Abstract
Objective
To document the activity profile of transcription factors following chondrocyte stimulation with hyaluronan (HA) hexasaccharides (HA6) and to determine the expression of genes whose transcriptional activation is tightly associated with the transcription factors.
Methods
Nuclear extracts from bovine articular chondrocytes treated with HA6 were subjected to transcription factor protein–DNA array analysis. Electrophoretic mobility shift assay (EMSA) analyses were performed to confirm the results of protein–DNA array. The gene expressions of matrix metalloproteinase 3 (MMP-3), type II collagen, and cartilage oligomeric matrix protein (COMP) were examined by quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR), and protease activity was assessed by casein zymography.
Results
In the protein–DNA array analysis, 12 transcription factors were up-regulated and 2 transcription factors were down-regulated in the chondrocytes treated with HA6. The transcription factors retinoic acid receptor (RAR), retinoid X receptor (RXR), and Sp-1 exhibited >2-fold increased activity by HA6 treatment, as confirmed by EMSA. RT-PCR analysis showed that the expression levels of MMP-3, type II collagen, and COMP messenger RNA, which are tightly associated with the activation of RAR, RXR, or Sp-1, were up-regulated by treatment with HA6. Addition of high molecular mass HA after HA6 treatment resulted in abrogation of the MMP-3 induction.
Conclusion
These results suggest that HA6 increase the activity of multiple transcription factors in chondrocytes and signal the enhanced expression of key genes involved in cartilage-matrix remodeling and turnover. The data also demonstrate that high molecular mass HA has a potential to suppress the signaling activated by HA6.
The glycosaminoglycan hyaluronan (HA) is a critical component of the extracellular matrix of articular cartilage. HA serves as a scaffold for the aggregation of cartilage proteoglycan (1) and, in this manner, forms a continuum of structure throughout the extracellular matrix. In addition, we have shown that HA facilitates the anchorage of these proteoglycan aggregates to the chondrocyte cell surface via its interaction with the membrane HA receptor CD44 (2–4). Thus, it is, in part, the interaction of chondrocytes directly with HA that constitutes the cell–matrix interaction that serves to regulate many aspects of chondrocyte metabolism (5). We have shown previously that one method of interference with the cell–matrix interactions of chondrocytes is through the use of excess small HA oligosaccharides, such as HA hexasaccharides (HA6) (2,4,6). HA6 compete with high molecular mass HA for CD44 binding; yet, they are too small in size to interfere with HA aggrecan or HA–link protein interactions (1,7,8).
The consequences of this loss of cell–matrix interactions can be dramatic. In a previous study, we demonstrated that the addition of small HA oligosaccharides to human or bovine articular cartilage slices resulted in the apparent activation of enhanced catabolism, including elevated release and activity of matrix metalloproteinases (MMPs), increases in aggrecanase neoepitope expression, and concomitant loss of proteoglycan from the tissue (6). This enhanced catabolic state mimicked experimental observations in which cartilage slices were incubated in the presence of interleukin-1 (IL-1) (9,10). However, unlike IL-1, chondrocytes treated with HA6 displayed an increase in aggrecan expression (6). These results suggest that whatever mechanism was responsible for the activation, the pathway differed significantly from pathways activated by IL-1. A similar catabolic response was also obtained in bovine articular cartilage slices when CD44 expression was reduced through the use of small phosphorothioate antisense oligonucleotides (11). Thus, it would appear that interference with cell–matrix interactions, by both perturbation of ligand binding and receptor-protein biosynthesis lead to similar changes in chondrocyte metabolism.
In recent years, HA has been used clinically, following injection into the intraarticular space of osteoarthritic joints, for the purpose of viscosupplementation. In addition to examining the possible biomechanical effects of viscosupplementation, investigators are attempting to determine whether there are chondroprotective effects on the cartilage itself and, if so, the mechanism for this protection. For example, in vitro experiments showed that high molecular mass HA inhibits the cartilage-matrix degradation caused by inflammatory cytokines (12,13). In contrast, the molecular mass of HA in cartilage is known to decrease with age (14), as does the average molecular mass of HA in the synovial fluid of patients with osteoarthritis (OA) (15). Therefore, there is at least a possibility that low molecular mass HA oligosaccharides are present and available to affect the metabolic state of articular cartilage chondrocytes.
The goal of this study was to use HA oligosaccharides as a model for changes in chondrocyte cell–matrix dysfunction and determine, in part, some of the underlying molecular mechanisms that become activated and give rise to the metabolic changes observed. Eukaryotic gene expression is regulated by protein transcription factors, which determine the frequency of transcriptional initiation by interacting with specific DNA binding sequences present in promoter/enhancer regions. The expression and activation of transcription factors are strictly regulated by various factors such as cytokines, growth factors, mechanical stress, cell–cell interactions, and cell–matrix interactions. Recently, a new technology, the protein–DNA array system, has been developed to analyze the activity profile of multiple transcription factors from a nuclear extract in a single assay. Using this system in combination with electrophoretic mobility shift assay (EMSA) analysis, we explored the transcriptional activation exerted by the stimulation of bovine articular chondrocytes with HA6. In addition, we examined the expression of genes whose transcriptional activation is tightly associated with the transcription factors found to be altered following HA6 treatment.
MATERIALS AND METHODS
Cell culture
Metacarpophalangeal joints from 18-month-old steers were obtained from a local slaughterhouse. Human articular cartilage was obtained from donors within 24 hours of death, through the Gift of Hope Organ and Tissue Donor Network (formerly, the Regional Organ Bank of Illinois). Articular cartilage was dissected from the talocrural ankle joint of donors, as previously described (16). To isolate chondrocytes, full-thickness slices of human or bovine noncalcified articular cartilage were subjected to sequential pronase/collagenase digestion (Calbiochem, San Diego, CA and Boehringer Mannheim, Indianapolis, IN, respectively). The immortalized human chondrocyte C-28/I2 cells were kindly provided by Dr. Mary Goldring (Harvard Institutes of Medicine, Boston, MA). The primary or immortalized chondrocytes were cultured as high-density monolayer (1 × 106 cells/100 mm2) in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Grand Island, NY) supplemented with 50 units/ml penicillin and 50 μg/ml streptomycin containing 10% fetal bovine serum (Summit Biotechnology, Fort Collins, CO) at 37°C in an atmosphere containing 5% CO2.
Application of HA hexasaccharides and HA
After gradual serum starvation with reduction of the concentration to 1% for 12 hours, and to 0% for 12 hours on day 2, the chondrocytes were treated with HA6 (Seikagaku, Tokyo, Japan) at 0–500 μg/ml for 0–48 hours. In some experiments, the cultures were washed excessively with DMEM after a 12-hour treatment with HA6, and then incubated in fresh media with or without 500 μg/ml high molecular mass HA (~1.9–3.9 × 106 daltons Healon from Pharmacia & Upjohn, Peapack, NJ, or ~5.0–7.3 × 105 daltons Hyalgan from Sanofi-Synthelabo, New York, NY).
