii) The perturbation of the Lys109 region observed for activating mutations is not detected in the phosphorylated protein. These results are consistent with a two-step model of activated CheY docking to the flagellar switch.
The ability to detect properties of interest in the external environment and respond appropriately is a fundamental function for most cells, whether of unicellular or multicellular organisms. One universal mechanism for signal transduction is the regulation of protein phosphorylation cascades, which in some cases can be constitutively activated by specific mutations in a critical component, leading to oncogenesis (1-4). The preceding paper (5) examined the phosphorylation-triggered activation of CheY, the terminal protein in the Escherichia coli chemotaxis phosphorylation cascade (reviewed in Refs. 6-9). Here we investigate activation and inactivation by specific mutations adjacent to the phosphorylation site of CheY. This study makes use of an experimentally important feature of the E. coli chemotaxis system: the signaling state of CheY can be assayed by its effect on motor rotation, since activated CheY is needed to switch the motor from counterclockwise to clockwise (CW)1 rotation.
Two classes of mutant CheY proteins have been generated for the purpose of analyzing the activation mechanism, (i) Activated mutants, CheY mutants that mimic the phosphorylated form have been sought, in order to augment the one known mutant of this type (10). Activated mutants are particularly useful because the short half-life of phospho-CheY (11, 12) precludes direct high resolution structural analysis. (ii) Docking site mutants, mutations which alter residues involved in the motor docking event have been pursued, and are identified as surface muations those which do not affect CheY phosphorylation or grossly affect CheY structure, yet block the CW signal normally transmitted by phospho-CheY. Such mutations help ascertain the location of the docking surface in the known structure of CheY (13, 31).
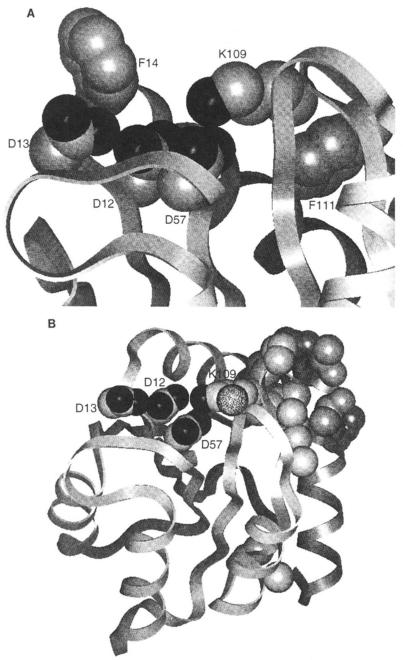
As in the preceding study (5), 19F NMR has been used to physically characterize the conformational effects of activation. 19F NMR is sensitive to perturbations in the local environment of fluorine labels and can be used to map out the regions of a protein affected by an activating mutation. Briefly, 4-fluorophenylalanine (4F-Phe) reporter groups were incorporated at each of the six Phe positions, chosen due to their useful locations in the molecule. Phe14 is adjacent to the highly conserved activation site carboxylate cluster (Fig. 1A). The cluster is composed of Asp57, the site of phosphorylation (14), which together with Asp12 and Asp13 forms the binding site for the essential Mg(II) ion in the phosphorylation and dephosphorylation reactions (15, 16). Phe111 is in van der Waals contact with the Lys109 residue, which in the crystal structure of the apoprotein forms a salt bridge with Asp57 (13) (Fig. 1A). Such a salt bridge could in principle act as a conformational “switch” in activation (12,13). The other four phenylalanines (Phe8, Phe30, Phe53, and Phe124) are on the opposite side of CheY from the activation site where they can monitor long range movements of eight of the ten secondary structural elements (Ref. 5, Fig. 1).
Fig. 1. Structure of the CheY activation site and location of selected residues.
A, the activation site includes Asp12, Asp13 (site of activating mutations), Asp57 (site of phosphorylation), and Lys109 (putative conformational switch). 4F-Phe reporter group locations for the carboxylate cluster (Phe14) and Lys109 (Phe111) in 19F NMR experiments are also indicated. B, the face of CheY that binds to the flagellar switch has been proposed to include residues Leu24, Glu27, Ala90, Ala99, Ser104, Val108, Pro110, Phe111, Thr112, Ala113, Thr115, and Glu117 (unlabeled side chains) (23, 32). In addition, Ala88 (stippled) and Lys109, which are contiguous with this surface, are implicated by the present study.
Activation of CheY (i.e. generation of CW flagellar rotation) begins with phosphorylation, then the phosphoprotein interacts with the flagellar switch. Phosphorylation of wild-type CheY triggers an allosteric conformational change extending from the activation site over much of the molecule, but has little effect on the 4F-Phel11 probe of the Lys109 region (5). Constitutively activated CheY mutants that function in the absence of phosphorylation offer an alternative window into the conformation of CheY that docks to the flagellar motor and induces CW rotation. The 19F NMR results described here suggest that perturbation of the Lys109 region is in fact a key feature of CheY activation by mutation. Furthermore, mutational activation triggers structural changes which are largely confined to the vicinity of the activation site. Together the 19F NMR studies suggest a multistep model for CheY activation.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Mutant Construction
The E. coli strains RP437 (wild-type for chemotaxis), K0641recA (ΔcheY ΔrecA), and RBB382 (ΔcheA ΔrecA) have been described (10, 17). RBB455 was made by transducing aux::Tn10 #43A phe from NK5138 (a gift from Nancy Kleckner) into HCB440 (a gift from Alan Wolfe) with P1. The complete genotype of RBB455 is Δtsr7021 Δ(cheA-cheY)1590::XhoI(Tn5) aux::Tn10#43A phe thr(am)l leuB6 his4 metF(am)159 eda50 rpsL136 thil ara14 lac Yl mtl1 xyl5 tonA31 tsx78. RBB455 is thus a phenylalanine auxotroph lacking all cytoplasmic chemotaxis proteins and all chemotaxis transducer proteins except Trg. RBB455 was used as the host strain to produce 4F-Phe-labeled CheY protein for 19F NMR. RBB838 was made by transducing cheB::rpsL::KanR::che Y377 from D345 (a gift from Steve Roman) (18) into RP437 with P1.
