Abstract
Mammalian lungs are branched networks containing thousands to millions of airways arrayed in intricate patterns that are crucial for respiration. How such trees are generated during development, and how the developmental patterning information is encoded, have long fascinated biologists and mathematicians. However, models have been limited by a lack of information on the normal sequence and pattern of branching events. Here we present the complete three-dimensional branching pattern and lineage of the mouse bronchial tree, reconstructed from an analysis of hundreds of developmental intermediates. The branching process is remarkably stereotyped and elegant: the tree is generated by three geometrically simple local modes of branching used in three different orders throughout the lung. We propose that each mode of branching is controlled by a genetically-encoded subroutine, a series of local patterning and morphogenesis operations, which are themselves controlled by a more global master routine. We show that this hierarchical and modular program is genetically tractable, and it is ideally suited to encoding and evolving the complex networks of the lung and other branched organs.
Many organs are composed of highly ramified tubular networks, each with a distinct architecture tailored to its physiological function. The bronchial tree of the human lung has over 105 conducting and 107 respiratory airways arrayed in an intricate pattern crucial for oxygen flow1–4. Classical studies of lung structure5–8 raise the question of how the information required to generate a tree of such complexity is biologically encoded9. Individually configuring thousands or millions of branches would require a tremendous amount of patterning information, far more than is biologically plausible, to specify when and where each branch forms during development, and the size, shape, and direction of outgrowth of each branch. One possibility is that the process is not precisely controlled, for example if branching occurs randomly to fill available space. Another is that control is precise but coding is simplified by repeated use of a branching mechanism, as in Mandelbrot’s fractal model and other elegant algorithms10–17.
Even with these attractive models and recent progress in identifying lung development genes18, understanding of the program that directs branching remains rudimentary. This is largely due to the complexity of the bronchial tree, which makes it difficult to follow branching dynamics beyond the earliest events19–21. Although branching of the lung and other organs can occur in culture22–25, it is unlikely these recapitulate the full pattern. Here, we have determined the complete in vivo pattern of branching and branch lineage of the mouse bronchial tree, and show that it is generated using three geometrically distinct local modes of branching coupled in three different sequences.
The branch lineage of the mouse bronchial tree
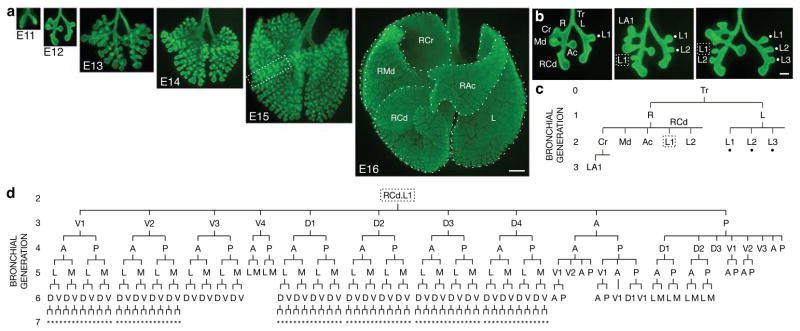
The bronchial tree develops by branching of airway epithelium into surrounding mesenchyme. Although the process cannot be visualized in living embryos with current techniques, we reasoned we could reconstruct the branching sequence from fixed specimens, provided the process is stereotyped. An immunostaining procedure was developed to visualize the full three-dimensional structure of the bronchial tree in fixed lungs (Fig. 1a). Examination of hundreds of wild-type CD1 specimens collected between embryonic day (E) 11 and E15 revealed that the branching pattern is remarkably stereotyped. This allowed us to reconstruct the sequence of events — where, when, and in what order, branches form — from finely staged specimens (Fig. 1b). This information was used to construct a lineage diagram representing the developmental history of the ~5000 branches of the bronchial tree (Fig. 1c, d; Supplementary Fig. 1). We found that there are three branching modes used repeatedly throughout the lung, which we call domain branching, planar bifurcation and orthogonal bifurcation.
Figure 1. Branching morphogenesis of the mouse bronchial tree.
a, Whole mount lungs (ventral view) at embryonic day indicated immunostained for E-Cadherin (green) to show airway epithelium. Dotted lines, right cranial (RCr), right middle (RMd), accessory (RAc), right caudal (RCd), and left (L) lobes. Bar, 500 μm. b, Reconstructing branching dynamics using three E12 specimens ~3 hours apart in age. Lateral 2° branches L1-3 (dots in b, c) sprout in proximal-to-distal order from left (L) 1° branch, as do lateral 2° branches L1 (box in b, c) and L2 from distal (RCd) portion of right (R) 1° branch. Bar, 200 μm. c, Branch lineage diagram for oldest lung in b. Branch names indicate lineage, e.g. RCd.L1 is first lateral 2° branch off RCd. d, Lineage diagram of RCd.L1 showing 250 descendant branches at E15 (box in a). A, anterior; D, dorsal; L, lateral; M, medial; P, posterior; V, ventral; *, orientation can vary.
Domain branching
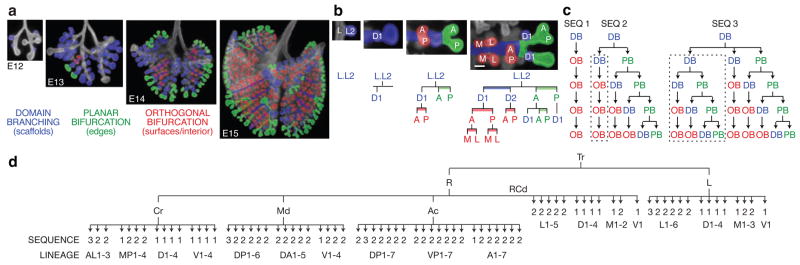
In domain branching, daughter branches form in rows (“domains”) at different positions around the circumference of the parent branch, like the rows of bristles on a bottle brush (Fig. 2f). In the left primary bronchus (L) lineage, the first secondary branch (L.L1, abbreviated L1) buds off the lateral aspect of the founder branch L late on E11 (Fig. 1b). Over the next two days additional branches sprout distal to L1, creating a row of lateral secondary branches numbered in the proximal-distal sequence in which they form (L1, L2, …; Fig. 1b, 2a). As these sprout, another row begins to form along the dorsal surface of L. The first dorsal branch (D1) buds just distal to the level of L1, and others bud sequentially in proximal to distal order (Fig. 2a, b). As this domain develops, a third row begins to sprout from the medial surface of L, then a fourth from the ventral surface (Fig. 2b, c). This ventral domain often consisted of just a single branch (V1) located distally, and sometimes there were none. Although rudimentary, this is a bona fide domain because we found rare wild-type variants and a mutant that form more complete rows (see below).
