Abstract
Background and objectives: Medullary sponge kidney (MSK) is a rare nephropathy characterized by cystic anomalies of precalyceal ducts, nephrocalcinosis, renal stones, and tubule dysfunctions. Its association with various malformations and cases of familial aggregation supports the conviction that genetic factors are involved, but no genetic studies have been conducted to date. It is hypothesized that MSK is due to a disruption at the “ureteric bud/metanephric blastema” interface caused by critical developmental genes functioning abnormally.
Design, setting, participants, & measurements: Fifty-five apparently sporadic MSK patients were analyzed by direct DNA sequencing of all exons and exon-intron boundaries of glial cell-derived neurotrophic factor (GDNF) gene and rearranged during transfection (RET) gene, which have a leading role in renal development.
Results: Two novel variants were found in heterozygosity in the MSK case population: GDNF{ENST00000344622}:c.−45G>C and c.−27+18G>A in a putative binding domain for paired-box 2 transcription factor. As a whole, eight patients showed these variations: four patients carried the c.[−45G>C; −27+18G>A] complex allele, and the others had the c.−27+18G>A alone. A case-control study revealed that these two alleles were significantly associated with MSK. Five of the eight cases were found to be familial, and the allele variants cosegregated with the disease in a seemingly dominant pattern of inheritance. Patients revealed no mutations in the RET gene.
Conclusions: This is the first report identifying GDNF gene sequence variations in patients with MSK and suggesting a role for this gene in the pathogenesis of some cases of the disease.
Medullary sponge kidney (MSK) is a rare nephropathy typically associated with nephrocalcinosis and renal stones, urinary acidification and concentration defects, and cystic anomalies in the precalyceal ducts. It is believed that it is a congenital disorder with late expression (1,2). MSK prevalence in the general population is uncertain, possibly 5 cases per 10,000 to 100,000 population. Approximately 3% to 5% of all renal stone formers have MSK, although much larger proportions (up to 20%) have been reported (3).
MSK has also been described in patients with various developmental disorders (e.g., congenital hemihypertrophy and Beckwith–Wiedemann syndrome) and in cases of renal developmental anomalies, including horse-shoe kidney, monolateral renal aplasia, contralateral congenital small kidney, and ureteral abnormalities (4–6). Although generally sporadic, familial cases have been reported seemingly with an autosomal dominant inheritance (7,8).
Although its association with malformations supports the idea that MSK is a developmental disorder (9), its pathogenesis is unknown, but it should explain the concomitant occurrence of alterations in the Wolffian duct-derived precalyceal and collecting ducts and in metanephric blastema-derived nephron, as well as the association of MSK with several malformative diseases. In 2000, a woman was described who had concomitant multiple endocrine neoplasia, MSK, and a RET proto-oncogene mutation; it was suggested that the rearranged during transfection (RET) gene mutation has a role in MSK (10).
RET and its ligand, the glial cell-derived neurotrophic factor (GDNF), are pivotal in renal development. Synthesis of GDNF by the metanephric blastema makes the ureteric bud branch away from Wolff's mesonephric duct to approach and invade the blastema. The tip of the bud expresses the GDNF receptor RET. The binding of RET-GDNF is essential not only for proper ureter and collecting duct formation, but also for the induction of nephrogenesis, morphogenesis, and kidney growth (11). In particular, the transition of the mesenchymal cells of the metanephros into nephronic cells, the proper polarization of the renal tubular cells, and the specialization of different tubular segments of the nephron all need differentiation “messages” originating from the “ureteric bud/metanephric blastema” interface (11,12).
In vivo experiments of GDNF or RET gene inactivation resulted in renal agenesis (13,14). On the other hand, animals heterozygous for GDNF or RET gene null allele develop unilateral renal agenesis or hypoplasia (13–15). Moreover, RET gene mutations were found in fetuses with bilateral or unilateral renal agenesis, and one of the latter cases also had a GDNF mutation (16).
