Abstract
The Escherichia coli d-galactose and d-glucose receptor is a two-domain structure with a sugar-binding site at the interface between domains. The structure of the closed cleft containing bound d-glucose has been determined crystallographically, but the open cleft remains to be characterized. The present study illustrates a generalizable approach that is used to detect and analyze both the open- and closed-cleft conformations in solution. A 19F nucleus located inside the cleft is monitored by 19F NMR. When the cleft is occupied by d-glucose, the 19F nucleus is found to be inaccessible to the aqueous paramagnetic probe Gd•EDTA, verifying that the occupied cleft is closed in solution and inaccessible to bulk solvent. When the cleft is empty, the 19F nucleus becomes accessible to the paramagnet such that the distance of closest approach is r ≤ 10 Å, indicating that the empty cleft opens at least transiently by an angle θ ≥ 18 ± 3°.
Substrate binding and dissociation is often controlled by a hinge mechanism that regulates access to a substrate site. When the hinge is closed, substrate is trapped in the site and inaccessible to bulk solvent, enabling active site chemistry to be carried out in a controlled environment where solvation may differ substantially from that provided by the aqueous phase. In addition, bound substrate can be retained for relatively long times, enabling sequestering of a substrate for prolonged activation of a receptor or for a slow enzymatic step. The most common type of hinge-regulated structure is the substrate site in a deep cleft between two protein domains; in this case, access is regulated by opening and closing the cleft (Bennett & Huber, 1984; Faber & Matthews, 1990). Such a hinged cleft is a common feature of lyases and binding proteins, including the serine proteases, the RNAses, the lysozymes, and the bacterial periplasmic binding proteins.
In the classic crystallographic study of cleft conformations in hexokinase, the substrate-occupied cleft is closed and shields the substrate from solvent, while the empty cleft opens to a hinge angle of 12° and allows bulk solvent to enter (Bennett & Steitz, 1980). Recent crystallographic studies of lysozyme present a more complex picture in which different crystal environments yield hinge angles ranging from 0° to 32° (Faber & Matthews, 1990; Dobson, 1990). While the crystallographic approach can provide strong evidence for hinge motions, it is important to consider that crystal packing forces could significantly perturb the hinge angle, particularly in the case of the empty cleft conformer, which is likely to be significantly more flexible than the substrate-occupied structure. Ideally the structural transitions of the cleft should also be studied in solution.
We are currently investigating the solution structural dynamics of the d-galactose and d-glucose receptor, a periplasmic receptor of Escherichia coli possessing a sugar-binding cleft between its two domains (Hazelbauer & Adler, 1971). The structure of this 36-kDa protein has been crystallographically determined to 1,9-Å resolution by Quiocho and co-workers (Vyas et al., 1987, 1988); in this conformation, the cleft contains bound d-glucose and is closed. 19F NMR studies have indicated that sugar binding is allosterically linked to other regions of the protein (Luck & Falke, 1991a,b): such allostery is required so that the buried sugar-binding site can control at least one surface site used in docking to transmembrane effector proteins. The receptor belongs to a structurally homologous class of periplasmic binding proteins (Quiocho et al., 1987), and it is generally believed that the entire class exhibits the hinged cleft mode of substrate binding (Mao et al., 1982; Miller et al., 1983; Sack et al., 1990), although the open cleft has not been observed in solution.
19F NMR provides a direct means to detect cleft opening and to probe the dimensions of the unoccupied cleft. As previously described, 19F can be incorporated into the receptor by substitution of 5-fluorotryptophan into its five tryptophan positions shown in Figure 1A (Luck & Falke, 1991a). The tryptophan at the 183 position is particularly useful because it lies in the sugar-binding cleft where its indole ring is in van der Waals contact with the carbon backbone of the bound d-glucose molecule, as illustrated in Figure 1B (Vyas et al., 1988). A testable prediction of the hinged cleft model is that Trp 183 will be fully shielded from bulk solvent in the presence of saturating substrate, which stabilizes the closed conformation of the cleft, but will be exposed to bulk water upon removal of substrate, which favors the open conformation. The present 19F NMR study uses an aqueous paramagnetic probe to test the solvent accessibility of 5F-Trp 183, which is verified to be inaccessible when d-glucose is bound and the cleft closed. In contrast, when the cleft is empty, the 5F-Trp 183 resonance is broadened by the close approach of the paramagnet, indicating that the cleft is at least transiently open to bulk solvent and enabling determination of the minimum angle of opening.
FIGURE 1.