Figure 2. Branching modes in lung development.
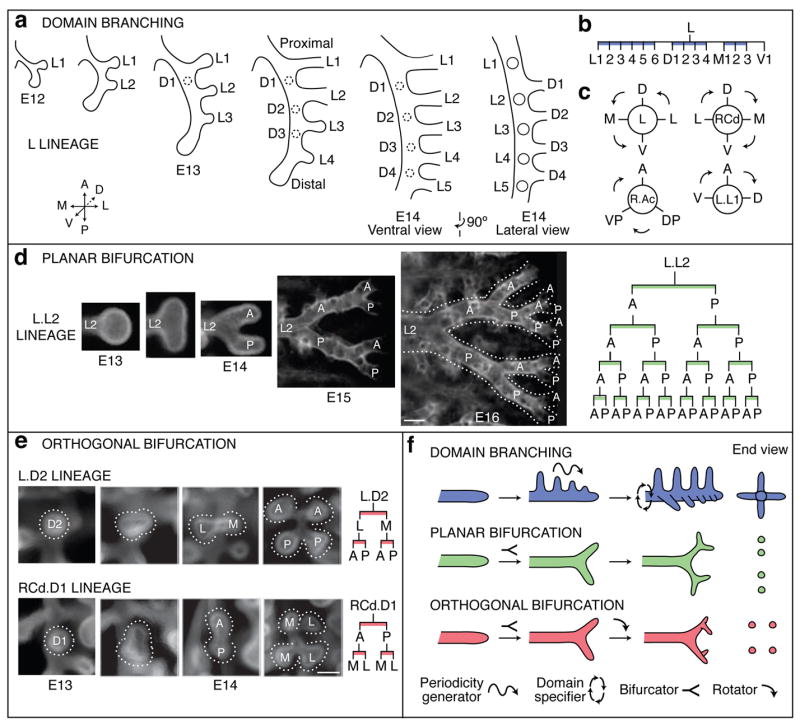
a–c, Domain branching. a, Schematics of lateral and dorsal 2° branches budding from L. Lateral 2° branches (L1-5) bud in proximal to distal order. Proximal-to-distal branching begins again in second domain (projecting into plane of figure) to form a row of dorsal 2° branches (D1-4; dashed circles). Right panel, E14 schematic rotated 90° to show dorsal branches. b, Lineage diagram of 2° branches from L. Branches form in four domains: lateral (L), dorsal (D), medial (M), and ventral (V), indicated by blue bars. c, Schematic cross sections through L and three other branches indicated showing positions of domains and order (arrows) domains are used. d, Planar bifurcation. Ventral view of branch L.L2 in series of fixed specimens from E13 to E16 showing sequential bifurcations along A-P axis. E15 and E16 specimens were stained with anti-Smooth Muscle α-Actin to highlight early branch generations, which are surrounded by smooth muscle. Dotted lines outline bifurcations. Right panel, lineage of L.L2 descendants formed by planar bifurcation; branches not yet formed in E16 specimen are in gray. Bar, 100 μm. e, Orthogonal bifurcation. End-on (dorsal) views of branches indicated in developmental series of E13 and E14 specimens. L.D2 bifurcates along L-M axis, and its daughters along A-P axis, whereas RCd.D1 bifurcates along A-P axis and its daughters along L-M axis. Bar, 100 μm. f, Schematics of branching modes. The first bifurcation in a series is classified retrospectively, based on the orientation of the subsequent bifurcation. Icons show patterning and morphogenesis operations inferred for each mode: proximal-distal periodicity generator, circumferential domain specifier, branch bifurcator, and bifurcation plane rotator.
Secondary branches off RCd (distal portion of the R primary branch) also arise by domain branching, beginning with a row of lateral branches (Fig. 1b). The spacing of branches in each row and the order in which rows trigger (Fig. 2c) are the same as in the L lineage, but the proximal-distal positions at which rows initiate and number of branches in each row are not. For example, the first dorsal branch (RCd.D1) forms proximal to the first lateral branch (RCd.L1), whereas in the L lineage the first dorsal branch (L.D1) forms distal to the first lateral branch (L.L1). Although the timing and spacing of branch budding within a row was regular and stereotyped, these parameters were not tightly coupled between rows. Thus, each row appears to comprise an independently patterned domain.
The results imply there are two patterning systems controlling domain branching: a proximal-distal system including a periodicity generator that controls the sequence of branching within each domain, and a circumferential system that specifies the positions of domains and the order domains are used (Fig. 2f). Also, because domains differ in number of branches and the position at which branching initiates, the proximal-distal system must also set the initiating position and register of each domain.
Planar and orthogonal bifurcation
Although many tertiary branches also form by domain branching (Fig. 2c and Supplementary Fig. 1), some tertiary and later generation branches form by a different mode in which the tip expands and bifurcates. In the L.L2 lineage (Fig. 2d), the founder branch bifurcates along the anterior-posterior axis to form a pair of 3° branches, which bifurcate again in a similar orientation to form four 4° branches. The process repeats, creating planar arrays of fourteen or more branches by E16. We call such series of two or more tip divisions, all of which occur in the same plane, planar bifurcation (Fig. 2f).
Other tertiary and most later generation branches form by a third branching mode called orthogonal bifurcation (Fig. 2e). Branches bifurcate at their tips, as in planar bifurcation. However, between each round of branching there is a ~90° rotation in the bifurcation plane, so that the four granddaughters are arranged in a rosette (Fig. 2f). Typically, this alternating sequence continues with the bifurcation plane rotating ~90° in each round, as in the RCd.L1.V1 lineage which undergoes at least four rounds and generates a cluster of 30 branches (Fig. 1d).
