Summary
During Bacillus subtilis sporulation, an endocytic-like process called engulfment results in one cell being entirely encased in the cytoplasm of another cell. The driving force underlying this process of membrane movement has remained unclear, although components of the machinery have been characterized. Here we provide evidence that synthesis of peptidoglycan, the rigid, strength bearing extracellular polymer of bacteria, is a key part of the missing force-generating mechanism for engulfment. We observed that sites of peptidoglycan synthesis initially coincide with the engulfing membrane and later with the site of engulfment membrane fission. Furthermore, compounds that block muropeptide synthesis or polymerization prevented membrane migration in cells lacking a component of the engulfment machinery (SpoIIQ), and blocked the membrane fission event at the completion of engulfment in all cells. In addition, these compounds inhibited bulge and vesicle formation that occur in spoIID mutant cells unable to initiate engulfment, as did genetic ablation of a protein that polymerizes muropeptides. This is the first report to our knowledge that peptidoglycan synthesis is necessary for membrane movements in bacterial cells and has implications for the mechanism of force generation during cytokinesis.
Introduction
A central question in cell biology is the nature of the forces driving membrane movement. During endocytosis, the cellular membrane invaginates and ultimately forms a lipid-bounded compartment that is distinct from the membrane. This event rarely occurs spontaneously and is dependent upon enzymes that facilitate this process. Proteins that polymerize into filaments, such as actin and tubulin, convert the chemical energy of polymerization into mechanical work (Theriot, 2000; Phillips et al., 2009). For example, actin polymerization mediates contractile ring formation during cytokinesis in Schizosaccharomyces pombe (Pelham and Chang, 2002) and facilitates deformations of the plasma membrane seen during endocytosis (Qualmann et al., 2000) or in protruding lamellipodia (Mitchison and Cramer, 1996). Microtubule polymerization drives the movement of organelles and chromosomes (Inoue and Salmon, 1995). However, while the polymerization of actin (Miyata et al., 1999) or tubulin (Elbaum et al., 1996) can generate a mechanical force that distorts membranes in vitro, it remains unclear whether polymerization is sufficient to explain in vivo membrane dynamics.
A process of membrane fission is seen in Bacillus subtilis that are responding to nutritional limitation. As part of this response, called sporulation, B. subtilis divides asymmetrically and undergoes engulfment, an endocytic-like process of membrane fission that results in one cell, the forespore, being entirely encased in the cytoplasm of the larger cell, the mother cell (Fig. 1A, top). Initially, an asymmetric division septum forms generating the two differently sized cells. The subsequent increasing curvature of this septum causes the forespore to assume a rounded shape comprised of a double membrane separated by a thin layer of peptidoglycan. SpoIID and SpoIIP are autolysins that hydrolyse cell wall and they have been proposed to drive this membrane movement by generating a force through a ratchet-like mechanism driven by the hydrolysis of peptidoglycan (Abanes-De Mello et al., 2002; Broder and Pogliano, 2006). Engulfment is also facilitated by the formation of the SpoIIQ–SpoIIIAH protein–protein zipper between the forespore and mother cell, which is required for membrane migration when the activity or levels of SpoIID and SpoIIP are reduced (Broder and Pogliano, 2006). During this movement, the forespore remains attached to the mother cell but in the final step, the forespore pinches off from the mother cell. However, the source of the force responsible for this final step remains unknown.
Fig. 1.
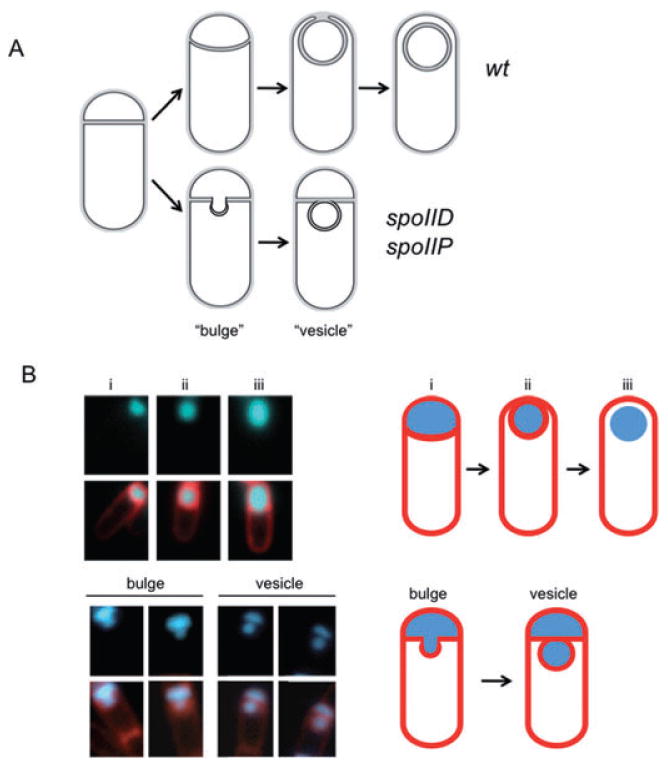
Membrane bulge and vesicle formation in engulfment mutants.
A. Sporulating wild-type cells undergo engulfment where the smaller forespore compartment becomes a free cell within the larger mother cell (top). Strains carrying spoIID or spoIIP mutations are blocked at the stage of asymmetric septation and undergo membrane bulging, eventually leading to the formation of membrane bound vesicles (bottom).
B. Wild-type (AES574) and spoIID strains (JDB2494) expressing CFP (blue) under control of a forespore-specific promoter (PspoIIQ) as a forespore marker were imaged at T4 in resuspension medium and stained with FM4-64 (red) to visualize membranes. Top, CFP is expressed in: (i) the forespore of wild-type cells undergoing asymmetric septation stained with FM4-64, (ii) engulfing cells stained with FM4-64 and (iii) engulfed cells not stained with FM4-64. Bottom, spoIID cells contained either a continuous CFP signal distribution between the forespore and the membrane protrusion (‘bulge’), or two distinguishable CFP signals separated by a membrane (‘vesicle’).
Membrane-bound compartments in other bacteria are thought to result from involutions of the cell membrane. For example, magnetosomes are membrane-associated organelles observed in magnetotactic bacteria composed of crystals of magnetite that are surrounded by a lipid bilayer (Komeili et al., 2006; Scheffel et al., 2006) and Escherichia coli carrying mutations in a gene necessary for proper cell shape produce membrane invaginations and cytoplasmic vesicles (Bendezu and de Boer, 2008). Sporulating B. subtilis strains carrying spoIID or spoIIP null mutations form asymmetric septa that fail to proceed with engulfment. Instead, membrane bulges appear gradually at the septa (Fig. 1A, bottom). Electron microscopy (Lopez-Diaz et al., 1986; Illing and Errington, 1991; Smith and Youngman, 1993; Bylund et al., 1994; Frandsen and Stragier, 1995) and staining with the membrane impermeable stain FM4-64 (Abanes-De Mello et al., 2002) indicate that the bulges in spoIIP and spoIID mutants are continuous with the forespore compartment. The origin of the forces which drive bulge formation are unknown, although by analogy with the roles of actin and tubulin polymerization in eukaryotic membrane fusion and fission, a protein or peptide capable of polymerizing could serve a similar role. Bulge formation in these mutants therefore offers a useful system to examine candidate force-generating mechanisms responsible for membrane movements in bacteria.