In light of the above, we hypothesize that MSK is due to a disruption at the ureteric bud/metanephric blastema interface caused by the GDNF or RET genes functioning abnormally.
Materials and Methods
Patients and Controls
MSK patients followed up at the nephrology divisions of the university hospitals in Padova and Verona were renal stone formers whose MSK was diagnosed during the workup for recurrent calcium nephrolithiasis on the strength of typical findings at intravenous urography and the exclusion of other causes of nephrocalcinosis. To be eligible for a diagnosis of MSK, patients had to have both kidneys involved, with typical nephrocalcinosis and/or cystic features in at least two papillae in each kidney. For the diagnosis of MSK, it is mandatory to demonstrate papillary precalyceal ectasias on films taken at least 10 minutes after injecting the contrast medium, with no compression maneuvers and no signs of obstruction. An independent radiologist reviewed the x-ray films of these patients to confirm the diagnosis. Fifty-five patients were considered for the purposes of this study. They were all apparently sporadic cases. On average, the number of affected papillae per kidney was 5 ± 3. Patients with full-blown MSK (i.e., severe, bilateral, all-papillae nephrocalcinosis) were excluded from the analysis presented here because of uncertainty in distinguishing MSK from other primary conditions [e.g., complete distal renal tubular acidosis (dRTA)] associated with nephrocalcinosis. Renal ultrasound; kidney, ureter, bladder x-ray; or intravenous urography were also performed in family members of five patients selected to look for any signs of MSK.
After obtaining informed written consent, venous blood was drawn for DNA studies. The local ethical committee approved the study.
Two different control populations were considered: 125 cord blood samples available in Padova and 85 blood samples from adult patients with idiopathic calcium nephrolithiasis (ICN) matched for age, gender, and geographical origin with the MSK patients. To be defined as ICN patients, the 85 control patients had to have no endocrine or other disorders in addition to calcium stone disease; normal serum creatinine and electrolyte concentrations; and no evidence of cystic kidney disorders, nephrocalcinosis, or obstructive nephropathy. The urinary pH measured in spot morning urine samples after overnight fasting had to be ≤5.5 to rule out tubular acidosis. All of these individuals were carefully examined for any signs of MSK, and it was ruled out.
Mutation Analysis
The coding and noncoding sequences of the RET and GDNF genes were analyzed in the MSK patients. Information on the GDNF and RET primers and PCR protocols used in this study are available upon request.
Genomic DNA was extracted from peripheral blood according to QIAamp DNA Blood Mini Kit standard procedures (QIAGEN, Valencia, CA). The BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) was used for direct PCR product sequencing. The purified amplicons were analyzed on a 3130 Genetic Analyzer (Applied Biosystems). The sequence variations identified were confirmed by two independent PCR reactions and sequencing both DNA strands and have been named according to the recent nomenclature for the description of human sequence variants (17) in relation to a cDNA reference sequence from the Ensembl Human Genome Browser (i.e., GDNF{ENST00000344622}). The genomic reference sequence is the Ensembl GDNF gene sequence ENSG00000168621. DNA samples from controls were analyzed for revealing the GDNF variations identified in patients.
Clinicobiochemical Characterization
The eight GDNF variant-positive and eight GDNF variant-negative, all unrelated, MSK patients that were one-to-one gender and age paired-matched underwent clinicobiochemical phenotyping. The patients were all administered a questionnaire on their personal and family history, particularly seeking information on urinary tract, glandular, and bone disorders; hyperuricemia and gout; cardiovascular and liver diseases; intestinal disturbances; nutritional habits; and any use of drugs affecting mineral and electrolyte metabolism. None of the women were in menopause. A comprehensive blood and urine biochemical characterization was performed twice in all patients, considering the means of the two values, and bone densitometry on dual-energy x-ray absorptiometry was always done at the Verona department to reduce variability. Hemogas analysis was performed in patients with a morning urine pH ≥5.5, and/or hypokalemia (≤3.5 mEq/L), and/or hypocitraturia to rule out classic dRTA.
