Abstract
Several quinone-based metabolites of industrial and environmental toxins are potent topoisomerase II poisons. These compounds act by adducting the protein, and previous studies suggest that they increase levels of enzyme-associated DNA strand breaks by at least two potential mechanisms. Quinones act directly on the DNA cleavage-ligation equilibrium of topoisomerase II by inhibiting the rate of ligation. They also block the N-terminal gate of the protein, thereby stabilizing topoisomerase II in its “closed clamp” form and trapping DNA in the central annulus of the enzyme. It has been proposed that this latter activity enhances DNA cleavage by increasing the population of enzyme molecules with DNA in their active sites, but a causal relationship has not been established. In order to more fully characterize the mechanistic basis for quinone action against topoisomerase II, the present study characterized the sensitivity of human topoisomerase IIα carrying a Cys455–>Ala mutation (top2αC455A) toward quinones. Cys455 was identified as a site of quinone adduction by mass spectrometry. The mutant enzyme was ~1.5–to2–fold hypersensitive to1,4-benzoquinone and the polychlorinated biphenyl quinone 4′Cl-2,5pQ, but displayed wild-type sensitivity to traditional topoisomerase II poisons. The ability of 1,4-benzoquinone to inhibit DNA ligation mediated by top2αC455A was similar to that of wild-type topoisomerase IIα. However, the quinone induced ~3 times the level of clamp closure with the mutant enzyme. These findings strongly support the hypothesis that the ability of quinones to block the N-terminal gate of the type II enzyme contributes to their actions as topoisomerase II poisons.
Introduction
Topoisomerase IIα is an essential enzyme that plays important roles in DNA replication and chromosome segregation. The enzyme relaxes, unknots, and untangles DNA by passing a double helix through a transient double-stranded break that it generates in a separate segment of DNA (1–7). To maintain genomic integrity during this process, topoisomerase IIα forms covalent bonds between active site tyrosyl residues and the 5′-DNA termini created by scission of the double helix (8–10). The covalent enzyme–cleaved DNA complex that results is known as the cleavage complex. When DNA tracking enzymes such as polymerases or helicases collide with these complexes, they convert them to permanent enzyme-linked double-stranded breaks in the genetic material (1–7). These breaks destabilize the genome, leading to illegitimate recombination and the formation of chromosomal aberrations. When present in sufficient concentrations, they trigger programmed cell death pathways (4, 11–17).
Agents that increase levels of topoisomerase IIα-mediated DNA strand breaks are called topoisomerase II poisons (4, 15, 18–20). Some topoisomerase II poisons, such as etoposide, doxorubicin, and mitoxantrone, are important anticancer drugs that are used to treat a wide variety of human malignancies. However, a small percentage of patients who receive therapy with these agents eventually develop secondary leukemias that feature aberrations in the mixed-lineage leukemia (MLL) gene at chromosomal band 11q23 (13, 14, 21–26). Other topoisomerase II poisons, such as the bioflavonoids (naturally found in fruits and vegetables), display chemopreventative properties in adults (27–30). In contrast, when ingested during pregnancy, they are believed to increase the risk of infant leukemias that include MLL rearrangements (31–35).
In addition to pharmaceutical agents and natural products, quinone metabolites of some industrial and environmental toxins, such as 1,4-benzoquinone [a reactive metabolite of benzene (36)] (37, 38), and a variety of PCB (polychlorinated biphenyl) quinone metabolites (39), act as topoisomerase II poisons. These highly reactive compounds are produced in the body as a result of detoxification or metabolism pathways (36, 40, 41). Cellular exposure to 1,4-benzoquinone or PCB quinones generates DNA strand breaks and other chromosomal aberrations, and has been linked to a variety of human health problems, including cancer (42–46). Furthermore, the accumulation of 1,4-benzoquinone in the bone marrow is believed to contribute to the leukemogenic properties of benzene, including leukemias that include MLL rearrangements (43).
Quinones are unique among characterized topoisomerase II poisons, in that their activity requires covalent attachment to the enzyme (37–39, 47–49). Although adduction inhibits the ability of topoisomerase IIα to ligate DNA molecules, this inhibition cannot completely account for the increase in enzyme-associated DNA strand breaks. During the strand passage event, topoisomerase II closes an N-terminal protein gate, and thereby forms a protein clamp that encircles the DNA within the central annulus of the enzyme. Quinone treatment blocks the N-terminal protein gate of topoisomerase IIα. It has been suggested that this latter effect contributes to the increase in topoisomerase II-associated strand breaks by trapping DNA in the active site of the enzyme (39, 49). However, evidence supporting this hypothesis is circumstantial.
Recently, four sites of quinone adduction on human topoisomerase IIα, Cys170, Cys392, Cys405, and Cys455, were identified by mass spectrometry (49). While mutation of Cys170 to Ala had no effect on quinone sensitivity, parallel mutation of Cys392 or Cys405 resulted in enzymes that were partially (~50%) resistant to 1,4-benzoquinone and the PCB quinone 4′Cl-2,5pQ (49). In both cases, mutation decreased the inhibitory effects of quinones on DNA ligation, but did not affect sensitivity to clamp closing. The present study characterized topoisomerase IIα that carried a C–>A mutation at residue C455 (top2αC455A). Top2αC455A was hypersensitive to 1,4-benzoquinone and the PCB quinone 4′Cl-2,5pQ. Mechanistic studies strongly suggest a causal link between the ability of quinones to close the N-terminal protein gate of topoisomerase IIα and their ability to increase levels of enzyme-generated double-stranded DNA breaks.
EXPERIMENTAL PROCEDURES
Enzymes and Materials
Human topoisomerase IIα was expressed in Saccharomyces cerevisiae and purified as described previously (50–52). Negatively supercoiled pBR322 DNA was prepared using a Plasmid Mega Kit (Qiagen) as described by the manufacturer. 1,4-Benzoquinone, etoposide, and genistein were obtained from Sigma, prepared as 20 mM stock solutions in 100% DMSO, and stored at 4 °C. The PCB quinone, 4′Cl-2,5pQ (the generous gift of Dr. Hans J. Lehmler and Dr. Larry W. Robertson, University of Iowa), was synthesized by coupling 4′-chloroaniline with 1,4-benzoquinone (53). The compound was prepared as a 20 mM stock in 100% DMSO, and stored at −20 °C. Amsacrine was obtained from Bristol-Myers Squibb, prepared as 20 mM stock solution in 100% DMSO, and stored at 4 °C. The quinolone CP-115,953 was the gift of Pfizer Global Research, dissolved as a 40 mM solution in 0.1 N NaOH, and stored at −20 °C. Immediately prior to use, the quinolone was diluted to 8 mM with 10 mM Tris, pH 7.9. All other chemicals were of analytical reagent grade.