Locations of tryptophan residues in the E. coli d-galactose and d-glucose receptor. (A) The α-carbon backbone structure of the receptor (Vyas et al., 1987). The structure contains a d-glucose molecule bound in the substrate cleft between the two domains and a Ca(II) ion (sphere) in the metal-binding site. The positions of five Trp residues that serve as fluorine-labeling sites in the present study are indicated. (B) Stereoview of the substrate cleft in the same structure from a slightly different angle. Indicated are the d-glucose molecule (lower ring) and the substrate-cleft residue Trp 183 (upper ring), the latter fluorinated in the present study and used as a probe. The isolated vertices represent the oxygen atoms of tightly bound water molecules. The cleft leads to bulk solvent, seen as the void on the right.
Materials and Methods
5-Fluorotryptophan-Labeled Receptor
The techniques used to generate and isolate 5F-Trp-labeled receptor were described in detail elsewhere (Luck & Falke, 1991a). Briefly, the plasmid pVB2 (Scholle et al., 1987) was expressed in the E. coli tryptophan auxotroph W3110 Trp A33 (Drapeau et al., 1968) by growth at 37 °C in the following media: 65 μg/mL 5F-Trp (Sigma), 16 μg/mL Trp (Sigma), M9 salts (Miller, 1972) with twice the standard phosphate salts, 2% (w/v) ca-samino acids (Difco Laboratories), 1% (v/v) glycerol as a carbon source, 20 μg/mL methionine, 10 μg/mL thiamine, 0.5 mM D-(+)-fucose [an inducer of the receptor promoter (Scholle et al., 1987)], and 50 μg/mL ampicillin. Cells were grown in this media at 37 °C, harvested, then standard procedures (Kellerman & Ferenci, 1982) were used to release the contents of the periplasm, predominantly the labeled receptor. The lysate was concentrated by ultrafiltration and dialyzed against a series of buffers: first a buffer containing 3.0 M guanidine HC1 to quantitatively release bound sugar by partially unfolding the receptor (Miller et al., 1980); then a refolding buffer containing 100 mM KC1, 10 mM Tris, pH 7.1 with HC1, and 0.5 mM CaCl2. Dialyzed samples were again concentrated to yield a final receptor concentration of 200–500 μM for NMR. Purity was checked by SDS–polyacrylamide electrophoresis with staining by Coomassie blue R-250; samples were typically >90% receptor.
19F NMR Measurements
NMR samples in the final dialysis buffer (above) contained also 10% D20 (v/v) as the lock solvent, 75 μM 3F-Phe as an internal frequency standard (−38.0 ppm relative to TFA at 0 ppm), and 5 mM CaCl2 to competitively exclude Gd(III) from the receptor Ca(II) site. Gd•EDTA was added as a stock solution (100 mM GdCl3, 500 mM EDTA, pH 7.1 with NaOH) to yield the final concentrations indicated in the figures. 19F NMR spectra were obtained at 470 MHz on a Varian VXR 500 fitted with a 5-mm 1H/19F probe. Standard coupled spectral parameters were a 12000-Hz spectral width, 16K data points, 60° pulse width, 0.68-s acquisition time, 1,0-s relaxation delay, 25-Hz line broadening, and temperature control at 25 °C. Other parameters were as previously described (Luck & Falke, 1991a).
Paramagnetic 19F NMR line broadenings of the 5F-Trp 183 resonance were determined as increases in the line width at half-height. Line width was graphically determined and was straightforward to measure in spectra of the receptor•d-glucose complex where the resonance was well resolved. In spectra of the sugar-empty receptor, the upfield side of the 5F-Trp 183 resonance overlaps the 5F-Trp 127 resonances, particularly at high Gd-EDTA concentrations. In this case the baseline was graphically determined by a line connecting the baselines 1 ppm upfield and downfield of the 5F-Trp 127/183 resonance group. The half-width at half-height was measured between the central vertical of the 5F-Trp 183 resonance and its downfield edge, which is well-resolved, then this half-width was doubled to yield the line width. Increases in line width were quantitatively consistent with the observed decreases in resonance height (Schmidt & Kuntz, 1984), providing evidence that the measurement procedure was accurate. The standard deviation of replicate line broadenings from different spectra was ±10 Hz (n = 3).
Molecular Graphics and Distance Measurements
Receptor coordinates provided by Quiocho and co-workers (Vyas et al., 1987) were displayed by Biosym Technologies insight software on an Evans and Sutherland PS-300 color graphics system driven by a VAXStation 3500. Gd-EDTA coordinates from the Cambridge Data Base (BIFZEV; Templeton et al., 1982) were viewed on the same system for distance measurements, while the software package SHELXTL from Siemens Analytical X-Ray Instruments was used to determine the least-squares plane and to plot perspective drawings of structures.