GDNF Expression Analysis
Individual I.2 in pedigree C (Figure 1)—an 80-yr-old woman with MSK ultimately found heterozygous for a GDNF variant—revealed a renal carcinoma and underwent open surgery for nephrectomy. A few papillae far away from the tumor were dissected from the healthy part of the kidney immediately after its removal. For control purposes, papillae were also obtained from three patients, approximately the same age as individual I.2, undergoing nephrectomy for renal cancer who had never had renal stones or other renal disease (as confirmed by histopathological examination of the healthy part of their kidneys).
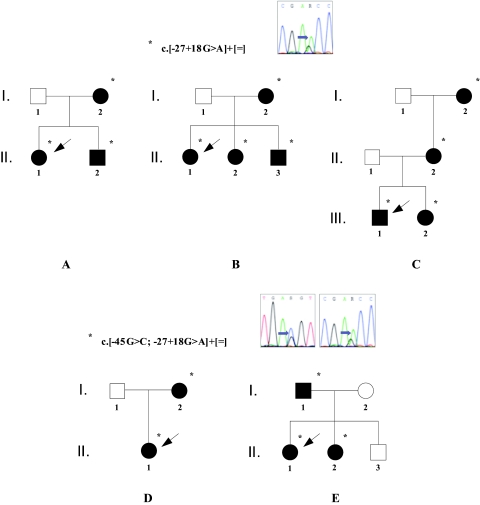
Figure 1.
Pedigrees of five MSK families with GDNF sequence variations, showing how the altered alleles cosegregate with the disease. Filled symbols identify individuals with MSK. Asterisks indicate heterozygous carriers of the variants (the simple allele with the c.−27+18G>A variant alone in families A, B, and C, and the complex allele comprising the c.−45G>C and the c.−27+18G>A sequence variations in families D and E). The arrows highlight the probands. Automated GDNF gene sequencing shows the nucleotide substitutions, identified by the arrow.
Total RNA was extracted from tissue fragments using the RNeasy Micro Kit (QIAGEN). A quantity of 200 ng of total RNA was retrotranscribed as reported (18). Because it was not possible to design a suitable couple of real-time PCR primers inside of the common region of the GDNF transcripts, quantitative comparative reverse transcriptase PCR was used to study GDNF gene expression (18). Data were normalized to the housekeeping 18S RNA subunit, with four repeats per experimental group. Information on the primers used to amplify the cDNAs and PCR protocols are available upon request. To obtain quantitative data, the number of PCR cycles was always selected within the exponential phase according to the kinetic strategy described elsewhere (18).
Statistical Analysis
The distributions of the sequence variations identified between MSK cases and controls were compared using Fisher's exact test. GDNF expression levels were expressed as mean ± SEM and analyzed using a t test. Patients' clinicobiochemical phenotypes were analyzed using a t test. Statistical significance was assumed when P < 0.05.
Results
Mutation Analysis
Two novel single nucleotide substitutions were found in heterozygosity by direct sequencing of the GDNF gene PCR products of the MSK case population as a whole (i.e., GDNF{ENST00000344622}:c.−45G>C in the 5′UTR exon 1 of the transcript and GDNF{ENST00000344622}:c.−27+18G>A in the genomic region downstream of this 5′UTR exon 1).
A complex allele consisting of the novel variations and a simple allele comprising the c.−27+18G>A variant alone were each identified in four patients. The allele frequency in the Veneto population was investigated in 250 consecutive cord blood sample chromosomes and was 0.008 for both, thus demonstrating these variants are rare. Instead, 170 blood sample chromosomes from adult patients with ICN were examined for the presence of the variants to estimate a case-control association. None of these controls showed them. There was a statistically significant difference in allele frequency between these adult controls and the MSK patients (P = 0.023).