Most bacteria contain peptidoglycan, a rigid polymer built from disaccharide peptide monomers that can be up to 5000 units in length (Hayhurst et al., 2008). Peptidoglycan monomers, also known as muropeptides, are synthesized in the bacterial cytoplasm by a series of essential and highly conserved enzymes that convert the sugar UDP-GlcNAc to the lipid-linked UDP-disaccharide pentapeptide (van Heijenoort, 2001) which is flipped across the membrane to the outside of the cell where cross-linking transpeptidation and polymerizing transglycosylation reactions link these muropeptides to mature peptidoglycan (Sauvage et al., 2008). While the glycan polymers of peptidoglycan are generated by transglycosylases, inhibition of the transpeptidation reaction by vancomycin also results in a loss of this polymerization activity in Staphylococcus aureus (Kim et al., 2008). Finally, since cell wall peptidoglycan has a Young's modulus of 1.7–25 MPa (Yao et al., 1999; Francius et al., 2008), it is up to 104 times stiffer than actin, so formation of mature peptidoglycan would produce mechanical work.
The requirement for peptidoglycan synthesis during bacterial growth complicates assessment of the hypothesis that it provides a driving force for membrane movement. We have therefore examined the role of peptidoglycan synthesis in three different membrane movements that occur during the non-essential process of B. subtilis sporulation. First, during the initial stages of engulfment, the membranes surrounding the forespore undergo a process of migration that results in the formation of curved membranes. We find that inhibition of peptidoglycan synthesis blocks this migration in a strain missing a secondary membrane migration system requiring SpoIIQ and we find that peptidoglycan biosynthesis is localized to the leading edge of the engulfing membrane in wild-type cells and in a mutant in which membrane migration occurs asymmetrically. Second, during the last stage of engulfment, peptidoglycan synthesis is localized to the last site of attachment between the two cells and antibiotics that inhibit peptidoglycan synthesis prevent separation of the two cells and completion of engulfment. Third, peptidoglycan synthesis occurs at sites of bulge formation in spoIID and spoIIP mutants and compounds that inhibit the synthesis of muropeptides block the initiation of these bulges and their ultimate formation as membrane-bounded, ‘vesicle’-like compartments. Consistent with the action of these compounds, a null mutation in a gene encoding a protein necessary for the synthesis of mature cross-linked peptidoglycan suppresses bulge formation. Together these results suggest that during engulfment, membrane migration and membrane fission require peptidoglycan biosynthesis.
Results
Peptidoglycan synthesis during engulfment
During the process of engulfment, the asymmetric septum becomes rounded and eventually pinches off from the mother cell, resulting in the formation of a membrane-bounded compartment (Fig. 1A, top). Sites of active peptidoglycan synthesis in engulfing cells were identified using a fluorescent ramoplanin that binds the reducing end of nascent glycan chains found at the initiation sites of peptidoglycan synthesis (Tiyanont et al., 2006). Sporulating cells showed a clear strong fluorescent signal at the sporulation septum during engulfment (Fig. 2A, i). This signal was due to active peptidoglycan synthesis because treatment of sporulating cells before polar septation with fosfomycin, an inhibitor of peptidoglycan synthesis, showed no ramoplanin labelling (Fig. S1, middle) and a derivative of ramoplanin that does not label lipid II (Tiyanont et al., 2006) did not produce similar patterns of staining (Fig. S1).
Fig. 2.
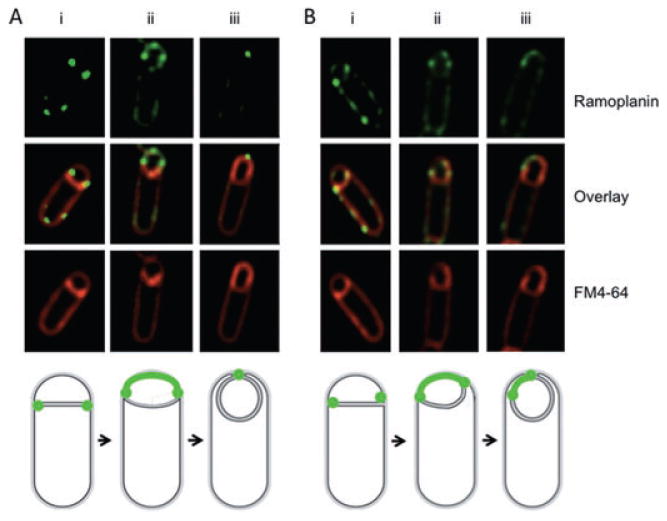
Peptidoglycan synthesis during engulfment. Engulfing cells contain sites of active peptidoglycan synthesis. Ramoplanin-FL (top) and FM4-64 (bottom) staining of (A) wild-type (PY79) cells and (B) spoIID(D210A) cells; centre panel are the overlay of the two images. (i) Ramoplanin-FL staining at septal tips, (ii) ramoplanin-FL staining around the forespore, (iii) ramoplanin-FL staining restricted to the point of contact between the advancing arms. Diagrams shown represent ramoplanin-FL (green) signals at different stages of engulfment corresponding to wild-type (left) and spoIID(D210A) (right) images.
Ramoplanin-FL fluorescence was enriched at the edges of the septal disk at early (Fig. 2A, i) and intermediate stages of engulfment (Fig 2A, ii and Fig. S2A) and eventually localized to the area of membrane fission (Fig. 2A, iii and Fig. S2B). The reduced signal in other regions of the forespore was likely not the result of reduced accessibility to the space between the two forespore membranes because forespore staining was similar in a spoIIQ mutant where the forespore membranes are less tightly associated (Broder and Pogliano, 2006) as compared with the wild type (Fig. S2). Once engulfment completed and the forespore had separated from the mother cell, the ramoplanin signal in the forespore disappeared (data not shown) likely because it is unable to cross the lipid bilayer (Hamburger et al., 2009). Next, we took advantage of a spoIID(D210A) mutant (KP1102; Gutierrez et al., 2010) where cells stained with FM4-64 show a clear asymmetry in membrane migration (80% of cells, 53 out of 66 cells) as exemplified by a single cell where one of the tips of the migrating membranes advances further than the other tip (Fig. 2B, cells ii and iii). Sites of peptidoglycan synthesis showed a similar asymmetrical staining pattern (Fig. 2B, top and middle), indicating that the pattern of peptidoglycan synthesis during engulfment reflects the asymmetrical migration of the membrane.
Inhibition of peptidoglycan synthesis blocks membrane migration
During engulfment, the septal membranes migrate around the forespore compartment ultimately resulting in the production of a cell within a cell. The observation that ramoplanin stained the leading edges of the engulfing forespore (Fig. 2A, ii) suggested that active peptidoglycan synthesis was involved in this process. We examined this possibility by performing time-lapse microscopy to follow single sporulating cells with FM4-64-stained membranes (Becker and Pogliano, 2007) and determining the effect of fosfomycin, an antibiotic that specifically inhibits MurAA, the first enzyme in the peptidoglycan biosynthetic pathway (Walsh, 2003). When 5 mM fosfomycin was added to wild-type cells at T1.5 after initiation of sporulation and cells were imaged starting at T2, no significant disruption could be detected in membrane migration (Fig. 3A and B and Fig. S3). However, membrane migration during engulfment is dependent on two partially redundant mechanisms, peptidoglycan hydrolysis by SpoIID and SpoIIP and the zipper-like interaction of SpoIIQ with SpoIIIAH. In mutants with reduced hydrolase activity, the SpoIIQ/SpoIIIAH system becomes essential (Broder and Pogliano, 2006), so a requirement for muropeptide synthesis for membrane migration could be similarly masked. Consistent with this hypothesis, time-lapse movies revealed that fosfomycin reduced membrane migration in spoIIQ mutant cells (Fig. 3C–E and Fig. S3A–C), as 54 out of 72 cells (75%) showed membrane migration in the spoIIQ mutant, but only 17 out of 55 (31%) in the spoIIQ mutant with fosfomycin. To ensure that fosfomycin did not have pleiotropic effects during sporulation of spoIIQ cells, we confirmed that a fluorescent marker under σF control was activated in spoIIQ cells that showed no membrane migration in the presence of fosfomycin (Fig. S3E).