pRBB40, which expresses cheYZ under the control of ptrp, has been described (10). pMClOO, a KanR derivative of pLCl–28 (19), was a gift from Steve Roman. The EcoRl-BstXI fragment of pRBB40 containing ptrp cheB′ was replaced with pMClOO fragments extending from the EcoRI site in cheA to the BstXI site in cheB using standard techniques. The resulting plasmid, pRBB42, carries ‘cheAW tar tap cheRBYZ flhB’ and was used as a donor to cross cheY alleles into the chromosome of RBB838 as described (18).
Single mutations in cheY, and double mutations at adjacent codons, were constructed by oligonucleotide-directed site-specific mutagenesis as previously described (10). Other double and triple cheY mutants were created by recombining appropriate restriction fragments of pRBB40 in vitro using standard techniques. All mutations were confirmed by DNA sequencing of the final plasmid.
Behavioral Assays
The ability of KO641recA/pRBB40 derivatives expressing various mutant CheY proteins to form chemotactic swarms on soft agar plates was assayed as previously described (10). The rotational behavior of tethered cells was assayed as previously described (20).
Protein Phosphorylation Assays
E. coli CheA and CheY proteins were purified to >95% homogeneity as described (21), except that the final gel filtration step was performed on a Superdex 75 FPLC column (Pharmacia LKB Biotechnology Inc.) equilibrated in TEDG buffer (5) with 0.75 M NaCl. Protein concentrations were determined using the BCA protein assay with bovine serum albumin as a standard (22).
Phosphorylation reactions, carried out as previously described (21), consisted of 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 0.1 mM [γ-32P]ATP (4,000–8,000 cpm/pmol), 1.2 μM CheA, and 12 μM CheY in a volume of 35–40 μl. Those reactions involving CheYD13K, CheYD13R, or the four double mutant proteins including these substitutions contained 8 μM CheA and 24 μ CheY, in order to favor formation of phospho-CheY and detect any phosphotransfer between CheA and these mutant CheY proteins. All reactions were performed at room temperature and were quenched at various times between 10 s and 2 h by adding 2 volumes of 2 × SDS-polyacrylamide gel electrophoresis sample buffer containing 25 mM EDTA. Samples were then analyzed by SDS-polyacrylamide gel electrophoresis and the level of phospho-CheY was determined by phosphorimaging (Molecular Dynamics).
19F NMR Procedures
CheY protein samples were prepared and 19F NMR spectra obtained as described in the preceding paper (5).
RESULTS
Genetic and Behavioral Characterization of Mutations Which Activate CheY
Replacement of Asp13 with Lys results in activation of E. coli CheY, even in the absence of the CheA kinase (10). To further explore the nature of the activation by the cheYD13K mutation, we constructed a series of additional amino acid substitutions at position 13. Among 15 amino acids tested to date, only Arg and Lys caused activation, whereas other large side chains such as Gin, Met, or Tyr did not (10, 14) (Table I). These results strongly suggest that a positive charge at position 13 is critical for CheY activation.
Table I. Amino acid substitutions in the CheY activation site.
Each of the 37 different amino acid substitutions tested at CheY positions 13, 57, or 109 abolish chemotaxis.
| Wild-type amino acid |
Speciesa | Mutant amino acid(s) |
Ref. |
|---|---|---|---|
| Activated mutantsb | |||
| Asp13 | E. coli | Lys | 10 |
| Arg | This work | ||
| Inactivated mutantsc | |||
| Asp13 | E. coli | Asn, Glu | 10 |
| Ala, Cys, Gin, Gly, His, Leu, Phe, Tyr | This work | ||
| S. typhimurium | Asn, Met, Ser, Thr | 14 | |
| Asp67 | E. coli | Asn, Glu, Lys | 10 |
| Ala, Gin, Leu, Ser | This work | ||
| S. typhimurium | Asn, Tyr | 14 | |
| Lys109 | E. coli | Arg, Cys, Gin, Glu, Gly, His, Leu, Met, Phe, Pro, Ser, Thr, Tyr, Val | This work |
| S. typhimurium | Arg | 12 |
The E. coli and S. typhimurium CheY proteins differ by only 3 out of 128 amino acids (19, 26) and are functionally interchangeable (36).
Activated mutant CheY proteins cause extensive CW flagellar rotation when expressed from pRBB40 in either the ΔcheY strain K0641recA or the ΔcheA strain RBB382.
Expression of inactivated mutant E. coli CheY proteins from pRBB40 in the ΔcheY strain K0641 recA resulted in exclusively CCW flagellar rotation, except for the D13Q substitution, which retained some CheA-dependent CW-generating activity. The related D13N substitution similarly retained slight activity in S. typhimurium (14). Expression of inactivated mutant S. typhimurium CheY proteins resulted in exclusively smooth swimming behavior (12, 14); thus these substitutions presumably also fail to support CW flagellar rotation.
A series of substitutions at positions 57 and 109 were next constructed to determine whether these activation site positions could generate additional activating mutations. No substitutions among a total of eight tested at position 57 were sufficient for activation, and each abolished chemotaxis (10, 14) (Table I). It follows that Asp57 is critical for CheY function, consistent with its role as the site of phosphorylation (14). Similarly, all 14 substitutions tested at position 109 abolished activity (12) (Table I), strongly suggesting that Lys109 is essential for wild-type CheY function.