Preparation of nuclear extract
Nuclear extracts were obtained from control and experimental cultures of bovine chondrocytes according to the instructions of the manufacturer, using the Nuclear Extraction kit (Panomics, Redwood, CA).
Protein–DNA array
Twenty-five micrograms of nuclear extract from control and experimental cultures was mixed with TransSignal probe mix (Panomics) containing a set of biotin-labeled DNA-binding oligonucleotides corresponding to the consensus sequences of 54 transcription factors, to allow the formation of protein–DNA complexes. Only protein–DNA complexes were isolated from the mixture, by agarose gel electrophoresis. The DNA was then separated from the protein, and finally, the separated DNA was hybridized to a TransSignal array (Panomics), which was spotted with consensus-binding sequences of 54 known transcription factors (see Figure 1). Detection was performed on chemiluminescence film (Hyperfilm ECL; Amersham Biosciences, Piscataway, NJ), based on a streptavidin–horseradish peroxidase (HRP) reaction. The films were scanned and the dot intensity (4 dots per transcription factor) was evaluated using Quantity One imaging software (Bio-Rad, Hercules, CA). The magnitude of the difference in the dot intensity on the membrane was calculated relative to that on control membrane. The experiment was performed twice using different nuclear extracts. Dot intensity was analyzed using Quantity One software (Bio-Rad).
Figure 1.
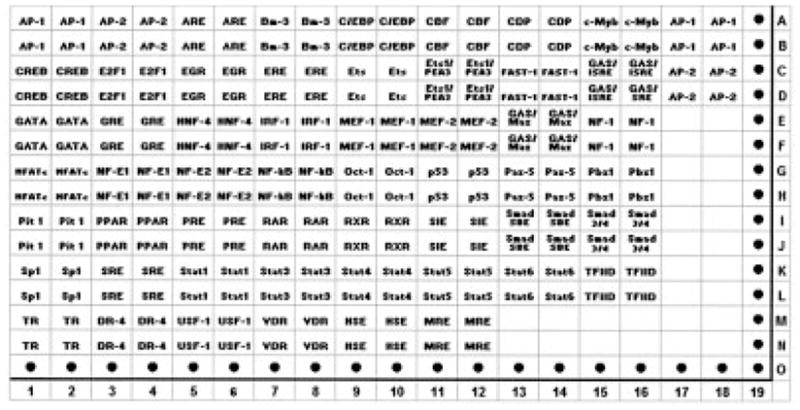
Schematic diagram of TransSignal protein–DNA array. The genes on the array are spotted in duplicate (the first row of each is DNA spotted at a standard concentration and the second row of each is DNA diluted 1:10).
EMSA analysis
Twenty-five micrograms of nuclear extracts from control or experimental cultures was mixed with biotin-labeled transcription factor probes (EMSA kit; Panomics) and incubated at 15°C for 30 minutes. Excess unlabeled double-stranded DNA was added for DNA–protein binding competition assay during this incubation time. The mixture was loaded onto a 6% polyacrylamide gel and electrophoresed at 120V in 0.5% Tris–borate–EDTA (TBE) for 1 hour. The sample was transferred in 0.5% TBE onto a nylon membrane at 300 mA for 30 minutes. After transfer, the sample was fixed on the membrane by ultraviolet crosslinking for 3 minutes (120,000 μJ). The bands were visualized after exposure to chemiluminescence film based on the streptavidin–HRP reaction. The reactions were analyzed in duplicate and performed twice using 2 separate nuclear extracts. Band intensities were quantified using Quantity One software (Bio-Rad). Statistical analyses were performed by Mann-Whitney U test to examine the differences in band intensity (mean ± SD pixel density from quadruplicate determinations) between control and experimental lanes.
Real-time reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was isolated from the bovine chondrocyte cultures according to the instructions of the manufacturer, using TRIzol reagent (Gibco BRL, Gaithersburg, MD), and was reverse transcribed with the GeneAmp RNA PCR kit (Perkin-Elmer, Roche Molecular Systems, Brachburg, NJ) with the PTC-100 Programmable Thermal Controller (MJ Research, Waltham, MA).
For real-time RT-PCR, the PCR products were detected by SYBR Green I Nucleic Acid gel stain (Molecular Probes, Eugene, OR). The primer-specific amplification and quantification cycles were run at the following temperatures: for GAPDH, amplification 57°C and quantification 74°C; for MMP-3, amplification 60°C and quantification 86°C; for type II collagen, amplification 60°C and quantification 74°C; and for cartilage oligomeric matrix protein (COMP), amplification 62°C and quantification 74°C. The quantification temperatures were set below the individual melting peak of each PCR product. The PCR primer sets were as follows: GAPDH, forward GTC-AAC-GGA-TTT-GGT-CGT-ATT-GGG, reverse TGC-CAT-GGG-TGG-AAT-CAT-ATT-GG; type II collagen, forward GCC-CTA-TGT-CCA-CAC-CGA-ATT, reverse ATG-ACA-ATC-TGG-CTC-CCA-AC; COMP, forward CAA-CTC-CGT-GTG-CAT-CAA-CAC-CC, reverse CAG-TCA-CGC-AGT-TGT-CCT-TAC-GGC; and MMP-3, forward CTC-ACA-GAC-CTG-ACT-CGG-TT, reverse CAC-GCC-TGA-AGG-AAG-AGA-TG.
Thermal cycling and fluorescence detection were performed in the Smart Cycler system (Cepheid, Sunnyvale, CA). Efficiency of the real-time PCR was calculated according to the equation provided by Rasmussen et al (17), as follows: E = 10[−1/slope] (for GAPDH, MMP-3, type II collagen, and COMP). The slope was determined from the graph depicting input of complementary DNA (in nanograms) (x axis) and cycle number at the crossing point (CP) (y axis). The CP is the PCR cycle number, which is the peak of the second derivative curve. The magnitude of the increase is calculated as a relative ratio of target gene (e.g., MMP-3) to GAPDH, in accordance with the new mathematical model and Equation 1 introduced by Pfaffl (18), as follows:
Promoter analysis for activator protein 1 (AP-1) in human immortalized chondrocytes
Human immortalized chondrocytes (C-28/I2) were plated 24 hours prior to transfection at a density of 1 × 106 cells/well in 6-well plates and transiently transfected with 2 μg of AP-1 promoter-reporter construct (pAP-1-Luc; Clontech, Palo Alto, CA [19]) using FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN) following the manufacturer’s instructions. The firefly luciferase promoterless vector, pGL2-Enhancer (Promega, Madison, WI), was used as a control. After 24 hours’ incubation, the cells were rinsed in phosphate buffered saline (PBS) and changed to serum-free conditions for 20–24 hours, followed by treatment with 250 μg/ml HA6 or 1 ng/ml IL-1β (R&D Systems, Minneapolis, MN). The cells were harvested after 24 hours, and luciferase activity was assayed using a luciferase reporter assay system (Promega). The data were normalized to the total protein levels in each well.
