Abstract
Objective
To conduct an investigation of clinical and genetic correlates of lipoprotein-associated phospholipase (Lp-PLA2) activity and mass in a large community-based cohort. Higher circulating Lp-PLA2 predicts cardiovascular disease risk, but sources of inter-individual variability are incompletely understood.
Methods
We conducted stepwise regression of clinical correlates of Lp-PLA2 in four Framingham Heart Study cohorts (n=8185; mean age 50±14 years, 53.8% women, 9.8% ethnic/racial minority cohort). We also conducted heritability and linkage analyses in Offspring and Generation 3 cohorts (n=6945). In Offspring cohort participants we performed association analyses (n=1535 unrelated) with 1943 common tagging SNPs in 233 inflammatory candidate genes.
Results
Sixteen clinical variables explained 57% of the variability in Lp-PLA2 activity; covariates associated with Lp-PLA2 mass were similar but only explained 27% of the variability. Multivariable-adjusted heritability estimates for Lp-PLA2 activity and mass were 41% and 25%, respectively. A linkage peak was observed for Lp-PLA2 activity (chromosome 6, LOD score 2.4). None of the SNPs achieved experiment-wide statistical significance, though 12 had q values <0.50, and hence we expect at least 50% of these associations to be true positives. The strongest multivariable-association with Lp-PLA2 activity was found for MEF2A (rs2033547; nominal p=3.20*10-4); SNP rs1051931 in PLA2G7 was nominally associated (p=1.26*10-3). The most significant association to Lp-PLA2 mass was in VEGFC (rs10520358, p=9.14*10-4).
Conclusions
Cardiovascular risk factors and genetic variation contribute to variability in Lp-PLA2 activity and mass. Our genetic association analyses need replication, which will be facilitated by web posting of our genetic association results.
Keywords: lipoprotein-associated phospholipase A2, inflammation, heritability, single nucleotide polymorphism
Introduction
Inflammation and oxidative stress contribute to atherogenesis. Circulating lipoprotein-associated phospholipase A2 (Lp-PLA2) has been scrutinized intensively as a marker of cardiovascular disease (CVD) risk because the enzyme exhibits pro-inflammatory and oxidative activities. A key feature is the transformation of oxidized low-density lipoprotein (LDL) in the arterial wall into highly proatherogenic reactants like lysophosphatidylcholine and oxidized fatty acids.
Research implicates Lp-PLA2 in multiple phases in the development of CVD. Circulating Lp-PLA2 concentrations predict the presence of coronary artery disease and correlate with endothelial dysfunction and early atherosclerosis in the coronary circulation.[1] Clinical and epidemiological studies consistently demonstrate associations with incident and recurrent coronary artery disease events.[2-4] Accounting for traditional CVD risk factors, higher blood Lp-PLA2 is associated with adverse long-term CVD outcomes.[2,5,6]
Prior reports have suggested that circulating Lp-PLA2 concentrations are related to both clinical and genetic factors, [7,8] but the determinants in the community are incompletely reported. We hypothesized that Lp-PLA2 activity and mass would be associated with CVD risk factors and genetic variation in the PLA2G7 gene coding for Lp-PLA2 and in other inflammatory SNPs. We report the association of Lp-PLA2 activity and mass with clinical factors. In addition, we describe heritability, genetic linkage, and the relation of variation in LpPLA2 activity and mass with variation in 13 common single nucleotide polymorphisms (SNPs) representing the PLA2G7 gene, and SNPs in inflammatory candidate genes in the community-based Framingham Heart Study.
Materials and Methods
Study Sample
Plasma Lp-PLA2 measurements were available from four Framingham Study cohort examinations, including: the Framingham Offspring cohort seventh follow-up examination (1998-2001; n=5124); the Third Generation cohort first examination enrolled from 2002-2005 (n=4085), and 804 Omni Study ethnic/racial minority participants (see Supplement). The study protocol was approved by the Boston University Medical Center Institutional Review Board and participants signed informed consent.
Lp-PLA2 Determination
Lp-PLA2 activity and mass were measured from overnight fasting plasma specimens that were stored at -80°C. Lp-PLA2 activity was measured using a colorimetric activity method (diaDexus CAM Kit, Inc., San Francisco, CA).[3] Lp-PLA2 mass was measured using a commercially available sandwich enzyme immunoassays (diaDexus PLAC® test, Inc., San Francisco, CA). Details of laboratory analysis are provided in the Supplement.
Genotyping
Genotyping was conducted by Perlegen Sciences, Inc., Mountain View, CA and the Broad Institute of Harvard and Massachusetts Institute of Technology in members of Offspring and Generation 3 cohorts (not Omni) cohorts. A total of 1943 SNPs in 233 inflammatory candidate genes passed quality control and entered analyses, more details on the methods are available in the Supplement. Linkage analyses were conducted using 640 polymorphic markers covering 22 autosomal chromosomes.