Generation of A Human Topoisomerase IIα Protein Carrying a Cys455–>A Mutation (top2αC455A)
The C455A mutation in the topoisomerase IIα PCR substrate was generated by cloning a SalI-KpnI fragment of YEpWob6 (54) that encoded the N-terminus of the human enzyme into pUC18. Site-directed mutagenesis was performed using the QuickChange II PCR system (Stratagene). The sequence of the forward and reverse primers used to generate the C455A mutation were CAGGGGGCCGAAACTCCACTGAGGCTACGCTTATCC and CCCTCAGTCAGGATAAGCGTAGCCTCAGTGGAGTTTCGGCCC. The mutagenized codons are underlined. The mutation was verified by sequencing and the SalI-KpnI fragment was cloned back into YEpWob6. The mutant human topoisomerase IIα enzyme (top2αC455A) was purified as described above.
Cleavage of Plasmid DNA
DNA cleavage reactions were carried out using the procedure of Fortune and Osheroff (55). Unless stated otherwise, assay mixtures contained 135 nM wild-type topoisomerase IIα or top2αC455A, and 10 nM negatively supercoiled pBR322 DNA in a total of 20 μL of 10 mM Tris-HCl, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0.1 mM NaEDTA, and 2.5% glycerol that contained 0–200 μM 1,4-benzoquinone, 4′Cl-2,5pQ, or etoposide; 50 μM genistein or amsacrine; or 5 μM CP-115,953. DNA cleavage was initiated by shifting mixtures to 37 °C and samples were incubated for 6 min to establish DNA cleavage-ligation equilibria. Enzyme-DNA cleavage intermediates were trapped by adding 2 μL of 5% SDS and 1 μL of 375 mM EDTA, pH 8.0. Proteinase K was added (2 μL of 0.8 mg/mL) and reaction mixtures were incubated for 30 min at 45 °C to digest the topoisomerase IIα. Samples were mixed with 2 μL of 60% sucrose in 10 mM Tris-HCl, pH 7.9, 0.5% bromophenol blue, and 0.5% xylene cyanol FF, heated for 15 min at 45 °C, and subjected to electrophoresis in 1% agarose gels in 40 mM Tris-acetate, pH 8.3, 2 mM EDTA that contained 0.5 μg/mL ethidium bromide. DNA cleavage was monitored by the conversion of negatively supercoiled plasmid DNA to linear molecules. DNA bands were visualized by ultraviolet light and quantified using an Alpha Innotech digital imaging system.
DNA Binding
Topoisomerase IIα binding to negatively supercoiled pBR322 DNA was assessed using an electrophoretic mobility shift assay (39, 56). Reaction mixtures contained 0–400 nM wild-type topoisomerase IIα or top2αC455A, and 5 nM plasmid DNA molecules in 20 μL of 10 mM Tris-HCl, pH 7.9, 30 mM KCl, 0.1 mM NaEDTA, and 2.5% glycerol. Samples were incubated for 6 min at 37 °C. Following the addition of 2 μL of 60% sucrose in 10 mM Tris-HCl, pH 7.9, samples were loaded directly onto a 1% agarose gel, and subjected to electrophoresis in 100 mM Tris-borate, pH 8.3, 2 mM EDTA. Gels were stained for 30 min with 0.5 μg/mL ethidium bromide and DNA was visualized as described above.
DNA Ligation
The DNA ligation reaction of wild-type human topoisomerase IIα or top2αC455A was monitored according to the procedure of Byl et al. (57). Topoisomerase IIα DNA cleavage/ligation equilibria were established using a plasmid substrate as described above in the absence or presence of 100 μM 1,4-benzoquinone. DNA ligation was initiated by shifting reaction mixtures from 37 °C to 0 °C, and reactions were stopped by the addition of 2 μL of 5% SDS followed by 1 μL of 375 mM NaEDTA, pH 8.0. Samples were processed and analyzed as described above for topoisomerase IIα cleavage reactions.
Protein Clamp Closing
Filter binding assays were used to analyze the salt-stable closed-clamp form of topoisomerase IIα (39, 58). Briefly, 5 nM wild-type human topoisomerase IIα or top2αC455A, and 2 nM pBR322 were incubated for 4 min at 37 °C in a total of 90 μL of clamp closing buffer (50 mM Tris-HCl, pH 8.0, 100 mM KCl, 1 mM EDTA, 8 mM MgCl2). 1,4-Benzoquinone (10 μL of 1 mM in 10% DMSO) or an equivalent amount of solvent was added, and mixtures were incubated for an additional 6 min at 37 °C.
Binding mixtures were loaded onto glass fiber filters (Millipore) pre-incubated in clamp closing buffer, and filtered in vacuo. Filters were washed 3 times with clamp closing buffer (low salt wash), followed by 3 washes with clamp closing buffer that contained 1 M NaCl (high salt wash), and 3 washes with 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 0.5% SDS. DNA in the eluates was precipitated with isopropanol and loaded onto a 1% agarose gel in 40 mM Tris-acetate, pH 8.3, 2 mM EDTA that contained 0.5 μg/mL ethidium bromide. DNA was visualized and quantified as described above.
RESULTS
Quinone metabolites of some industrial and environmental toxins, including 1,4-benzoquine and PCB quinones, are potent topoisomerase II poisons in vitro and in cultured human cells (42–46). In contrast to “traditional” topoisomerase II poisons, such as etoposide and other anticancer drugs, the mechanism by which quinones increase levels of topoisomerase II-associated DNA breaks is not well understood. Quinones require adduction to the enzyme in order to act as topoisomerase II poisons and modification of Cys392 and Cys405 inhibits the ability of topoisomerase IIα to ligate cleaved DNA molecules (37–39, 47–49). However, this inhibition does not completely account for the increase in enzyme-associated DNA strand breaks.