For five of eight patients, mutational screening was performed in the parents to investigate the origin of the sequence variations and it was confirmed that the alleles were inherited from an apparently healthy heterozygous mother or father. A familial occurrence of MSK was surprisingly revealed by accurate clinical and, in all patients, radiologic investigation. In fact, five relatives had had renal colics before; two of these had passed one stone each. In one relative, microhematuria was disclosed. In the remaining three subjects, only the radiologic investigation disclosed MSK. Direct DNA sequencing of all members of the families showed the GDNF alleles cosegregating with the disease in a seemingly autosomal dominant pattern (Figure 1).
We analyzed the RET gene in MSK patients, but no disease-causing mutations or unknown variants were detected.
Clinicobiochemical Characterization
All subjects had an estimated creatinine clearance (Cockcroft–Gault formula) >80 ml/min/1.73 m2. No evident abnormality in renal size in patients with the GDNF variants was observed. The questionnaire revealed no particular differences between the two groups of MSK patients. None of them had classic dRTA or hyperparathyroidism. Table 1 shows the results of urine and blood tests and bone densitometry. GDNF variant-positive patients had marginally lower blood potassium concentration and higher fasting spot urine pH levels. They also had markedly higher urinary calcium and lower citraturia, both outside of the normal range. Bone densitometry disclosed a significantly reduced T-score in the lumbar spine.
Table 1.
Clinicobiochemical phenotype of MSK patients positive or negative for GDNF variants
| Parameters | GDNF Variant + | GDNF Variant − |
|---|---|---|
| 24-hour urine | ||
| pH (fasting, morning sample) | 5.92 ± 0.27 | 5.61 ± 0.67 |
| potassium (mEq) | 58.22 ± 18.78a | 61.64 ± 23.00 |
| calcium (mg) | 315.00 ± 38.42a | 184.90 ± 54.38 |
| phosphate (mg) | 920.73 ± 59.12 | 810.87 ± 68.38 |
| citrate (mg) | 238.44 ± 77.05a | 331.90 ± 81.44 |
| Blood | ||
| calcium (mg/dl) | 9.23 ± 0.49 | 9.63 ± 0.46 |
| phosphorus (mg/dl) | 3.08 ± 0.65 | 3.23 ± 0.58 |
| potassium (mEq/L) | 3.50 ± 0.41 | 3.92 ± 0.55 |
| chloride (mEq/L) | 105.8 ± 2.89 | 104.6 ± 3.44 |
| Bone serum alkaline phosphatase (IU/L) | 23.92 ± 5.39 | 30.30 ± 6.03 |
| Total vertebral (L1 to L4) T-score | −2.04 ± 0.20a | −1.13 ± 0.26 |
Statistically significant differences versus GDNF variant-negative values.
GDNF Expression Analysis
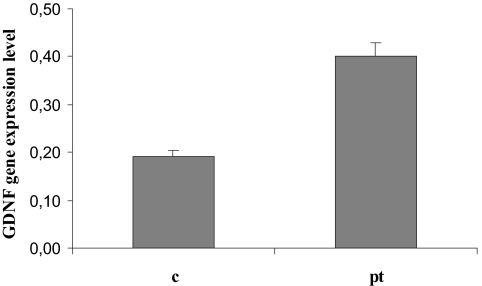
Adult kidney mRNA samples from a patient with MSK carrying the c.−27+18G>A GDNF gene variant and three control samples were analyzed by reverse transcriptase PCR. The GDNF expression level was significantly higher in the MSK patient's kidney (P = 0.01) (Figure 2).
Figure 2.
GDNF expression analysis using reverse transcriptase PCR in a fragment of renal papillary tissue from a patient with MSK (pt), ultimately found heterozygous for the c.−27+18G>A variant, and in papillary specimens from control individuals who had never had renal stones or any other renal disease (c). Relative gene expression was calculated as the ratio between the quantity (ng/μl) of the PCR product of the target gene and of the housekeeping gene. The result represents a total of four reverse transcriptase PCR reactions from each RNA sample.