Results
Use of 19F NMR To Detect Cleft Opening: General Approach
The 19F nucleus is a simple I = 1/2 spin system exhibiting high sensitivity (0.833 relative to 1H) and frequency dispersion (>10-fold that of 1H). As a result 19F is well suited as an observation nucleus in NMR studies of macromolecules too large or too insoluble for multidimensional NMR techniques (Sykes et al., 1974; Pratt & Ho, 1975; Lu et al., 1976; Kooistra & Richards, 1978; Post et al., 1984; Wilson & Dahlquist, 1985; Peerson et al., 1990; Li et al., 1990; Luck & Falke, 1991a,b). We have previously incorporated 5F-Trp into the D-galactose and D-glucose receptor by overexpressing a plasmid containing the receptor gene in the E. coli tryptophan auxotroph W3110 Trp A33 with addition of 5F-Trp to the growth medium (Luck & Falke, 1991a). The resulting labeling efficiency is 65 ± 10% 5F-Trp at each of the five tryptophan positions in the receptor. The subtle F/H substitution leaves the receptor structure essentially intact as judged by substrate binding equilibria and kinetics: the affinity of the fluorine-labeled sugar-binding cleft for D-galactose is decreased <3.0-fold, while the rate of Tb3+ dissociation from the receptor's EF-hand-like Ca2+-binding site (Vyas et al., 1987; Snyder et al., 1990) is increased <2.5-fold. All 5F-Trp resonances have been assigned in the primary structure by replacement of individual tryptophans with tyrosine or phenylalanine via oligonucleotide-directed mutagenesis (Luck & Falke, 1991a). As previously discussed, the 5F-Trp 127 and 133 residues at the base of the Ca(II)-binding loop each give rise to a pair of resonances (Figures 3A and 4A), indicating that this region of the protein possesses two distinct conformational populations that exchange slowly on the NMR time scale, probably representing two different covalent structures (Luck & Falke, 1991a,b).
FIGURE 3.
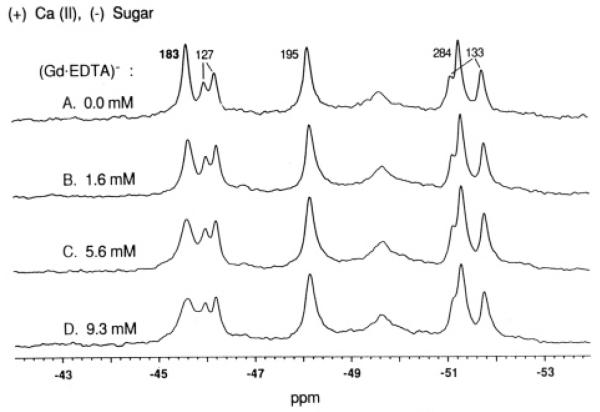
Effect of paramagnetic probe on the l9F NMR resonances of the receptor: substrate cleft occupied by d-glucose, Shown are a series of l9F NMR spectra for the 5F-Trp-labeled receptor saturated with d-glucose. Spectra were obtained after adding the indicated concentrations of Gd(HI) and a 5-fold larger concentration (4-fold molar excess) of EDTA, The assignment of each resonance to a specific 5F-Trp residue is given; that of the 5F-Trp 183 resonance from the sugar-binding cleft is shown in bold. All samples contained 100 mM KC1, 10 mM Tris, pH 7.1, 10% D2O, 5.0 mM CaCl2, 1.0 mM d-glucose, and 75 μM free 3F-Phe as an internal frequency reference. Spectra were at 470 MHz, 25 °C.
FIGURE 4.
Effect of paramagnetic probe on the 19F NMR resonances of the receptor: substrate cleft empty. Shown are a series of 19F NMR spectra for 5F-Trp-labeled receptor from which sugar was quantitatively removed. Other details are as in Figure 3.
The present study focuses on the 19F nucleus at the 5-pos-ition of the indole ring of Trp 183 in the sugar-binding cleft. The crystal structure of the closed cleft containing bound D-glucose indicates that the bound D-glucose and Trp 183 are fully buried near the center of the cleft (Figure 1B) (Vyas et al., 1987). Also trapped is a layer of bound water molecules extending from the sugar to the bulk solvent. These ordered waters are distinct from the disordered bulk solvent and they fill a cavity ~4 Å wide between the two parallel surfaces defining the cleft (for example 3.8 Å between the van der Waals surfaces of Met 182 and Asn 43). Clearly, the cleft must open to admit probes with larger diameters. The minimum angle by which the hinge must open to admit a probe is a simple function of how far the probe penetrates into the cleft and the probe radius. The present goal is to detect the entry of a paramagnet into the open cleft by its effect on the spin—spin relaxation of the observe 19F nucleus, then to use this line broadening to calculate the penetration distance and hinge angle.