Quinone adduction also blocks the N-terminal protein gate of topoisomerase IIα (39, 48, 49). However, the relationship between this activity and effects on DNA cleavage has not been established. Two lines of circumstantial evidence suggest that this latter effect contributes to the increase in enzyme-mediated DNA strand breaks. First, the bisdioxopiperazine, ICRF-193, is a mixed function inhibitor of topoisomerase II that traps DNA in the central annulus of the enzyme by closing the N-terminal protein gate (59). Although this action inhibits overall catalytic activity, it leads to a modest increase in enzyme-mediated DNA cleavage (<2–fold) (60). Second, quinones such as plumbagin, which do not significantly effect gate closure, are poor topoisomerase II poisons (49).
In an effort to more fully define the mechanistic basis by which quinones act as topoisomerase II poisons, four sites of quinone adduction on human topoisomerase IIα, Cys170, Cys392, Cys405, and Cys455, were identified by mass spectrometry and mutated to alanine residues (49). This study previous analyzed the effects of Cys–>Ala mutations at residues 170, 392, and 405, which are located in the N-terminal domain of human topoisomerase IIα (49, 61, 62). Mutation of residues Cys392 or Cys405 (or both) in the enzyme (top2αC392A, top2αC405A, or top2αC392/405A) resulted in enzymes that displayed wild-type DNA cleavage activity and sensitivity to etoposide, but were partially (~50%) resistant to quinones. Although these mutations decreased the effects of 1,4-benzoquinone and PCB quinones on DNA ligation, they did not alter the ability of quinones to close the N-terminal protein gate of the enzyme.
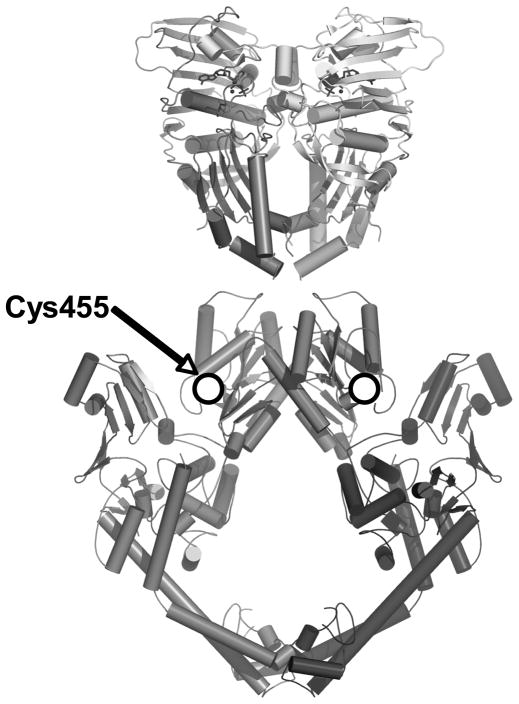
The fourth adducted residue, Cys455, is located in the catalytic core of human topoisomerase IIα rather than the N-terminal domain (Figure 1). Furthermore, as described below, mutation of this residue to an alanine altered the basal DNA cleavage activity of the enzyme. Therefore, the effects of the C455A mutation on quinone sensitivity were not analyzed in the earlier study.
Figure 1.
Cys455 in human topoisomerase IIα was identified by mass spectrometry as a site of quinone adduction. A composite of the crystal structures of the yeast catalytic core and N-terminal domain is shown and the location of the homologous cysteine residue that is adducted in human topoisomerase IIα is indicated by shaded circles. Adapted from Refs. (61, 62).
Human Topoisomerase IIα Carrying a Cys455–>Ala Mutation (top2αC455A) is Hypersensitive to Quinones
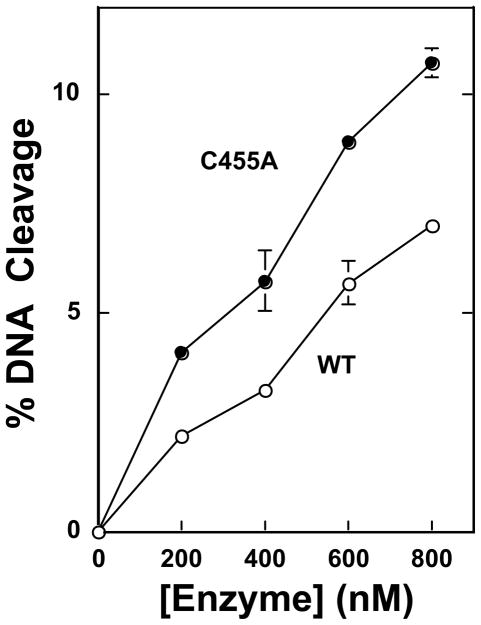
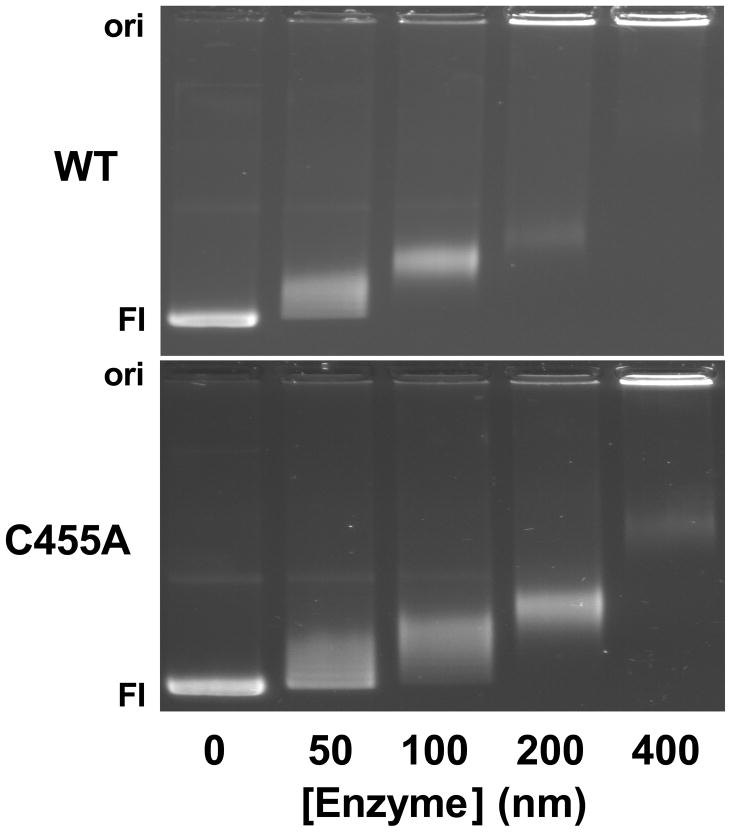
The previously characterized Cys–>Ala mutants (top2αC392A, top2αC405A, or top2αC392/405A) displayed wild-type DNA cleavage activity in the absence of topoisomerase II poisons (49). In contrast, as seen in Figure 2, top2αC455A exhibits a cleavage activity with negatively supercoiled plasmid DNA that is ~70% higher than that of the wild-type enzyme. This enhancement of DNA cleavage was not due to an increase in DNA binding by the mutant enzyme. In fact, top2αC455A displayed a lower binding affinity for negatively supercoiled plasmid DNA than did wild-type topoisomerase IIα (Figure 3). In addition, as determined by nitrocellulose filter binding experiments (39), the mutant and wild-type enzymes bound similar levels of linear plasmid DNA or oligonucleotide (data not shown).