The anatomical orientation of each round of orthogonal bifurcation is generally stereotyped and can be distinct for different branches. For example, L.D2 first bifurcates along the lateral-medial and then the anterior-posterior axis, whereas RCd.D1 does the reverse (Fig. 2e). However, orientation control appears to deteriorate over time because the orientation of late generations in the RCd.L1.V1 and other lineages were less stereotyped, although they were always oriented orthogonal to the preceding bifurcation.
Pattern of deployment of the local branching modes
The three branching modes described above are used at many different times and positions and account for nearly all branching events in the first five days of lung development (Supplementary Fig. 1). Pseudocoloring branches according to the mode by which they form revealed that each mode is associated with a specific aspect of lung design (Fig. 3a). Domain branching is used first and generates the central scaffold of each lobe, setting its overall shape (e.g., trigonal pyramidal for RAc). Planar bifurcation forms the thin edges of lobes, and orthogonal bifurcation creates lobe surfaces and fills the interior.
Figure 3. Deployment of branching modes.
a, Lungs from Fig. 1a with branches pseudocolored blue (domain branching), green (planar bifurcation), or red (orthogonal bifurcation) to indicate branching mode used to form the branch. b, Close-up of L.L2 (dorsal view, lateral at right) pseudocolored as in a. Lineage diagrams are also colored to indicate branching mode. Branching mode can switch between generations and a single branch (e.g. L.L2) can form daughters by more than one mode. Bar, 100 μm. c, Sequences of branching mode use. DB, domain branching; OB, orthogonal bifurcation; PB, planar bifurcation. Sequence 2 includes a lineage formed by Sequence 1 (dotted box) and Sequence 3 includes a lineage formed by Sequence 2 (dotted box). d, Diagram showing sequence in c used to generate each lineage (named after the founder branch, see Supplementary Fig. 1) off the lobar branches (R.Cr, R.Md, R.Ac, RCd, L). Most or all lineages within a domain use the same sequence.
There is no global transition from one branching mode to another. At many developmental stages all three modes are used concurrently (Fig. 3a), and even individual branches can use more than one mode (Fig. 3b). Furthermore, branching proceeds at different and somewhat variable rates in different lineages (see below). Thus, deployment of the branching modes is not controlled by a global developmental or generational clock. Rather, each lineage proceeds independently through a characteristic sequence of branching modes.
With three branching modes and seven or more generations of branches, there are thousands of possible sequences in which branching modes could be used. However, only three were observed (Fig. 3c). In sequence 1, a founder branch (e.g., L.D2) formed by domain branching switches immediately and permanently to orthogonal bifurcation (Fig. 2e). In sequence 2, a founder branch formed by domain branching (e.g. L.L2, Fig. 3b) forms some daughters (e.g. L.L2.D1) by domain branching, which switch permanently to orthogonal bifurcation, as in Sequence 1 (dotted box in Fig. 3c). However, the founder forms other daughters by planar bifurcation (e.g. L.L2.A, L.L2.P). These daughters continue to undergo planar bifurcation at their tips, and also form domain branches along their length. These domain branches switch permanently to orthogonal bifurcation. In sequence 3, a founder and some of its descendants recapitulate Sequence 2 (right half of Sequence 3, Fig. 3c). However, the founder also forms daughters that themselves follow Sequence 2 (dotted box, left half of Sequence 3).
Figure 3d shows where each of the three sequences are used and the lineages they generate. Remarkably, these three sequences of deployment of the three branching modes describe the complete lineage of the bronchial tree.
Variability and errors in the branch lineage
Although much of the branching process is stereotyped, it is not invariant. There was local variability in the temporal sequence of branching so that some lineages get ahead of, or fall behind, other lineages (Supplementary Fig. 2). There was also spatial variability including subtle differences in register between branches in different domains of a parent branch, and relaxation in the absolute orientation of later rounds of orthogonal bifurcation. None of this temporal or subtle spatial variability altered branch lineage. However, we also found variants that did affect lineage, which we call branching “errors.”
Errors were identified in specimens with branch patterns that could not be reconciled with the canonical lineage unless an anomalous branching event had occurred. For example, there were specimens in which a branch originated off the wrong parent branch, a “branch displacement” error (Fig. 4a, b), or a branch was missing and daughter branches sprang directly from the grandparent (“skipping a generation”; Fig. 4c, d). Despite their inappropriate origins, such branches went on to branch in their usual manner (Fig. 4a′-d′), demonstrating that the information controlling a branch’s subsequent branching is not encoded within the parental branch, and that continuation of the branching program is not contingent upon completion of each prior step. We also found lungs in which a founder branch and all its descendants were missing, such as the L.V1 and RCd.V1 lineages (Fig. 2b and Supplementary Fig. 1). In each of these “optional” lineages, the founder forms by domain branching and is typically the only branch formed in the last domain used.
Figure 4. Branching errors.