Statistical Analysis
Clinical Correlates
Skewed distributions led us to employ natural logarithmic transformation of both markers. Lp-PLA2 mass and activity stepwise linear regression models were performed with forwards selection (inclusion p<0.05). Age, sex and cohort were forced into the model. The model selected from the following clinical variables: current smoking, alcohol consumption, body mass index, waist circumference, systolic and diastolic blood pressures, fasting biomarkers (calculated low density lipoprotein[LDL]- and high density lipoprotein[HDL]-cholesterol, triglycerides, glucose), diabetes, medications (hypertension, lipid therapy, hormone replacement in women, aspirin [≥3 per week]), prevalent CVD, and season. R2 for the overall model and partial R2 for individual variables were assessed. In secondary analyses, we tested the interactions among age, sex, cohort, LDL- and HDL-cholesterol with respect to association with Lp-PLA2. A two-sided p<0.05 was considered statistically significant for clinical correlates analysis. SAS version 8.1 (http://www.sas.com/presscenter/guidelines.html, Cary, NC) was used for clinical analyses and creation of phenotype residuals for genetic analyses adjusting for age, sex, cohort, smoking, alcohol consumption, body mass index, waist, systolic and diastolic blood pressure, total/HDL-cholesterol, triglycerides, glucose, diabetes, the four medication classes listed above, prevalent CVD, and season.
Heritability, Linkage and Association
Heritability analyses were restricted to Offspring and Generation 3 individuals in families with ≥2 phenotyped individuals (n=6945 individuals, 782 families). Multivariable-adjusted Lp-PLA2 residuals were examined in association with inflammatory SNPs. For each association, multiple testing was accounted for by computing the q value. See Supplement for details on genetic analyses.
Results
Participant Characteristics
The clinical and laboratory characteristics of the study participants available for phenotype, linkage, and candidate gene analyses are presented in Table 1; Omni participants were unavailable for genetic analyses. The clinical characteristics by study cohort are displayed in Supplement Table 1. The SNP study sample had an older mean age (62 vs. 49 years in the phenotype and linkage samples, respectively). Pearson's correlation coefficient between Lp-PLA2 activity and mass was 0.46 (95% confidence interval 0.45, 0.48).
Table 1. Study Participants Characteristics by Analysis Sample.
| Phenotype (N=8185) |
Heritability (N=6945) |
Candidate Gene (N=1535) |
|
|---|---|---|---|
| Cohort | Omni, Generation 3, Offspring | Generation 3, Offspring | Offspring |
| Age, years | 49.4±13.8 | 49.0±13.8 | 61.8±9.3 |
| Women, % | 53.8 | 53.2 | 51.3 |
| Ethnic minority, % | 9.8 | -- | -- |
| Current smoking, % | 14.7 | 15.2 | 13.0 |
| Alcohol consumption, drinks/week,% none (women/men) | 32.1/22.7 | 29.2/21.2 | 35.7/27.7 |
| 1-7 (women)/1-14 (men) | 53.2/60.5 | 56.3/62.1 | 46.0/52.8 |
| >7 (women)/>14 (men) | 14.7/16.9 | 14.6/16.7 | 18.3/19.5 |
| Body mass index, kg/m2 | 27.5±5.5 | 27.5±5.5 | 28.2±5.2 |
| Waist, cm | 96±15 | 96±15 | 100±14 |
| Systolic blood pressure, mm Hg | 121±17 | 121±17 | 128±19 |
| Diastolic blood pressure, mm Hg | 75±10 | 75±10 | 74±10 |
| Triglycerides, mmol/L | 1.4±1.0 | 1.4±1.0 | 1.6±1.0 |
| LDL-cholesterol, mmol/L | 3.0±0.8 | 3.0±0.8 | 3.1±0.8 |
| HDL-cholesterol, mmol/L | 1.4±0.4 | 1.4±0.4 | 1.4±0.4 |
| Total/HDL-cholesterol, ratio | 3.9±1.4 | 3.9±1.4 | 4.0±1.3 |
| Fasting glucose, mmol/L | 5.5±1.4 | 5.5±1.3 | 5.9±1.6 |
| Diabetes, % | 8.0 | 7.4 | 15.6 |
| Hypertension treatment, % | 20.1 | 19.5 | 35.2 |
| Lipid treatment, % (% statin users) | 13.0 (12) | 13.0 (12) | 22.4 (20) |
| Aspirin ≥3 per week, % | 18.4 | 18.0 | 34.6 |
| Hormone replacement, % | 8.6 | 8.5 | 16.8 |
| Prevalent cardiovascular disease, % | 6.5 | 6.4 | 13.7 |
| Individuals, per Season | |||
| Spring, % | 27.6 | 28.4 | 22.4 |
| Summer, % | 26.1 | 26.4 | 29.1 |
| Fall, % | 26.2 | 25.4 | 28.5 |
| Winter, % | 20.2 | 19.9 | 20.0 |
| Lp-PLA2 | |||
| Mass, ng/mL | 257±79 | 260±79 | 302±97 |
| Activity, nmol/mL/min | 152±39 | 156±38 | 144±36 |
Numbers are presented as mean±SD for continuous, or percent) for categorical variables.
Clinical measures were available in Offspring cohort (n=3296), Omni cohort wave 1 (n=400), wave 2 (n=404), and Generation 3 cohort (n=4085).
Multivariable Clinical Correlates of Lp-PLA2
As displayed in Table 2, in stepwise multivariable linear regression models with age, sex and cohort forced in, Lp-PLA2 activity and mass concentrations were positively associated with higher mean age, smoking, and LDL-cholesterol, and inversely associated with being a woman or a minority, alcohol consumption category, medications (hypertension, lipid-lowering, and hormone replacement therapy). Lp-PLA2 activity was strongly inversely associated with HDL-cholesterol; however, mass was only weakly associated and the direction was positive. Both mass and activity were associated with season, though not with a consistent pattern. In addition, Lp-PLA2 activity was positively associated with CVD, and inversely associated with body mass index, whereas Lp-PLA2 mass was positively associated with diastolic blood pressure. Triglycerides were positively correlated with Lp-PLA2 activity. Clinical correlates explained 57.4% of the inter-individual variability in Lp-PLA2 activity, with LDL-cholesterol alone explaining 14.3% of the total variance, HDL-cholesterol explaining 6.4%. The overall model R2 for Lp-PLA2 mass was 27.5%.