Discussion
This study investigates the possibility of a genetic basis for MSK that generally appears as a sporadic condition but has also occasionally been observed in families with an apparent autosomal dominant inheritance. We have enrolled the biggest MSK population on a national level and conducted a mutation analysis on the genes that we identified as likely candidates for the disease's etiology. Two GDNF sequence variations never described before in heterozygosity were identified in eight patients. Variant frequency was investigated in consecutive cord blood sample chromosomes, but we could not be sure these individuals were really healthy rather than affected with MSK with clinical manifestations presenting late. Thus, a true estimate of variant-phenotype association was carried out using a more carefully selected adult control population (i.e., patients with ICN, chosen according to definite criteria that clearly excluded patients with MSK). The resulting score was significant, thus pointing to a likely role for these sequence variations in the onset of the disease.
In fact, we demonstrated that the GDNF variants were inherited from one of the parents in five of the eight MSK patients and that all of the family members of these pedigrees, who were ultimately found heterozygous for these variants, had MSK. This also confirmed the dominant single trait hypothesis suggested by pedigree analyses of the few familial cases that have been reported in literature. A founder effect for this disorder in the Veneto region should be considered, according to which the sequence variants should also be inherited in the other three patients carrying an altered GDNF allele.
According to our findings, we hypothesize that an underestimation of familial cases of MSK might exist, and that the prevalence of the MSK condition in the general population might be higher than previously estimated.
Sequence variations in noncoding regions usually cause quantitative alterations in gene expression product. To estimate the c.−27+18G>A GDNF variant's ability to alter transcription, we had the exceptional chance to collect renal tissue from a patient suffering from MSK and carrying this variant. Such a variation might be the most likely culprit responsible for MSK because it was the only one that was always identified in the altered alleles. We found the GDNF gene expression in the patient's papillary tissue significantly higher than in controls. This finding is consistent with a regulatory quantitative genetic effect of the GDNF variant and with our hypothesis that a GDNF gene dysregulation is responsible for MSK.
GDNF expression is high in the developing kidney (19), but few studies have addressed the role of the GDNF in human pathology. In agreement with our findings, these studies indicate that GDNF expression is very low in the adult human kidney (20) and that GDNF gene reactivation may occur in the tubules in dysplastic/cystic renal disorders (21,22).
Because it was not possible to obtain a renal tissue specimen from a patient affected with MSK and carrying the other novel GDNF variant identified, we used an in silico approach utilizing the Automated Splice Site Analyses, the ESEfinder, and RESCUE-ESE web interfaces to evaluate the transcriptional consequences of this variation. A new cryptic splice site created by the c.−45G>C substitution, associated with an increase of 6 times in the percentage of splicing factor recruitment, was predicted by all three software programs. This change might lead to an aberrantly spliced transcript. The c.−45G>C allele variant was never found alone in controls or patients: we suggest that it might represent a crucial deleterious mutation that has no lethal phenotypic effect when associated with the c.−27+18G>A sequence variation. Alternatively, the c.−27+18G>A substitution might be an ancestral mutation in disease-bearing chromosomes on which the functional variant arises. The bioinformatic approach needs to be validated using experimental methods; nevertheless, it suggests that the c.−45G>C sequence variation is also not neutral.
