Abstract
Background
Yersinia pestis, the agent of plague, has caused many millions of human deaths and still poses a serious threat to global public health. Timely and reliable detection of such a dangerous pathogen is of critical importance. Lysis by specific bacteriophages remains an essential method of Y. pestis detection and plague diagnostics.
Methodology/Principal Findings
The objective of this work was to develop an alternative to conventional phage lysis tests – a rapid and highly sensitive method of indirect detection of live Y. pestis cells based on quantitative real-time PCR (qPCR) monitoring of amplification of reporter Y. pestis-specific bacteriophages. Plague diagnostic phages ϕA1122 and L-413C were shown to be highly effective diagnostic tools for the detection and identification of Y. pestis by using qPCR with primers specific for phage DNA. The template DNA extraction step that usually precedes qPCR was omitted. ϕA1122-specific qPCR enabled the detection of an initial bacterial concentration of 103 CFU/ml (equivalent to as few as one Y. pestis cell per 1-µl sample) in four hours. L-413C-mediated detection of Y. pestis was less sensitive (up to 100 bacteria per sample) but more specific, and thus we propose parallel qPCR for the two phages as a rapid and reliable method of Y. pestis identification. Importantly, ϕA1122 propagated in simulated clinical blood specimens containing EDTA and its titer rise was detected by both a standard plating test and qPCR.
Conclusions/Significance
Thus, we developed a novel assay for detection and identification of Y. pestis using amplification of specific phages monitored by qPCR. The method is simple, rapid, highly sensitive, and specific and allows the detection of only live bacteria.
Introduction
Yersinia pestis is a Gram-negative nonsporulating bacterium belonging to the family Enterobacteriaceae. Y. pestis is the causative agent of bubonic and pneumonic plague, a primarily zoonotic infection. The bacterium is usually transmitted from rodents and lagomorphs to humans by flea bite. Pneumonic plague is a severe infection transmissible from person to person by respiratory droplets and thought to be responsible for about 200 million human deaths during three historic pandemics. Nowadays, natural plague foci exist in Asia, Eastern Europe, Africa and both Americas, and about 2,000 cases of human plague are registered by the World Health Organization every year [1]–[3]. Y. pestis is classified by the CDC as a category A biothreat agent due to its easy person-to-person dissemination via aerosol, high lethality, and wide recognition as a biowarfare agent that is likely to cause mass casualties [4]. The problem is aggravated by the fact that multidrug-resistant strains of Y. pestis have been isolated from humans including a strain resistant to all drugs currently used for plague treatment and prophylaxis [5], [6].
In the classic clinical scenario, a confirmed diagnosis of plague includes the isolation of a pure culture of Y. pestis and its identification, or observing a 4-fold difference in titers of antibodies against F1 (capsule antigen) in two serum specimens from the same patient [7], [8]. This process usually takes at least 48–72 hours, which is unacceptable due to the rapid or fulminant course of plague. Therefore, numerous rapid tests for the detection of Y. pestis have been developed. Most of these include different variants of polymerase chain reaction, PCR [9]–[15]. Disadvantages of conventional PCR tests include the need to analyze the PCR products by gel electrophoresis and frequent contamination of the laboratory by the amplicons. Real-time PCR, allowing researchers to see an ongoing reaction by using fluorescent reporters has significantly improved the rapid detection of Y. pestis [16]–[26]. This approach has been successfully used for testing simulated [18], [19], [22] and actual [16] clinical specimens, environmental samples [22], food [26], and experimentally infected fleas [16], [18]. The shortest assay time described was 4 h [22], and the maximum sensitivity reached one bacterial cell per sample [18], [22], [25]. However, real-time PCR using bacterial DNA as template also has some disadvantages: it requires a preceding step of DNA extraction and cannot discriminate between live, dead and/or dormant bacteria present in diagnostic specimens.