Western blot for IL-1 in human primary chondrocytes
Conditioned media (protein content 15 μg) from human primary chondrocyte cultures that were treated with HA6 for 24 hours were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electro-blotted onto nitrocellulose membrane. After blocking with 5% bovine serum albumin, the membrane was incubated with mouse anti-human IL-1β monoclonal antibody (clone CZD; Biological Resources Branch, National Cancer Institute, Frederick, MD). The membrane was washed extensively in PBS–Tween (0.2%) and incubated with anti-mouse biotinylated secondary antibody. The HRP activity was detected on radiographic film using the enhanced chemiluminescence system.
Casein zymography
Serum-free conditioned medium of bovine articular chondrocytes treated with or without HA6 for 24 hours was assayed for protease activity using casein zymography (20). Ten times–concentrated samples (30 μl) were separated by 10% SDS-PAGE containing casein (0.5 mg/ml; Sigma, St. Louis, MO). The gels were washed twice in 2.5% Triton X-100 in 50 mM Tris HCl, 5 mM CaCl2, 1 μM ZnCl2 (pH 7.6), for 30 minutes each, and then rinsed in the same buffer without Triton X-100 for 20 minutes. The gels were incubated overnight at 37°C with 50 mM Tris HCl (pH 7.6), 5 mM CaCl2, 1 μM ZnCl2, 1 mM aminophenylmercuric acetate (APMA), 1% Triton X-100, and 0.02% NaN3. Staining was performed for 1 hour at room temperature with 1% Coomassie brilliant blue in 50% methanol and 25% acetic acid, and destaining was performed with 30% methanol and 1% formic acid until clear bands over a dark background were observed.
RESULTS
Protein–DNA array findings
The particular protein–DNA array that we used documents the activation state of 52 of the most common nuclear transcription factors (Figure 1). Bovine articular chondrocytes were incubated without (Figure 2A) or with (Figure 2B) an optimal concentration of HA6 for 6 hours, and the nuclear extracts were isolated and analyzed for activated transcription factors within that compartment. The first observation to note concerning the array analysis is that, in general, the pixel intensities for most (38 of 52) of the transcription factors were below detectable limits or were unchanged between control and HA6-treated chondrocytes. Nonetheless, 12 transcription factors were up-regulated and 2 were down-regulated in the chondrocytes treated with HA6 (Table 1). The transcription factors RAR, RXR, and Sp-1 were increased more than 2-fold (Table 1).
Figure 2.
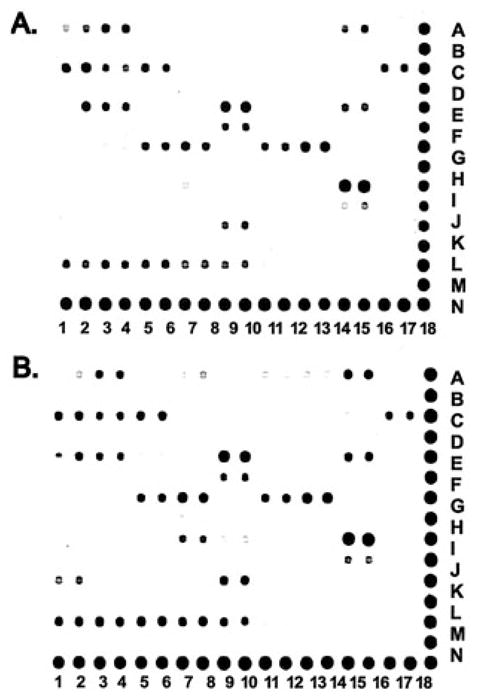
Transcription factor protein–DNA array analysis results obtained from bovine chondrocytes without (A) or with (B) treatment with hyaluronan hexasaccharides.
Table 1.
Summary of transcription factors whose activity was reduced or enhanced following exposure of bovine articular chondrocytes to hyaluronan hexasaccharides (HA6)
| Fold difference* | Transcription factors |
|---|---|
| 0.5–0.9† | CREB, CRE |
| 1.5–2.0 | BM3, CBF, CDF, cMyb, HSE, MRE, NF-κB, Pbx1, VDR (DR3) |
| >2.0 | RAR (DR5), RXR (DR1), Sp-1 |
Values are the fold difference in pixel intensity of hybridization dots derived from nuclear extracts of HA6-treated cells versus untreated control chondrocytes. The activity of the remaining 38 transcription factors (see Figure 1) was unchanged by treatment.
Indicative of inhibition.
EMSA findings
To confirm the results of the protein–DNA array, EMSA analyses of the transcription factors retinoic acid receptor (RAR), retinoid X receptor (RXR), and Sp-1 were performed. As shown in Figure 3, EMSA demonstrated enhanced activation of RAR (Figure 3A), RXR (Figure 3B), and Sp-1 (Figure 3C) present in nuclei of chondrocytes treated with HA6. Treatment of chondrocytes with HA6 for 6 hours resulted in a significant (P < 0.05) 2-fold increase in band intensity for RAR, RXR, and Sp-1 as compared with that for untreated control cells (mean ± SD pixel density 2.156 ± 0.711, 2.246 ± 0.062, and 2.022 ± 0.221, respectively). Specificity was determined by control experiments in which the same nuclear extracts isolated from control and HA6-treated cells were incubated with excess competitive unlabeled probes for each transcription factor (Figure 3, lanes 3 and 4). For the Sp-1 transcription factor, the target transcription factor band was decreased but not totally abolished by excess competitive probe. For the RAR and RXR transcription factors, competitive unlabeled probes completely eliminated the shifted target band. In the absence of nuclear extract (Figure 3, lane 5), no shifted bands were detected.
Figure 3.

Electrophoretic mobility shift assay (EMSA) analysis of RAR (A), RXR (B), and Sp-1 (C). EMSA reactions were performed using biotin-labeled probes that were incubated with 25 μg of nuclear extracts from chondrocytes treated with hyaluronan hexasaccharides (lanes 2 and 4) or untreated chondrocytes (lanes 1 and 3). Arrows denote the band location of the relevant transcription factor. For competition analysis, an EMSA reaction was performed in the presence of excess unlabeled probe (lanes 3 and 4). Lane 5 shows the probes without addition of nuclear extracts. The reactions were analyzed in duplicate and performed twice using 2 separate nuclear extracts. Band intensities were quantified using Quantity One software.
Changes in gene expression of MMP-3, type II collagen, and COMP
As a functional approach to determining whether HA6 treatment gives rise to a substantial activation of RAR, RXR, and Sp-1, the expression of cartilage genes known to be up-regulated by these transcription factors in situ was investigated. Activation of retinoid-related signaling pathways has previously been shown to enhance the induction of MMP-3 messenger RNA (mRNA) in articular chondrocytes (21,22). As shown in Figure 4A, the expression of MMP-3 mRNA was markedly enhanced by HA6 treatment, especially at concentrations of ≥250 μg/ml, as determined by real-time RT-PCR analysis. Induction was observed at 12 hours after treatment, and expression remained elevated throughout the experimental period (Figure 4B). Enhanced caseinolytic activity was also detected in the conditioned medium of experimental samples in the absence of activation by APMA (results not shown).