Table 2. Stepwise Linear Regression: Multivariable correlates of log Lp-PLA2 Activity and Mass (n=7989).
| Activity | Mass | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable, unit | β | 95% CI | P value | Partial R2, % | β | 95% CI | P value | Partial R2, % |
| Covariates forced in | ||||||||
| Age, per 10 years | 0.007 | 0.002, 0.01 | 0.003 | 0.05 | 0.02 | 0.01, 0.02 | <0.0001 | 0.28 |
| Women vs. men | -0.11 | -0.12, -0.10 | <0.0001 | 3.02 | -0.06 | -0.08, -0.05 | <0.0001 | 0.83 |
| Offspring Cohort (reference) | -- | -- | <0.0001 | 11.53 | -- | -- | <0.0001 | 8.4 |
| Generation 3 | 0.17 | 0.16, 0.18 | -0.21 | -0.23, -0.19 | ||||
| Omni wave 1 | -0.18 | -0.20, -0.16 | -0.28 | -0.31, -0.26 | ||||
| Omni wave 2 | -0.13 | -0.15, -0.11 | -0.29 | -0.32, -0.26 | ||||
| Covariates selected by model | ||||||||
| Current smoking, yes/no | 0.04 | 0.02, 0.05 | <0.0001 | 0.21 | 0.08 | 0.06, 0.09 | <0.0001 | 0.76 |
| Alcohol drinks/week | ||||||||
| 1-7 women; 1-14 men | -0.009 | -0.02, 0.00004 | 0.0008 | 0.08 | -0.03 | -0.04, -0.01 | 0.0004 | 0.14 |
| >7 women; >14 men | -0.02 | -0.04, -0.01 | -0.03 | -0.04, -0.009 | ||||
| Body mass index, per 5kg/m² | -0.01 | -0.01, -0.007 | <0.0001 | 0.16 | -- | -- | -- | -- |
| Diastolic blood pressure, per 20 mm Hg | -- | -- | -- | -- | 0.006 | 0.0003, 0.01 | 0.04 | 0.04 |
| Log-triglycerides | -0.03 | -0.04, -0.03 | <0.0001 | 0.76 | -- | -- | -- | -- |
| Log-LDL-cholesterol | 0.37 | 0.35, 0.38 | <0.0001 | 14.32 | 0.20 | 0.18, 0.21 | <0.0001 | 3.54 |
| Log-HDL-cholesterol | -0.31 | -0.33, -0.29 | <0.0001 | 6.40 | 0.02 | -0.0004, 0.05 | 0.0004 | 0.11 |
| Cardiovascular disease, yes/no | 0.02 | 0.002, 0.04 | 0.03 | 0.02 | -- | -- | -- | -- |
| Hypertension medications, yes/no | -0.02 | -0.03, -0.009 | 0.0003 | 0.07 | -0.02 | -0.04, -0.007 | 0.004 | 0.08 |
| Lipid medications, yes/no | -0.03 | -0.04, -0.01 | <0.0001 | 0.10 | -0.08 | -0.10, -0.06 | <0.0001 | 0.74 |
| Hormone replacement therapy, yes/no | -0.06 | -0.07, -0.04 | <0.0001 | 0.30 | -0.08 | -0.10, -0.06 | <0.0001 | 0.57 |
| Season (vs. Winter) | ||||||||
| Spring | -0.009 | -0.021, 0.001 | 0.0005 | 0.09 | -0.002 | -0.02, 0.01 | <0.0001 | 0.25 |
| Summer | -0.0004 | -0.01, 0.01 | 0.03 | 0.01, 0.05 | ||||
| Fall | 0.01 | 0.002, 0.02 | -0.004 | -0.02, 0.01 | ||||
| Model R2 | 57.4 | 27.5 | ||||||
See methods section for candidate covariates for stepwise selection. Cholesterol concentrations were log-transformed, triglycerides were log-normalized for analysis.
Secondary clinical analyses
Because different investigators may vary in their approach to building multivariable models we provide age-, sex- and cohort-adjusted models in the Supplement Table 2.
In the multivariable model, we observed effect modification by sex, age and cohort for the relation of LDL- and HDL-cholesterol with Lp-PLA2 activity (data not shown). LDL-cholesterol was more strongly associated with Lp-PLA2 activity (p=0.02) and mass (p=0.001) in women than in men, and less associated in older than in younger individuals for both activity (p=0.03) and mass (p=0.02). Activity was strongly inversely associated with HDL-cholesterol but there was an absence of an association with mass. For both LDL- and HDL-cholesterol we observed statistical evidence for interaction with cohort with Lp-PLA2 activity (p=0.002) and mass (p<0.0001). Individuals with diabetes had lower Lp-PLA2 activity and mass, although the inverse association was only borderline significant for activity. Diabetes was not included in the final stepwise models. No associations with glucose lowering medications or interactions with diabetes were observed. The exclusion of individuals on lipid treatment did not change the final model (Supplement Table 3).
Heritability and Genetic Linkage
Multivariable-adjusted heritability estimates were 41% (standard error [SE]=0.02) for Lp-PLA2 activity and 25% (SE=0.02) for Lp-PLA2 mass. No genome-wide significant linkage peak was detected for Lp-PLA2 mass or activity. A linkage peak for Lp-PLA2 activity was observed at 79 cM on chromosome 6 with a LOD score of 2.4 (nominal p=4.3×10-4; genome-wide[9] p= 0.21, Figure 1).