Thus, the known CheY activating mutations each introduce a positive charge at position 13. A key question regarding these mutants is whether phosphorylation is required for their activity. The observation that the cheYD13K and cheYD13R mutations cause tumbling behavior in a ΔcheA host suggests that phosphorylation of CheY may not be necessary for CW flagellar rotation. However, CheA can phosphorylate the CheYD13K and CheYDl3R proteins in vitro, although at a dramatically reduced rate in comparison to wild-type CheY (Ref. 23; see also Table III below). Furthermore, it is conceivable that a kinase related to CheA, or a small molecule phosphodonor, could phosphorylate these mutant CheY proteins in vivo (9, 24). To determine if phosphorylation is necessary for CheYD13K or CheYD13R function, we constructed a series of double mutants containing an activating mutation at position 13 and a second mutation at position 57, the site of phosphorylation (Table II). The CheYD13K and CheYD13R mutants retained activity even upon replacement of Asp57 with either Ala, which cannot be phosphorylated, or Glu, which is phosphorylated very slowly (10) (Tables II and III). The CheYDl3K D57E, CheYD13K D57A, CheYD13R D57E, and CheYD13R D57A proteins were not detectably phosphorylated by CheA in vitro (Table III), and mutation of Asp67 has previously been shown to eliminate phosphorylation by small molecule phosphodonors (5, 24), ruling out the possibility of phosphorylation occurring at a site other than Asp57. Thus CheY phosphorylation is not essential for CW flagellar rotation, and the phosphoryl group must play an indirect role in the activation process. It follows that phosphorylation is likely to activate wild-type CheY by a conformational change; a change in multimeric state was ruled out previously (10, 23).
Table III.
Characterization of selected CheY mutants
| CheY mutant | Swarma | Bias in ΔcheA hostb |
Phe111 shift |
In vitro phosphorylationc |
|---|---|---|---|---|
| ppmd | ||||
| Wild-type | Che+ | CCW | 0 | ++ |
| Salt bridge mutants (see Fig. 2) | ||||
| D57N | Che− | CCW | −0.3 | + |
| D57Q | Che− | CCW | −0.3 | 0 |
| D57A | Che− | CCW | −0.5 | 0 |
| K109Q | Che− | CCW | −0.6 | ++ |
| K109R | Che− | CCW | −0.9 | ++ |
| Activation site mutants (see Fig. 3) | ||||
| D13N | Che− | CCW | −0.1 | ++ |
| F14Y | Che+ | CCW | 0 | NDe |
| N59Q | Che+, Sm | CCW | 0 | ++ |
| A88S | Che−, Lg | CCW | −0.1 | ++ |
| D57E | Che− | CCW | −0.1 | + |
| Activated mutants (see Fig. 5) | ||||
| D13K | Che−, Lg | CW | −0.3 | + |
| D13K D57E | Che−, Lg | CW | −0.4 | 0 |
| D13K D57A | Che−, Lg | CW | −0.7 | 0 |
| D13R | Che−, Lg | CW | −0.4 | + |
| D13RD57E | Che−, Lg | CW | −0.5 | 0 |
| D13R D57A | Che−, Lg | CW | −0.9 | 0 |
| Phe cluster mutants (see Fig 6) | ||||
| K4R | Che+, Sm | CCW | −0.1 | ND |
| L6C | Che+ | CCW | 0 | ND |
| L84I | Che+ | CCW | −0.1 | ND |
| N121L | Che+ | CCW | 0 | ND |
| A113P | Che− | Reversing | −0.1 | ++ |
Swarms of K0641 rec4/pRBB40 derivatives carrying the various mutatations are classified according to their rate of growth in comparison to wild-type: ≥50% of wild-type = Che+; 25–50% of wild-type = Che+, Sm; 10–25% of wild type = Che−, Lg; ≤l0% of wild-type = Che−. In addition, rings are observed on swarms in the two “Che+” classifications, but not on the “Che−” swarms.
Rotational behavior of tethered RBB382/pRBB40 derivatives carrying the various cheY mutations. “CCW” denotes exclusively counter-clockwise flagellar rotation, whereas “CW” denotes varying degrees of predominantly clockwise flagellar rotation.
In vitro phosphorylation of CheY by phosphotransfer from CheA. Relative amount of phospho-CheY observed after a 30-min reaction: 25–100% of wild-type = ++; 1–25% of wild-type = +, not detectably phosphorylated = 0.
The error in determining resonance frequency inherent in the 19F NMR technique is ~0.1 ppm; thus only frequency shifts larger than this are significant (5).
ND, not determined.
Table II.
Dependence of CheY activated mutants on Asp51 and Lys109
| cheY Mutant | Bias in ΔcheA Hosta |
|---|---|
| Wild-type | CCW |
| Activated Asp13 single mutants | |
| D13K | CW |
| D13R | CW |
| Asp13/Asp57 double mutants | |
| D13K D57A | CW |
| D13K D57E | CW |
| D13K D57N | Mostly CCW |
| D13K D57Q | CCW |
| D13R D57A | CW |
| D13R D57E | CW |
| D13R D57N | CCW |
| D13R D57Q | CCW |
| Asp13/Lys109 double mutants | |
| D13K K109E | CCW |
| D13K K109L | CCW |
| D13K K109Q | CCW |
| D13K K109R | CCW |
| D13R K109E | CCW |
| D13R K109L | CCW |
| D13R K109Q | CCW |
| D13R K109R | CCW |
Rotational behavior of tethered RBB382/pRBB40 derivatives carrying the various cheY mutations. “CCW” denotes exclusively counterclockwise flagellar rotation unless otherwise noted, whereas “CW” denotes varying degrees of predominantly clockwise flagellar rotation. Note that each of the single CheY mutants at positions 57 and 109 (D57A, D57E, D57N, D57Q, K109E, K109L, K109Q, K109R) are CCW by themselves.