It is noteworthy that both novel GDNF variants are located in a putative binding domain for paired-box 2 transcription factor (PAX2) that is known to have a major role in nephrogenesis (23,24), thus supporting the importance of this region in gene regulation. An altered GDNF response to PAX2 might affect interaction with RET, disrupting the differentiation “messages” originating from the interface between the ureteric bud and the metanephric blastema. Because this step is fundamental to nephrogenesis, renal tubular cell polarization might be altered, leading to a mistargeting of carriers involved in acidification and ion handling and consequently to functional abnormalities. The suggestion that the GDNF sequence variations exert their influence on the disease is supported by the finding of a specific clinicobiochemical phenotype in patients carrying them in comparison with patients without the GDNF variants (i.e., a more severely disturbed distal tubular acidification and a strongly negative calcium balance). Both of these conditions are very common in MSK patients but are generally subclinical (3,9). They were definitely worse in the patients positive for GDNF variants, as demonstrated by their higher urinary pH, lower serum potassium levels, subnormal citraturia, higher calciuria, and, most importantly, more severe osteopenia.
In this study, characteristics and limitations should be mentioned. First, our study individuals came from a well defined geographical area, thus eliminating the possibility of confounding factors due to population stratification, but, on the other hand, limiting the possibility to perform crosspopulation comparison. In addition, the number of cases and controls is low, but both groups were accurately selected on the basis of specific clinical and biochemical investigations.
We suggest that our genetic findings may help to clarify the pathogenesis of MSK, and that the analysis of other genes belonging to the same developmental pathway in MSK patients might help to confirm this genetic hypothesis. In conclusion, we report here on the discovery of two novel GDNF gene variants that may have a role in the pathogenesis of MSK. The data suggest that there is a subset of MSK patients that acquires this disease as a genetic developmental disorder.
Disclosures
None.
Acknowledgments
We sincerely thank the patients and their families for participation. We thank Monica Ceol and Emilia Tiralongo for technical support. This study was supported by grants 2002061783_002 and 2006062453_002 from the Italian Ministry of Education, University, and Research.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1..Kasap B, Soylu A, Oren O, Türkmen M, Kavukçu S: Medullary sponge kidney associated with distal renal tubular acidosis in a 5-year-old girl. Eur J Pediatr 165: 648–651, 2006 [DOI] [PubMed] [Google Scholar]
- 2..Gupta S, Shanbag P, Vaidya M: Medullary sponge kidney. Indian J Pediatr 69: 1091–1092, 2002 [DOI] [PubMed] [Google Scholar]
- 3..Cameron S: Medullary sponge kidney. In: Oxford Textbook of Clinical Nephrology, 3rd Ed., edited by Davison AM, Cameron JS, Grunfeld J-P, Ponticelli C, Ritz E, Winearls CG, van Ypersele C.Oxford University Press, Oxford, United Kingdom, 2004, pp 2495–2501 [Google Scholar]
- 4..Lambrianides AL, John DR: Medullary sponge disease in horseshoe kidney. Urology 29: 426–427, 1987 [DOI] [PubMed] [Google Scholar]
- 5..Gambaro G, Fabris A, Citron L, Tosetto E, Anglani F, Bellan F, Conte M, Bonfante L, Lupo A, D'Angelo A: An unusual association of contralateral congenital small kidney, reduced renal function and hyperparathyroidism in sponge kidney patients: On the track of the molecular basis. Nephrol Dial Transplant 20: 1042–1047, 2005 [DOI] [PubMed] [Google Scholar]