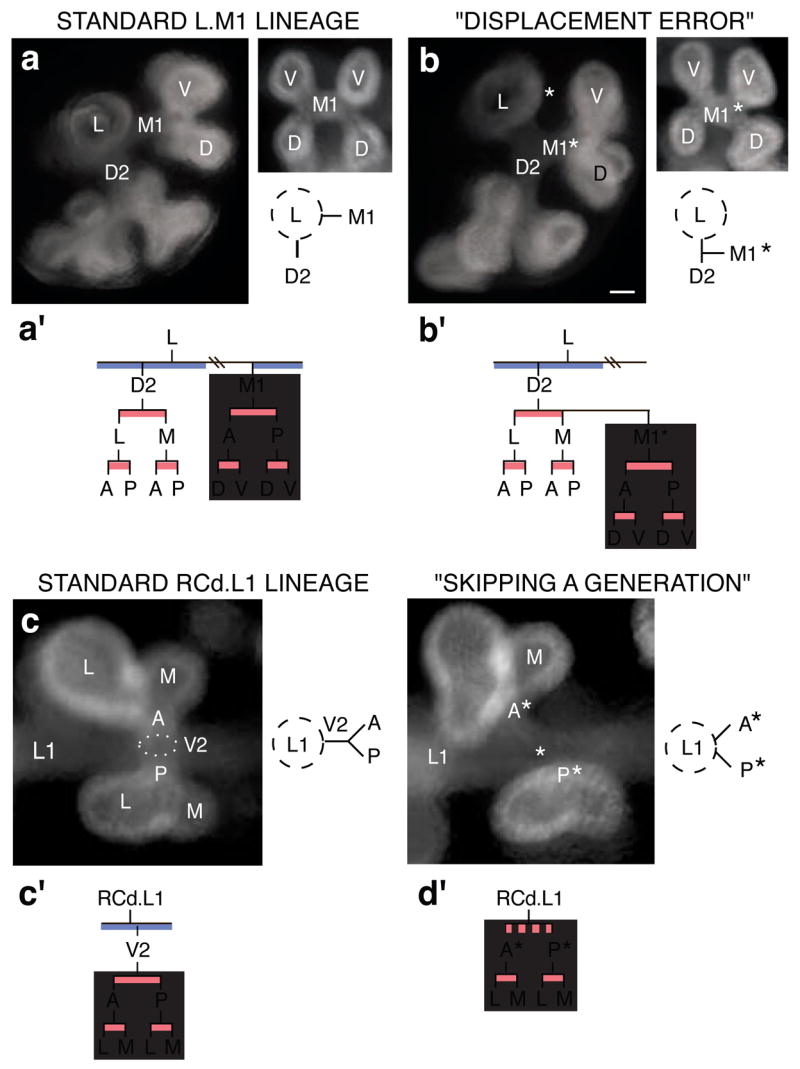
(a, b) “Displacement error”. a, Standard arrangement of D2 and M1 2° branches off L. Left panel, end-on (anterior) view of L and descendants. Right panel, end-on (medial) view of M1 and descendants; below, schematic of anterior view. a′, Standard lineage of L, D2 and M1. Grey box, M1 lineage. b, Same views as a of lung in which M1* arises off D2, normally a sister branch. Bar (for a and b), 50 μm. b′, M1* follows the normal M1 lineage (grey box). (c, d) “Skipping a generation”. c, Standard arrangement of V2 arising off RCd.L1. Left panel, end-on (ventral) view of V2 and its descendants, which form by orthogonal bifurcation. Right panel, schematic depicting side (lateral) view of V2 and its daughters. c′, Standard lineage of RCd.L1 and V2. Grey box, descendants of V2. d, Same view (left) and schematic (right) of lung that skipped V2 generation. Bar (for c and d), 50 μm. Anterior (A*) and posterior (P*) branches sprout directly from RCd.L1, normally their grandparent. d′, Descendants of missing V2 (grey box) follow normal lineage.
Some errors, such as branch displacement, were rare, occurring at <1% of all branching events scored. Others were more common, and some, like skipping generations in the RCd.L1 lineage, approached the frequency of the “normal” event. Errors were not randomly distributed but tended to occur at specific sites and times in lung development. A more limited analysis of five inbred strains (A/J, C3H/HeJ, C57BL/6J, DBA/2J, and FVB/NJ) showed the same pattern of branching and same types of errors and other variation as in the outbred CD1 strain, with one exception detailed below. Thus, errors are not due to genetic heterogeneity. Rather, they identify intrinsically imprecise steps in the branching program.
Genetic control of branch lineage and pattern
To begin to elucidate the genetic basis of the branching program, we investigated branch lineage and pattern in two mutants and an inbred strain with airway patterning defects. In inversus viscerum (iv) mutants in the Dnahc11 dynein heavy chain gene26, left-right axis specification is randomized and in about half the animals the positions and gross structures of organs is reversed27. We found that the lung branching pattern and lineage was completely reversed in some mutant embryos (Supplementary Fig. 3), demonstrating that deployment and coupling of the branching modes is under global genetic control, downstream of Dnahc11 and the left-right asymmetry pathway.
In contrast to this global effect, null mutations in sprouty2 (Spry2)28, encoding a Sprouty family receptor tyrosine kinase inhibitor29,30, had local and subtle effects on branch pattern and lineage: there were extra branches in the ventral domains off L and RCd (Fig. 5a, b; Supplementary Fig. 4). The extra branches sprouted earlier and more proximally than the normal branches in these domains, expanding the domains toward the base of the parent branch. The ectopic branches formed additional generations, creating ectopic lineages indistinguishable from those of normal branches in the domain. Thus, Spry2 restricts the number of branches in the two ventral domains, and in its absence normally non-branching regions along the parent branches acquire the branching identity of more distal regions.
Figure 5. Genetic control of branch pattern and lineage.