It is worth noting that although Ala or Glu substitutions at position 57 were functionally tolerated in the activated mutants, Asn or Gin substitutions were not (Table II). Furthermore, the Ala substitution at position 57 slightly reduced CW flagellar rotation caused by the cheYD13K mutation and actually enhanced activity of the cheYD13R mutation (data not shown). These observations suggest that the precise structure of the activation site region is important for CheY activation.
The behavioral assays described so far were performed using cells that expressed the mutant CheY proteins from a multicopy plasmid. To determine if the activated mutant proteins retained function when expressed at single copy levels, the cheYD13K, cheYD13K D57A, cheYD13R, and cheYD13R D57A alleles were crossed into the chromosome of an otherwise wild-type host. Each mutant retained the ability to generate CW rotation, although it was significantly less pronounced than in the multicopy case (data not shown). Wild-type phospho-CheY (in a ΔcheZ host) was more efficient at generating CW rotation than any of the four mutants tested.
To examine the role of Lys109 in the activated cheYmutants, double mutants containing Arg or Lys activating substitutions at position 13 and Arg (basic), Gln (polar), Glu (acidic), or Leu (hydrophobic) substitutions at position 109 were constructed. All eight combinations completely abolished CW rotation in a Δche A strain (Table II). This indicates that Lys109 is essential for the CW signaling activity of the activated mutants, just as it is for wild-type phospho-CheY.
Mutations Suggested by Other Response Regulator Proteins
The chemotaxis signal transduction network belongs to a diverse family of bacterial two-component regulatory systems defined by amino acid sequence similarity (8, 25–27). The residues corresponding to Asp13, Asp67, and Lys109 of CheY are almost universally conserved among the response regulator proteins (9, 25). Two exceptions are Caulobacter crescentus FlbD and Myxococcus xanthus FrzG, which carry Lys at position 13 and Arg or Leu at position 109 (28, 29). The unusual activation site residues found in FlbD or FrzG might provide insight into the mechanism of CheY activation and thus were explored further. The combinations of Lys at position 13 with Arg or Leu at position 109, which are presumably active in FlbD and FrzG, were not functional in CheY (Table II). However, FlbD and FrzG also contain Gly at position 12, rather than a very highly conserved acidic (Asp or Glu) residue (25, 28, 29). The analysis was therefore extended to examine the activity of cheYD12G, cheYD12G D13K, cheYD12G D13K K109L, and cheYD12G D13KK109R mutations. All four mutations eliminated CW rotation in KO641recA/pRBB40 (data not shown). It follows that the Gly12 substitution does not compensate for the absence of Lys109.
19F NMR Analysis of the Activated Conformation: An Assay for Perturbations of Lys109
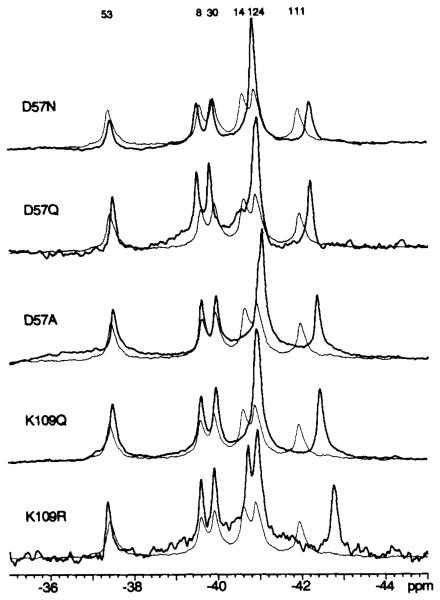
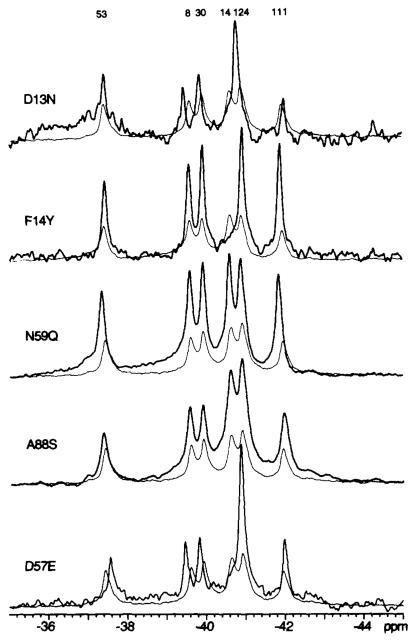
The effect of activation on Lys109 is of particular interest due to its putative function as a conformational switch (12,13). Thus, protein engineering was employed to determine the effect of perturbations in the Lys109 region on the 19F NMR resonances of 4F-Phe-labeled CheY. 19F NMR spectra were obtained for mutant CheY proteins bearing substitutions at positions 57 or 109 that would be expected to prevent formation of the Asp57-Lys109 salt bridge observed in the crystal structure of apo-CheY (13) (Che-YD57N, CheYD57Q, CheYD57A, CheYKl09Q). A key focus was the effect of such substitutions on the chemical shift of the 4F-Phe111 resonance, since this residue is in van der Waals contact with the Lys109 side chain and is located in the same β35-α5 loop as Lys109. An upfield frequency shift (0.3–0.6 ppm) of the 4F-Phe111 resonance was observed for each of the indicated electrostatic substitutions (Fig. 2); these shifts are significantly larger than the 0.1 ppm measurement error (see “Materials and Methods” of Ref. 5). A complementary test used a steric substitution at the 109 position (K109R) which, due to the critical dependence of salt bridges on favorable steric interactions (30), might also prevent salt bridge formation. A large upfield frequency shift (0.9 ppm) was observed for this mutant (Fig. 2, Table III), even larger than the shift observed for the electrostatic substitutions. In contrast, alteration of other activation site residues that are not expected to be directly coupled to the Lys109 region (CheYD13N, CheYFHY, CheYN59Q, and CheYA88S) did not perturb the 4F-Phe111 resonance (Fig. 3, Table III). Therefore, the chemical shift of the 4F-Phe111 resonance can be utilized as a specific spectroscopic signature to monitor perturbations of the Lys109 region in the activated CheY mutants.