Fig. 3.
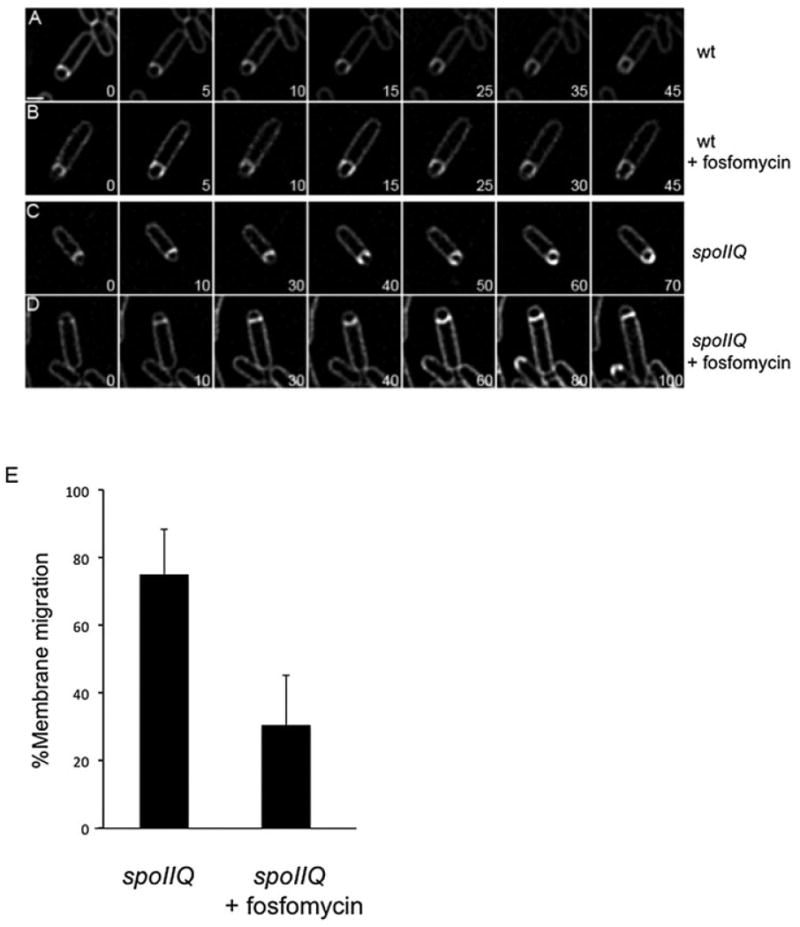
Inhibition of muropeptide synthesis blocks membrane migration in a spoIIQ mutant. All cultures were grown on agarose pads composed of A+B medium at 30°C and stained with FM4-64. The initial image in each sequence was taken at approximately T2 after sporulation initiation and arbitrarily set to t = 0 min. The time of subsequent images of the membrane stain is indicated in minutes in the lower right corner.
A. Wild type (PY79).
B. Wild type + 5 mM fosfomycin added at T1.5 after initiation of sporulation.
C. spoIIQ (KP575).
D. spoIIQ + 5 mM fosfomycin added at T1.5.
E. Histogram showing the percentage of cells where membrane migrated in ΔspoIIQ cells with (17 out of 55 cells or 75 ± 5%) or without (54 out of 72 cells or 30 ± 6%) 5 mM fosfomycin added at T1.5 after initiation of sporulation. Means are significantly different by t-test (P < 10−5). For complete movies, see Fig. S3. Scale bar = 1 μm.
A fluorescence-based assay to monitor completion of engulfment
The presence of a strong ramoplanin signal at the point of contact between the two engulfing membrane arms (Fig. 2A, iii) suggested that peptidoglycan synthesis was occurring at an appropriate time and place to play a role in the separation of the outer forespore membrane from the mother cell membrane that marks completion of membrane fission during engulfment. We examined this possibility using a strain that expresses CFP only in the forespore along with visualization of membranes by the lipophilic fluorescent dye FM4-64 that was added just prior to microscopy. We expressed CFP under control of a forespore specific promoter (PspoIIQ), which is active only upon completion of the septum and thus CFP-positive cells must have progressed past this point in sporulation. When asymmetric septation was completed in this strain, the forespore contained a CFP signal that was surrounded by an FM4-64 signal (Fig. 4A, panels ‘T3.5’, ‘T4’; numbers indicate the time in hours after the initiation of sporulation). Once engulfment ends, the forespore has detached from the mother cell and since FM4-64 does not cross membranes (Sharp and Pogliano, 1999), the forespore is no longer accessible to the dye. Thus, an engulfed forespore had a CFP signal but no FM4-64 staining (Fig. 4A, panel ‘T5’, yellow arrow), whereas a forespore that has not completed engulfment and remains attached to the mother cell was stained with FM4-64 (Fig. 4A, panel ‘T4.5’, green arrow). This assay therefore distinguishes cells at the pre-separation step and cells where the membranes had completed fission and separated.
Fig. 4.
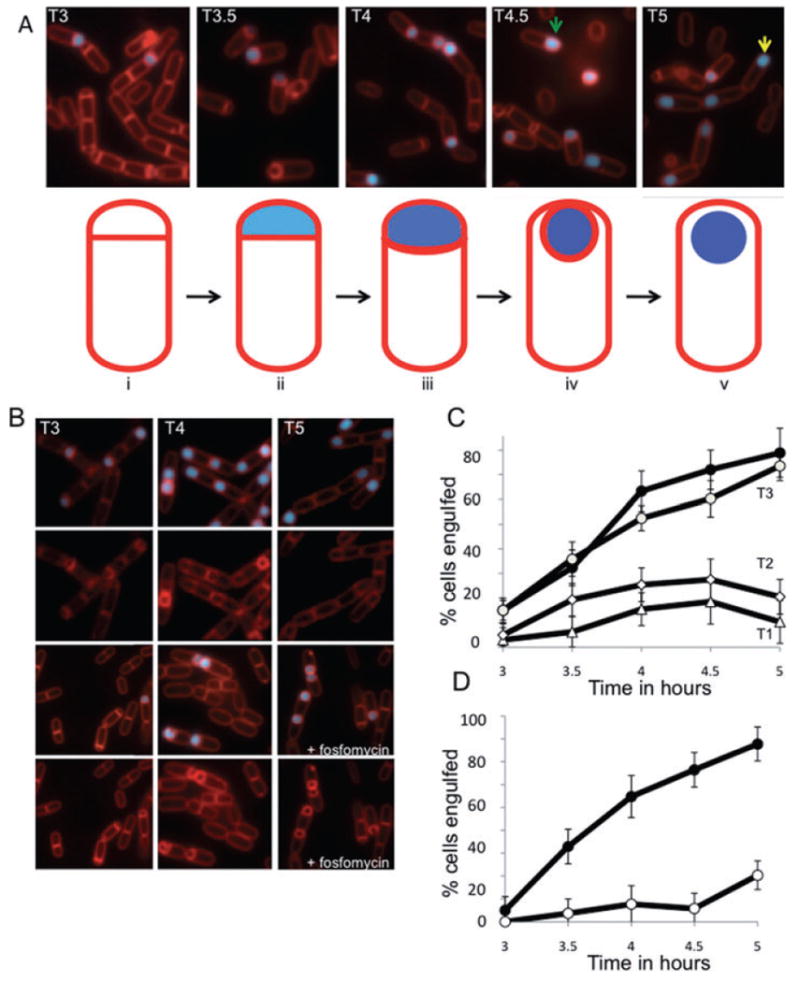
Inhibition of muropeptide synthesis blocks membrane fission.