Viruses of bacteria (bacteriophages, or phages) have been used for the diagnostics of bacterial infections since the 1930s [27]. Bacteriophage-based detection exploits fundamental properties of lytic phages: specific targeting, infection and lysis of host cells with simultaneous amplification of phage particles. Routine phage lysis assays including phage typing are still actively used for the detection and identification of various bacterial pathogens [28]–[32] but several new phage-based technologies have been developed for more rapid and efficient detection of medically important bacteria. One group of methods is based on monitoring the release of bacterial ATP [33] or such enzymes as adenylate kinase [34] or β-D-galactosidase [35] from lysed cells. Another group utilizes the indirect detection of bacteria via the propagation of specific phages engineered to express luciferase reporter genes [36]–[39], green fluorescent protein [40], or biotin ligase with subsequent biotinylation and conjugation to quantum dots [41], [42]. Finally, the technically simplest and yet efficient method of phage-based bacterial detection is the monitoring of genome amplification of a wild-type phage in the presence of a sensitive bacterium using quantitative real-time PCR (qPCR). This approach has been used first as an alternative to a standard plating test for bacteriophage λ quantitation [43] and then successfully applied for the indirect detection of Bacillus anthracis [44] and the plant pathogen Ralstonia solanacearum [45], as well as for phage-based determination of different kinds of bacterial fecal pollution of water [46].
The bacteriophage lysis test has been an integral part of Y. pestis detection and identification and bacteriological diagnosis of plague for about 80 years [7], [8], [47]–[50]. There are three well-studied and widely used Y. pestis-specific plague diagnostic phages. Two of them, ϕA1122 [7], [48], [49], [51] and the Pokrovskaya phage [47], [52]–[54], are similar to enterobacteriophage T7, display highly lytic activities and very broad lytic spectra towards Y. pestis strains of different origin but also lyse some strains of Yersinia pseudotuberculosis, the closest phylogenetic relative of Y. pestis. This insufficient specificity in the case of ϕA1122 can be overcome by reduction of growth temperature from 26–37°C to 20–25°C [7], [51]. ϕA1122 is recommended as an important plague diagnostic tool by the CDC [7], [49]. The third plague diagnostic phage, L-413C, is a lytic mutant of a temperate phage. L-413C is active only against Y. pestis (as shown on 6,000 global isolates) and rare restrictionless strains of Escherichia coli and does not lyse any of the 2,000 strains of Y. pseudotuberculosis tested [50], [52]–[54]. We have recently sequenced the genome of L-413C and shown its high homology to coliphage P2 and a mosaic structure of its tail fiber protein H, which is responsible for high specificity of L-413C [50].
The phage lysis assay usually requires the isolation of a pure Y. pestis culture, which takes at least 48 h. The test itself takes additional 18–24 h [7], [8]. In a recent publication, Schofield and colleagues [39] have used a fluorescently labeled ϕA1122 to monitor the early steps of its propagation and expedite the indirect detection of Y. pestis. A genetically modified phage was engineered to express luciferase reporter genes luxAB in targeted Y. pestis cells and allowed the detection of 820 or more bacterial cells 60 min after adding the phage. However, there was also a signal observed with 2 (of 10) Y. pseudotuberculosis strains and 1 (of 10) Yersinia enterocolitica isolates. Moreover, the prolonged incubation of the reporter phage with Y. pestis cells, longer than 90 min, resulted in a gradual decline in signal strength [39].
In the present study, we further explored the diagnostic capabilities of not only ϕA1122 but also L-413C and designed a simple (employing native, non-modified phages) rapid (4 h), highly sensitive (up to 103 CFU/ml, equivalent to 1 cell per 1-µl sample) and specific assay for the indirect detection of live Y. pestis cells by qPCR monitoring of the reporter phage burst. Bacteriophage ϕA1122 provided the maximum sensitivity and displayed a significant titer rise in simulated blood specimens. This phage did not lyse any of the 17 Y. enterocolitica strains tested but showed some degree of amplification on 4 of the 20 Y. pseudotuberculosis strains tested at 28°C. The specificity of this assay was increased to practically 100% by incubating at 24°C. The L-413C-based assay was less sensitive (≥100 bacteria per sample) but more specific. We propose to use a parallel 2-phage qPCR assay for rapid and definitive identification of Y. pestis.