Figure 4.
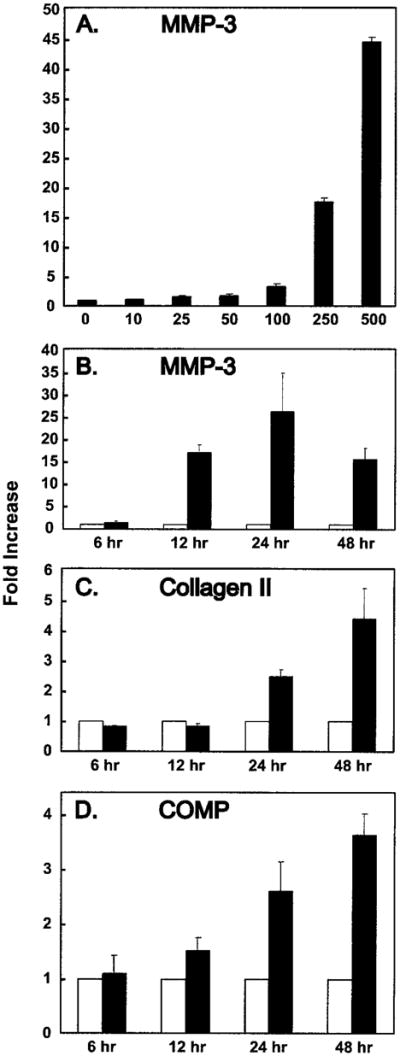
Effects of hyaluronan hexasaccharides (HA6) on the expression of matrix metalloproteinase 3 (MMP-3), type II collagen (collagen II), and cartilage oligomeric matrix protein (COMP) mRNA in bovine articular chondrocytes. Reverse transcription products of total RNA from chondrocytes with or without HA6 treatment were amplified by using a specific primer set for each target gene. The ratio of increase in the sample relative to the untreated control was calculated from the crossing point using the modification of Equation 1 as described by Rasmussen et al (17). A, MMP-3 induction by HA6 at the various doses (0–500 μg/ml) for 24 hours of treatment. B, C, and D, Time-course response (from 6 to 48 hours) to 250 μg/ml HA6 by MMP-3, type II collagen, and COMP mRNA, respectively. Open bars = no HA6; solid bars = addition of HA6. Bars show the mean and SD.
Sp-1 is a crucial transcription factor in the regulation of type II collagen and COMP expression. Both type II collagen and COMP promoter/enhancer regions have several Sp-1 binding sites, and overexpression of Sp-1 has been shown to stimulate type II collagen and COMP protein synthesis (23,24). The addition of 250 μg/ml of HA6 to bovine articular chondrocytes increased the expression of both type II collagen and COMP mRNA by 2.5-fold at 24 hours and by ~4-fold at 48 hours (Figures 4C and D, respectively) as compared with untreated control chondrocytes.
Reversibility by HA of HA6 effects on chondrocytes
If HA oligosaccharides such as HA6 exert their effects on chondrocytes through interaction with cell surface receptors, these effects should be reversible and completely abrogated by the presence of excess high molecular mass HA. To examine this possibility, chondrocytes were incubated without or with HA6 for 12 hours. The medium was then changed to fresh medium without HA6 and the cells incubated for an additional 12 hours in the presence or absence of 500 μg/ml high molecular mass HA. As shown in Figure 5A, incubation of chondrocytes with HA6 resulted in a 30-fold increase in MMP-3 mRNA expression as compared with that in untreated control chondrocytes. When the HA6 was removed for the last 12 hours of incubation, the level of expression of MMP-3 mRNA decreased by 75%, to ~7 times that of control cells. However, following washout of HA6 and subsequent incubation in the presence of high molecular mass HA, the expression of MMP-3 was again equivalent to control values. Furthermore, high molecular mass HA also had a reductive effect (~60%) on the basal MMP-3 expression level in articular chondrocytes.
Figure 5.
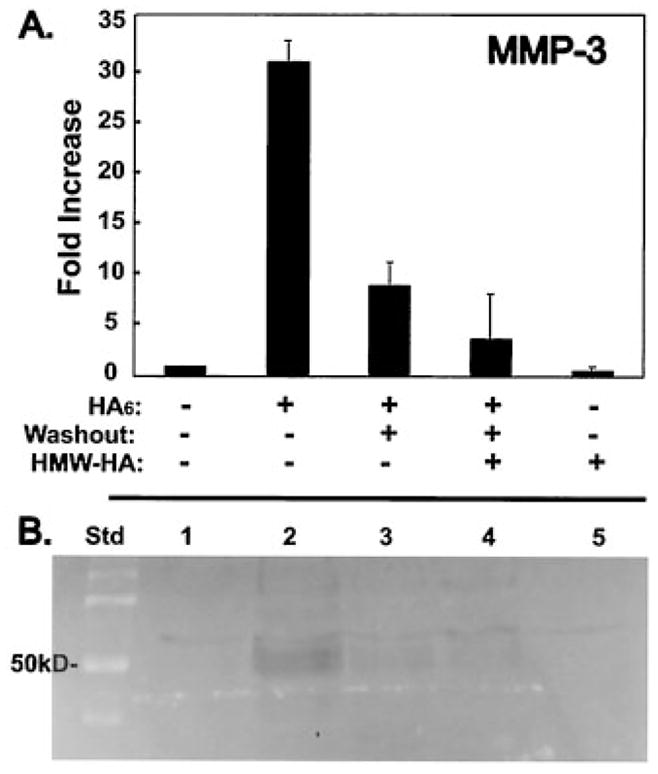
Reversibility of HA6 effects on chondrocytes by high molecular mass (HMW) HA. A, MMP-3 mRNA expression was determined by amplification of reverse transcription products from total RNA using a specific primer set for MMP-3. The ratio of increase in the sample relative to the untreated control was calculated from the crossing point using Equation 1. Chondrocytes were analyzed after no treatment, after treatment with HA6 only, after treatment with HA6 followed by a 12-hour washout, and after treatment with HA6 followed by a 12-hour washout with incubation media containing HMW HA. The first experiment utilized Healon (average molecular weight 1,900–3,000 kd; ratio of treated to control 0.766) and the second experiment utilized Hyalgan (average molecular weight 500–730 kd; ratio of treated to control 6.790). Chondrocytes were also incubated for 12 hours in media containing HMW HA only. Bars show the mean and SD of 2 experiments. B, In separate experiments, the conditioned medium of chondrocytes treated in a manner similar to that described in A was collected and was assayed by casein zymography. In this reverse image, a clearing band at 50 kd is apparent and indicative of MMP-3 in the conditioned medium of chondrocyte cultures treated with HA6 (lane 2). Analysis of conditioned media after a 12-hour washout showed reduction in the intensity of the 50-kd band (lane 3), more so in the 12-hour washout media containing HMW HA (lane 4). Chondrocytes were also incubated for 12 hours in media containing HMW HA only (lane 5), which showed no activity, as in the control untreated condition (lane 1). The results are representative of 3 different experiments. Std = standard (see Figure 4 for other definitions).