Paramagnetic T2 Relaxation: Theory
Previous studies have illustrated the application of paramagnetic broadening to macromolecular systems and have discussed the physics of the experiment [reviewed by Dwek (1973); Mildvan and Gupta (1978); Schmidt and Kuntz (1984); Bertini and Luchinat (1986); Kosen (1989); Saterlee (1990); and Petros et al. (1990)]. In the present application, a paramagnet rapidly exchanges between solution and one or more sites near an NMR nucleus. The resulting spin–spin relaxation of the nucleus (R2) will include additive contributions from each of the paramagnet sites:
| (1) |
where R2o is the nuclear spin–spin relaxation in the absence of paramagnet, R2p is the additional relaxation due to the paramagnet, fi, is the fractional occupancy of the ith paramagnet site, and R2pi is the maximum relaxation contribution of the ith site. This maximum contribution is realized when the site is fully occupied by paramagnet, in which case the magnetic dipole–dipole relaxation of the nucleus due to a paramagnet at distance r is given by the simplified Solomon and Bloembergen (1956) equation:
| (2) |
where γX is the magnetogyric ratio for nuclear spin X, g is the Landé g factor for a free electron spin, β is the Bohr magneton, S is the spin quantum number of the paramagnetic center, and ω is the NMR resonance frequency of the observe nucleus I in rad s−1. The net correlation time τci for this dipole–dipole interaction is given by
| (3) |
and thus contains contributions from the electron spin–lattice relaxation time Tlei of the paramagnet, the rotational correlation time τr of the interdipole vector (generally the rotational correlation time of the protein and thus site independent), and the time τbi the paramagnet remains bound in the ith site (Dwek, 1973). Equations 2 and 3 assume that (1) there is no contact between the electronic and nuclear spins (zero hyperfine coupling); (2) the resonance frequency of the electron spin is large enough that ωS ⪢ ωI and ωs ⪡ τc−1; and (3) the T1e and T2e relaxation times of the paramagnetic center are similar. Together eq 2 and 3 provide information on paramagnet–nuclear distances when relaxation data are available.
Experimentally, the paramagnetic relaxation is generally measured from NMR line widths. For the 19F NMR resonance of 5F-Trp, for example, the line shape is non-Lorentzian but the linewidth (△1/2) can be decomposed into a spin–spin relaxation rate (R2 = T2−1) and a constant component due to nuclear coupling with the nearby aromatic protons (J ~ 30 Hz):
| (4) |
Then the excess relaxation due to the paramagnet (R2p’ eq 1) can be simply determined from the paramagnetic line broadening, which is the difference between the line widths in the presence (△1/2+) and absence (△1/2− = R2o/π) of the paramagnet:
| (5) |
When the paramagnetic is mobile, the number, locations, and occupancies of its sites near the nucleus will generally be unknown, so that paramagnetic relaxation will not uniquely determine the distance of closest approach to the nucleus. However, such data do place an upper bound on the distance of closest approach: this limit is calculated with eq 2 and the assumption that the observed paramagnetic relaxation stems soley from one fully occupied paramagnet binding site. In reality the site occupancy will generally be much less than the assumed unity, in which case the paramagnet must visit sites closer to the nucleus to yield the same paramagnetic relaxation. It follows that the distance calculated for full occupancy will always overestimate the distance of closest approach.
Paramagnetic Probe Complex: Gd•EDTA
The paramagnetic center chosen for the present study if Gd(III), a trivalent lanthanide possessing a large magnetic moment (S = 7/2) and a long relaxation time, which together account for its usefulness as an NMR line-broadening agent (Saterlee, 1990). The EDTA complex is used to provide a paramagnet of large size requiring a correspondingly large angle of hinge opening for entry into the cleft (the diameter of the unhydrated complex calculated perpendicular to the least squares plane is d = 9.7 Å; see Figure 2 legend); in addition, the chelate significantly reduces the dependence of electronic relaxation rates on environment (T1e−1 and T2e−1, eq 3). The structure of Gd-EDTA in Figure 2 (Templeton et al., 1982) reveals that EDTA provides four coordinating oxygens and two nitrogens, while three additional coordinating oxygens are provided by water molecules to yield a coordination number of nine. The most rapid fluctuation in the magnetic dipole–dipole interaction is the electronic relaxation of Gd(III), which thereby controls the correlation time (eq 3; τc−1 ≃ T1e−1 ≃ 2 × 109 Hz; Dwek et al., 1975). Sources of slower fluctuations include the tumbling of the protein–paramagnet complex (τr−1 = 108 Hz for a spherical protein of 36 kDa) and the chemical exchange of paramagnet between the substrate cleft and bulk solution (τb−1 = 108 Hz for diffusion-limited exchange of 10 mM complex), All 19F NMR experiments were conducted at 470 MHz, corresponding to ωI = 3.0 × 109 rad•Hz in eq 2.
FIGURE 2.