A. Assay of membrane fission. Top, sporulating wild-type cells (AES574) express CFP (blue) under control of a forespore specific promoter (PspoIIQ). Membranes of cells at intermediate stages of engulfment (T3–T4.5, green arrow) are also stained by a membrane impermeant dye (FM4-64, red) just before imaging; upon completion of membrane fission, cells are no longer stained by this dye (yellow arrow). Time after resuspension is indicated. Bottom, cells at different stages of sporulation. Prior to the completion of asymmetric septation (i), no CFP (blue) signal is observed. Following completion of asymmetric septation, a faint CFP signal is observed in the forespore (ii). As the cells proceed through engulfment (iii), the CFP signal increases. When membrane movement is complete (iv), the forespore remains attached to the mother cell and stained with FM4-64 (red). When the forespore is released from the mother cell (v), the CFP signal is no longer surrounded by an FM4-64 signal.
B. Fosfomycin blocks engulfment before membrane separation. Sporulating wild-type cells were either treated with fosfomycin (5 mM) at T2 (bottom panels) or not treated (top panels). Time after resuspension is indicated.
C. Inhibition of membrane fission by fosfomycin is dependent on time of addition. Wild-type cells (AES574) were untreated (filled circles) or fosfomycin (5 mM) was added at T1 (triangles), T2 (diamonds) or T3 (open circles) and the fraction of cells with a CFP signal not surrounded by an FM4-64 signal was determined.
D. Fosfomycin-resistant murAA mutant engulfs normally in the presence of fosfomycin. The effect of fosfomycin on engulfment of a merodiploid strain (JDB2426) expressing an IPTG-inducible C117D murAA allele and YFP under a forespore-specific promoter (PspoIIQ-yfp) was determined. Filled circles, both 1 mM IPTG was added at T0 and 5 mM fosfomycin was added at T2; open circles, only 5 mM fosfomycin at T2.
Inhibition of muropeptide synthesis blocks engulfment membrane fission
This fusion assay was used to determine the effect of inhibiting peptidoglycan synthesis on completion of engulfment. Addition of fosfomycin (5 mM) after asymmetric septation at T2 of sporulation allowed membrane migration but blocked membrane fission (i.e. the detachment of the forespore from the mother cell; Fig. 4B, bottom images). When fosfomycin was added at different times after the start of sporulation, fosfomycin blocked engulfment membrane fission only if it was added before T3, but had no effect when added at T3 (Fig. 4C). A decreasing fraction of cells containing a forespore that had detached from the mother cell was observed at fosfomycin concentrations ranging from 1 mM to 10 mM (Fig. S4).
When 5 mM fosfomycin was added at T2 to a strain that expressed an inducible fosfomycin-resistant allele of MurAA (C117D), the fraction of sporulating cells that completed engulfment was significantly higher than a culture where this allele was not expressed (Fig. 4D, Fig. S5). Since the presence of this MurAA mutant allele by itself did not effect engulfment (Fig. S6), fosfomycin must be blocking engulfment membrane fission by directly inhibiting muropeptide synthesis.
Inhibition of peptidoglycan synthesis blocks completion of engulfment
The requirement of muropeptide synthesis to complete engulfment suggests that polymerization of these monomers during formation of mature peptidoglycan was also necessary for engulfment. Vancomycin inhibits transpeptidation and thereby blocks the transglycosylation step necessary for polymerization. Addition of vancomycin at T2 after resuspension blocked engulfment membrane fission (Fig. 5A). Importantly, this was not an effect of blocking septal synthesis since the assay uses a fluorescent reporter whose activity is dependent on the completion of asymmetric septation. This inhibition did depend on when vancomycin was added in sporulation although it still blocked completion of engulfment in 50% of cells when it was added as late as T3.5 (Fig. 5B). Thus, muropeptide polymerization is necessary for completion of engulfment defined as the release of the forespore from the mother cell.
Fig. 5.
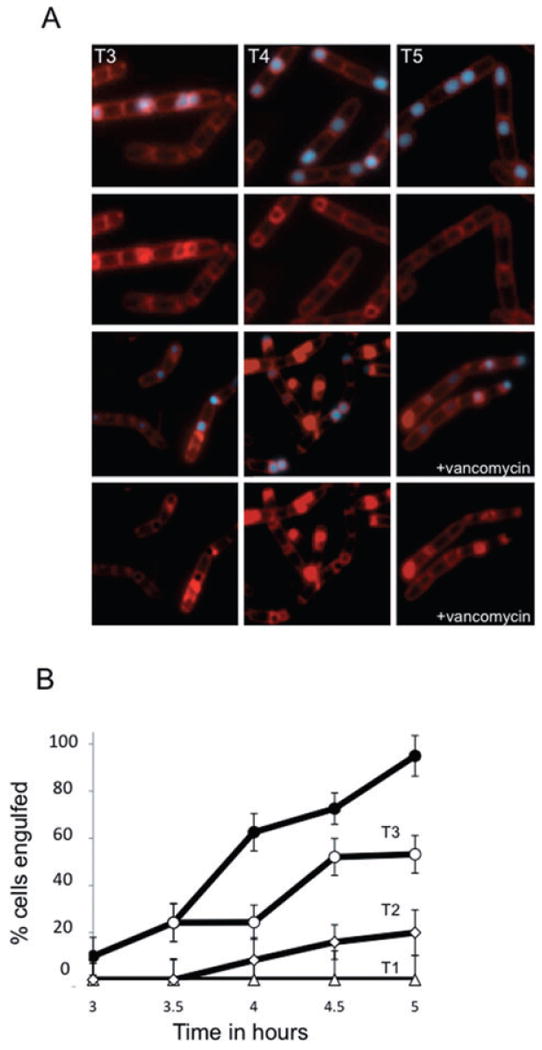
Inhibition of muropeptide transpeptidation blocks completion of engulfment.
A. Vancomycin blocks engulfment before membrane fission. At T2 of sporulation, cells (AES574) expressing CFP (blue) under control of a forespore-specific promoter (PspoIIQ) were either treated with vancomycin (0.5 μg ml−1; bottom rows) or not treated (top rows). Membranes were visualized with FM4-64 (red). Time after resuspension is indicated.
B. Inhibition of membrane fission depends on the time of addition. The percentage of sporulating cells completing membrane fission was determined for untreated cells (filled circle) or cells incubated with vancomycin (0.5 μg ml−1) at T1 (triangles), T2 (diamonds) or T3.5 (open circles) of sporulation by resuspension.
Topology of membrane bulges
Sporulating cells lacking either the SpoIIP or the SpoIID autolysins fail to initiate engulfment and instead produce septal membrane bulges (Fig. 1A, bottom, Fig. S7; Lopez-Diaz et al., 1986; Illing and Errington, 1991; Smith and Youngman, 1993; Bylund et al., 1994; Frandsen and Stragier, 1995). Time-lapse microscopy of single cells revealed that bulge formation was continuous and occurred over a period of ∼30 min (Fig. 6A, Fig. S7). To characterize the topology and origin of these bulges in living cells, we expressed CFP under control of a forespore specific promoter (PspoIIQ). We observed a CFP signal in membrane bulges of a spoIID null mutant indicating that they had been, at least transiently, contiguous with the forespore (Fig. 1B, ‘bulge’). In some cells, the CFP signal in the forespore and the bulge was not continuous, and the presence of an FM4-64 signal surrounding these discontinuous bulges suggests that the bulge was transformed into a physically distinct membrane-bounded vesicle (Fig. 1B, ‘vesicle’; Fig. S8).