We also performed casein zymography to assess the protease activity in the serum-free conditioned medium of bovine chondrocyte cultures without or with HA6 or high molecular mass HA. As shown in the reverse image in Figure 5B, casein zymography revealed enhanced caseinolytic activity determined by the appearance of a cleared zone of ~50 kd in the conditioned medium from the cells treated with HA6 (lane 2) as compared with untreated controls (lane 1). Removal of HA6 at 12 hours resulted in reduced caseinolytic activity (lane 3), which is consistent with the changes in MMP-3 mRNA levels. Addition of high molecular mass HA during the washout of HA6 resulted in further reduction of caseinolytic activity (lane 4), and incubation of chondrocytes with high molecular mass HA alone did not stimulate caseinolytic activity above control levels (lane 5). Altogether, the results show that changes in the 50-kd caseinolytic activity paralleled the changes in expression of MMP-3 mRNA observed in Figure 5A. Thus, the effects of HA6 are reversible and compete with interactions of high molecular mass HA at the surface of chondrocytes.
Effect of HA6 on AP-1 and IL-1β expression
To investigate whether the effects of HA6 on chondrocytes, particularly the dramatic increases in MMP-3 expression, were related to the activation of IL-1–mediated signaling pathways, the effects of HA6 on AP-1 expression were examined. The results illustrated in Figures 2A and B demonstrated that there was minimal increase in AP-1 expression, as measured using the protein–DNA array method of analysis. To explore this further, human immortalized chondrocytes (C-28/I2 cells) transfected with an AP-1–responsive luciferase vector were incubated without or with IL-1β, HA6, HA4, or high molecular mass HA for 24 hours. As shown in Figure 6A, there was a 4.4-fold increase in luciferase activity following exposure of these cells to IL-1β, as expected (19); however, there was no change in luciferase activity in response to HA6. Similarly, there was no change in luciferase activity in response to HA4, and only a small change (1.3-fold increase) occurred due to the inclusion of high molecular mass HA (results not shown).
Figure 6.

Lack of activation of interleukin-1β (IL-1β) signaling path-ways by HA6. A, Activator protein 1 (AP-1) promoter activity in C-28/I2 immortalized human chondrocytes was determined using the AP-1 promoter-reporter construct pAP-1-Luc, which contains 3 tandem repeats of the AP-1 consensus sequences, transiently transfected into immortalized human chondrocytes. The chondrocytes were treated with phosphate buffered saline (PBS) (control), IL-1β, or HA6 as indicated, and relative luciferase activity was determined. Luciferase activity was increased by IL-1β but unchanged by HA6 treatment. B, IL-1β production by human articular chondrocytes treated with HA6 was determined when normal human chondrocytes from 4 different donors were exposed to HA6 for 24 hours and the IL-1β in the conditioned media was measured by Western blot analysis using IL-1β antibodies. Although chondrocytes from the different donors produced different levels of IL-1β, no difference in IL-1β following treatment with HA6 (+) as compared with untreated cells (−) from the same donor was detected in the conditioned media. See Figure 4 for other definitions.
Addition of fibronectin fragments to chondrocytes has also been shown to enhance MMP-3 activity (this mechanism is, in part, mediated by the induced release of endogenous IL-1 [25]). To investigate whether the effects of HA6 on MMP-3 expression were related to a release of endogenous IL-1, changes in the accumulation of released IL-1β protein in the medium of chondrocyte cultures were examined. In order to perform this study using anti–IL-1β antibodies and an immunoblotting approach, articular chondrocytes derived from the talocrural joint of 4 human donors were utilized. As shown in Figure 6B, chondrocytes from 3 of the 4 donors exhibited release and accumulation of IL-1β in the culture medium following 24 hours of incubation. However, in cultures from all 4 donors there was no change (increase or decrease) in the level of IL-1β present in the medium following exposure of the cells to HA6. Thus, it is unlikely that the effects of HA6 are dependent on release of endogenous IL-1, at least during the same time frame evaluated in these experiments.
DISCUSSION
In pathologic musculoskeletal conditions, including OA, fragmentation of HA is induced, thus increasing the amount of low molecular mass HA in the synovial fluid (15). This increase in low molecular mass HA leads to a reduction in viscosity of the synovial fluid. Furthermore, various functions of low molecular mass HA have been reported, particularly with regard to the inflammatory process (26,27). The HA receptor CD44 is responsible for endocytosis-mediated catabolism of HA; however, other mechanisms of signal transduction exerted by HA via CD44 have not yet been clarified in chondrocytes.
Regulation of transcription factors is tightly associated with frequency of gene expression, and consequently, is also associated with protein synthesis, reflecting the changes in cell metabolism. Conventionally, transcription factor activation was examined by EMSA analysis, which uses consensus-binding probes to a single target transcription factor. Newly developed protein–DNA arrays provide a means for rapid screening of multiple transcription factors present in activated versus control cell samples. This approach is highly useful in the case of HA oligosaccharide–mediated signaling, since little is known about the signaling pathways involved, what transcription factors are activated, or even which genes are affected. Nonetheless, protein–DNA arrays are only semiquantitative and can be used only as an initial screen to focus attention on the likely candidate transcription factors. The activation of each of these candidate transcription factors must be validated by subsequent gel-shift assays.
By using this array, we found that the activity of RAR and RXR was remarkably up-regulated in chondrocyte nuclei by the stimulation of HA6. EMSA analysis also showed the activation of RAR and RXR in nuclear extracts of chondrocytes treated with HA6. The nuclear retinoic acid receptors (i.e., RAR) and retinoid X receptor (i.e., RXR) classes are each composed of α, β, and γ subtypes with more than 1 isoform for each receptor subtype (28). RARs associate with RXRs to form RAR-RXR heterodimers, which bind target DNA sequences, called retinoic acid response elements, consisting of directly repeated hexameric half-sites with the consensus sequence 5′-PuG(G/A)(T/A)CA-3′, separated by 1–5 nucleotides depending on the target gene.
It is well established that retinoid signals are required for normal skeletal development. For example, repression of retinoid signaling is a major requirement for chondrogenesis because RAR-mediated repression is required for induction of SOX9, which plays an essential role in establishing the precartilaginous condensations and in initiating chondroblast differentiation (29). Weston et al (30) demonstrated a tight inverse correlation between retinoid receptor activity and SOX9 transactivation of Colα1(II) and showed that inhibition of RARα activity was sufficient to induce chondrogenesis in vitro. In contrast, the stimulation of retinoid signals results in dedifferentiation of articular chondrocytes (31). Retinoic acid also induces the gene expression of MMP-3 (stromelysin 1), which cleaves proteoglycans, collagens, gelatin, and the link protein of the proteoglycan aggregate synthesized by human articular chondrocytes (21). In addition, analysis of mRNA from mice in the collagen-induced arthritis model showed that an RAR antagonist prevented the overexpression of MMP-3 in the arthritic joints (22). These findings indicate that retinoid signals exert deleterious effects on articular chondrocytes by phenotypic modulation or induction of MMP-3.