Structure of Gd•EDTA and determination of diameter. The structure of the sodium salt of the Gd•EDTA complex shows the Gd(III) ion to have a coordination number of nine: six coordinating atoms provided by EDTA and three by water molecules. Noncoor-dinating waters of hydration are removed in the figure and not considered in diameter calculations. The diameter was determined as follows. The least-squares plane, which is placed to minimize the rms distances of atoms above and below the plane, was found to lie perpendicular to the page approximately along the O10–N1 axis. This plane defines the orientation of the unhydrated molecule most likely to slide into a narrow cleft. The atoms most distant from the plane are O7 and O8 at 3.6 and 3.3 Å above and below the plane, respectively; thus the thickness of the molecule perpendicular to the plane is 9.7 Å, including the van der Waals radii of O7 and O8. This diameter was used in hinge-angle calculations (see text). If the hydrating waters are included in the structure, the largest diameter is 11.8 Å from the H1 hydrogen on C1 to the H12’ hydrogen on a water molecule hydrated to O10.
Effect of Gd•EDTA on Receptor 19F NMR Resonances
To quantitate the broadening due to the paramagnetic complex, 19F NMR spectra have been obtained for several probe concentrations. Figure 3 summarizes the spectra obtained when the receptor contains bound D-glucose. In this case the crystal structure provides an internuclear distance of ≥ 15 Å between the l9F observe nucleus in the closed cleft and the paramagnetic center, yielding a predicted line broadening of △△υ1/2 ≤ 16 Hz (from eq 2 for a fully occupied paramagnet site). The observed broadening of the 5F-Trp 183 resonance is insignificant even at the maximum probe concentration (△△υ1/2 = 7 ± 10 Hz for 9.3 mM Gd•EDTA), confirming that this residue is buried deep within the closed cleft where it is in-accessible to bulk solvent and the probe. Other residues closer to the surface exhibit more accessibility. For example, the l9F nucleus of 5F-Trp 284 is ~4 Å from bulk solvent and is broadened significantly (ΔΔν1/2 = 40 ± 10 Hz).
Figure 4 summarizes the effects of the paramagnet on the empty cleft. In this case, the 5F-Trp 183 resonance exhibits a large paramagnetic broadening that is >2-fold greater then that of any other resonance, indicating that this residue becomes selectively exposed to solvent when sugar is removed. The maximum paramagnet concentration illustrates the greatly increased accessibility of 5F-Trp 183: the broadening of the 5F-Trp 183 resonance caused by 9.3 mM Gd•EDTA is ΔΔν1/2 = 130 ± 10 Hz for the empty cleft, or 15-fold larger than for the d-glucose-containing cleft. This increased line broadening is due to rapid exchange of paramagnet between the cleft and solution, since the 5F-Trp 183 resonance is uniformly broadened (in contrast a slowly exchanging paramagnet would yield two resonances, one broadened by the presence of paramagnet and the other unbroadened).
When the data of Figures 3 and 4 are used to analyze the relationship between the 5F-Trp 183 broadening and the Gd•EDTA concentration, a linear dependence is observed (not shown; the 5F-Trp 183 slopes are 14 ± 1 Hz of line broadening per millimolar Gd•EDTA for the empty cleft and 0.7 ± 0.1 Hz of linebroadening per millimolar Gd•EDTA for the d-glucose-occupied cleft). Such linearity indicates that the binding of paramagnet to the cleft is far from saturation and rules out the existence of a high-affinity paramagnet-binding site. Instead the paramagnet must either diffuse freely within the cleft or weakly associate with one of the low-affinity sites there; the possibility that low-affinity sites exist is supported by the suggestion that Gd-diethylenetriaminepentaacetate, a similar complex, binds weakly to carboxylates (Petros et al., 1990) of which there are three in the sugar-binding site alone [Asp 14, Asp 154, and Asp 236; Quiocho et al. (1987)]. There is no detectable structural change when Gd•EDTA is added to the empty cleft; in particular, there is no frequency shift for 5F-Trp 183 or any other 5F-Trp resonance upon addition of paramagnet (Figure 4), while these frequencies have previously been shown to be highly sensitive to structural changes in the receptor (Luck & Falke, 1991a,b). Together the evidence indicates that Gd•EDTA interacts weakly with the open cleft without perturbing its structure.
Control experiments summarized in Figure 5 rule out specific alternative interpretations of the broadening. The paramagnetic Gd•EDTA complex is required for the broadening as illustrated by the absence of broadening when the diamagnetic La•EDTA is substituted (Figure 5B). And the broadening is not caused by a small concentration of free Gd(III) ion released by dissociation of the Gd•EDTA complex (this concentration would be [Gd(III)free] ~ 10−17 M when [Gd(III)total] = 9 mM and [EDTAtotal] = 45 mM at pH 7.1), since 10−5 M free Gd(III) ion causes no detectable broadening (Figure 5C). These results confirm that the species causing the broadening is an EDTA complex of Gd(III).