Figure 2.
DNA cleavage activity of wild-type human topoisomerase IIα and top2αC455A. Cleavage activity was assessed using 0–800 nM enzyme. Assay mixtures contained wild-type enzyme (WT, open circles) or top2αC455A (C455A, closed circles). Error bars represent the standard deviation of at least three independent experiments.
Figure 3.
Binding of negatively supercoiled plasmid DNA by wild-type human topoisomerase IIα and top2αC455A. DNA Binding was assessed using 0–400 nM wild-type (WT) or mutant (C455A) enzyme. DNA products were analyzed by gel electrophoresis and visualized by staining with ethidium bromide. The presence of enzyme-DNA complexes is indicated by a shift in the electrophoretic mobility of negatively supercoiled DNA. Topoisomerase II-bound DNA exhibited a slower mobility or remained at the gel origin (Ori). The position of negatively supercoiled plasmid (form I, FI) is indicated. Gels are typical of at least three independent experiments.
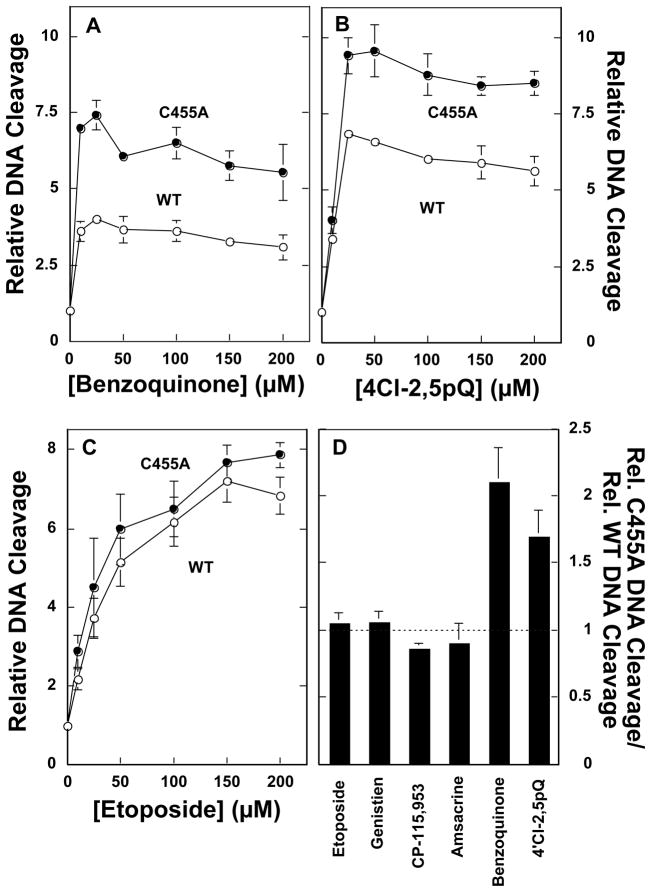
To further analyze the properties of top2αC455A, its sensitivity toward topoisomerase II poisons was compared to that of wild-type human topoisomerase IIα. It should be noted that the data shown in Figure 4 are plotted as relative DNA cleavage. Thus, the data account for the enhanced scission activity of top2αC455A by setting the initial level of DNA cleavage in the absence of topoisomerase II poisons to 1.0 for both the mutant and wild-type enzymes.
Figure 4.
Effects of quinones and traditional topoisomerase II poisons on the DNA cleavage activity of wild-type human topoisomerase IIα and top2αC455A. Assay mixtures contained wild-type topoisomerase IIα (WT, open circles) or top2αC455A (C455A, closed circles). Relative levels of DNA cleavage are shown. For both the wild-type and mutant enzymes, the level of DNA cleavage in the absence of topoisomerase II poisons was set to 1.0. Panel A: DNA cleavage was assessed in the presence of 0–200 μM benzoquinone. Panel B: DNA cleavage was assessed in the presence of 0–200 μM 4′Cl-2,5pQ. Panel C: DNA cleavage was assessed in the presence of 0–200 μM etoposide. Panel D: DNA cleavage was assessed in the presence of 50 μM etoposide, genistein, amsacrine, benzoquinone, or 4′Cl-2,5pQ, or 5 μM CP-115,953. Values represent the ratio of DNA cleavage generated by top2αC455A divided by that generated by the wild-type enzyme. The dotted line represents equal sensitivities for the indicated compound by both enzymes. Values below the line indicate resistance, while those above the line indicate hypersensitivity. Error bars represent the standard deviation of at least three independent experiments.
As seen in Figure 4A, B, and D, the mutant enzyme was hypersensitive (~1.5– to 2–fold) to two quinone-based topoisomerase II poisons, 1,4-benzoquinone and the PCB quinone 4′Cl-2,5pQ. The enhanced sensitivity did not result from an increased affinity for quinones, because at saturating (i.e., plateau) concentrations of 1,4-benzoquinone or 4′Cl-2,5pQ, levels of DNA cleaved by the mutant enzyme were always higher than those observed for the wild-type enzyme.