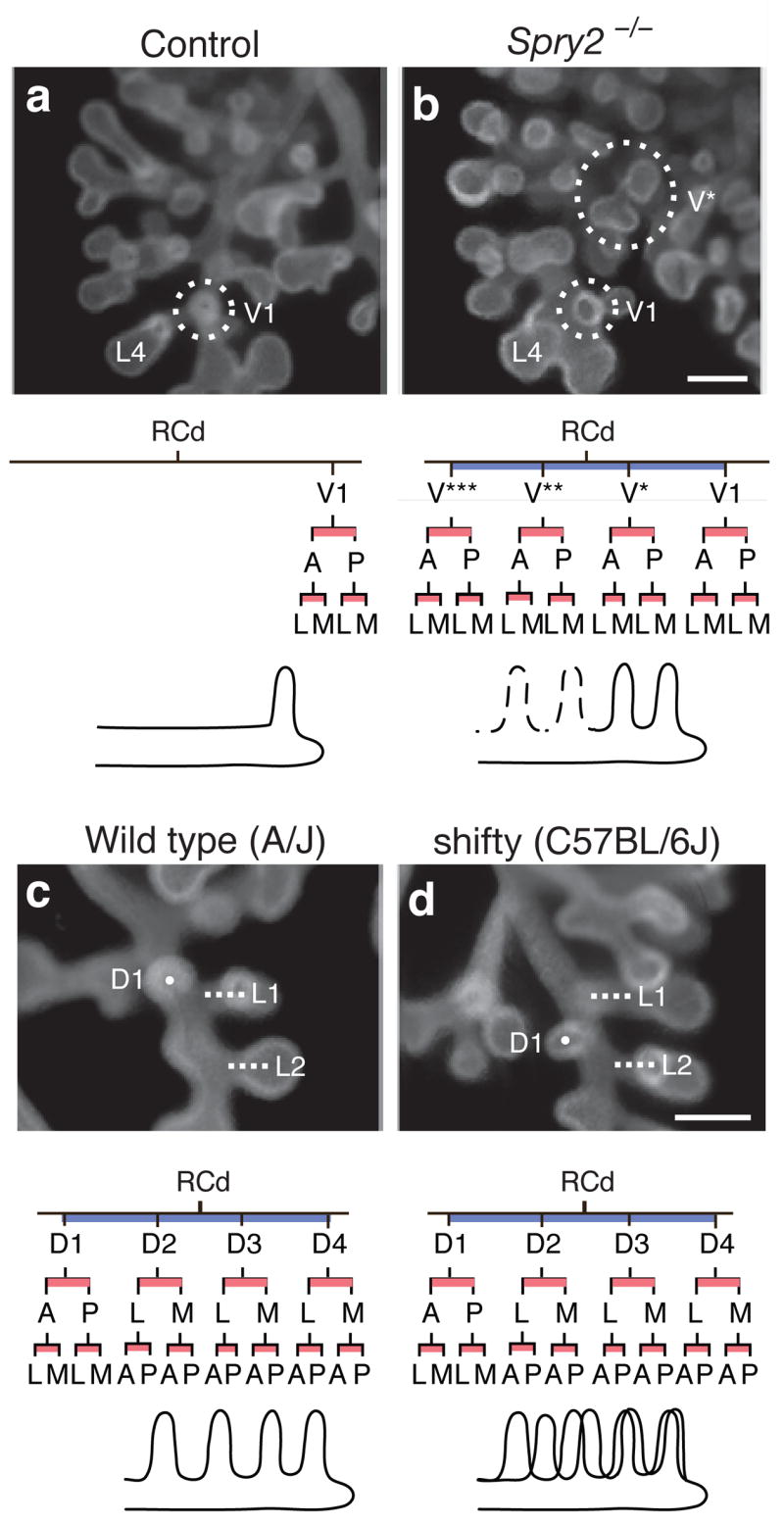
(a, b) Ectopic domain branching in Spry2−/− mutants. a, RCd lobe (ventral view) of E12.5 control (Spry2+/−) lung with a single 2v branch (V1, circled) off RCd, at level of RCd.L4 (L4). Below, RCd.V1 lineage and schematic of RCd with single ventral 2° branch (V1). b, Same view of E12.5 Spry2−/− lung showing the normal ventral 2° branch (V1) and an ectopic branch (V*) that forms earlier and proximal to V1. V* has already sprouted additional generations of branches. Below, lineage and schematic show V* plus additional ectopic ventral branches (V**, V***; dashed lines in schematic) seen in other Spry2−/− lungs (Supplementary Fig. 4). Bar (for a and b), 200 μm. (c, d) Shifted domains in strain C57BL/6J. c, RCd lobe (dorsal view) of E12.5 lung from control strain A/J. Dorsal 2° branch RCd.D1 (D1; white dot) forms just proximal to lateral 2° branch RCd.L1 (L1, white line). Below, lineage and schematic of RCd.D1 and other branches in D domain. d, Same view of E12.5 lung from C57BL/6J mouse with shifty phenotype. RCd.D1 forms distal to RCd.L1. In shifty lungs, entire dorsal domain (black line in schematic) is shifted distally along RCd relative to wild type (grey line in schematic) but lineage is unaffected (except when there is a full unit shift and RCd.D4 lineage is missing, as indicated in gray; see Supplementary Fig. 5). Bar (for c and d), 200 μm.
One inbred strain (C57BL/6J) had a subtle defect in branch positioning: branches in the dorsal domains of L and RCd were shifted distally along the parent branch (Fig. 5c, d; Supplementary Fig. 5). Despite this shift, the subsequent branch pattern and lineage of the displaced branches were unperturbed. We named the phenotype shifty and propose that the shifty locus encodes a modulator of the pathway that sets the proximal-distal register of domains.
DISCUSSION
The branching pattern and lineage of the mouse bronchial tree reveals the logic of the lung branching program. Three local modes of branching (Fig. 2f) are used in three different sequences in the developing lung (Fig. 3). Each branching mode (domain branching, planar bifurcation, and orthogonal bifurcation) creates a different arrangement of branches (bottle-brush, planar array, rosette) and serves a specific function in lung design (scaffold, edge, surface/interior). The repeated use of these branching modes, along with a hierarchical control and coupling scheme, allows genetic encoding of the complex but stereotyped bronchial tree.
A formal model of the airway branching program
All three branching modes are geometrically simple and easy to encode. We propose that each is controlled by a locally operative, genetic subroutine, a series of discrete patterning and morphogenesis events (Figs. 2f, 6). Domain branching requires a proximal-distal “periodicity generator” that sets the timing and spacing of branches within a domain, and a circumferential “domain specifier” that dictates the positions of domains around the parent branch and the order domains trigger. Planar and orthogonal bifurcation require a branch “bifurcator”, and orthogonal bifurcation requires a “rotator” that reorients the bifurcation plane by 90° between events. All subroutines require a “branch generator.” Some of these steps may themselves be modular and shared among subroutines (Fig. 6).
Figure 6. A formal model of the lung branching program.