FIGURE 5.
Controls for the paramagnetic broadening caused by Gd•EDTA. Shown are a series of 19F NMR spectra comparing the effects of different aqueous species on the 5F-Trp 183 resonance of the empty cleft. (A) 5F-Trp 183 resonance of the empty cleft, as in Figure 4A. (B) Effect of 9.3 mM La•EDTA (4-fold molar excess of EDTA). (C) Effect of 10 nM free Gd(III) (introduced by dialysis against 10 μM GdCl3 in final dialysis buffer). (D) Effect of 5.6 mM Gd•EDTA (4-fold molar excess of EDTA). Other sample and spectral parameters are as in Figure 3.
It is worth considering in detail a specific alternative mechanism in which the broadening of 5F-Trp 183 in the empty cleft does not stem from cleft opening. In principle a modest paramagnetic broadening could result from a simple 180° rotation of the 5F-Trp 183 indole ring about the Cβ–Cγ bond, which would place the 19F observe nucleus closer to bulk solvent even if the cleft remained closed. However such a picture is disfavored by the following points. Indole rotation is likely to be restricted by the nearby protein charges; for example, a 180° rotation would place the indole within 1 Å of two carboxylates, Asp 14 and Glu 149. Such a rotation would still leave the observe 19F nucleus 5 Å from the bulk solvent, too far to yield the observed paramagnetic broadening unless the paramagnet is fixed nearby on the protein surface. And 180° ring flips often yield characteristic double resonances (Wagner & Wutrich, 1986) that have not been observed for the 5F-Trp 183 19F resonance even at 4 °C (not shown). Evidence that the cleft must spend a significant fraction of its time in an open conformation is provided by the rapid binding and dissociation of sugar at its deeply buried site within the cleft (see below). Together the structural constraints argue that paramagnet closely approaches the observe nucleus by entering the open cleft.
Calculation of the Distance of Closest Approach and the Angle of Cleft Opening
Substitution of the maximum line broadening observed for the empty cleft into eq 2 enables calculation of the upper limit on the distance of closest approach between the nucleus and the paramagnet. The result is rc ≤ 10 Å, which requires at least half of the paramagnet diameter to penetrate into the cleft as illustrated in Figure 6. It is worth reemphasizing that the true closest approach is likely shorter than this upper limit that is calculated for a fixed nucleus-paramagnet separation, since the true separation is likely to be heterogeneous and the paramagnet concentration is far from saturating (see above). In fact, the closest approach may well be a collision of the hydrated probe with the nucleus, which from the minimum radius of the probe (5 Å) and the van der Waals radius of fluorine (1.35 Å) would yield rc ~ 6 Å.
FIGURE 6.
Schematic depiction of the hinged cleft model and the structural parameters determined for the open con former. (A) In the closed conformer containing trapped d-glucose, the hinge angle of the substrate cleft is defined as θ = 0° and the hinge point X is assumed to be located on the central hinge strand in the same plane as the upper cleft surface. The 4-Å cavity between the surfaces of the closed cleft is filled with substrate and tightly bound water molecules (Figure 1B) but is inaccessible to bulk solvent and the Gd•EDTA probe. The minimum internuclear distance FGd from the 19F observe nucleus (F) on 5F-Trp 183 to the probe (Gd) is the distance from F to the nearest solvent molecule outside the cleft (10 Å from F to H20 317) summed with the radius of Gd (5 Å). (B) When substrate is removed, the cleft opens, at least transiently, so that the probe can now enter the cleft and approach the observe nucleus F, The following distances were used to calculate the open hinge angle: FX = 9 Å, the distance between F and the Cα of ser 112 in the hinge. FGd = 10 Å, the maximum internuclear distance between F and the paramagnetic center as calculated from the observed paramagnetic line broadening (see text). XY ≃ Fx + FGd = 19 Å, the hypotenuse in the hinge angle calculation. YZ = d − 4 Å = 6 Å, the dimension that a cleft of width 4 Å must open to admit a probe of diameter d = 10 Å. Finally θ = sin−1 (YZ/XY) ≃ 18°, the angle by which the closed cleft must open to admit the probe. When the location of the hinge point is moved to other positions on the hinge strand closest to the observe nucleus, always within the 4-Å cavity of the cleft, the uncertainty in the minimum hinge angle is θ = 18 ± 3°.