To determine whether the enhanced susceptibility of top2αC455A was unique to quinones, the sensitivity of the enzyme toward a series of traditional (i.e., non-covalent) topoisomerase II poisons was assessed. Drugs from four different classes were employed for these experiments, including etoposide (a demethylepipodophyllotoxin), genistein (an isoflavone), CP-115,953 (a quinolone), and amsacrine (an anilinoacridine). As seen in Figure 4C and D, the sensitivity of the mutant enzyme toward these traditional topoisomerase II poisons was similar to that of wild-type topoisomerase IIα. Therefore, the hypersensitivity of top2αC455A appears to be specific to quinone-based topoisomerase II poisons.
Basis for the Quinone Hypersensitivity of top2αC455A
As discussed above, quinones have two effects on human topoisomerase IIα that may contribute to their actions as topoisomerase II poisons: they inhibit the ability of the enzyme to ligate cleaved DNA and they block the N-terminal gate of the protein (49). As a first step in determining the mechanistic basis for the quinone hypersensitivity of top2αC455A, the ability of the enzyme to ligate DNA was characterized. In the absence of quinones, the mutant enzyme ligated DNA ~1/3 slower than did wild-type topoisomerase IIα (Figure 5). This decreased ligation rate probably accounts (at least in part) for the higher levels of DNA cleavage generated by top2αC455A in the absence of topoisomerase II poisons.
Figure 5.
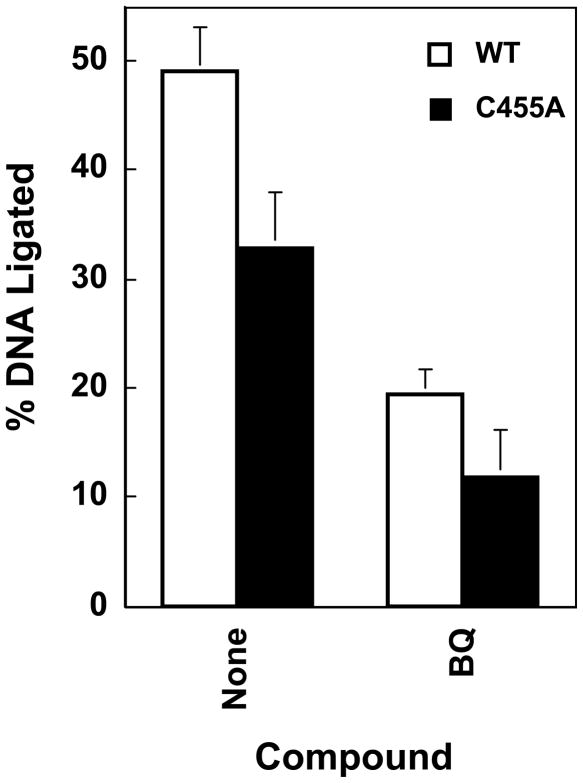
DNA ligation mediated by wild-type human topoisomerase IIα and top2αC455A in the absence and presence of 1,4-benzoquinone. Samples contained wild-type topoisomerase IIα (WT, open bars) or top2αC455A (C455A, closed bars) and were incubated at 37 °C to establish DNA cleavage/ligation equilibria. Reactions were shifted to 0 °C for 10 s, and DNA ligation was quantified by the loss of linear cleaved molecules. DNA cleavage/ligation equilibria were established in the absence (None) or presence (BQ) of 100 μM 1,4-benzoquinone. Error bars represent the standard deviation of at least three independent experiments.
Addition of 1,4-benzoquinone to reaction mixtures decreased the ability of top2αC455A to ligate DNA (Figure 5). Levels of ligation mediated by top2αC455A dropped ~2.8–fold (from 33.1% to 11.9%) in the presence of the quinone. This drop was similar to the 2.5–fold decrease observed for wild-type topoisomerase IIα (from 49.0% to 19.3%). Therefore, the heightened sensitivity of top2αC455A to quinones does not appear to be related to effects on DNA ligation.
To further explore the basis for quinone hypersensitivity, the ability of 1,4-benzoquinone to block the N-terminal protein gate of top2αC455A was characterized (Figure 6). In these experiments, the mutant and wild-type enzymes were incubated with circular DNA substrates prior to the addition of 1,4-benzoquinone. Blocking the N-terminal gate traps the circular substrate in the central annulus of topoisomerase IIα, generating a non-covalent protein-DNA complex that is stable in 1 M NaCl (39, 58).
Figure 6.
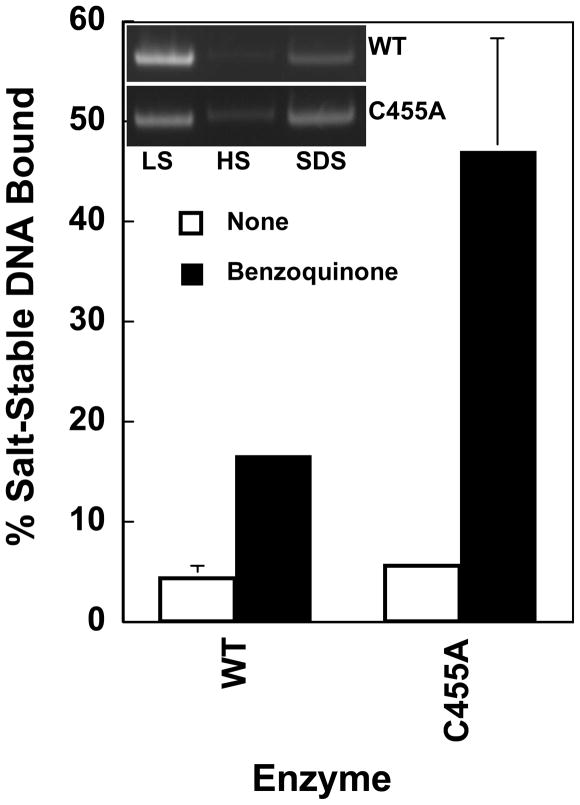
Ability of quinones to close the N-terminal gate of wild-type human topoisomerase IIα (WT) and top2αC455A (C455A). Filter binding assays were used to analyze the salt-stable closed-clamp form of topoisomerase II. Enzyme-DNA complexes were incubated in the absence (None, open bars) or presence (filled bars) of 100 μM 1,4-benzoquinone. Samples were applied to glass fiber filters, and eluted by sequential washes in low salt (LS), high salt (HS), and SDS. Eluted DNA was subjected to electrophoresis in an agarose gel. Representative gels are shown in the inset. Salt-stable non-covalent enzyme-DNA complexes were monitored by quantifying the amount of plasmid in the SDS wash relative to the total plasmid eluted in all three washes. Error bars represent the standard deviation of at least three independent experiments.