Fig. 6.
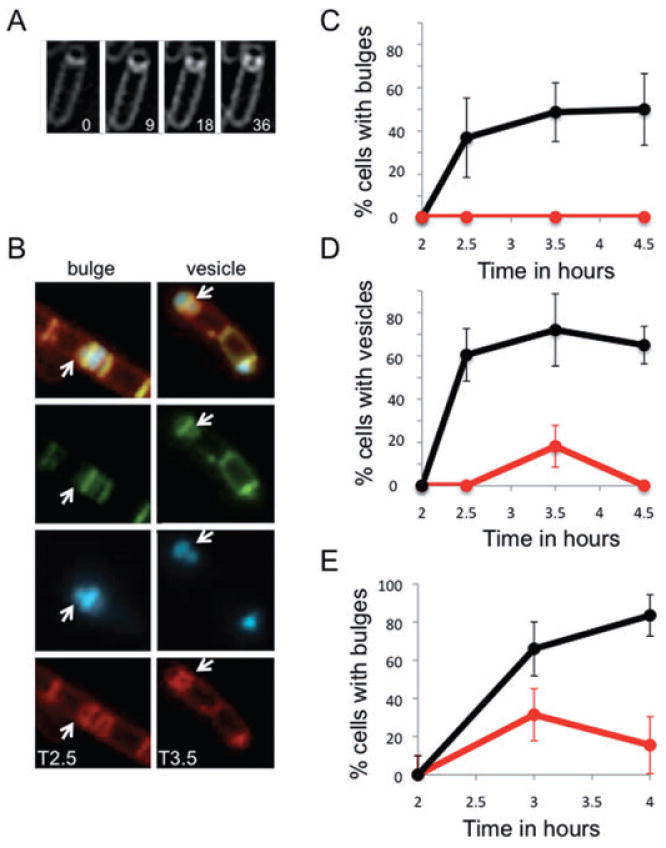
Muropeptide synthesis and polymerization are required for bulge and vesicle formation.
A. Time-lapse images of a bulge progressively forming in a spoIID mutant strain (JDB2395) sporulating in a pad at 30°C and stained with FM4-64. Initial image was taken at approximately T2 after sporulation initiation and arbitrarily set to t = 0 min. The time of subsequent images is indicated in minutes in the lower right corner. See movie Fig. S7.
B. In a spoIID mutant strain (JDB2494) expressing CFP (blue) in the forespore, bulge and vesicles (white arrows) are sites of peptidoglycan synthesis. Membranes were visualized with FM4-64 (red) and newly synthesized peptidoglycan identified by ramoplanin-FL, a fluorescent ramoplanin derivative (green). Time after start of sporulation is indicated.
C. Fosfomycin blocks bulge formation. Bulges do not form in a strain (JDB2494) lacking spoIID when 5 mM fosfomycin was added at T2 of sporulation (red line) compared with cells where no fosfomycin was added (black line).
D. Fosfomycin blocks vesicle formation. Percentage of cells with vesicles in strains carrying mutations in spoIID and expressing CFP at the forespore (JDB2494) in the absence (black) or in the presence of 5 mM fosfomycin added at T2 of sporulation (red).
E. Vancomycin blocks bulge formation. Percentage of spoIID cells (JDB2494) with bulges in the absence (black) or in the presence of 0.5 μg ml−1 vancomycin (red) added at T2 of sporulation. Bulges and vesicles were identified as defined in Fig. 1B.
Peptidoglycan synthesis in membrane bulges
What is the driving force underlying the formation of these bulges and vesicles? They originated from the septum, a site of prior peptidoglycan synthesis. Bulge-forming spoIID cells stained with ramoplanin-FL at T4 of sporulation had a fluorescent signal associated with the septum, indicating the presence of active peptidoglycan synthesis (Fig. 6B). We examined whether peptidoglycan synthesis was necessary for bulge formation by blocking it using fosfomycin. When 5 mM fosfomycin was added at T2 of sporulation to either spoIID (Fig. 6C, red) or spoIIP (Fig. S9) mutants, many fewer bulges were observed as compared with untreated cells (Fig. 6C, Fig. S9, black). Furthermore, addition of fosfomycin at T2 also blocked formation of vesicles that presumably originated as bulges (Fig. 6D). This effect of fosfomycin was due to a direct inhibition of peptidoglycan synthesis because a spoIID strain carrying the MurAA(C117D) allele exhibited bulge formation during sporulation even in the presence of 5 mM fosfomycin (Fig. S10).
While the inhibition of muropeptide synthesis results indirectly in decreased levels of mature, polymerized peptidoglycan, the experiments using fosfomycin do not address the possibility that direct inhibition of synthesis of peptidoglycan polymers from muropeptide monomers would similarly block formation of bulges and vesicles. We used vancomycin to answer this question, and observed that addition of vancomycin (0.5 μg ml−1) at T2 of sporulation blocked bulge (Fig. 6E) and vesicle formation (Fig. S11) in a strain lacking spoIID, indicating that synthesis of mature peptidoglycan is necessary for bulge formation. This inhibition by vancomycin could be through an indirect effect on lipid synthesis if reducing lipid synthesis blocked bulge formation. We addressed this possibility by using cerulenin, an inhibitor of an essential enzyme (FadD) in the fatty acid biosynthetic pathway. However, addition of cerulenin had no effect on bulge formation when it was added to spoIID cells at the same time in sporulation (T3.5) where the inhibitory effect of vancomycin was observed (Fig. S12). While addition of cerulenin at earlier time points (T2; data not shown) prevented bulge formation in spoIID cells, this may be due to the membrane growth that is evident in time-lapse movies of bulge formation (Fig. S7).
A peptidoglycan transpeptidase is necessary for bulge formation
The muropeptide monomer is incorporated into mature peptidoglycan by enzymes mediating transpeptidation and transglycosylation reactions. During sporulation, the transpeptidase SpoVD is necessary for the production of mature peptidoglycan (Vasudevan et al., 2007). A complementing GFP–SpoVD fusion localizes to septal membrane bulges and vesicles (Fig. 7A), similar to the pattern observed with ramoplanin-FL (Fig. 6B). This similarity suggested that SpoVD is involved, and perhaps necessary, for the peptidoglycan synthesis occurring in the bulges. In fact, bulge formation is reduced in a spoIIP strain lacking SpoVD (Fig. 7B, red) as compared with the parent spoIIP strain (black). The result is consistent with the inhibition of bulge formation by the transpeptidase inhibitor vancomycin in spoIID (Fig. 6E) and spoIIP cells (data not shown) and demonstrates that a protein necessary for the synthesis of mature peptidoglycan is also necessary for bulge formation.
Fig. 7.
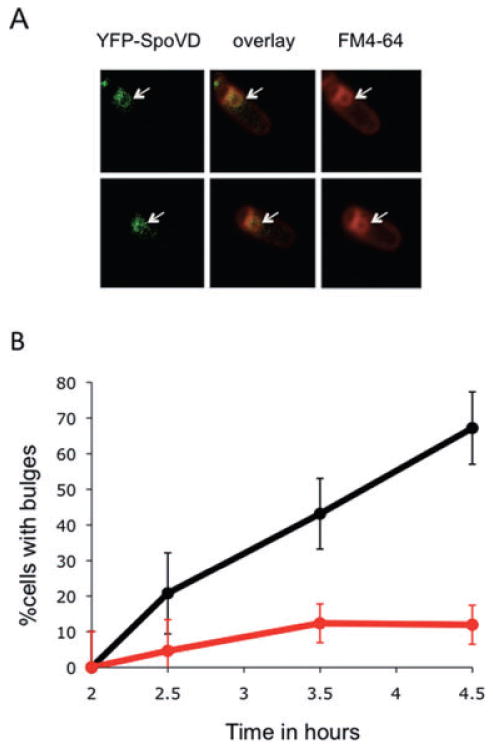
Muropeptide-polymerizing enzyme is required for bulge formation.