The distance of closest approach can in turn be used to calculate the angle by which the cleft must open to admit the probe. This calculation is based on the simple picture of cleft geometry illustrated in Figure 6 and discussed in its legend. The resulting angle of opening is θ ≥ 18 ± 3° for a closest approach of rc = 10 Å. The 3° uncertainty in this minimum hinge angle stems from uncertainty in the position of the hinge. The true angle is likely to be significantly greater than 18° because the calculation assumes the unhydrated paramagnet diameter and the minimum penetration into the cleft. Furthermore, spectroscopic evidence suggests that under the relevant experimental conditions (pH 7.1; [EDTA]/[Gd(III)] = 5) the paramagnet may be Gd•(EDTA)2, a complex of unknown structure which must be significantly larger than Gd•EDTA.
Discussion
19F NMR paramagnetic broadening measurements have enabled both the open- and closed-cleft conformations of the d-galactose and d-glucose receptor to be detected in solution and have shown that the open cleft, for which the hinge angle is θ ≥ 18 ± 3°, allows close approach of a large probe to the substrate site inside the cleft. These results fully support the hinged cleft model of substrate binding (Mao et al., 1982; Miller et al., 1983). The findings are also consistent with a crystallographic study of the empty-cleft conformer of a structurally homologous periplasmic binding protein: the leucine/isoleucine/valine-binding protein has an open cleft hinge angle of θ = 18° in the crystal (Sack et al., 1990); in solution, the cleft may be open by an even larger angle since the NMR-determined angle is a lower limit.
Direct observation of the open cleft in solution provides a molecular explanation for two previously reported findings. First, the closing of a large open cleft would cause shifts of secondary structural elements contacting the cleft or hinge, thereby generating structural changes in regions distant from the cleft as observed by 19F NMR (Luck & Falke, 1991a). Second, an open cleft would provide an unhindered diffusion pathway for substrate between its binding site and solvent, explaining the relatively large rate constant for sugar binding (kon = 4 × 107 M−1 s−1 for d-glucose; Miller et al., 1980). If sugar binding is in the diffusion-controlled limit when the cleft is open, then the fraction of time the empty cleft is open is Popen = 1/100 since the observed kon is 100-fold smaller than the diffusion-controlled limit. However, additional factors are likely to contribute to the reduction of kon including the failure of substrate trajectories to enter the narrow cleft and the orientation requirements for binding within the cleft; thus the open conformer probability is a lower limit such that 0.01 ≤ Popen ≤ 1. The kon data also place constraints on the frequency of opening. The largest d-glucose concentration utilized in the kinetic study was 3.2 mM (Miller et al., 1980); for this concentration, the on-rate (kon•3.2 mM = 105 s−1) requires that the empty cleft be open at least once every 10−5 s.
The present results illustrate the usefulness of paramagnetic broadening NMR in the analysis of induced structural changes. The approach is especially well suited to systems where enough structural information is available to formulate predictions regarding the structural change; in an ideal case, the approach combines the complementary strengths of crystallography (high-resolution structural information) and NMR (solution information on structure and dynamics) to test a specific structural model.
Acknowledgments
We sincerely thank Dr. Florante Quiocho and co-workers for providing protein coordinates and Drs, Gareth Eaton, Sandra Eaton, Curt Haltiwanger, and Florante Quiocho for helpful discussions.
Footnotes
Support of this work by the National Institutes of Health (R01-GM40731) and by a University of Colorado CRCW Junior Faculty Development Award (J.J.F.) is gratefully acknowledged.
Registry No. Trp, 73-22-3; Ca, 7440-70-2; d-glucose, 50-99-7; d-galactose, 59-23-4.