In the absence of quinone, top2αC455A and wild-type topoisomerase IIα trapped similar levels of DNA in a salt-stable non-covalent complex (5.6% vs. 4.5%, respectively). However, in the presence of 1,4-benzoquinone, a dramatic difference was observed (Figure 6). Whereas the level of DNA trapped by the wild-type enzyme rose to 16.8%, the level of DNA trapped by top2αC455A was ~3 times higher (47.1%). These data strongly suggest that the enhanced ability of quinones to block the N-terminal gate of top2αC455A contributes to the hypersensitivity of the enzyme toward these topoisomerase II poisons.
Discussion
Quinone metabolites of a variety of industrial and environmental toxins are potent topoisomerase II poisons (37, 38, 47, 48). These compounds act by adducting the protein, and previous studies suggest that they increase levels of enzyme-DNA cleavage complexes by at least two potential mechanisms (37, 38, 47, 48). Quinones act directly on the DNA cleavage-ligation equilibrium of topoisomerase II by inhibiting the rate of ligation. Quinones also block the N-terminal gate of the protein, thereby stabilizing the enzyme in its “clamp-closed” form (39, 49). It has been proposed that this latter activity raises levels of cleavage complexes by increasing the population of enzyme molecules with DNA in their active sites, but a causal relationship has not been established.
The present study characterized the sensitivity of top2αC455A toward quinones. Cys455 was identified as a site of quinone adduction by mass spectrometry (49). The mutant enzyme was ~1.5– to 2–fold hypersensitive to 1,4-benzoquinone and 4′Cl-2,5pQ, but displayed wild-type sensitivity to traditional topoisomerase II poisons. The ability of 1,4-benzoquinone to inhibit DNA ligation mediated by top2αC455A was similar to that of wild-type topoisomerase IIα. However, the quinone induced ~3 times more clamp closure with the mutant enzyme. These findings strongly support the hypothesis that quinones increase levels of topoisomerase II-associated DNA strand breaks, at least in part, by blocking the N-terminal gate of the enzyme.
Since Cys455 is located in the catalytic core of topoisomerase IIα as opposed to the N-terminal domain (61, 62), it is unlikely that the residue plays a direct role in clamp closure. In this regard, the loss of Cys455 has little effect on the rate of subunit crosslinking (data not shown).
How then does the Cys455–>Ala mutation, which removes a site for quinone adduction, enhance the effects of quinones on the N-terminal gate of topoisomerase IIα? At least three possibilities exist. First, Cys455 may simply represent a “non-productive sink” for quinone adduction. This would diminish levels of compound available to modify amino acid residues involved in clamp closure. We believe that this possibility is unlikely. If the sole role of Cys455 in quinone action was to draw these compounds away from other residues, levels of DNA cleavage mediated by wild-type topoisomerase IIα would eventually approach those seen with top2αC455A as quinone concentrations reached saturation. As seen in Figure 2, this was not the case.
Second, adduction of Cys455 may attenuate the ability of quinones to block the N-terminal gate of topoisomerase II by a yet to be defined mechanism. Therefore, removal of the residue would enhance the actions of quinones on clamp closure.
Third, the majority of Cys455 exists in a disulfide bridge with Cys427 (63); at any given time, only 10 to 15% of the residue exists as a free sulfhydryl (63). The existence of the Cys427-Cys455 disulfide bridge may impede closure of the N-terminal gate of topoisomerase IIα. In this case, adduction of Cys455 by one quinone molecule would prevent the disulfide bridge from reforming, thus enhancing the ability of other quinone molecules to block the N-terminal gate. In a parallel fashion, mutation of Cys455–>Ala would prevent the formation of the disulfide bridge and enhance clamp closing by mimicking the effects of quinone adduction.
Since top2αC455A displayed altered basal DNA cleavage activity, it is most likely that the Cys455–>Ala mutation increases the sensitivity of topoisomerase IIα to quinones by an allosteric mechanism such as those discussed in points two and three. However, it is not possible to distinguish between these possibilities (or an even more complicated scenario) at the present time. Further mechanistic and structural studies most likely will be required to address this complex issue.
In conclusion, mutation of Cys455–>Ala in human topoisomerase IIα results in an enzyme that is hypersensitive to quinones. The increased sensitivityof top2αC455Ato 1,4-benzoquinone correlates with an enhanced ability of this compound to block the N-terminal gate of the protein. These findings provide strong evidence that the effects of quinones on the N-terminal gate play an important role in the actions of these compounds as topoisomerase II poisons.
Acknowledgments
We are grateful to Dr. H. J. Lehmler and Dr. Larry W. Robertson, University of Iowa, for the generous gift of 4′Cl-2,5pQ (synthesized under the auspices of National Institute of Superfund Basic Research Program grant P42 ES013661), and to Omari J. Bandele and Joseph E. Deweese for critical reading of the manuscript.
Abbreviations
- PCB
Polychlorinated biphenyl
- 4′Cl-2
5pQ, 2-(4-chloro-phenyl)-[1,4]benzoquinone
Footnotes
This work was supported by National Institutes of Health research grants GM33944 (NO). RPB was a trainee under grant 5 T32 CA09582 from the National Institutes of Health.