In the present study, the expression of MMP-3 mRNA and caseinolytic activity were markedly enhanced in the articular chondrocytes treated with HA6. It can therefore be speculated that the resultant catabolic activity of chondrocytes caused by MMP-3 induction is achieved by the activation of retinoid signals through HA6 stimulation. In addition, the results of protein–DNA array showed increased activity of NF-κB, which is also closely associated with MMP-3 induction in articular chondrocytes (32,33), suggesting another possible mechanism for the increases in the MMP-3 levels by the stimulation with HA6. Taken together, our results demonstrate that the enhancement of cartilage degradation in OA joints by low molecular mass HA is a consequence of induction of MMP-3 via retinoid and/or NF-κB signaling pathway(s).
Interestingly, the induction of MMP-3 by HA6 was completely abrogated by treatment with high molecular mass HA in the middle of the experimental period. Recently, Julovi et al (34) demonstrated that direct interaction of high molecular mass HA with CD44 inhibited the induction of MMP-1, MMP-3, and MMP-13 by IL-1β in articular chondrocytes. Furthermore, Spessotto et al (35) showed that the HA–CD44 engagement directly inhibited the expression of MMP-9 in osteoclasts. These results support the possibility that the interaction of high molecular mass HA with CD44 exerts anabolic actions, thus abrogating the induction of MMPs. However, this notion also gives rise to an argument concerning the diversity of CD44 signaling depending on the size of HA.
HA6 is the minimum size for binding CD44 (36), and it can therefore be speculated that one HA hexasaccharide binds to one CD44, whereas high molecular mass HA is able to bind to multiple CD44 receptors. This function of high molecular mass HA may lead to clustering of CD44 in the cell membrane, and result in the stabilization of the cortical cytoskeletal conformation. Varedi et al (37) reported that reorganization of the cytoskeleton induced expression of mRNA for collagenases in dermal fibroblasts. It has also been reported that gene expression of MMP-1, MMP-3, and MMP-9 is regulated by the activation of protein kinase C in chondrocytes (38,39). Protein kinase C was shown to be a downstream effector of CD44 during the activation of NF-κB by HA fragments in T-24 carcinoma cells (40). Thus, HA6 treatment seems to disturb the stabilization of cytoskeletal conformation accomplished by high molecular mass HA–CD44 interactions, consequently leading to the activation of transcription factors affecting catabolism.
In this study, the basal MMP-3 mRNA level was recovered considerably by the washout of HA6 in the middle of the experimental period. New synthesis of endogenous HA by chondrocytes may function to reduce the effect of small HA oligosaccharides on MMP-3 induction by the reconstruction of HA–CD44 interactions, CD44 clustering, and cytoskeletal reorganization. This also emphasizes the importance of interaction of high molecular mass HA and CD44 in chondrocyte homeostasis. Although we have found CD44 to be the primary HA receptor expressed by articular chondrocytes, the potential participation of other HA receptors, such as receptor for HA-mediated motility (RHAMM), Toll-like receptor 4, or LYVE-1, will need to be determined in future studies. Other extracellular matrix receptors, such as integrins, also play a role in the activation of MMPs. For example, extracellular fibronectin fragments activate MMPs through an IL-1–mediated pathway (41,42). Moreover, the transcription factor AP-1 is involved in the induction of MMPs by IL-1 (33). In the present study, stimulation of chondrocytes by HA6 did not result in the activation of AP-1. In addition, the expression level of IL-1β was not changed by HA6 treatment of human articular chondrocytes. Thus, several lines of evidence suggest that the HA oligosaccharide–induced events differ substantially from those induced by IL-1 or fibronectin fragments.
Because of its dual ability to cleave collagen and aggrecan, MMP-13 is also considered a major enzyme responsible for matrix resorption of OA and rheumatoid arthritis cartilage. MMP-13 has a particular affinity for type II collagen and selectively enhances cleavage and denaturation of this type of collagen (43), and can also cleave aggrecan at specific sites (44). Interestingly, recent studies have also shown that retinoid-related signals strongly enhance the induction of MMP-13 in chondrocytic cells through the action of RAR-RXR heterodimers (45). In addition, reduced expression of MMP-13 is observed in cytokine-stimulated cells when NF-κB is maintained in the cytoplasm by constitutive levels of IκB (46). Thus, it can be speculated that MMP-13 is also induced by HA6 through these transcription factors. In our preliminary studies using the promoter assay with MMP-13 constructs, HA6 up-regulated the promoter activity of MMP-13 (Ohno S, et al: unpublished observations). We are currently investigating the inductive mechanism of MMP-13 by HA oligosaccharides.
In this study, Sp-1 was also remarkably activated in chondrocytes by the treatment with HA6. Type II collagen and COMP have several Sp-1 sites in their promoter regions, and Sp-1 specifically activates type II collagen and COMP synthesis (23,24). In RT-PCR analysis, the gene expressions of type II collagen and COMP were up-regulated by the treatment with HA6. Furthermore, expression of HA synthase 2 mRNA was increased 2.1-fold by HA oligosaccharides, and aggrecan mRNA was increased 1.8-fold by HA oligosaccharides in human articular chondrocytes (47). These results also suggest that anabolic responses, as evidenced by matrix synthesis, take place simultaneously with the catabolic responses induced by small HA oligosaccharides. In OA cartilage, chondrocytes are subjected to rapid matrix turnover with both anabolic and catabolic activities elevated, and the subsequent imbalance between matrix synthesis and degradation advances the pathologic development. Although the direct links between transcription factors and genes have not been clarified, it can be supposed that the specific transcription factors activated via HA signal transduction modulate cartilage-matrix metabolism.
In summary, we have demonstrated with the use of transcription factor protein–DNA array and EMSA analyses that treatment with HA oligosaccharide increases the activity of RAR, RXR, and Sp-1 in nuclei of articular chondrocytes. Furthermore, we have shown the transcriptional up-regulation of the genes for MMP-3, type II collagen, and COMP, which is tightly associated with the activation of RAR, RXR, and Sp-1. The links between these genes and transcription factors should be clarified in future studies.
Acknowledgments
We thank the Seikagaku Corporation (Japan) for providing us with the hyaluronan hexasaccharide. We thank the Gift of Hope Organ and Tissue Donor Network of Chicago, Illinois for providing the human donor tissues. The generosity and beneficence of the donor families for access to the human tissues are greatly appreciated.
Supported by grants from the NIH (R01-AR-43384, R01-AR-39507, and P50-AR-39239) and the Arthritis Foundation.