A. The transpeptidase SpoVD localizes to the bulges. A strain lacking spoIIP and expressing a YFP–SpoVD fusion (JDB2553) was sporulated by resuspension and images acquired at T4. Two cells are shown. Left, YFP–SpoVD signal; middle is overlay of the YFP–SpoVD and FM4-64 signals; right, FM4-64. White arrows show the bulge.
B. Bulge formation is suppressed by a spoVD mutation. Percentage of spoIIP (JDB2396; black) and spoIIP spoVD (JDB2537; red) cells with bulges was determined. Bulge formation was assessed as defined in Fig. 1B.
Discussion
Muropeptide polymerization during engulfment
Our results allow historical experiments concerning the role of peptidoglycan biosynthesis in engulfment to be put into a mechanistic context. Among the earliest hints of such a role was the observation that removal of the cell wall by lysozyme treatment leads to a block in sporulation, but only when this treatment occurs before engulfment (Fitz-James, 1964). Consistent with this observation, B. subtilis exposed to vancomycin during engulfment do not reach a phase bright state, suggesting that peptidoglycan synthesis is required towards the end of engulfment (Dancer, 1979). During engulfment, a thin layer of peptidoglycan known as the germ cell wall is generated adjacent to the inner forespore membrane (Tipper and Linnett, 1976; Meador-Parton and Popham, 2000). Thus, the inhibitory effect of vancomycin on engulfment (Fig. 5) could be due to an inhibition of germ cell wall synthesis.
Consistent with this possibility, our observation that sites of active peptidoglycan synthesis are distributed around the engulfing forespore (Figs 2A and 8A, top) suggests that muropeptide polymerization could be mediating earlier stages of engulfment. Protoplasts of B. subtilis that lack peptidoglycan are still able to undergo this membrane movement in a mechanism that depends on the SpoIIQ–SpoIIIAH zipper, which is dispensable for engulfment in intact cells, but which is independent of the SpoIID–SpoIIP peptidoglycan hydrolase proteins that are essential in intact cells (Broder and Pogliano, 2006). Our results demonstrate that the inhibition of muropeptide synthesis blocks membrane migration in intact cells lacking the SpoIIQ–SpoIIIAH zipper (Fig. 3), suggesting that peptidoglycan polymerization comprises a mechanism for membrane migration during engulfment that is essential in the absence of the Q–AH zipper. We therefore propose that peptidoglycan polymerization and the SpoIIQ–SpoIIIAH zipper comprise two redundant mechanisms for force generation (Fig. 8B). Peptidoglycan hydrolysis is required early in engulfment in order to release the asymmetric septum from the cellular cross-wall, and proteins that mediate this event are therefore required for the initiation of engulfment (Abanes-De Mello et al., 2002; Morlot et al., 2010). Peptidoglycan hydrolysis is required throughout membrane migration (Abanes-De Mello et al., 2002; Gutierrez et al., 2010), but it remains unclear if it provides force for membrane migration or plays some other role, such as removing steric barriers to the advancing membranes.
Fig. 8.
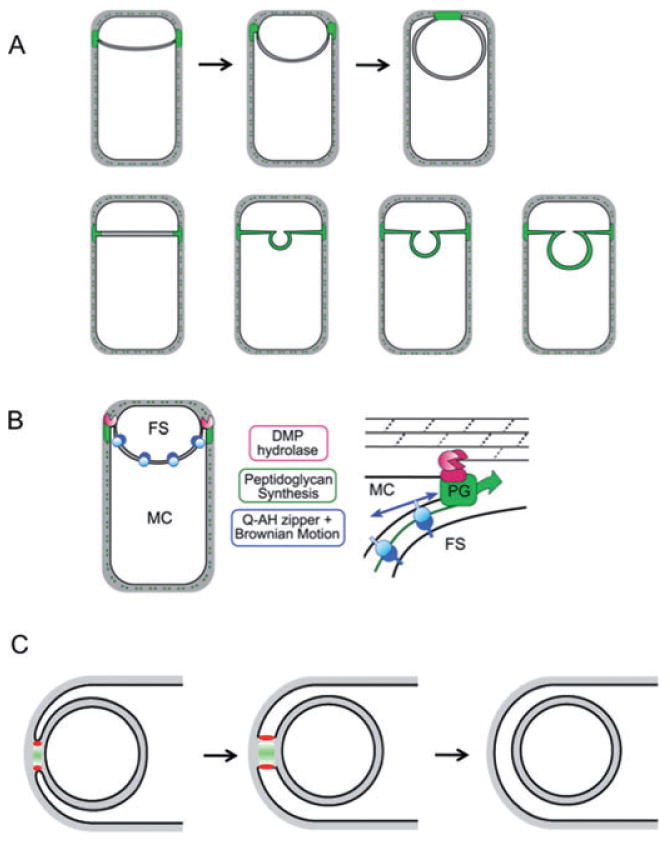
Role of peptidoglycan polymerization in engulfment.
A. Diagram shows distribution of active peptidoglycan synthesis (green) during membrane migration and before detachment of the forespore from the mother cell (top) as well as during bulge formation (bottom).
B. The polymerization of these newly synthesized muropeptides partially drives membrane movement and results in the separation of the two cells. Our data suggest that membrane movement can be mediated either by peptidoglycan synthesis (PG; green) or by the SpoIIQ–SpoIIIAH proteins (blue) and that peptidoglycan hydrolysis mediated by the DMP proteins (red) plays a necessary role in detaching the septal peptidoglycan from the transverse peptidoglycan. FS, forespore; MC, mother cell.
C. Polymerization of newly synthesized muropeptides into peptidoglycan (green) at the point of contact between the migrating arms of the membranes pushes the septal membranes apart. Following detachment, the forespore becomes an independent membrane-bounded compartment in the cytoplasm. This polymerization is mediated by an unidentified transpeptidase-transglycosylase (red).
During the last step of engulfment, membrane fission, peptidoglycan synthesis was restricted to the zone of contact between the advancing membranes (Fig. 2, panel iii) and inhibition of muropeptide synthesis (Fig. 4) or polymerization (Fig. 5) prevented detachment of the forespore from the mother cell (engulfment membrane fission). The production of a rigid peptidoglycan polymer in this zone of contact could generate a mechanical force that drives elongation of the bud neck, distorting it into a tube, that eventually breaks, leading to cell separation. Then, localized peptidoglycan hydrolysis would allow the membranes to meet and rearrange (Fig. 8C). A membrane protein, SpoIIIE, is necessary for mediating this fission event during engulfment (Sharp and Pogliano, 2003) and our results suggest that SpoIIIE could interact with membrane proteins involved in peptidoglycan synthesis and organize their spatial distribution at the point of membrane fission. Consistent with this possibility, the membrane domain of SpoIIIE is necessary for its localization to the septum and its function in membrane fission (Sharp and Pogliano, 2003) and for the final steps of cytokinesis at the asymmetric septum (Liu et al., 2006).