References
- 1.Wang JC. DNA Topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim Biophys Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Quart Rev Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 4.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 5.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Velez-Cruz R, Osheroff N. DNA Topoisomerases: Type II. In: Lennarz W, Lane MD, editors. Encyclopedia of Molec Biol. Elsevier Science; San Diego: 2004. pp. 806–811. [Google Scholar]
- 8.Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 9.Liu LF, Rowe TC, Yang L, Tewey KM, Chen GL. Cleavage of DNA by mammalian DNA topoisomerase II. J Biol Chem. 1983;258:15365–15370. [PubMed] [Google Scholar]
- 10.Zechiedrich EL, Christiansen K, Andersen AH, Westergaard O, Osheroff N. Double-stranded DNA cleavage/religation reaction of eukaryotic topoisomerase II: evidence for a nicked DNA intermediate. Biochemistry. 1989;28:6229–6236. doi: 10.1021/bi00441a014. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann SH. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim Biophys Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann SH, Gore SD, Miller CB, Jones RJ, Zwelling LA, Schneider E, Burke PJ, Karp JE. Topoisomerase II and the response to antileukemic therapy. Leukemia Lymph. 1998;29:217–237. doi: 10.3109/10428199809068560. [DOI] [PubMed] [Google Scholar]
- 13.Rowley JD. The critical role of chromosome translocations in human leukemias. Ann Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 14.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 15.Wilstermann AM, Osheroff N. Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr Top Med Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 16.Sordet O, Khan QA, Kohn KW, Pommier Y. Apoptosis induced by topoisomerase inhibitors. Curr Med Chem Anti-Canc Agents. 2003;3:271–290. doi: 10.2174/1568011033482378. [DOI] [PubMed] [Google Scholar]
- 17.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst) 2006;5:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 18.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 19.Walker JV, Nitiss JL. DNA topoisomerase II as a target for cancer chemotherapy. Cancer Invest. 2002;20:570–589. doi: 10.1081/cnv-120002156. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anti-Canc Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 21.DeVore R, Whitlock J, Hainsworth T, Johnson D. Therapy-related acute nonlymphocytic leukemia with monocytic features and rearrangement of chromosome 11q. Ann Intern Med. 1989;110:740–742. doi: 10.7326/0003-4819-110-9-740. [DOI] [PubMed] [Google Scholar]
- 22.Ratain MJ, Rowley JD. Therapy-related acute myeloid leukemia secondary to inhibitors of topoisomerase II: from the bedside to the target genes. Annals Oncol. 1992;3:107–111. doi: 10.1093/oxfordjournals.annonc.a058121. [DOI] [PubMed] [Google Scholar]
- 23.Smith MA, Rubinstein L, Anderson JR, Arthur D, Catalano PJ, Freidlin B, Heyn R, Khayat A, Krailo M, Land VJ, Miser J, Shuster J, Vena D. Secondary leukemia or myelodysplastic syndrome after treatment with epipodophyllotoxins. J Clin Oncol. 1999;17:569–577. doi: 10.1200/JCO.1999.17.2.569. [DOI] [PubMed] [Google Scholar]
- 24.Pui CH, Relling MV. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br J Haematol. 2000;109:13–23. doi: 10.1046/j.1365-2141.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 25.Felix CA. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med Pediatr Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 26.Leone G, Voso MT, Sica S, Morosetti R, Pagano L. Therapy related leukemias: susceptibility, prevention and treatment. Leuk Lymphoma. 2001;41:255–276. doi: 10.3109/10428190109057981. [DOI] [PubMed] [Google Scholar]
- 27.Adlercreutz H, Markkanen H, Watanabe S. Plasma concentrations of phyto-oestrogens in Japanese men. Lancet. 1993;342:1209–1210. doi: 10.1016/0140-6736(93)92188-y. [DOI] [PubMed] [Google Scholar]
- 28.Lamartiniere CA. Protection against breast cancer with genistein: a component of soy. Am J Clin Nutr. 2000;71:1705S–1709S. doi: 10.1093/ajcn/71.6.1705S. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar FH, Adsule S, Padhye S, Kulkarni S, Li Y. The role of genistein and synthetic derivatives of isoflavone in cancer prevention and therapy. Mini Rev Med Chem. 2006;6:401–407. doi: 10.2174/138955706776361439. [DOI] [PubMed] [Google Scholar]
- 30.Siddiqui IA, Adhami VM, Saleem M, Mukhtar H. Beneficial effects of tea and its polyphenols against prostate cancer. Mol Nutr Food Res. 2006;50:130–143. doi: 10.1002/mnfr.200500113. [DOI] [PubMed] [Google Scholar]
- 31.Ross JA, Potter JD, Robison LL. Infant leukemia, topoisomerase II inhibitors, and the MLL gene. J Natl Cancer Inst. 1994;86:1678–1680. doi: 10.1093/jnci/86.22.1678. [DOI] [PubMed] [Google Scholar]
- 32.Ross JA, Potter JD, Reaman GH, Pendergrass TW, Robison LL. Maternal exposure to potential inhibitors of DNA topoisomerase II and infant leukemia (United States): a report from the Children’s Cancer Group. Cancer causes & control: CCC. 1996;7:581–590. doi: 10.1007/BF00051700. [DOI] [PubMed] [Google Scholar]
- 33.Ross JA. Dietary flavonoids and the MLL gene: A pathway to infant leukemia? Proc Natl Acad Sci U S A. 2000;97:4411–4413. doi: 10.1073/pnas.97.9.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc Natl Acad Sci U S A. 2000;97:4790–4795. doi: 10.1073/pnas.070061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spector LG, Xie Y, Robison LL, Heerema NA, Hilden JM, Lange B, Felix CA, Davies SM, Slavin J, Potter JD, Blair CK, Reaman GH, Ross JA. Maternal diet and infant leukemia: the DNA topoisomerase II inhibitor hypothesis: a report from the children’s oncology group. Cancer Epidemiol Biomarkers Prev. 2005;14:651–655. doi: 10.1158/1055-9965.EPI-04-0602. [DOI] [PubMed] [Google Scholar]
- 36.Ross D. The role of metabolism and specific metabolites in benzene-induced toxicity: evidence and issues. J Toxicol Environ Health A. 2000;61:357–372. doi: 10.1080/00984100050166361. [DOI] [PubMed] [Google Scholar]
- 37.Lindsey RH, Jr, Bromberg KD, Felix CA, Osheroff N. 1,4-Benzoquinone is a topoisomerase II poison. Biochemistry. 2004;43:7563–7574. doi: 10.1021/bi049756r. [DOI] [PubMed] [Google Scholar]