Figure 1.
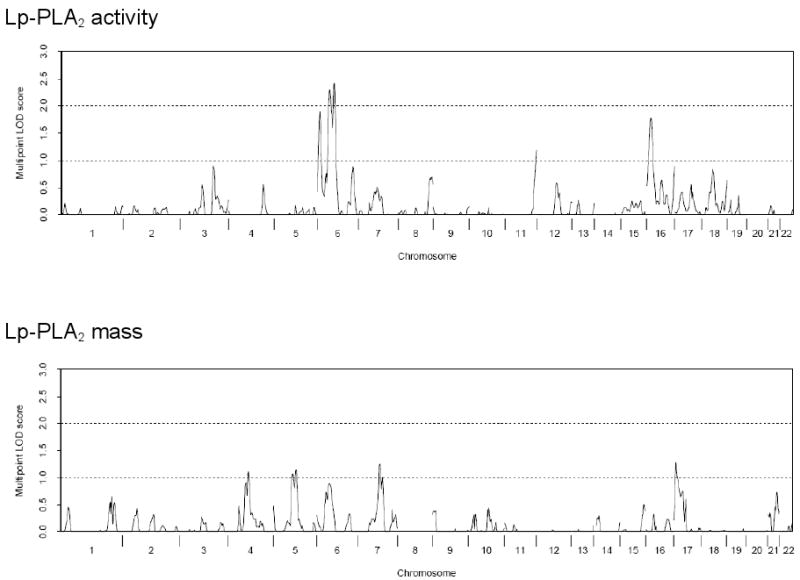
Multivariable-adjusted multipoint linkage of Lp-PLA2 activity and mass on chromosome 6. The observed linkage peak for Lp-PLA2 activity had a LOD score of 2.4, p=4.3×10-4. The PLA2G7 gene is located at approximately 73 cM on chromosome 6.
Inflammatory Candidate Genotype Associations
The top associations from the inflammatory candidate gene approach are tabulated in Table 3; upon publication complete results will be available at http://www.framinghamheartstudy.org/research/resresults.html. Accounting for multiple statistical testing, none of the SNPs achieved experiment-wide significance, but 12 had q values <0.50. The top SNP in relation to Lp-PLA2 activity in multivariable-adjusted analysis was observed in MEF2A (rs2033547, p=3.20*10-4). For Lp-PLA2 mass the most statistically significant SNP was in VEGFC (p=9.14*10-4). A SNP in the gene coding for Lp-PLA2, PLA2G7 (rs1051931), was nominally associated with Lp-PLA2 activity (p=1.26*10-3), but not mass (p=0.45). The other genotyped PLA2G7 SNPs did not reach nominal significance (p≥0.0057) for either activity or mass. The overall variability in Lp-PLA2 activity and mass concentrations explained by each SNP that was associated was ≤1.0%. Supplement Table 4 provides top results for the age- and sex-adjusted models relating the inflammatory SNPs to Lp-PLA2 concentrations.
TABLE 3. Association of inflammatory candidate gene SNPs and multivariable-adjusted log Lp-PLA2 in Offspring cohort (n=1535)*.
| Allelic Variant | Gene | Protein Gene Codes for | Chr* | Location | Major→ Minor Allele | MAF | Beta heterozygotes | SE | Beta homozygotes minor allele | SE | R2 | P-Value | q-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2033547 | MEF2A | myocyte enhancer factor 2A | 15 | 980324 | A|T | 46.1 | 0.12 | 0.06 | -0.15 | 0.08 | 1.0 | 3.20*10-4 | 0.25 |
| rs1029153 | CXCL12 | chemokine (C-X-C motif) ligand 12 | 10 | 441871 | A|G | 26.7 | -0.20 | 0.06 | 0.12 | 0.11 | 1.0 | 3.44*10-4 | 0.25 |
| rs10520358 | VEGFC | vascular endothelial growth factor C | 4 | 1779630 | C|T | 10.8 | 0.13 | 0.07 | -0.90 | 0.27 | 1.0 | 5.12*10-4 | 0.25 |
| rs6019910 | PTGIS | prostaglandin I2 synthase | 20 | 476230 | C|T | 6.7 | -0.08 | 0.08 | -1.16 | 0.31 | 1.0 | 5.14*10-4 | 0.25 |
| rs193276 | CD44 | CD44 molecule (Indian blood group) | 11 | 35140414 | T|C | 23.6 | -0.02 | 0.06 | 0.47 | 0.13 | 0.9 | 1.21*10-3 | 0.31 |
| rs1051931 | PLA2G7 | phospholipase A2, group VII | 6 | 46780902 | A|G | 19.4 | 0.21 | 0.06 | 0.21 | 0.15 | 0.9 | 1.26*10-3 | 0.31 |
| rs10520358 | VEGFC | vascular endothelial growth factor C | 4 | 1779630 | C|T | 10.8 | -0.01 | 0.08 | -1.17 | 0.31 | 0.9 | 9.14*10-4 | 0.96 |
| rs4335431 | TGFB2* | transforming growth factor, β 2 | 1 | 2149460 | T|C | 7.5 | 0.26 | 0.09 | -0.85 | 0.38 | 0.9 | 1.26*10-3 | 0.96 |
| rs12058490 | TGFB2* | 1 | 2149476 | A|G | 7.9 | 0.24 | 0.09 | -0.85 | 0.38 | 0.9 | 1.45*10-3 | 0.96 | |
| rs10128001 | PTGER3 | prostaglandin E receptor 3 | 1 | 712568 | C|T | 8.6 | -0.26 | 0.09 | -0.58 | 0.34 | 0.8 | 2.89*10-3 | 0.99 |
| rs17038663 | CX3CR1 | chemokine (C-X3-C motif) receptor 1 | 3 | 392803 | C|T | 5.9 | 0.31 | 0.10 | -0.29 | 0.70 | 0.7 | 3.29*10-3 | 0.99 |
Lp-PLA2 activity and mass adjusted for 19 covariates (see methods for details).