Fig. 2. Effect of mutations that perturb the Asp57-Lys109 interaction on the 19F NMR spectra of 4F-Phe-labeled CheY.
Each spectral pair consists of the spectrum from wild-type CheY (fine line) overlaid on the spectrum from the indicated mutant (bold line). Sample parameters were: 2 mM CheY, 2 mM MgCl2, 50 mM KCl, 50 mM NaCl, 50 mM Tris-HCl, pH 7.0, 10% D20, 50 μm 5-fluorotryptophan as internal frequency standard, 25 °C. Assignments are from the preceding paper (Ref. 5, Fig. 3).
Fig. 3. Effect of control activation site mutations on the 19F NMR spectra of 4F-Phe-labeled CheY.
Each spectral pair consists of the spectrum from wild-type CheY (fine line) overlaid on the spectrum from the indicated mutant (bold line). Sample parameters were as described in the legend to Fig. 2.
Assignment of 19F NMR Resonances in Activated CheY Mutants
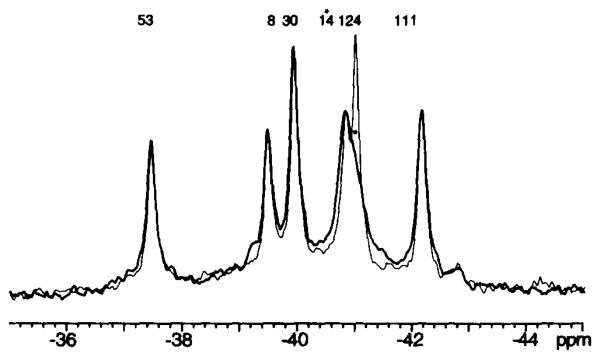
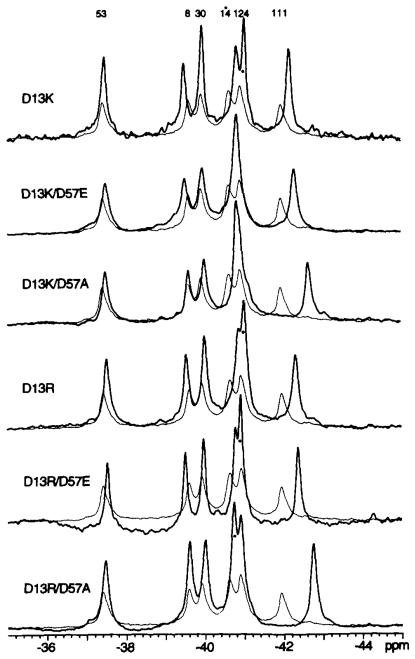
In order to interpret 19F NMR spectra of the various activated mutant CheY proteins, it is necessary to eliminate ambiguities in the resonance assignments. The six 4F-Phe resonances in the 19F NMR spectrum of wild-type CheY were assigned as described in the preceding paper (5). Most resonances in mutant CheY proteins can be straightforwardly assigned by comparison with the wild-type resonances. However, it was important to directly assign the 4F-Phe14 resonance, since the activated mutants all contain an altered residue at the adjacent position 13, which could yield dramatic frequency shifts. The 4F-Phe14 residue provides the most solvent exposed probe side chain in CheY (5, 13, 31), such that the 4F-Phe14 resonance is significantly broadened by collision with the water-soluble paramagnetic probe Gd(III)·EDTA, via a magnetic dipole-dipole interaction between the 19F nucleus and the Gd(III) unpaired electrons (S = 7/2) (5). Spectra of each of the six activated mutant CheY proteins were thus obtained in the presence and absence of Gd(III)·EDTA, enabling resolution of the overlapping 4F-Phe14 and 4F-Phe124 resonances. The most broadened 4F-Phe14 resonance was found to be upfield of the less broadened 4F-Phe124 resonance in the CheYD13K mutant, rather than downfield as in wild-type CheY (Fig. 4). The relative positions of the 4F-Phe14 and 4F-Phe124 resonances were similarly reversed in the CheYD13R and CheYD13R D57E spectra (Fig. 5).
Fig. 4. Use of Gd(III) EDTA to assign the 4F-Phe14 19F NMR resonance from 4F-Phe-labeled CheYD13K.
Spectra taken in the absence (fine line) or presence (bold line) of the aqueous paramagnetic probe Gd(III)·EDTA are displayed. Sample parameters were as described in the legend to Fig. 2, except MgCl2 was omitted, and additional divalent-free KCl was substituted for the NaCl.
Fig. 5. Effect of activating mutations on the 19F NMR spectra of 4F-Phe-labeled CheY.
Each spectral pair consists of the spectrum from wild-type CheY (fine line) overlaid with the spectrum from the indicated mutant (bold line). Assignment of the 4F-Phel4 resonance, determined with Gd(III)·EDTA, is indicated by the asterisk. Sample parameters were as described in the legend to Fig. 2.