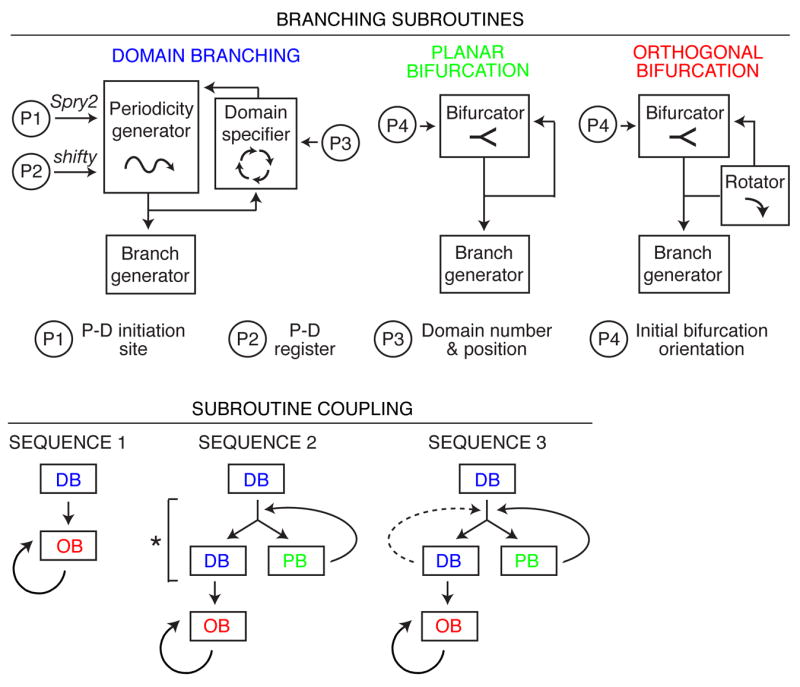
(Top) Representation of branching modes as subroutines using different combinations of patterning and morphogenesis operations (boxed; see Fig. 2f). Subroutines are locally modified by input parameters P1–P4 that regulate variables indicated. P-D, proximal-distal. (Bottom) Three subroutine coupling schemes generate the three observed sequences of subroutine use (Fig. 3c). The schemes are related: bypass of step marked by asterisk in Sequence 2 generates Sequence 1, and repeat of domain branching step (dashed line) generates Sequence 3. DB, domain branching; PB, planar bifurcation; OB, orthogonal bifurcation.
Because the pattern of deployment of the branching modes is complex but stereotyped, we infer that there is higher order, perhaps emergent, patterning information, “the master routine”, which calls subroutines at specific times and positions in the lung development program. The three different sequences in which branching modes are combined (Fig. 3d) are each simple variants of a general coupling scheme (Fig. 6). Thus, the master routine need only encode the three variants, and where each is used. Coding is further simplified because most or all branches within a domain use the same scheme (Fig. 3d).
Although the sequences of subroutine use are rigidly specified, temporal variability in the program (Supplementary Fig. 2) implies that the master routine does not function as a control center that calls subroutines individually in a fixed global order. Rather, it appears to set the coupling scheme for each lineage early and then allow each lineage to proceed through its sequence independently. The program is also regulative because branching continues normally after suffering errors (Fig. 4).
Because there are stereotyped local differences in the branching modes, such as the number of branches in a domain and the absolute orientation of orthogonal bifurcation, the master routine must also encode position-specific modifications in the subroutines, which we represent as local input parameters (P) (Fig. 6). Setting these local parameters may be the most computationally intensive part of the program.
Elucidating the genetic basis of the branching program
A critical challenge ahead is to determine the genetic and molecular basis of the master routine, three subroutines, and the local parameters. With the lineage in hand, functions can now be assigned with unprecedented precision to the dozens of extant lung development genes18,31–34. We found that Spry2 regulates the site of initiation and number of branches in specific domains (P1, Fig. 6), and shifty controls the proximal-distal register of entire domains (P2, Fig. 6). It will be particularly important to identify genes that underlie the periodicity generator, domain specifier, bifurcator, and rotator, because they are central to the distinctive geometries of the branching modes and likely to involve novel patterning processes.
Airway branching is one of many processes required to build a lung. Others include airway size control, airway cell differentiation, alveolus formation, and patterning pulmonary blood vessels. Because airways appear to set arterial and smooth muscle pattern35, and signals from the airways likely direct morphogenesis of other tissues in the lung too36, parsing the airway branching program is a critical step towards elucidating the full program of lung development.
Evolution of branching networks
Branching networks come in many sizes, cellular architectures, and branching complexities that differ between organs and species37–39. For example, the human bronchial tree contains millions of branches, several orders of magnitude more than in mouse, whereas the lobes of frog lung are unbranched sacs. Human and mouse lungs also differ in lobation and branch pattern.
The modular logic of the mouse lung branching program suggests how such structural diversity is created during evolution40–42. New branching patterns can arise by reiterative use of subroutines or new patterns of their deployment. Indeed, the limited developmental data available for human and pig provides evidence that at least domain branching and orthogonal bifurcation are used in other animals20,21,43. New branching patterns can also arise by local modifications of subroutines, like the increased number and altered positions of domain branches in Spry2 and shifty mutants, and the reduction of the standard four-domain structure to three domains for R.Ac. More extreme modifications could create entirely new subroutines: orthogonal bifurcation may have evolved from planar bifurcation by acquisition of the rotator function. Branching subroutines controlled by a master routine may represent a general biological strategy for encoding and evolving complex branch patterns.
METHODS SUMMARY
Outbred CD1 embryos, inbred A/J, C3H/HeJ, C57BL/6J, DBA/2J, and FVB/NJ embryos, and iv (ref. 27) and Spry2ΔORF/ΔORF (ref. 28) null and littermate control embryos were dissected in PBS. Noon of the day a vaginal plug was detected was considered ~E0.5. Lungs were fixed and airway epithelium was visualized by indirect immunofluorescence after staining with rat anti-E-Cadherin primary antibody (clone ECCD-2)44, biotinylated secondary antibody, and avidin-peroxidase and tyramide histochemistry. Some lungs were double stained with Cy3-conjugated mouse anti-Smooth Muscle α-Actin antibody (clone 1A4) to enhance visualization of early branch generations. See online Methods and Supplementary Methods for details.
METHODS
Mice
CD1 mice (Charles River Laboratories) were used for the wild type analysis. Inbred strains (Jackson Laboratory) were: A/J (n=21 lungs analyzed), C3H/HeJ (n=8), C57BL/6J (n=20), DBA/2J (n=10), and FVB/NJ (n=13) mice. iv/iv mice27 were from Jackson Laboratory. Spry2−/− embryos and their Spry2+/− and Spry2−/− littermates were obtained from crosses of Spry2 mutant alleles28 on three different mixed genetic backgrounds; similar results were obtained for all three.
Imaging
All specimens were imaged on Leica MZFLIII, Leica MZ16FA or M2 Bio (Kramer Scientific) fluorescence stereomicroscopes. Images were captured with a Spot RT slider (Diagnostic Instruments) camera with Spot software or a Retiga 2000R (Q Imaging) camera with Image-Pro (Media Cybernetics) software. Adobe Photoshop software was used to adjust image levels and to pseudocolor the images in Fig. 3a.