Q values were calculated within phenotype.
Partial R2 refers to the proportion of multivariable-adjusted Lp-PLA2 value explained by the specific SNP.
TGFB2 LD r2=0.95
Discussion
Principal Findings
Although LpPLA2 mass and activity were moderately correlated with each other, we found important differences in their relations to key lipid traits, amount of variability explained by clinical correlates, heritability and genetic associations. These data suggest that LpPLA2 mass and activity may provide complementary biological information. Of note, circulating concentrations were lower in women and in ethnic minorities (compared to men and whites, respectively) and increased with age, consistent with recent reports.[2-4,6] Clinical covariates explained 57.4% of the inter-individual variability of Lp-PLA2 activity, and 25% in Lp-PLA2 mass. Both, Lp-PLA2 activity and mass were heritable phenotypes. For activity a linkage peak was observed on chromosome 6 in the genomic region of the PLA2G7 gene (∼73 cM; LOD 2.4). Whereas correcting for multiple testing, none of our findings achieved experiment-wide significance, we observed several suggestive associations (q value<0.50). Among SNPs from 233 inflammatory candidate genes, rs22033547 in MEF2A was associated with Lp-PLA2 activity in multivariable-adjusted analyses. For Lp-PLA2 mass, the most statistically significant association was noted for SNP rs10520358 in VEGFC gene. A SNP in the gene coding for Lp-PLA2 protein, PLA2G7 was nominally significantly associated with Lp-PLA2 concentrations (p=1.26*10-3). Each SNP that was associated explained less than 1% of the variability of circulating Lp-PLA2 activity or mass. We provide a full-disclosure web-based resource so that other investigators can replicate our findings.
Clinical Correlates of Lp-PLA2
Prior studies have observed that mean Lp-PLA2 concentrations are higher in men, with advancing age, and with smoking.[10-12] Further, previous experimental and epidemiological investigations noted that Lp-PLA2 activity and mass are associated positively with clinical variables such as LDL-cholesterol, and systolic blood pressure,[13,14] and inversely with HDL-cholesterol, lipid treatment,[15] and alcohol consumption.[16] A weak or no relation to Lp-PLA2 measures was seen for other classical cardiovascular risk factors like body mass index and diabetes.[3,4] We confirmed most of the relations observed in prior reports. For body mass index, we found a statistically significant, but clinically small association with Lp-PLA2 activity but not mass. Whereas HDL-cholesterol was the second highest explanatory variable for activity, HDL-cholesterol was only borderline significantly associated with mass. Similar to these recent reports, in our study, Lp-PLA2 activity was more closely related to clinical variables and classical cardiovascular risk factors compared to mass, and clinical correlates explained a higher amount of variability in Lp-PLA2 activity.
The correlation between Lp-PLA2 mass and activity that we found in the current study was 0.46, and is within the range of correlation coefficients for other large samples previously reported (r=0.36[17], r=57[13,18], r=0.57[3]) . The study that reported a substantially higher correlation included only n=148 individuals.[19]
The variation in clinical and genetic factors associated with mass and activity suggests that they convey different clinical information. Hence, it is not surprising that mass and activity are only moderately correlated. The assays for activity and mass have been successfully used in large-scale studies. However, we concede that different measurement approaches may affect the correlations between mass and activity. Caslake et al. used a different immunoassay, and found a higher correlation betweenLp-PLA2 activity and mass.[19] The mass assay utilized in our study may not retrieve all available Lp-PLA2.[15]
We observed differences between LpPLA2 mass and activity in their correlation and their association with clinical covariates including LDL- and HDL-cholesterol concentrations, and different results for heritability and genetic association analyses. We observed that more than 20% of the variability in Lp-PLA2 activity was explained by cholesterol concentrations, whereas less than 4% of Lp-PLA2 mass was accounted for by common lipid measurements. Previous reports have hypothesized that Lp-PLA2 activity is a marker for small dense LDL, a well-established atherosclerotic risk factor, whereas the correlation with Lp-PLA2 mass is low. In addition, Lp-PLA2 associated with plasma lipoprotein particles may be present in a partially inactive state.[20]
There is controversy regarding which marker is more prognostically robust.[2,3] A recent meta-analysis of 14 studies (activity measurements: n=5, mass measurements: n=9) with 20,549 participants concluded that the measurement did not make a difference in the relation to outcome.[21] The similarities and differences between mass and activity will be further elucidated by future clinical and genetic meta-analyses.
Genetics of Lp-PLA2
Our family-based data add novel information on the heritability and linkage of SNPs with Lp-PLA2 concentrations. Substantial heritability for Lp-PLA2 activity measures between 0.55 and 0.62 has been reported before.[22,23] We observed a linkage peak for Lp-PLA2 activity 6cM away from the PLA2G7 gene.