References
- 1.Hascall VC, Heinegard D. Aggregation of cartilage proteoglycans. II. Oligosaccharide competitors of the proteoglycan-hyaluronic acid interaction. J Biol Chem. 1974;249:4242–9. [PubMed] [Google Scholar]
- 2.Knudson CB. Hyaluronan receptor-directed assembly of chondrocyte pericellular matrix. J Cell Biol. 1993;120:825–34. doi: 10.1083/jcb.120.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knudson W, Bartnik E, Knudson CB. Assembly of pericellular matrices by COS-7 cells transfected with CD44 homing receptor genes. Proc Natl Acad Sci U S A. 1993;90:4003–7. doi: 10.1073/pnas.90.9.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudson W, Aguiar DJ, Hua Q, Knudson CB. CD44-anchored hyaluronan-rich pericellular matrices: an ultrastructural and biochemical analysis. Exp Cell Res. 1996;228:216–28. doi: 10.1006/excr.1996.0320. [DOI] [PubMed] [Google Scholar]
- 5.Knudson W, Loeser RF. CD44 and integrin matrix receptors participate in cartilage homeostasis. Cell Mol Life Sci. 2002;59:36–44. doi: 10.1007/s00018-002-8403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knudson W, Casey B, Nishida Y, Eger W, Kuettner KE, Knudson CB. Hyaluronan oligosaccharides perturb cartilage matrix homeostasis and induce chondrocytic chondrolysis. Arthritis Rheum. 2000;43:1165–74. doi: 10.1002/1529-0131(200005)43:5<1165::AID-ANR27>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 7.Solursh M, Hardingham TE, Hascall VC, Kimura JH. Separate effects of exogenous hyaluronic acid on proteoglycan synthesis and deposition in pericellular matrix by cultured chick embryo limb chondrocytes. Dev Biol. 1980;75:121–9. doi: 10.1016/0012-1606(80)90148-7. [DOI] [PubMed] [Google Scholar]
- 8.Kimura JH, Hardingham TE, Hascall VC, Solursh M. Biosynthesis of proteoglycans and their assembly into aggregates in cultures of chondrocytes from the Swarm rat chondrosarcoma. J Biol Chem. 1979;254:2600–9. [PubMed] [Google Scholar]
- 9.Ratcliffe A, Tyler JA, Hardingham TE. Articular cartilage cultured with interleukin 1. Biochem J. 1986;238:571–80. doi: 10.1042/bj2380571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clements KM, Price JS, Chambers MG, Visco DM, Poole AR, Mason RM. Gene deletion of either interleukin-1β, interleukin-1β–converting enzyme, inducible nitric oxide synthase, or stromelysin 1 accelerates the development of knee osteoarthritis in mice after surgical transection of the medial collateral ligament and partial medial meniscectomy. Arthritis Rheum. 2003;48:3452–63. doi: 10.1002/art.11355. [DOI] [PubMed] [Google Scholar]
- 11.Chow G, Nietfeld JJ, Knudson CB, Knudson W. Antisense inhibition of chondrocyte CD44 expression leading to cartilage chondrolysis. Arthritis Rheum. 1998;41:1411–9. doi: 10.1002/1529-0131(199808)41:8<1411::AID-ART10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 12.Shimazu A, Jikko A, Iwamoto M, Koike T, Yan W, Okada Y, et al. Effects of hyaluronic acid on the release of proteoglycan from the cell matrix in rabbit chondrocyte cultures in the presence and absence of cytokines. Arthritis Rheum. 1993;36:247–53. doi: 10.1002/art.1780360217. [DOI] [PubMed] [Google Scholar]
- 13.Stove J, Gerlach C, Huch K, Gunther KP, Puhl W, Scharf HP. Effects of hyaluronan on proteoglycan content of osteoarthritic chondrocytes in vitro. J Orthop Res. 2002;20:551–5. doi: 10.1016/S0736-0266(01)00141-3. [DOI] [PubMed] [Google Scholar]
- 14.Bayliss MT, Dudhia J. Hyaluronan synthesis in human articular cartilage. In: Kennedy J, Phillips G, Williams P, Hascall V, editors. Hyaluronan. Cambridge: MFK Group Ltd; 2002. pp. 297–302. [Google Scholar]
- 15.Dahl L, Dahl IM, Engstrom-Laurent A. Concentration and molecular weight of sodium hyaluronate in synovial fluid from patients with rheumatoid arthritis and other arthropathies. Ann Rheum Dis. 1985;44:817–22. doi: 10.1136/ard.44.12.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishida Y, Knudson CB, Eger W, Kuettner KE, Knudson W. Osteogenic protein 1 stimulates cell-associated matrix assembly by normal human articular chondrocytes: up-regulation of hyaluronan synthase, CD44, and aggrecan. Arthritis Rheum. 2000;43:206–14. doi: 10.1002/1529-0131(200001)43:1<206::AID-ANR25>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen TB, Uttenthal A, de Stricker K, Belak S, Storgaard T. Development of a novel quantitative real-time RT-PCR assay for the simultaneous detection of all serotypes of foot-and-mouth disease virus. Arch Virol. 2003;148:2005–21. doi: 10.1007/s00705-003-0145-2. [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Im HJ, Pacione C, Chubinskaya S, van Wijnen AJ, Sun Y, Loeser RF. Inhibitory effects of insulin-like growth factor-1 and osteogenic protein-1 on fibronectin fragment- and interleukin-1β-stimulated matrix metalloproteinase-13 expression in human chondrocytes. J Biol Chem. 2003;278:25386–94. doi: 10.1074/jbc.M302048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Resa P, Mira E, Quesada AR. Enhanced detection of casein zymography of matrix metalloproteinases. Anal Biochem. 1995;224:434–5. doi: 10.1006/abio.1995.1063. [DOI] [PubMed] [Google Scholar]
- 21.Flannery CR, Little CB, Caterson B, Hughes CE. Effects of culture conditions and exposure to catabolic stimulators (IL-1 and retinoic acid) on the expression of matrix metalloproteinases (MMPs) and disintegrin metalloproteinases (ADAMs) by articular cartilage chondrocytes. Matrix Biol. 1999;18:225–37. doi: 10.1016/s0945-053x(99)00024-4. [DOI] [PubMed] [Google Scholar]
- 22.Beehler BC, Hei YJ, Chen S, Lupisella JA, Ostrowski J, Starrett JE, et al. Inhibition of disease progression by a novel retinoid antagonist in animal models of arthritis. J Rheumatol. 2003;30:355–63. [PubMed] [Google Scholar]
- 23.Ghayor C, Chadjichristos C, Herrouin JF, Ala-Kokko L, Suske G, Pujol JP, et al. Sp3 represses the Sp1-mediated transactivation of the human COL2A1 gene in primary and de-differentiated chondrocytes. J Biol Chem. 2001;276:36881–95. doi: 10.1074/jbc.M105083200. [DOI] [PubMed] [Google Scholar]
- 24.Deere M, Rhoades Hall C, Gunning KB, LeFebvre V, Ridall AL, Hecht JT. Analysis of the promoter region of human cartilage oligomeric matrix protein. Matrix Biol. 2001;19:783–92. doi: 10.1016/s0945-053x(00)00127-x. [DOI] [PubMed] [Google Scholar]