References
- Anglister J, Frey T, McConnell HM. Biochemistry. 1984;23:5372–5375. doi: 10.1021/bi00317a041. [DOI] [PubMed] [Google Scholar]
- Bennett WS, Steitz TA. J. Mol. Biol. 1980;140:211–230. doi: 10.1016/0022-2836(80)90103-5. [DOI] [PubMed] [Google Scholar]
- Bennett WS, Huber R. Crit. Rev. Biochem. 1984;15:291–384. doi: 10.3109/10409238409117796. [DOI] [PubMed] [Google Scholar]
- Bertini I, Luchinat C. NMR of Paramagnetic Molecules in Biological Systems. Benjamin/Cummins Publishing Co., Inc.; Menlo Park, CA: 1986. [Google Scholar]
- Dobson CM. Nature. 1990;348:198–199. doi: 10.1038/348198a0. [DOI] [PubMed] [Google Scholar]
- Drapeau GR, Brammer WJ, Yanofsky C. J. Mol. Biol. 1968;35:357–367. doi: 10.1016/s0022-2836(68)80030-0. [DOI] [PubMed] [Google Scholar]
- Dwek RA. NMR in Biochemistry. Clarendon Press; Oxford, U.K.: 1973. [Google Scholar]
- Dwek RA, Levy HR, Radda GK, Seeley PJ. Biochim. Biophys. Acta. 1975;337:26–33. doi: 10.1016/0005-2744(75)90282-x. [DOI] [PubMed] [Google Scholar]
- Faber JR, Matthews BW. Nature. 1990;348:263–266. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
- Gueron M. J. Magn. Reson. 1975;19:58–66. [Google Scholar]
- Hazelbauer GL, Adler J. Nature (London), New Biol. 1971;230:101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- Kellermann OK, Ferenci T. Methods Enzymol. 1982;90:459–463. doi: 10.1016/s0076-6879(82)90171-9. [DOI] [PubMed] [Google Scholar]
- Kooistra DA, Richards JH. Biochemistry. 1978;17:345–351. doi: 10.1021/bi00595a024. [DOI] [PubMed] [Google Scholar]
- Kosen PA. Methods Enzymol. 1989;177:86–121. doi: 10.1016/0076-6879(89)77007-5. [DOI] [PubMed] [Google Scholar]
- Li E, Qian S, Yang NC, d'Avignon A, Gordon JI. J. Biol. Chem. 1990;265:11549–11554. [PubMed] [Google Scholar]
- Lu P, Jarema MC, Mosar K, Daniel WE. Proc. Natl. Acad. Sci. U.S.A. 1976;73:3471–3475. doi: 10.1073/pnas.73.10.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. Biochemistry. 1991a;30:4248–4256. doi: 10.1021/bi00231a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. Biochemistry. 1991b;30:4257–4261. doi: 10.1021/bi00231a022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Pear MR, McCammon JA, Quiocho FA. J. Biol. Chem. 1982;257:1131–1133. [PubMed] [Google Scholar]
- Mildvan A, Gupta RK. Methods Enzymol. 1978;49:322–359. doi: 10.1016/s0076-6879(78)49017-2. [DOI] [PubMed] [Google Scholar]
- Miller DM, Olson JS, Quiocho FA. J. Biol. Chem. 1980;255:2465–2471. [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Press, Cold Spring Harbor; NY: 1972. [Google Scholar]
- Peerson OB, Pratt EA, Truong HTN, Ho C, Rule GS. Biochemistry. 1990;29:3256–3262. doi: 10.1021/bi00465a017. [DOI] [PubMed] [Google Scholar]
- Petros AM, Mueller L, Kopple KD. Biochemistry. 1990;29:10041–10048. doi: 10.1021/bi00495a005. [DOI] [PubMed] [Google Scholar]
- Pratt EA, Ho C. Biochemistry. 1975;14:3035–3040. doi: 10.1021/bi00684a037. [DOI] [PubMed] [Google Scholar]
- Post JFM, Cottam PF, Simplaceau V, Ho C. J. Mol. Biol. 1984;179:729–743. doi: 10.1016/0022-2836(84)90164-5. [DOI] [PubMed] [Google Scholar]
- Quiocho FA, Vyas NK, Sack JS, Vyas MN. Cold Spring Harbor Symp. Quant. Biol. LII. 1987:453–463. doi: 10.1101/sqb.1987.052.01.052. [DOI] [PubMed] [Google Scholar]
- Sack JS, Saper MA, Quiocho FA. J. Mol. Biol. 1989;206:171–191. doi: 10.1016/0022-2836(89)90531-7. [DOI] [PubMed] [Google Scholar]
- Satterlee JD. Concepts Magn. Reson. 1990;2:119–129. [Google Scholar]
- Schmidt PG, Kuntz ID. Biochemistry. 1984;23:4261–4266. doi: 10.1021/bi00313a038. [DOI] [PubMed] [Google Scholar]
- Scholle A, Vreemann J, Blank V, Nold A, Boos W, Manson MD. Mol. Gen. Genet. 1987;208:247–253. doi: 10.1007/BF00330450. [DOI] [PubMed] [Google Scholar]
- Snyder EE, Buoscio BW, Falke JJ. Biochemistry. 1990;29:3937–3943. doi: 10.1021/bi00468a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon I, Bloembergen N. J. Chem. Phys. 1956;25:261–266. [Google Scholar]
- Sykes BD, Wingarten HJ, Schlesinger M. Proc. Natl. Acad. Sci. U.S.A. 1974;71:469–473. doi: 10.1073/pnas.71.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton LK, Templeton DH, Zalkin A, Ruben HW. Acta Crystallogr., Sect B. 1982:2155–2157. [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. Nature. 1987;327:635–638. doi: 10.1038/327635a0. [DOI] [PubMed] [Google Scholar]
- Vyas NK, Vyas MN, Quiocho FA. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- Wilson ML, Dahlquist FW. Biochemistry. 1985;24:1920–1928. doi: 10.1021/bi00329a018. [DOI] [PubMed] [Google Scholar]