Earlier candidate gene association analyses have been undertaken to characterize frequent polymorphisms in the PLA2G7 gene area in Europeans.[7] The most interesting SNP is Ala379Val (rs1051931), which has been related to circulating Lp-PLA2 and atherosclerotic disease.[7,8,24] We confirmed the association with higher Lp-PLA2 activity in homozygotes of the minor allele and extended them to a large community-based cohort. None of the SNPs in the PLA2G7 gene was significantly related to Lp-PLA2 mass in our study. For the functional variant rs1051931, decreased activity of the secreted enzyme has been shown and may explain differences in association between Lp-PLA2 activity and mass.[25] However, for this genetic variant, as well as for the other top SNPs, our power to demonstrate experiment-wide significance was moderate, and conclusions about null findings need to be drawn cautiously. Only replication studies will show whether genetic association is different for Lp-PLA2 activity compared to mass.
Top associations of SNPs in inflammatory genes comprised the MEF2A gene for multivariable-adjusted Lp-PLA2 activity and the VEGFC gene for Lp-PLA2 mass. MEF2A encodes an important transcription factor, myocyte enhancer factor 2. Mutations in this gene have been related to myocardial infarction and coronary artery disease.[26] The protein encoded by VEGFC belongs to the vascular endothelial growth factor family and is abundantly expressed in carcinomatous tissue.[27] VEGFC promotes inflammation and angiogenesis,[28] and may have an impact on pressure-induced endothelial cell proliferation.[29] Whereas it is biologically plausible that both SNPs are related to Lp-PLA2 concentrations, we acknowledge that our findings will need to be replicated. The candidate genes were chosen based on prior evidence from experimental or epidemiological data on their relevance in the inflammatory process. The present study is not suitable to demonstrate whether the candidate genes have a direct, potentially causal, or indirect relation to Lp-PLA2. If our genetic association findings are validated, further experimental work will be needed to uncover the exact mechanisms.
Strengths and Limitations
The measurement of Lp-PLA2 with a strict quality control protocol, well-characterized routinely ascertained CVD risk factors, enabling multivariable models, the large community-based cohort limiting referral bias, the dense family relations, facilitating heritability and genetic linkage analyses, the broad panel of inflammatory candidate genes examined, all constitute study strengths. Known SNPs were chosen to adequately cover the PLA2G7 genomic region. However, several limitations must be noted. The covariates were assessed cross-sectionally; hence, we cannot determine directionality and causal nature of clinical factors associated with Lp-PLA2 concentrations. The study sample was predominantly white and middle-aged to elderly. Because of the Omni cohorts' small size, we lacked statistical power to examine specific ethnic/racial subgroups in clinical models, and we did not have genetic data in the Omni cohort. The generalizability of genetic findings in other ethnicities is unknown.
We had adequate statistical power to demonstrate heritability, but our power to find significant linkage was modest. Although genome-wide significance was not achieved, we report the Lp-PLA2 activity linkage peak on chromosome 6 because the LOD score was suggestive and in the PLA2G7 genomic region. The adjustment of Lp-PLA2 variables for 16 covariates, which are partially correlated with Lp-PLA2 may have lead to over-adjustment and diminished the strength of association. We acknowledge that because of multiple testing inherent in examining 1943 inflammatory SNPs, the reported associations may represent false positive findings; our novel genetic findings will need to be replicated by others. Conversely, because of the need to account for multiple testing, we also may have false negative Lp-PLA2-SNP findings for associations with modest effect sizes.
Clinical Implications
Our study represents a comprehensive approach to examining the clinical and genetic factors influencing Lp-PLA2 activity and mass in a large community-based cohort. The current data strengthen the evidence that Lp-PLA2 activity and mass represent different pathophysiological entities and thus may convey different clinical information. Compared to mass, our results clearly demonstrate a stronger relation of Lp-PLA2 activity with cardiovascular risk factors, especially the known association with HDL-cholesterol and triglycerides, which indicate potential targets for Lp-PLA2 activity modification. Both traits are heritable. Linkage and association data suggest that Lp-PLA2 activity is, at least in part, influenced by variation in the genomic region of PLA2G7. Our study suggests that for Lp-PLA2 mass trans-acting genetic factors might be of importance. Whereas multiple associations of diverse inflammatory biomarkers with known cardiovascular risk factors and outcome have been reported, it generally is not clear whether they are surrogate markers of an underlying pathophysiological process or active agents in the disease process. Understanding the clinical and genetic contributors to Lp-PLA2 variation may set the stage for prevention and interventional strategies.
Supplementary Material
Acknowledgments
Lp-PLA2 activity measurements were provided by GlaxoSmithKline and mass measurements by diaDexus at no cost to the FHS.
Supported by NIH/NHLBI contract N01-HC-25195 and NIH grants HL64753 & HL076784 AG028321 (E.J.B.), HL71039 (R.S.V). NIH Research career award HL04334 (R.S.V.), NIH grant HG000848 (J.D.); Deutsche Forschungsgemeinschaft (German Research Foundation) Research Fellowship SCHN 1149/1-1 (RS). Portion of these analyses were conducted using the Boston University Linux Cluster for Genetic Analysis (LinGA) funded by the NIH NCRR (National Center for Research Resources) Shared Instrumentation grant (1S10RR163736-01A1).