Constructing the branch lineage of the mouse bronchial tree
For each branch, the lineage was reconstructed by assembling the branch patterns of fixed, immunostained lungs from E11 up to E15 (as described in the text) into local, self-consistent orders from which we could infer the dynamic sequence and pattern of branching. For most branches, the lineage is based on groups of specimens that included all or most intermediates. However, for some branches that form during E14, we did not obtain every intermediate but were nevertheless able to reconstruct the lineage and branching dynamics based on the morphological similarity of the intermediates obtained to those of positions where we were able to reconstruct the lineage in detail. Lineages L.M2-3, RCd.M2, R.Ac.A1-4, and R.RMid.V1-4 were the most difficult to reconstruct. They form relatively late and, because of the shape of the lobes, become difficult to visualize clearly as branching proceeds.
Representing the branch lineage in the lineage diagram
Because there is variability among specimens in the local rate of progression through the lineage (see text and Supplementary Fig. 2), and because branching in most positions continues beyond E15, the extent of the lineage shown in Supplementary Figure 1 was, with two exceptions, based on a single, representative E15 lung. One exception was the number of branches in each domain off L and RCd. The number of 2° branches in these eight domains appears complete by E14 because the number of branches did not increase between E14 and E15 as it did for other domains. Hence, for these eight domains, we show in the lineage diagram the maximum number of 2° branches observed in five different E15 lungs. The other exception was the number of rounds of orthogonal bifurcation shown for each sublineage. In the lineage diagram, we show the same number of rounds of orthogonal bifurcation for each daughter branch even though sister branches do not always bifurcate synchronously, and we show a maximum of four rounds because it was difficult to ascertain the branching pattern beyond that. The R.Md.D1 “optional” lineage (see text) was present in the E15 specimen used and hence is included in the lineage diagram but other optional lineages were not, including R.Ac.P1, and anterior or posterior 3° branches off L.L2-6 and RCd.L1-5. Branching “errors” (see text and Fig. 4b–e) are also not included in the diagram.
Because there is some variability in the absolute orientation of orthogonal bifurcations (see text), particularly late in a series of orthogonal bifurcations, in the lineage diagram we did not name the branches produced by the fourth round of orthogonal bifurcations by their orientation, as we do for the earlier rounds, but instead use an asterisk. For the earlier rounds, the orientation given is the one most frequently observed.
Assigning branching modes
Assigning the mode of formation to each branch in the lineage was obvious for most branches from the branching dynamics inferred from the series of developmental intermediates. However, the following branches warrant special comment.
Some branches that appear to form by domain branching (e.g., RCd.V1 and L.V1; see text) were “singletons,” i.e. they were the only branch found in a particular domain. We assume that these are unusual domains in which just a single branch typically forms or domains in °which additional branches form later in development, hence we assigned these branches as the first branch in the domain (e.g. L.L2.A.D1). Likewise, some entire domains that are missing in the lineage presumably form later in development. However there may also be more variation in the number and position of domains that form late.
Because bifurcations were identified as planar or orthogonal based on the orientation of the subsequent round of bifurcation, for the final generation in lineages in which we did not collect information on subsequent branching events, we designated the bifurcation as orthogonal or planar based on morphology and analogy to neighboring positions in the lineage. For example, L.L2.A.A.V1.A and .P which form off a branch that forms by domain branching, look like the first round of orthogonal bifurcation as seen off other branches that form by domain branching.
In several positions in the lineage there were potential ambiguities in assigning branching modes. One was at 1° branch tips where several branches we assigned as forming by domain branching (RCd.L4, RCd.L5, L.5, and L.6) could also be interpreted as planar bifurcations based on branching dynamics and morphology. However, this alternative interpretation is unlikely because elsewhere in the developing lung branches that form by planar bifurcation undergo domain branching in the same domains used by the parent, whereas these branches use only a subset of the domains used by the parent branches. It is also possible based on branching morphology to interpret RCd.L5 and L.L6 as simply the continuation of the 1° branch. However, this too is unlikely because they only form daughter branches in two of the four domains of the 1° branch.
In lineages using Sequence 3 (e.g., L.L1), where a domain (e.g., L.L1.A1 and .2) and daughter branches that form by planar bifurcation (e.g., L.L1.A and .P) lie in the same plane, domain branches could alternatively be interpreted as forming by highly asymmetric planar bifurcations in which one daughter branch of the bifurcation appears to form part of the domain and the other appears to be a continuation of the parent branch. However, we do not prefer this interpretation because all other planar bifurcations are symmetric. Rather, we suggest this relatedness indicates how domain branching may have evolved from planar bifurcation.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank members of the Krasnow lab and Pat Brown, Doug Brutlag, Nir Hacohen, David Kingsley, Lars Mündermann, Jim Spudich, and Elaine Storm for advice and discussion, and Maya Kumar and Maria Petersen for help preparing figures. This work was funded by grants from National Institutes of Health (to M.A.K. and G.R.M.). M.A.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions R.J.M. and M.A.K. conceived the experiments. R.J.M. designed and performed experiments and collected data. O.K. and G.R.M. contributed to conception and design of Spry2 experiments and provided genotyped Spry2 embryos. R.J.M. and M.A.K. analyzed the data and wrote the manuscript. All authors discussed results and edited the manuscript.
Author Information Correspondence and requests for materials should be addressed to R.J.M. (ross.metzger@ucsf.edu) or M.A.K. (krasnow@cmgm.stanford.edu).