- 38.Lindsey RH, Jr, Bender RP, Osheroff N. Effects of benzene metabolites on DNA cleavage mediated by human topoisomerase II alpha: 1,4-hydroquinone is a topoisomerase II poison. Chem Res Toxicol. 2005;18:761–770. doi: 10.1021/tx049659z. [DOI] [PubMed] [Google Scholar]
- 39.Bender RP, Lehmler HJ, Robertson LW, Ludewig G, Osheroff N. Polychlorinated Biphenyl Quinone Metabolites Poison Human Topoisomerase IIalpha: Altering Enzyme Function by Blocking the N-Terminal Protein Gate. Biochemistry. 2006;45:10140–10152. doi: 10.1021/bi0524666. [DOI] [PubMed] [Google Scholar]
- 40.Oakley GG, Devanaboyina U, Robertson LW, Gupta RC. Oxidative DNA damage induced by activation of polychlorinated biphenyls (PCBs): implications for PCB-induced oxidative stress in breast cancer. Chem Res Toxicol. 1996;9:1285–1292. doi: 10.1021/tx960103o. [DOI] [PubMed] [Google Scholar]
- 41.Srinivasan A, Lehmler HJ, Robertson LW, Ludewig G. Production of DNA strand breaks in vitro and reactive oxygen species in vitro and in HL-60 cells by PCB metabolites. Toxicol Sci. 2001;60:92–102. doi: 10.1093/toxsci/60.1.92. [DOI] [PubMed] [Google Scholar]
- 42.Ludewig G, Dogra S, Glatt H. Genotoxicity of 1,4-benzoquinone and 1,4-naphthoquinone in relation to effects on glutathione and NAD(P)H levels in V79 cells. Environ Health Perspect. 1989;82:223–228. doi: 10.1289/ehp.8982223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith MT. The mechanism of benzene-induced leukemia: a hypothesis and speculations on the causes of leukemia. Environ Health Perspect. 1996;104(Sup 6):1219–1225. doi: 10.1289/ehp.961041219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sze CC, Shi CY, Ong CN. Cytotoxicity and DNA strand breaks induced by benzene and its metabolites in Chinese hamster ovary cells. J Appl Toxicol. 1996;16:259–264. doi: 10.1002/(SICI)1099-1263(199605)16:3<259::AID-JAT342>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 45.Nakayama A, Koyoshi S, Morisawa S, Yagi T. Comparison of the mutations induced by p-benzoquinone, a benzene metabolite, in human and mouse cells. Mutat Res. 2000;470:147–153. doi: 10.1016/s1383-5718(00)00099-1. [DOI] [PubMed] [Google Scholar]
- 46.Ludewig G. Cancer Initiation by PCBs. In: Robertson LW, Hansen LG, editors. PCBs, Recent Advances in Environmental Toxicology and Health Effects. University Press of Kentucky; Lexington, KY: 2001. pp. 337–354. [Google Scholar]
- 47.Wang H, Mao Y, Chen AY, Zhou N, LaVoie EJ, Liu LF. Stimulation of topoisomerase II-mediated DNA damage via a mechanism involving protein thiolation. Biochemistry. 2001;40:3316–3323. doi: 10.1021/bi002786j. [DOI] [PubMed] [Google Scholar]
- 48.Bender RP, Lindsey RH, Jr, Burden DA, Osheroff N. N-acetyl-p-benzoquinone imine, the toxic metabolite of acetaminophen, is a topoisomerase II poison. Biochemistry. 2004;43:3731–3739. doi: 10.1021/bi036107r. [DOI] [PubMed] [Google Scholar]
- 49.Bender RP, Ham AJ, Osheroff N. Quinone-Induced Enhancement of DNA Cleavage by Human Topoisomerase IIalpha: Adduction of Cysteine Residues 392 and 405. Biochemistry. 2007 doi: 10.1021/bi062017l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J Biol Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 51.Elsea SH, Hsiung Y, Nitiss JL, Osheroff N. A yeast type II topoisomerase selected for resistance to quinolones. Mutation of histidine 1012 to tyrosine confers resistance to nonintercalative drugs but hypersensitivity to ellipticine. J Biol Chem. 1995;270:1913–1920. doi: 10.1074/jbc.270.4.1913. [DOI] [PubMed] [Google Scholar]
- 52.Kingma PS, Greider CA, Osheroff N. Spontaneous DNA lesions poison human topoisomerase IIα and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 53.Amaro AR, Oakley GG, Bauer U, Spielmann HP, Robertson LW. Metabolic activation of PCBs to quinones: reactivity toward nitrogen and sulfur nucleophiles and influence of superoxide dismutase. Chem Res Toxicol. 1996;9:623–629. doi: 10.1021/tx950117e. [DOI] [PubMed] [Google Scholar]
- 54.Wasserman RA, Austin CA, Fisher LM, Wang JC. Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase II alpha in yeast. Cancer Res. 1993;53:3591–3596. [PubMed] [Google Scholar]
- 55.Fortune JM, Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase IIα by blocking DNA cleavage. J Biol Chem. 1998;273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- 56.Fortune JM, Lavrukhin OV, Gurnon JR, Van Etten JL, Lloyd RS, Osheroff N. Topoisomerase II from Chlorella virus PBCV-1 has an exceptionally high DNA cleavage activity. J Biol Chem. 2001;276:24401–24408. doi: 10.1074/jbc.M101693200. [DOI] [PubMed] [Google Scholar]
- 57.Byl JA, Fortune JM, Burden DA, Nitiss JL, Utsugi T, Yamada Y, Osheroff N. DNA topoisomerases as targets for the anticancer drug TAS-103: primary cellular target and DNA cleavage enhancement. Biochemistry. 1999;38:15573–15579. doi: 10.1021/bi991791o. [DOI] [PubMed] [Google Scholar]
- 58.Vaughn J, Huang S, Wessel I, Sorensen TK, Hsieh T, Jensen LH, Jensen PB, Sehested M, Nitiss JL. Stability of the topoisomerase II closed clamp conformation may influence DNA-stimulated ATP hydrolysis. J Biol Chem. 2005;280:11920–11929. doi: 10.1074/jbc.M411841200. [DOI] [PubMed] [Google Scholar]
- 59.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci U S A. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang KC, Gao H, Yamasaki EF, Grabowski DR, Liu S, Shen LL, Chan KK, Ganapathi R, Snapka RM. Topoisomerase II poisoning by ICRF-193. J Biol Chem. 2001;276:44488–44494. doi: 10.1074/jbc.M104383200. [DOI] [PubMed] [Google Scholar]
- 61.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 62.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc Natl Acad Sci U S A. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hasinoff BB, Wu X, Krokhin OV, Ens W, Standing KG, Nitiss JL, Sivaram T, Giorgianni A, Yang S, Jiang Y, Yalowich JC. Biochemical and proteomics approaches to characterize topoisomerase IIalpha cysteines and DNA as targets responsible for cisplatin-induced inhibition of topoisomerase IIalpha. Mol Pharmacol. 2005;67:937–947. doi: 10.1124/mol.104.004416. [DOI] [PubMed] [Google Scholar]