Abbreviations
- CVD
cardiovascular disease
- HDL
high-density lipoprotein
- LD
linkage disequilibrium
- LDL
low-density lipoprotein
- Lp-PLA2
lipoprotein-associated phospholipase A2
- LOD
logarithm of the odds
- SE
standard error
- SNP
single nucleotide polymorphism
Reference List
- 1.Lavi S, McConnell JP, Rihal CS, Prasad A, Mathew V, Lerman LO, Lerman A. Local production of lipoprotein-associated phospholipase A2 and lysophosphatidylcholine in the coronary circulation: association with early coronary atherosclerosis and endothelial dysfunction in humans. Circulation. 2007;115:2715–21. doi: 10.1161/CIRCULATIONAHA.106.671420. [DOI] [PubMed] [Google Scholar]
- 2.Packard CJ, O'Reilly DS, Caslake MJ, McMahon AD, Ford I, Cooney J, Macphee CH, Suckling KE, Krishna M, Wilkinson FE, Rumley A, Lowe GD. Lipoprotein-associated phospholipase A2 as an independent predictor of coronary heart disease. West of Scotland Coronary Prevention Study Group. N Engl J Med. 2000;343:1148–55. doi: 10.1056/NEJM200010193431603. [DOI] [PubMed] [Google Scholar]
- 3.Koenig W, Twardella D, Brenner H, Rothenbacher D. Lipoprotein-associated phospholipase A2 predicts future cardiovascular events in patients with coronary heart disease independently of traditional risk factors, markers of inflammation, renal function, and hemodynamic stress. Arterioscler Thromb Vasc Biol. 2006;26:1586–93. doi: 10.1161/01.ATV.0000222983.73369.c8. [DOI] [PubMed] [Google Scholar]
- 4.Ballantyne CM, Hoogeveen RC, Bang H, Coresh J, Folsom AR, Heiss G, Sharrett AR. Lipoprotein-associated phospholipase A2, high-sensitivity C-reactive protein, and risk for incident coronary heart disease in middle-aged men and women in the Atherosclerosis Risk in Communities (ARIC) study. Circulation. 2004;109:837–42. doi: 10.1161/01.CIR.0000116763.91992.F1. [DOI] [PubMed] [Google Scholar]
- 5.Blake GJ, Dada N, Fox JC, Manson JE, Ridker PM. A prospective evaluation of lipoprotein-associated phospholipase A(2) levels and the risk of future cardiovascular events in women. J Am Coll Cardiol. 2001;38:1302–6. doi: 10.1016/s0735-1097(01)01554-6. [DOI] [PubMed] [Google Scholar]
- 6.Daniels LB, Laughlin GA, Sarno MJ, Bettencourt R, Wolfert RL, Barrett-Connor E. Lipoprotein-associated phospholipase A2 is an independent predictor of incident coronary heart disease in an apparently healthy older population: the Rancho Bernardo Study. J Am Coll Cardiol. 2008;51:913–9. doi: 10.1016/j.jacc.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ninio E, Tregouet D, Carrier JL, Stengel D, Bickel C, Perret C, Rupprecht HJ, Cambien F, Blankenberg S, Tiret L. Platelet-activating factor-acetylhydrolase and PAF-receptor gene haplotypes in relation to future cardiovascular event in patients with coronary artery disease. Hum Mol Genet. 2004;13:1341–51. doi: 10.1093/hmg/ddh145. [DOI] [PubMed] [Google Scholar]
- 8.Abuzeid AM, Hawe E, Humphries SE, Talmud PJ. Association between the Ala379Val variant of the lipoprotein associated phospholipase A2 and risk of myocardial infarction in the north and south of Europe. Atherosclerosis. 2003;168:283–8. doi: 10.1016/s0021-9150(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 9.Tang HK, Siegmund D. Mapping quantitative trait loci in oligogenic models. Biostatistics. 2001;2:147–62. doi: 10.1093/biostatistics/2.2.147. [DOI] [PubMed] [Google Scholar]
- 10.Brilakis ES, McConnell JP, Lennon RJ, Elesber AA, Meyer JG, Berger PB. Association of lipoprotein-associated phospholipase A2 levels with coronary artery disease risk factors, angiographic coronary artery disease, and major adverse events at follow-up. Eur Heart J. 2005;26:37–44. doi: 10.1093/eurheartj/ehi010. [DOI] [PubMed] [Google Scholar]
- 11.Koenig W, Twardella D, Brenner H, Rothenbacher D. Lipoprotein-associated phospholipase A2 predicts future cardiovascular events in patients with coronary heart disease independently of traditional risk factors, markers of inflammation, renal function, and hemodynamic stress. Arterioscler Thromb Vasc Biol. 2006;26:586–93. doi: 10.1161/01.ATV.0000222983.73369.c8. [DOI] [PubMed] [Google Scholar]
- 12.Zhang SY, Shibata H, Karino K, Wang BY, Kobayashi S, Masuda J, Nabika T. Comprehensive evaluation of genetic and environmental factors influencing the plasma lipoprotein-associated phospholipase A2 activity in a Japanese population. Hypertens Res. 2007;30:403–9. doi: 10.1291/hypres.30.403. [DOI] [PubMed] [Google Scholar]
- 13.Persson M, Nilsson JA, Nelson JJ, Hedblad B, Berglund G. The epidemiology of Lp-PLA(2): distribution and correlation with cardiovascular risk factors in a population-based cohort. Atherosclerosis. 2007;190:388–96. doi: 10.1016/j.atherosclerosis.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Furberg CD, Nelson JJ, Solomon C, Cushman M, Jenny NS, Psaty BM. Distribution and Correlates of Lipoprotein-Associated Phospholipase A(2) in an Elderly Cohort: The Cardiovascular Health Study. J Am Geriatr Soc. 2008 doi: 10.1111/j.1532-5415.2008.01667.x. [DOI] [PubMed] [Google Scholar]