Effect of cheY-activating Mutations on the Asp57-Lys109 Salt Bridge
The 19F NMR spectra of each of the six available activated CheY mutants are displayed in Fig. 5. As expected, the position of the 4F-Phe14 resonance, adjacent to the activating mutation at position 13, was in each case shifted relative to its position in the spectrum of wild-type CheY. However, the most striking difference between the spectra of the mutant and wild-type proteins was that the 4F-Phe111 resonance exhibited large upfield frequency shifts of 0.3–0.9 ppm in each of the activated mutants (Table III). These were similar to the shifts observed for 4F-Phe111 when Lys109 was perturbed by disrupting the Asp57-Lys109 interaction (Fig. 2). Such results strongly suggest that the activating mutations generate a conformational change in the vicinity of Lys109. Moreover, the activated mutants CheYDl3K D57A and CheYD13R D57A provide further evidence that the electrostatic interaction between the 57 and 109 positions is not required in the activated state, since these proteins are able to regulate the motor without any possibility of a Lys109 salt bridge to the 57 position (Table III). Together these results are consistent with the proposal that the electrostatic interaction between the 57 and 109 positions is weakened or broken in the activated state.
Long Range Conformational Effects in CheY Mutants
In contrast to the 4F-Phe14 and 4F-Phe111 resonances, the 4F-Phe8, 4F-Phe30, 4F-Phe53, and 4F-Phe124 resonances in the spectra of the activated mutants exhibited small or undetectable frequency shifts (Fig. 5). In general, the shifts were in the same direction (upfield for 4F-Phe30 and 4F-Phe53; down-field for 4F-Phe8 and 4F-Phe124) as observed for phospho-CheY, but smaller in magnitude (5). However, the failure to consistently observe these resonance shifts in all six mutants suggests the structural changes that cause them are not essential for activation. For example, the 4F-Phe8 resonance did not shift in CheYD13K D57A and CheYD13R D57A, but shifted downfield in the other four mutants.
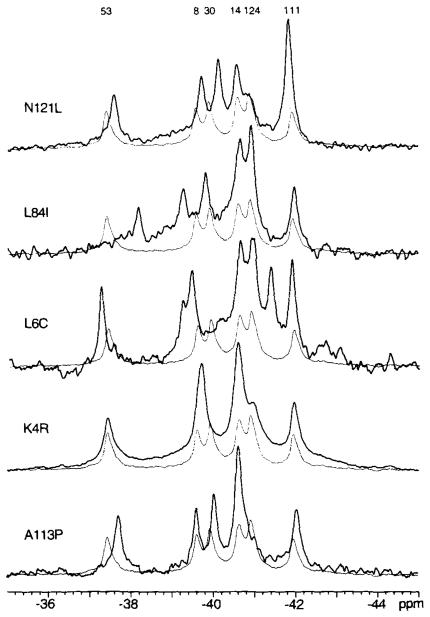
The long range conformational change triggered by phosphorylation (5) indicates that it is possible to couple structural changes in the activation site of CheY to changes on the opposite side of the molecule. However, the mechanism by which phosphorylation affects the global structure of CheY appears to be highly specific, since structural changes are not easily transmitted within the molecule. Specifically, the resonances of the Phe cluster were not significantly perturbed by 10 of 16 mutations in the activation site (Figs. 2, 3, and 5). Similarly, amino acid substitutions in the van der Waals environment of the Phe cluster (CheYK4R, CheYL6C, CheYL84I, CheYN121L) resulted in large frequency shifts for the resonances from the cluster, but had little effect on the activation site probes 4F-Phe14 and 4F-Phe111 (Fig. 6).
Fig. 6. Effect of mutations perturbing the Phe cluster on the 19F NMR spectra of 4F-Phe-labeled CheY.
Each spectral pair consists of the spectrum from wild-type CheY (fine line) overlaid with the spectrum from the indicated mutant (bold line). Sample parameters were as described in the legend to Fig. 2.
Ala88 May Interact with the Flagellar Switch
Identification of the CheY docking surface that interacts with the flagellar switch is necessary for a complete understanding of the mechanism of activation. Progress has been made in this regard from analysis of second site suppressor mutations that restore interaction between CheY and the switch proteins (23, 32) (Fig. 1B). A complementary approach is to identify residues whose alteration specifically reduces or abolishes the CheY/switch interaction. Ala88 appears to have these properties. The cheYA88S mutation has a minimal effect on CheY structure, but a large effect on signaling function. The 19F NMR spectrum of the CheYA88S mutant is indistinguishable from that of wild-type CheY (Fig. 3), and spectra of the two proteins obtained during phosphorylation reactions were also the same (data not shown). However, the CheYA88S mutant has a reduced ability to generate CW flagellar rotation. Virtually all rotating tethered KO641recA/pRBB40 cells expressing wild-type CheY exhibited reversing flagellar rotation with no strong rotational bias (data not shown). In contrast, half of the rotating tethered cells expressing CheYA88S exhibited exclusively counterclockwise rotation. These observations are consistent with the proposal that Ala88 interacts with the flagellar switch.
DISCUSSION
Structural Features of CheY Activation
We have undertaken a combined physical, genetic, and biochemical investigation of the CheY activation mechanism. The following key conclusions are suggested by the results described here and in the accompanying paper (5).
The Inactive Conformation
(a) Mg(II) binding to the activation site of CheY does not perturb the Lys109 residue proposed to be involved in the activation switch (Ref. 5, Fig. 5). (b) Larger divalent cations (Ca(II), Sr(II), Ba(II)) do perturb Lys109 (Ref. 5, Fig. 5). (c) Divalent cation binding generates a conformational change localized predominantly in the activation site region (Ref. 5, Fig. 5).
The Phosphorylated Conformation
(a) Phosphorylation of Asp57 does not perturb Lys109 (Ref. 5, Fig. 8). (b) Phosphorylation triggers an allosteric conformational change detected up to 20 A from Asp57 (Ref. 5, Fig. 8).