- 25.Homandberg GA, Meyers R, Xie DL. Fibronectin fragments cause release of IL-1 from human cartilage in vitro. Clin Physiol. 1993;10:1–6. [Google Scholar]
- 26.McKee CM, Penno MB, Cowman M, Burdick MD, Strieter RM, Bao C, et al. Hyaluronan (HA) fragments induce chemokine gene expression in alveolar macrophages: the role of HA size and CD44. J Clin Invest. 1996;98:2403–13. doi: 10.1172/JCI119054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodge-Dufour J, Noble PW, Horton MR, Bao C, Wysoka M, Burdick MD, et al. Induction of IL-12 and chemokines by hyaluronan requires adhesion-dependent priming of resident but not elicited macrophages. J Immunol. 1997;159:2492–500. [PubMed] [Google Scholar]
- 28.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–54. [PubMed] [Google Scholar]
- 29.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–9. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 30.Weston AD, Chandraratna RA, Torchia J, Underhill TM. Requirement for RAR-mediated gene repression in skeletal progenitor differentiation. J Cell Biol. 2002;158:39–51. doi: 10.1083/jcb.200112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishihara A, Fujii M, Sampath TK, Miyazono K, Reddi AH. Bone morphogenetic protein signaling in articular chondrocyte differentiation. Biochem Biophys Res Commun. 2003;301:617–22. doi: 10.1016/s0006-291x(02)03068-1. [DOI] [PubMed] [Google Scholar]
- 32.Sylvester J, Liacini A, Li WQ, Dehnade F, Zafarullah M. Tripterygium wilfordii Hook F extract suppresses proinflammatory cytokine-induced expression of matrix metalloproteinase genes in articular chondrocytes by inhibiting activating protein-1 and nuclear factor-κB activities. Mol Pharmacol. 2001;59:1196–205. doi: 10.1124/mol.59.5.1196. [DOI] [PubMed] [Google Scholar]
- 33.Liacini A, Sylvester J, Li WQ, Zafarullah M. Inhibition of interleukin-1-stimulated MAP kinases, activating protein-1 (AP-1) and nuclear factor κB (NF-κB) transcription factors down-regulates matrix metalloproteinase gene expression in articular chondrocytes. Matrix Biol. 2002;21:251–62. doi: 10.1016/s0945-053x(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 34.Julovi SM, Yasuda T, Shimizu M, Hiramitsu T, Nakamura T. Inhibition of interleukin-1β–stimulated production of matrix metalloproteinases by hyaluronan via CD44 in human articular cartilage. Arthritis Rheum. 2004;50:516–25. doi: 10.1002/art.20004. [DOI] [PubMed] [Google Scholar]
- 35.Spessotto P, Rossi FM, Degan M, di Francia R, Perris R, Colombatti A, et al. Hyaluronan-CD44 interaction hampers migration of osteoclast-like cells by down-regulating MMP-9. J Cell Biol. 2002;158:1133–44. doi: 10.1083/jcb.200202120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohda D, Morton CJ, Parkar AA, Hatanaka H, Inagaki FM, Campbell ID, et al. Solution structure of the link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell. 1996;86:767–75. doi: 10.1016/s0092-8674(00)80151-8. [DOI] [PubMed] [Google Scholar]
- 37.Varedi M, Ghahary A, Scott PG, Tredget EE. Cytoskeleton regulates expression of genes for transforming growth factor-β1 and extracellular matrix proteins in dermal fibroblasts. J Cell Physiol. 1997;172:192–9. doi: 10.1002/(SICI)1097-4652(199708)172:2<192::AID-JCP6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 38.Ogata Y, Pratta MA, Nagase H, Arner EC. Matrix metalloproteinase 9 (92-kDa gelatinase/type IV collagenase) is induced in rabbit articular chondrocytes by cotreatment with interleukin 1β and a protein kinase C activator. Exp Cell Res. 1992;201:245–9. doi: 10.1016/0014-4827(92)90271-9. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka S, Hamanishi C, Kikuchi H, Fukuda K. Factors related to degradation of articular cartilage in osteoarthritis: a review. Semin Arthritis Rheum. 1998;27:392–9. doi: 10.1016/s0049-0172(98)80019-x. [DOI] [PubMed] [Google Scholar]
- 40.Fitzgerald KA, Bowie AG, Skeffington BS, O’Neill LA. Ras, protein kinase C ζ, and I κ B kinases 1 and 2 are downstream effectors of CD44 during the activation of NF-κB by hyaluronic acid fragments in T-24 carcinoma cells. J Immunol. 2000;164:2053–63. doi: 10.4049/jimmunol.164.4.2053. [DOI] [PubMed] [Google Scholar]
- 41.Saito S, Yamaji N, Yasunaga K, Saito T, Matsumoto S, Katoh M, et al. The fibronectin extra domain A activates matrix metalloproteinase gene expression by an interleukin-1-dependent mechanism. J Biol Chem. 1999;274:30756–63. doi: 10.1074/jbc.274.43.30756. [DOI] [PubMed] [Google Scholar]
- 42.Yasuda T, Poole AR. A fibronectin fragment induces type II collagen degradation by collagenase through an interleukin-1–mediated pathway. Arthritis Rheum. 2002;46:138–48. doi: 10.1002/1529-0131(200201)46:1<138::AID-ART10051>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell PG, Magna HA, Reeves LM, Lopresti-Morrow LL, Yocum SA, Rosner PJ, et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J Clin Invest. 1996;97:761–8. doi: 10.1172/JCI118475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fosang AJ, Last K, Knauper V, Murphy G, Neame PJ. Degradation of cartilage aggrecan by collagenase-3 (MMP-13) FEBS Lett. 1996;380:17–20. doi: 10.1016/0014-5793(95)01539-6. [DOI] [PubMed] [Google Scholar]
- 45.Jimenez MJ, Balbin M, Alvarez J, Komori T, Bianco P, Holmbeck K, et al. A regulatory cascade involving retinoic acid, Cbfa1, and matrix metalloproteinases is coupled to the development of a process of perichondrial invasion and osteogenic differentiation during bone formation. J Cell Biol. 2001;155:1333–44. doi: 10.1083/jcb.200106147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mengshol JA, Vincenti MP, Coon CI, Barchowsky A, Brinckerhoff CE. Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor κB: differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum. 2000;43:801–11. doi: 10.1002/1529-0131(200004)43:4<801::AID-ANR10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 47.Nishida N, Knudson CB, Knudson W. Extracellular matrix recovery by human articular chondrocytes after treatment with hyaluronan hexasaccharides or Streptomyces hyaluronidase. Mod Rheumatol. 2003;13:62–8. doi: 10.3109/s101650300009. [DOI] [PubMed] [Google Scholar]