References
- 1.Weibel ER. The pathway for oxygen. Harvard University Press, Cambridge; Massachusetts: 1984. [Google Scholar]
- 2.West GB, Brown JH, Enquist BJ. A general model for the origin of allometric scaling laws in biology. Science. 1997;276:122–126. doi: 10.1126/science.276.5309.122. [DOI] [PubMed] [Google Scholar]
- 3.Bejan A. Shape and structure, from engineering to nature. Cambridge University Press; Cambridge: 2000. [Google Scholar]
- 4.Mauroy B, Filoche M, Weibel ER, Sapoval B. An optimal bronchial tree may be dangerous. Nature. 2004;427:633–636. doi: 10.1038/nature02287. [DOI] [PubMed] [Google Scholar]
- 5.Aeby C. Der Bronchialbaum der Säugethiere und des Menschen, nebst Bemerkungen über den Bronchialbaum der Vögel und Reptilien. Engelmann; Leipzig: 1880. [Google Scholar]
- 6.Boyden EA. Segmental anatomy of the lungs; a study of the patterns of the segmental bronchi and related pulmonary vessels. McGraw-Hill; New York: 1955. [Google Scholar]
- 7.Weibel ER, Gomez DM. Architecture of the human lung. Science. 1962;137:577–585. doi: 10.1126/science.137.3530.577. [DOI] [PubMed] [Google Scholar]
- 8.Weibel ER. Morphometry of the human lung. Academic Press Inc.; New York: 1963. [Google Scholar]
- 9.Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science. 1999;284:1635–1639. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- 10.Meinhardt H. Morphogenesis of lines and nets. Differentiation. 1976;6:117–123. doi: 10.1111/j.1432-0436.1976.tb01478.x. [DOI] [PubMed] [Google Scholar]
- 11.Mandelbrot BB. The fractal geometry of nature. W. H. Freeman; New York: 1983. [Google Scholar]
- 12.Nelson TR, Manchester DK. Modeling of lung morphogenesis using fractal geometries. IEEE Transactions on Medical Imaging. 1988;7:321–327. doi: 10.1109/42.14515. [DOI] [PubMed] [Google Scholar]
- 13.Kitaoka H, Takaki R, Suki B. A three-dimensional model of the human airway tree. J Appl Physiol. 1999;87:2207–2217. doi: 10.1152/jappl.1999.87.6.2207. [DOI] [PubMed] [Google Scholar]
- 14.Tawhai M, Pullan A, Hunter P. Generation of an anatomically based three-dimensional model of the conducting airways. Ann Biomed Eng. 2000;28:793–802. doi: 10.1114/1.1289457. [DOI] [PubMed] [Google Scholar]
- 15.Prusinkiewicz P, Lindenmayer A. The algorithmic beauty of plants. Springer-Verlag; New York: 1990. [Google Scholar]
- 16.Miura T. Modeling lung branching morphogenesis. Curr Top Dev Biol. 2007;81:291–310. doi: 10.1016/S0070-2153(07)81010-6. [DOI] [PubMed] [Google Scholar]
- 17.Tebockhorst S, Lee D, Wexler AS, Oldham MJ. Interaction of epithelium with mesenchyme affects global features of lung architecture: A computer model of development. J Appl Physiol. 2007;102:294–305. doi: 10.1152/japplphysiol.00665.2006. [DOI] [PubMed] [Google Scholar]
- 18.Cardoso WV, Lu J. Regulation of early lung morphogenesis: Questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 19.His W. Zur Bildungsgeschichte der Lungen beim menschlichen Embryo. Archiv für Anatomie und Entwicklungsgeschichte. 1887;1887:89–106. [Google Scholar]
- 20.Flint JH. Development of the lungs. Am J Anat. 1906;6:1–138. [Google Scholar]
- 21.Heiss R. Zur Entwicklung und Anatomie der menschlichen Lunge. Archiv für Anatomie und Physiologie, anatomische Abteilung. 1919:1–129. [Google Scholar]
- 22.Borghese E. The development in vitro of the submandibular and sublingual glands of Mus musculus. J Anat. 1950;84:287–302. [PMC free article] [PubMed] [Google Scholar]
- 23.Alescio T. Osservazioni su culture organotipiche di polmone embrionale di topo. Arch Ital Anat Embriol. 1960;65:323–363. [PubMed] [Google Scholar]
- 24.Massoud EA, et al. In vitro branching morphogenesis of the fetal rat lung. Pediatr Pulmonol. 1993;15:89–97. doi: 10.1002/ppul.1950150205. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe T, Costantini F. Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev Biol. 2004;271:98–108. doi: 10.1016/j.ydbio.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 26.Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature. 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hummel KP, Chapman DB. Visceral inversion and associated anomalies in the mouse. J Hered. 1959;50:9–13. [Google Scholar]
- 28.Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Hacohen N, et al. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 30.Minowada G, et al. Vertebrate sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–4475. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- 31.Eppig JT, et al. Mouse genome database. 2005 Available at www.informatics.jax.org.
- 32.Hogan BL. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 33.Warburton D, et al. Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr Res. 2005;57:26R–37R. doi: 10.1203/01.PDR.0000159570.01327.ED. [DOI] [PubMed] [Google Scholar]
- 34.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 35.Metzger RJ, Tang N, Martin GR, Krasnow MA. Airway control of pulmonary artery patterning in mouse lung development. (In preparation) [Google Scholar]
- 36.Espinoza FH, Firkus C, Fish G, Krasnow MA. Genomic analysis of signaling and receiving centers in the embryonic mouse lung. (In preparation) [Google Scholar]
- 37.al-Awqati Q, Goldberg MR. Architectural patterns in branching morphogenesis in the kidney. Kidney Int. 1998;54:1832–1842. doi: 10.1046/j.1523-1755.1998.00196.x. [DOI] [PubMed] [Google Scholar]
- 38.Lu P, Sternlicht MD, Werb Z. Comparative mechanisms of branching morphogenesis in diverse systems. J Mammary Gland Biol Neoplasia. 2006;11:213–228. doi: 10.1007/sl0911-006-9027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomson AA, Marker PC. Branching morphogenesis in the prostate gland and seminal vesicles. Differentiation. 2006;74:382–392. doi: 10.1111/j.1432-0436.2006.00101.x. [DOI] [PubMed] [Google Scholar]
- 40.Shubin NH, Alberch P. A morphogenetic approach to the origin and basic organization of the tetrapod limb. Evol Biol. 1986;20:319–387. [Google Scholar]
- 41.Shubin N, Tabin C, Carroll S. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–648. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- 42.Prusinkiewicz P, et al. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- 43.Wells LJ, Boyden EA. The development of the bronchopulmonary segments in human embryos of horizons XVII to XIX. Am J Anat. 1954;95:163–201. doi: 10.1002/aja.1000950202. [DOI] [PubMed] [Google Scholar]
- 44.Hirai Y, Nose A, Kobayashi S, Takeichi M. Expression and role of E- and P-cadherin adhesion molecules in embryonic histogenesis. I. Lung epithelial morphogenesis. Development. 1989;105:263–270. doi: 10.1242/dev.105.2.263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.