The Mutation-activated Conformation
(a) Activation of CheY can occur in the absence of phosphorylation (Tables II and III). (b) Positively charged Arg or Lys side chains at position 13 activate CheY (Table I), (c) The Lys109 region is significantly perturbed in the activated mutant CheY proteins (Fig. 5). (d) Activation of CheY by mutation results in structural changes that appear to be primarily localized to the activation site (Fig. 5).
Docking to the Motor
(a) Activating mutations give rise to a different conformation than phosphorylation, indicating that one or both conformations must change upon docking to the motor (Fig. 5; and Ref. 5, Fig. 8). (b) Lys109 is essential for activation of CheY either by phosphorylation or mutation (Tables I and II), (c) An electrostatic interaction between Lys109 and the 57 position is unnecessary for docking, since the CheYD13K D57A and CheYDl3R D57A mutants can signal (Table II). However, breaking the Asp57-Lys109 salt bridge is not sufficient for activation (Table III), (d) Ala88 appears to lie on the motor docking surface (Figs. 1B and 3; Table III).
Together these observations may be synthesized into a multistep model of CheY activation in which phosphorylation and the activating mutations have similar structural consequences when docked to the motor, but not in solution.
Two-step Working Model of CheY Activation
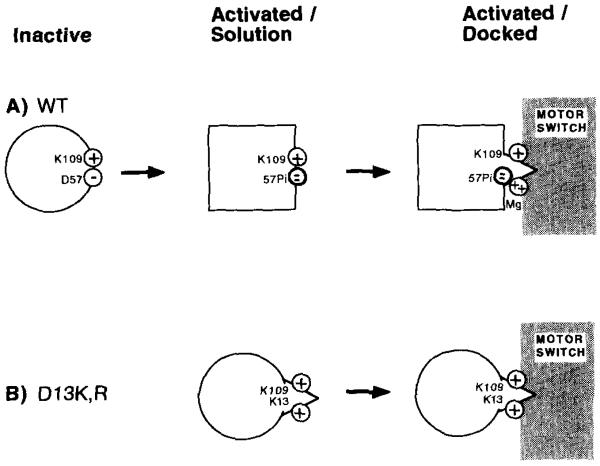
The following working model of CheY activation is the simplest model consistent with all available information (Fig. 7). In the absence of a divalent metal ion, there is a salt bridge between Asp57 and Lys109 (13).
Fig. 7. Two-step working model of CheY activation.
The indicated schematic model accounts for observations described in this and the preceding paper (5). See text for detailed discussion. A, activation by phosphorylation. Shown are the Asp57 and Lys109 side chain charges in the activation site of wild-type CheY. Upon phosphorylation of Asp57, the Lys109 side chain retains its original conformation, perhaps forming a salt bridge with the phosphoryl group. When phospho-CheY docks to the flagellar switch, a Mg(II) ion binds to the phosphoryl group and breaks any remaining electrostatic interaction between Lys109 and the acylphosphate. The Mg(II) and repositioned Lys109 interact with the switch to cause CW flagellar rotation. B, activation by mutation. A positively charged Lys or Arg side chain at position 13 mimics the position of the surface Mg(II) in phosphorylated wild-type CheY. This positive charge also disrupts the Asp57-Lys109 electrostatic interaction, together yielding partial activation.
In the initial inactive state, Asp57 coordinates Mg(II) bound in the activation site (16). The lack of a 4F-Phe111 frequency shift upon Mg(II) binding (Ref. 5, Fig. 5) indicates that the position of Lys109 is not significantly perturbed. Thus, either the Asp57 carboxylate group bridges the Lys and Mg(II) charges, as has been observed in other phospho-chemistry enzymes (33–35), or the electrostatic interaction between Asp57 and Lys109 is broken, but Lys109 does not relocate.
The observation that diverse substitutions at the 57 and 109 positions cause similar 4F-Phe111 frequency shifts in the inactive state (Fig. 2) can be explained in two ways. The simplest hypothesis is that the salt bridge between Asp67 and Lys109 in the apo-site remains intact when Mg(II) binds, but is broken by specific mutations at these positions, thereby yielding similar perturbations of the 4F-Phe111 probe. Alternatively, if the Asp57-Lys109 salt bridge is already broken in the Mg(II)-occupied conformer, one must propose that the indicated substitutions generate structural perturbations which fortuitously yield similar 4F-Phe111 frequency shifts. Additional studies are needed to characterize the status of the salt bridge in the solution structure of CheY.
In the first activation step, Asp57 is phosphorylated. The Lys109 side chain repositions only slightly (Fig. 7A), thereby explaining the lack of perturbation of the 4F-Phe111 resonance. In this conformation Lys109 might form a salt bridge with the phosphoryl group. An analogous lack of perturbation is observed for this resonance in the D57E mutant (Fig. 3) which, like phosphorylation, is proposed to move the negative charge of the 57 position to a more distal location on its side chain. Phosphorylation of Asp57 causes a long range conformational change in CheY (5); however, this global change does not occur in the activated CheY mutants (Figs. 5 and 7B), and thus must be of secondary importance for generating CW flagellar rotation.
The second activation step occurs when phospho-CheY docks to the flagellar switch complex. A Mg(II) ion is proposed to bind to the phosphoryl group and thus break any remaining Lys109-phosphoryl group electrostatic interaction (Fig. 7A). It is unclear whether the original Mg(II) ion is repositioned or a second Mg(II) ion enters the activation site. The essential Lys109 side chain and Mg(II) ion are proposed to interact directly with the flagellar switch to cause CW flagellar rotation. This second activation step has not been observed experimentally, but is postulated in order to account for the required (i) Lys13 or Arg13 and (ii) Lys109 positive charges in the activated mutants.