- 6..Iwig J, Thiemann KJ: Medullary sponge kidney with megapyelon and primary congenital megaureter in unilateral renal aplasia. Fortschr Geb Rontgenstr Nuklearmed 111: 137–140, 1969 [PubMed] [Google Scholar]
- 7..Copping GA: Medullary sponge kidney: Its occurrence in a father and daughter. CMAJ 96: 608–611, 1967 [PMC free article] [PubMed] [Google Scholar]
- 8..Kuiper JJ: Medullary sponge kidney in three generations. NY State J Med 71: 2665–2669, 1971 [PubMed] [Google Scholar]
- 9..Gambaro G, Feltrin GP, Lupo A, Bonfante L, D'Angelo A, Antonello A: Medullary sponge kidney (Lenarduzzi-Cacchi-Ricci disease): A Padua Medical School discovery in the 1930s. Kidney Int 69: 663–670, 2006 [DOI] [PubMed] [Google Scholar]
- 10..Diouf B, Ka EH, Calender A, Giraud S, Diop TM: Association of medullary sponge kidney disease and multiple endocrine neoplasia type IIA due to RET gene mutation: Is there a causal relationship?. Nephrol Dial Transplant 15: 2062–2066, 2000 [DOI] [PubMed] [Google Scholar]
- 11..Schedl A, Hastie ND: Cross-talk in kidney development. Curr Opin Genet Develop 10: 543–549, 2000 [DOI] [PubMed] [Google Scholar]
- 12..Huber SM, Braun GS, Segerer S, Veh RW, Horster MF: Metanephrogenic mesenchyme-to epithelium transition induces profound expression changes of ion channels. Am J Renal Physiol 279: F65–F76, 2000 [DOI] [PubMed] [Google Scholar]
- 13..Sánchez MP, Silos-Santiago I, Frisén J, He B, Lira SA, Barbacid M: Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382: 70–73, 1996 [DOI] [PubMed] [Google Scholar]
- 14..Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, Pachnis V: Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor RET. Nature 367: 380–383, 1994 [DOI] [PubMed] [Google Scholar]
- 15..Manie S, Santoro M, Fusco A, Billaud M: The RET receptor: Function in development and dysfunction in congenital malformation. Trends Genet 17: 580–589, 2001 [DOI] [PubMed] [Google Scholar]
- 16..Skinner MA, Safford SD, Reeves JG, Jackson ME, Freemerman AJ: Renal aplasia in humans is associated with RET mutations. Am J Hum Genet 82: 344–351, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17..den Dunnen JT, Antonarakis SE: Nomenclature for the description of human sequence variations. Hum Genet 109: 121–124, 2001 [DOI] [PubMed] [Google Scholar]
- 18..Ceol M, Forino M, Gambaro G, Sauer U, Schleicher ED, D'Angelo A, Anglani F: Quantitation of TGF-beta1 mRNA in porcine mesangial cells by comparative kinetic RT/PCR: comparison with ribonuclease protection assay and in situ hybridization. J Clin Lab Anal 15: 215–222, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19..Suter-Crazzolara C, Unsicker K: GDNF is expressed in two forms in many tissues outside the CNS. Neuroreport 5: 2486–2488, 1994 [DOI] [PubMed] [Google Scholar]
- 20..Orth SR, Ritz E, Suter-Crazzolara C: Glial cell line-derived neurotrophic factor (GDNF) is expressed in the human kidney and is a growth factor for human mesangial cells. Nephrol Dial Transplant 15: 589–595, 2000 [DOI] [PubMed] [Google Scholar]
- 21..El-Ghoneimi A, Berrebi D, Levacher B, Nepote V, Infante M, Paris R, Simonneau M, Aigrain Y, Peuchmaur M: Glial cell line derived neurotrophic factor is expressed by epithelia of human renal dysplasia. J Urol 168: 2624–2628, 2002 [DOI] [PubMed] [Google Scholar]
- 22..Lee DC, Chan KW, Chan SY: RET receptor tyrosine kinase isoforms in kidney function and disease. Oncogene 21: 5582–5592, 2002 [DOI] [PubMed] [Google Scholar]
- 23..Brophy PD, Ostrom L, Lang KM, Dressler GR: Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development 128: 4747–4756, 2001 [DOI] [PubMed] [Google Scholar]
- 24..Zhang Z, Quinlan J, Grote D, Lemire M, Hudson T, Benjamin A, Roy A, Pascuet E, Goodyer M, Raju C, Houghton F, Bouchard M, Goodyer P: Common variants of the glial cell-derived neurotrophic factor gene do not influence kidney size of the healthy newborn. Pediatr Nephrol 24: 1151–1157, 2009 [DOI] [PubMed] [Google Scholar]