Muropeptide polymerization during bulge and vesicle formation
Membrane bulges in a spoIID or spoIIP strain form initially as a deformation of the membrane and then develop into a clear invagination that, at least in some cases, becomes a membrane-bounded vesicle whose cytoplasmic contents are visibly separated from that of the forespore (Fig. 1A, bottom; Fig. 1B). These bulges are sites of active peptidoglycan formation (Fig. 6B) and inhibiting synthesis of the muropeptide precursor using fosfomycin or of mature peptidoglycan using vancomycin blocks formation of the bulges (Fig. 6C and E). The effect of fosfomycin was on MurAA since a merodiploid strain expressing a resistant mutant protein (MurAAC117D) generated vesicles even in the presence of fosfomycin (Fig. S10). The effect of vancomycin was likely on an inhibition of polymerization directly and not on an inhibition of peptidoglycan hydrolases since inhibition of polymerization in E. coli leads to increased muramidase activity (Kohlrausch and Holtje, 1991). Thus, septal peptidoglycan polymerization could exert a mechanical force on the septal membrane that, by analogy to actin polymerization during lamellipodial motility (Mogilner and Oster, 1996), would drive membrane deformations (Fig. 8A, bottom).
The requirement of the SpoVD transpeptidase (Fig. 7A and B) is consistent with the observed inhibition of bulge formation by vancomycin (Fig. 6E) and demonstrates that a protein necessary for the synthesis of mature peptidoglycan is also necessary for bulge formation. In addition, the presence of bulges only on the mother cell side can be explained by this result as SpoVD is under control of σE, which is only active in the mother cell. If the forespore had a higher osmotic pressure than the mother cell because of the presence of equally sized chromosomes in the very differently sized compartments, this pressure could push the bulges from the middle of the septum (Perez et al., 2000). However, we did not observe a significant difference in forespore size between the spoIIP and the spoIIP spoVD strains despite their observed differences in bulge formation (Fig. S13).
Rod-shaped E. coli depleted for proteins necessary for cell shape transform into spherical cells that contain large intracellular, membrane-bounded vesicles (Bendezu and de Boer, 2008). The inability of these mutant cells to couple the rate of phospholipid synthesis to changes in shape suggests that vesicle production is caused by overproduction of phospholipids. In B. subtilis, a similar coupling is not known to exist (Paoletti et al., 2007), consistent with our observation that fatty acid synthesis was not required at later time points where peptidoglycan synthesis was required for bulge and vesicle formation (Fig. S12) even though it was necessary for initial bulge formation.
Peptidoglycan polymerization as a force-generating mechanism during cytokinesis
A basic question in microbiology is the origin of the force underlying cell division. Although there are well-conserved proteins known to be involved, including the tubulin homologue FtsZ that forms constricting polymeric rings at mid-cell, the source of the force generating these constrictions remains unknown (Weiss, 2004; Margolin, 2005). It has been suggested on theoretical grounds (Lan et al., 2007; 2009; Ghosh and Sain, 2008; Allard and Cytrynbaum, 2009) as well as from electron microscopic observations of FtsZ polymer structure in vivo (Li et al., 2007) that the energetics of FtsZ ring depolymerization could provide such a force. In tubular vesicles, FtsZ forms rings that produce visible constrictions (Osawa et al., 2008) consistent with the ability of the constricting Z ring to generate a force. The absence of complete septal closure despite formation of physically distinct compartments seen in some E. coli amidase mutants suggests that, at least in these cells, membrane invagination could be mediated solely by constriction of the divisome (Priyadarshini et al., 2007).
Gradients of peptidoglycan synthesis generate cell curvature in the bacterium Caulobacter crescentus (Cabeen et al., 2009), supporting the notion that spatially differentiated peptidoglycan synthesis produces physical force sufficient to generate changes in cell shape that occur during cytokinesis (Huang et al., 2008). High-magnification images of dividing S. aureus demonstrated the presence of peptidoglycan lying in tight apposition to the membrane. In fact, careful measurement of the diameter of the very tip of the in-growing septum indicated that this site was the locus of peptidoglycan synthesis (Matias and Beveridge, 2007). Thus, this synthesis could provide a driving force for the ‘iris-like’ movement of the septum from the initial stages of membrane invagination to the completion of septation.
Cell wall polymerization could be used in other systems to generate active and locally controlled forces. Although peptidoglycan is only found in bacteria [and perhaps in the chloroplasts of some plant cells (Machida et al., 2006)], yeast and other fungi have rigid cell walls composed of long polysaccharides (Cabib et al., 2001) that, similar to peptidoglycan, are polymerized by transglycosylases (Cabib et al., 2008). In fact, growing S. pombe exerts a large mechanical force at its cell tips that is independent of actin cables and sufficient to play a role in piercing membranes during host invasion (Minc et al., 2009). Finally, a variety of cell types such as budding yeasts can undergo cytokinesis in the absence of an actin–myosin contractile ring (Bi et al., 1998). Thus, polymerization of fungal polysaccharides could serve a similar function as muropeptide polymerization in providing a force-generating mechanism for cytokinesis.
Experimental procedures
Microbiological methods
Bacillus subtilis strains are derivatives of PY79 (Table S1) and details of their construction are described in Supporting information. Standard procedures were used to prepare and handle recombinant DNA and to transform E. coli. B. subtilis was transformed using competent cells made by the two-step method (Cutting and Vander Horn, 1990). Sporulation for microscopy was conducted at 30°C and used CH medium or 25% LB for growth and A+B medium for resuspension (Sterlini and Mandelstam, 1969). To measure completion of engulfment, membranes from sporulating cells expressing CFP under control of a forespore-specific promoter (PspoIIQ) were stained with FM4-64 (1 μg ml−1). Every half hour starting at T3 a sample of cells was observed under the microscope. Cells expressing CFP were counted and the fraction of those not exhibiting FM4-64 staining of the forespore was determined.
Reagents
Fosfomycin and vancomycin were obtained from Sigma, cerulenin was obtained from Cayman Chemical, and FM4-64 was from Invitrogen. Ramoplanin-FL and ramoplanin-2c were gifts of Dr Suzanne Walker (Harvard Medical School).
Fluorescence microscopy
One microlitre of FM4-64 (Molecular Probes; 100 μg ml−1) was added to each sample of 100 μl of sporulating cells that were taken at designated times after resuspension, immediately prior to collection by centrifugation. The pellet was resuspended in 10 μl of PBS, and added to a poly-l-lysine pre-treated coverslip. All microscopy (except Fig. 2 and Fig. S2) was performed on a Nikon Eclipse 90i with a 100× objective using phase contrast and captured by a Hamamatsu Orca-ER camera using Nikon Elements BR software. CFP, YFP, FITC (Ramoplanin-FL) and TRITC (FM4-64) exposures were 400 ms. Other microscopy was performed using an Applied Precision Spectris microscope.
Dynamics of engulfing forespores
Time-lapse microscopy was performed as described (Becker and Pogliano, 2007). Briefly, FM4-64 was added to a final concentration of 0.5 μg ml−1 in a 1.2% solution of molten agar/media (A+B) and added to the well of a culture slide and covered with a glass slide. After cooling, the slide was removed and two air pockets were cut out of the agar leaving a 3–5 mm agar bridge in the centre of the well. Sporulating cells suspended in A+B with FM4-64 (0.5 μg ml−1) were added at T2 to the agar bridge and covered by a glass coverslip. To prevent drying during the experiment, 50% glycerol was applied to the region of contact between the slide and the coverslip. The slide equilibrated in an environmentally controlled chamber at 30°C (Precision Control Weather Station) for at least 10 min prior to visualization. Images were acquired using an Applied Precision Spectris microscope.