This model can account for the phenotypes of numerous cheY mutations. The positively charged Arg or Lys side chain at position 13 in the activated mutants is proposed to mimic the Mg(II) that interacts with the switch in phospho-CheY (Fig. 7B). The presence of the basic residue at position 13 is also proposed to disrupt the Asp57-Lys109 electrostatic interaction, allowing Lys109 to bind to the switch. Evidence consistent with this proposed disruption of the Asp57-Lys109 interaction is provided by the cheYD13K D57A and cheYD13R DS7A mutations, which prevent salt bridge formation yet retain signaling (Table II). Finally, although CheYK109R is stably phosphorylated (12), it is proposed to be incapable of generating CW flagellar rotation because the Arg109 side chain cannot interact properly with the switch (Fig. 7B). All mutants lacking Lys109 would similarly be defective (Table I).
Interaction of CheY and the Flagellar Switch
A face of the CheY molecule that interacts with the flagellar switch has been tentatively identified, based on the location of che Y mutations that suppress mutations in genes encoding switch proteins (23, 32), as illustrated in Fig. 1B. We have made three further observations consistent with the proposal that this region is responsible for interaction of CheY with the flagellar switch. First, the region includes Lys109, which is essential for generation of CW flagellar rotation by either phospho-CheY or the activated mutant CheY proteins (Tables I and II). Second, the putative switch interaction region is adjacent to the phosphorylation site, and activation of CheY can occur in the absence of a long range conformational change (Fig. 5). Third, evidence has been presented that Ala88, which is located on the border of the proposed docking region, could directly interact with the switch (Fig. 1B).
Acknowledgments
We are indebted to Karl Volz for access to the crystal structure of E. coli CheY prior to publication. We thank Nancy Kleckner, Steve Roman, and Alan Wolfe for providing bacterial strains and plasmids, and Lisa Alex, Stephan Schuster, and Ron Swanson for comments on the manuscript.
Footnotes
This work was supported by National Research Service Award Fellowship AI07798 (to R. B. B.) and National Institutes of Health Grants AI19296 (to M. I. S.) and GM40731 (to J. J. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: CW, clockwise; 4F-Phe, 4-fluoro-phenylalanine.
REFERENCES
- 1.Cantley LC, Auger KR, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 2.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Nature. 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 3.Milburn MV, Tong L, DeVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Science. 1991;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 4.Kjelsberg MA, Cotecchia S, Ostrowski J, Caron MG, Lefkowitz RJ. J. Biol. Chem. 1992;267:1430–1433. [PubMed] [Google Scholar]
- 5.Drake SK, Bourret RB, Luck LA, Simon MI, Falke JJ. J. Biol. Chem. 1993;268:13081–13088. [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart RC, Dahlquist FW. Chem. Rev. 1987;87:997–1025. [Google Scholar]
- 7.Macnab RM. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt FC, editor. American Society of Microbiology; Washington, D. C.: 1987. pp. 732–759. [Google Scholar]
- 8.Bourret RB, Borkovieh KA, Simon MI. Annu. Rev. Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- 9.Stock JB, Lukat GS, Stock AM. Annu. Rev. Biophys. Biophys. Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- 10.Bourret RB, Hess JF, Simon MI. Proc. Natl. Acad. Sci. U. S. A. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess JF, Oosawa K, Kaplan N, Simon MI. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 12.Lukat GS, Lee BH, Mottonen JM, Stock AM, Stock JB. J. Biol. Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- 13.Volz K, Matsumura P. J. Biol. Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 14.Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL, Koshland DE., Jr. J. Biol. Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 15.Lukat GS, Stock AM, Stock JB. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- 16.Stock JB, Surette MG, McCleary WR, Stock AM. J. Biol. Chem. 1992;267:19753–19756. [PubMed] [Google Scholar]
- 17.Parkinson JS, Houts SE. J. Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell CB, Dahlquist FW. J. Bacteriol. 1989;171:2614–2618. doi: 10.1128/jb.171.5.2614-2618.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumura P, Rydel JJ, Linzmeier R, Vacante D. J. Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray D, Bourret RB, Simon MI. Mol. Biol. Cell. 1993 doi: 10.1091/mbc.4.5.469. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess JF, Bourret RB, Simon MI. Methods Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- 22.Stoscheck CM. Methods Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 23.Roman S, Meyers M, Volz K, Matsumura P. J. Bacteriol. 1992;174:6247–6255. doi: 10.1128/jb.174.19.6247-6255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukat GS, McCleary WR, Stock AM, Stock JB. Proc. Natl. Acad. Sci. U. S. A. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stock JB, Ninfa AJ, Stock AM. Microbiol. Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stock A, Koshland DE, Jr., Stock J. Proc. Natl. Acad. Sci. U. S. A. 1985;82:7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stock A, Chen T, Welsh D, Stock J. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramakrishnan G, Newton A. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2369–2373. doi: 10.1073/pnas.87.6.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCleary WR, McBride MJ, Zusman DR. J. Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dao-pin S, Sauer U, Nicholson H, Matthews BW. Bio-chemistry. 1991;30:7142–7153. doi: 10.1021/bi00243a015. [DOI] [PubMed] [Google Scholar]
- 31.Stock AM, Mottonen JM, Stock JB, Schutt CE. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 32.Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM. J. Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight S, Anderson I, Branden CI. J. Mol. Biol. 1990;215:113–160. doi: 10.1016/S0022-2836(05)80100-7. [DOI] [PubMed] [Google Scholar]
- 34.Lebioda L, Stec B. Biochemistry. 1991;30:2817–2822. doi: 10.1021/bi00225a012. [DOI] [PubMed] [Google Scholar]
- 35.Lundqvist T, Schneider G. Biochemistry. 1991;30:904–908. doi: 10.1021/bi00218a004. [DOI] [PubMed] [Google Scholar]
- 36.DeFranco AL, Parkinson JS, Koshland DE., Jr. J. Bacteriol. 1979;139:107–114. doi: 10.1128/jb.139.1.107-114.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]