- 15.Saougos VG, Tambaki AP, Kalogirou M, Kostapanos M, Gazi IF, Wolfert RL, Elisaf M, Tselepis AD. Differential effect of hypolipidemic drugs on lipoprotein-associated phospholipase A2. Arterioscler Thromb Vasc Biol. 2007;27:2236–43. doi: 10.1161/ATVBAHA.107.147280. [DOI] [PubMed] [Google Scholar]
- 16.Oei HH, van dM I, Hofman A, Koudstaal PJ, Stijnen T, Breteler MM, Witteman JC. Lipoprotein-associated phospholipase A2 activity is associated with risk of coronary heart disease and ischemic stroke: the Rotterdam Study. Circulation. 2005;111:570–5. doi: 10.1161/01.CIR.0000154553.12214.CD. [DOI] [PubMed] [Google Scholar]
- 17.O'Donoghue M, Morrow DA, Sabatine MS, Murphy SA, McCabe CH, Cannon CP, Braunwald E. Lipoprotein-associated phospholipase A2 and its association with cardiovascular outcomes in patients with acute coronary syndromes in the PROVE IT-TIMI 22 (PRavastatin Or atorVastatin Evaluation and Infection Therapy-Thrombolysis In Myocardial Infarction) trial. Circulation. 2006;113:1745–52. doi: 10.1161/CIRCULATIONAHA.105.612630. [DOI] [PubMed] [Google Scholar]
- 18.Persson M, Hedblad B, Nelson JJ, Berglund G. Elevated Lp-PLA2 levels add prognostic information to the metabolic syndrome on incidence of cardiovascular events among middle-aged nondiabetic subjects. Arterioscler Thromb Vasc Biol. 2007;27:1411–6. doi: 10.1161/ATVBAHA.107.142679. [DOI] [PubMed] [Google Scholar]
- 19.Caslake MJ, Packard CJ, Suckling KE, Holmes SD, Chamberlain P, Macphee CH. Lipoprotein-associated phospholipase A(2), platelet-activating factor acetylhydrolase: a potential new risk factor for coronary artery disease. Atherosclerosis. 2000;150:413–9. doi: 10.1016/s0021-9150(99)00406-2. [DOI] [PubMed] [Google Scholar]
- 20.Gazi I, Lourida ES, Filippatos T, Tsimihodimos V, Elisaf M, Tselepis AD. Lipoprotein-associated phospholipase A2 activity is a marker of small, dense LDL particles in human plasma. Clin Chem. 2005;51:2264–73. doi: 10.1373/clinchem.2005.058404. [DOI] [PubMed] [Google Scholar]
- 21.Garza CA, Montori VM, McConnell JP, Somers VK, Kullo IJ, Lopez-Jimenez F. Association between lipoprotein-associated phospholipase A2 and cardiovascular disease: a systematic review. Mayo Clin Proc. 2007;82:159–65. doi: 10.4065/82.2.159. [DOI] [PubMed] [Google Scholar]
- 22.Guerra R, Zhao B, Mooser V, Stafforini D, Johnston JM, Cohen JC. Determinants of plasma platelet-activating factor acetylhydrolase: heritability and relationship to plasma lipoproteins. J Lipid Res. 1997;38:2281–8. [PubMed] [Google Scholar]
- 23.Diego VP, Rainwater DL, Wang XL, Cole SA, Curran JE, Johnson MP, Jowett JB, Dyer TD, Williams JT, Moses EK, Comuzzie AG, Maccluer JW, Mahaney MC, Blangero J. Genotype x adiposity interaction linkage analyses reveal a locus on chromosome 1 for lipoprotein-associated phospholipase A2, a marker of inflammation and oxidative stress. Am J Hum Genet. 2007;80:168–77. doi: 10.1086/510497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu PY, Li YH, Wu HL, Chao TH, Tsai LM, Lin LJ, Shi GY, Chen JH. Platelet-activating factor-acetylhydrolase A379V (exon 11) gene polymorphism is an independent and functional risk factor for premature myocardial infarction. J Thromb Haemost. 2006;4:1023–8. doi: 10.1111/j.1538-7836.2006.01895.x. [DOI] [PubMed] [Google Scholar]
- 25.Kruse S, Mao XQ, Heinzmann A, Blattmann S, Roberts MH, Braun S, Gao PS, Forster J, Kuehr J, Hopkin JM, Shirakawa T, Deichmann KA. The Ile198Thr and Ala379Val variants of plasmatic PAF-acetylhydrolase impair catalytical activities and are associated with atopy and asthma. Am J Hum Genet. 2000;66:1522–30. doi: 10.1086/302901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Fan C, Topol SE, Topol EJ, Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003;302:1578–81. doi: 10.1126/science.1088477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su JL, Yen CJ, Chen PS, Chuang SE, Hong CC, Kuo IH, Chen HY, Hung MC, Kuo ML. The role of the VEGF-C/VEGFR-3 axis in cancer progression. Br J Cancer. 2007;96:541–5. doi: 10.1038/sj.bjc.6603487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cha HS, Bae EK, Koh JH, Chai JY, Jeon CH, Ahn KS, Kim J, Koh EM. Tumor necrosis factor-alpha induces vascular endothelial growth factor-C expression in rheumatoid synoviocytes. J Rheumatol. 2007;34:16–9. [PubMed] [Google Scholar]
- 29.Shin HY, Smith ML, Toy KJ, Williams PM, Bizios R, Gerritsen ME. VEGF-C mediates cyclic pressure-induced endothelial cell proliferation. Physiol Genomics. 2002;11:245–51. doi: 10.1152/physiolgenomics.00068.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