Finally, the following metric was used for defining membrane migration: ‘No migration’ is defined as: 40 min of observation with no advancement of the membranes (curvature is acceptable as long as the distance between septum edge and cell pole does not decrease). ‘Migration’ is defined as: advancement that is maintained for at least 20 min of observation. In addition, cells must be observed for 40 min without going off the screen or lysing.
Supplementary Material
Acknowledgments
We thank Suzanne Walker (Harvard Medical) for generously supplying fluorescent derivatives of ramoplanin, Avigdor Eldar and Michael Elowitz (Caltech) for strains and Allison Fay for constructing pAF54. This work was supported by NIH R01GM83468 (J.D.) and R01 R01GM57045 (K.P.). P.M. is a Helen Hay Whitney Fellow and J.D. is an Irma T. Hirschl Scholar.
Footnotes
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard JF, Cytrynbaum EN. Force generation by a dynamic Z-ring in Escherichia coli cell division. Proc Natl Acad Sci USA. 2009;106:145–150. doi: 10.1073/pnas.0808657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EC, Pogliano K. Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol Microbiol. 2007;66:1066–1079. doi: 10.1111/j.1365-2958.2007.05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, de Boer PA. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol. 2008;190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund JE, Zhang L, Haines MA, Higgins ML, Piggot PJ. Analysis by fluorescence microscopy of the development of compartment-specific gene expression during sporulation of Bacillus subtilis. J Bacteriol. 1994;176:2898–2905. doi: 10.1128/jb.176.10.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Roh DH, Schmidt M, Crotti LB, Varma A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J Biol Chem. 2001;276:19679–19682. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- Cabib E, Farkas V, Kosik O, Blanco N, Arroyo J, McPhie P. Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J Biol Chem. 2008;283:29859–29872. doi: 10.1074/jbc.M804274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting SM, Vander Horn PB. Genetic analysis. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. New York, NY: John Wiley & Sons; 1990. pp. 24–74. [Google Scholar]
- Dancer BN. Requirement for peptidoglycan synthesis during sporulation of Bacillus subtilis. J Bacteriol. 1979;140:786–797. doi: 10.1128/jb.140.3.786-797.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum M, Kuchnir Fygenson D, Libchaber A. Buckling microtubules in vesicles. Phys Rev Lett. 1996;76:4078–4081. doi: 10.1103/PhysRevLett.76.4078. [DOI] [PubMed] [Google Scholar]
- Fitz-James PC. Sporulation in protoplasts and its dependence on prior forespore development. J Bacteriol. 1964;87:667–675. doi: 10.1128/jb.87.3.667-675.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francius G, Domenech O, Mingeot-Leclercq MP, Dufrene YF. Direct observation of Staphylococcus aureus cell wall digestion by lysostaphin. J Bacteriol. 2008;190:7904–7909. doi: 10.1128/JB.01116-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen N, Stragier P. Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol. 1995;177:716–722. doi: 10.1128/jb.177.3.716-722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B, Sain A. Origin of contractile force during cell division of bacteria. Phys Rev Lett. 2008;101:178101. doi: 10.1103/PhysRevLett.101.178101. [DOI] [PubMed] [Google Scholar]
- Gutierrez J, Smith R, Pogliano K. SpoIID peptidoglycan hydrolase activity is required throughout engulfment during Bacillus subtilis sporulation. J Bacteriol. 2010 doi: 10.1128/JB.00127-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger JB, Hoertz AJ, Lee A, Senturia RJ, McCafferty DG, Loll PJ. A crystal structure of a dimer of the antibiotic ramoplanin illustrates membrane positioning and a potential Lipid II docking interface. Proc Natl Acad Sci USA. 2009;106:13759–13764. doi: 10.1073/pnas.0904686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc Natl Acad Sci USA. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Cegelski L, Stueber D, Singh M, Dietrich E, Tanaka KS, et al. Oritavancin exhibits dual mode of action to inhibit cell-wall biosynthesis in Staphylococcus aureus. J Mol Biol. 2008;377:281–293. doi: 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlrausch U, Holtje JV. Analysis of murein and murein precursors during antibiotic-induced lysis of Escherichia coli. J Bacteriol. 1991;173:3425–3431. doi: 10.1128/jb.173.11.3425-3431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Lan G, Wolgemuth CW, Sun SX. Z-ring force and cell shape during division in rod-like bacteria. Proc Natl Acad Sci USA. 2007;104:16110–16115. doi: 10.1073/pnas.0702925104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci USA. 2009;106:121–126. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NJ, Dutton RJ, Pogliano K. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol. 2006;59:1097–1113. doi: 10.1111/j.1365-2958.2005.05004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S, Mandelstam J. spoIID operon of Bacillus subtilis: cloning and sequence. J Gen Microbiol. 1986;132:341–354. doi: 10.1099/00221287-132-2-341. [DOI] [PubMed] [Google Scholar]
- Machida M, Takechi K, Sato H, Chung SJ, Kuroiwa H, Takio S, et al. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc Natl Acad Sci USA. 2006;103:6753–6758. doi: 10.1073/pnas.0510693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias VR, Beveridge TJ. Cryo-electron microscopy of cell division in Staphylococcus aureus reveals a mid-zone between nascent cross walls. Mol Microbiol. 2007;64:195–206. doi: 10.1111/j.1365-2958.2007.05634.x. [DOI] [PubMed] [Google Scholar]
- Meador-Parton J, Popham DL. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J Bacteriol. 2000;182:4491–4499. doi: 10.1128/jb.182.16.4491-4499.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc N, Boudaoud A, Chang F. Mechanical forces of fission yeast growth. Curr Biol. 2009;19:1096–1101. doi: 10.1016/j.cub.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Miyata H, Nishiyama S, Akashi K, Kinosita K., Jr Protrusive growth from giant liposomes driven by actin polymerization. Proc Natl Acad Sci USA. 1999;96:2048–2053. doi: 10.1073/pnas.96.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti L, Lu YJ, Schujman GE, de Mendoza D, Rock CO. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J Bacteriol. 2007;189:5816–5824. doi: 10.1128/JB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R, Kondev J, Theriot J. Physical Biology of the Cell. New York: Garland Science; 2009. [Google Scholar]
- Priyadarshini R, de Pedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM, Kelly RB. Molecular links between endocytosis and the actin cytoskeleton. J Cell Biol. 2000;150:F111–F116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. The membrane domain of SpoIIIE is required for membrane fusion during Bacillus subtilis sporulation. J Bacteriol. 2003;185:2005–2008. doi: 10.1128/JB.185.6.2005-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Youngman P. Evidence that the spoIIM gene of Bacillus subtilis is transcribed by RNA polymerase associated with sigma E. J Bacteriol. 1993;175:3618–3627. doi: 10.1128/jb.175.11.3618-3627.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA. The polymerization motor. Traffic. 2000;1:19–28. doi: 10.1034/j.1600-0854.2000.010104.x. [DOI] [PubMed] [Google Scholar]
- Tipper DJ, Linnett PE. Distribution of peptidoglycan synthetase activities between sporangia and forespores in sporulating cells of Bacillus sphaericus. J Bacteriol. 1976;126:213–221. doi: 10.1128/jb.126.1.213-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc Natl Acad Sci USA. 2006;103:11033–11038. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, Popham DL. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol. 2007;65:1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x. [DOI] [PubMed] [Google Scholar]
- Walsh CT. Antibiotics. Washington, DC: ASM; 2003. [Google Scholar]
- Weiss DS. Bacterial cell division and the septal ring. Mol Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- Yao X, Jericho M, Pink D, Beveridge T. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J Bacteriol. 1999;181:6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