Materials and Methods
Ethics statement
Human blood from anonymous donors was commercially purchased from Biological Specialty Corp. (Colmar, Philadelphia). All experiments with blood artificially contaminated with Y. pestis, phage particles or phage DNA were performed under the human subjects use protocol approved by the Walter Reed Army Institute of Research Institutional Review Board (Protocol WRAIR #1119).
Bacterial strains, bacteriophages and growth media
Bacterial strains used in this work are listed in Table 1. All bacteria and bacteriophage L-413C were taken from the laboratory strain collection. Phages ϕA1122, P2 vir1, and T7 were kindly provided by Dr. Martin E. Schriefer (Bacterial Diseases Branch, Division of Vector-Borne Infectious Disease, National Center for Zoonotic, Vector-Borne and Enteric Diseases, Centers for Disease Control and Prevention, Ft. Collins, Colorado), Dr. Richard Calendar (Department of Molecular and Cell Biology, University of California, Berkeley, California), and Dr. Ian J. Molineux (Section of Molecular Genetics and Microbiology, and Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, Texas), respectively. High-concentration stocks of ϕA1122 were prepared using attenuated Y. pestis strain CO92 pgm− cultured at 28°C by the low multiplicity of infection method [55]. The same method was used for large-scale isolation of T7 phage but it was grown on E. coli C600 at 37°C. L-413C and P2 vir1 phage stocks were prepared on Y. pestis CO92 pgm− as described previously [50] using different temperatures of incubation, 28 or 37°C, respectively. Liquid Brain Heart Infusion (BHI) medium (Becton-Dickinson, Franklin Lakes, NJ) or BHI plates containing 1.5% Bacto Agar (Becton-Dickinson) and these with a 0.7% agar overlay were used for growing bacteria and phages. BHI for the experiments with L-413C and P2 vir1 phages was supplemented with 1.6 mM MgCl2, 0.5 mM CaCl2, and 0.1% glucose. SM buffer [55] was used for phage storage and dilutions. All bacterial strains were grown at 28°C unless specifically indicated. Phage plaque assays were performed by the double-layer agar method as described earlier [55] with overnight incubation for L-413C and P2 vir1 phages and 5–6 h incubation for ϕA1122. Bacterial test cultures in case of P2 vir1 were grown at 37°C.
Table 1. Bacterial strains used and results of phage propagation as detected by a standard lysis procedure and by the described qPCR assay.
| Species | Strain | ϕA1122 Growth/qPCR | L-413C Growth/qPCR | Species | Strain | ϕA1122 Growth/qPCR | L-413C Growth/qPCR |
| Yersinia pestis | A1122 | + | + | Y. enterocolitica | 2516-87 (O:9) | − | − |
| Y. pestis | CO92 pgm− | + | + | Y. enterocolitica | YF194 (O:18) | − | − |
| Y. pestis | KIM | + | + | Y. enterocolitica | YE330 (O:20) | − | − |
| Yersinia pseudotuberculosis | 1 (IA)* | − | − | Y. enterocolitica | YF315 (O:22) | − | − |
| Y. pseudotuberculosis | IB (IB) | − (24°C)** | − | Y. enterocolitica | Y240 (O:25) | − | − |
| Y. pseudotuberculosis | PB1/+ (I) | − (24°C) | − | Y. enterocolitica | 661-83 (O:27) | − | − |
| Y. pseudotuberculosis | EP2/+ (I) | − | − | Y. enterocolitica | YE312 (O:34) | − | − |
| Y. pseudotuberculosis | MD67 (I) | − | − | Y. enterocolitica | Y219 (O:44) | − | − |
| Y. pseudotuberculosis | 7 (IIA) | − | − | Y. enterocolitica | ATCC 49397 | − | − |
| Y. pseudotuberculosis | 1779 (IIB) | − | − | Yersinia aldovae | ATCC 35237 | − | − |
| Y. pseudotuberculosis | 43 (III) | − | − | Y. aldovae | 669-83 | − | − |
| Y. pseudotuberculosis | III(65) (III) | − | − | Yersinia bercovieri | ATCC 43970 | − | − |
| Y. pseudotuberculosis | MD31 (III) | − | − | Yersinia frederiksenii | ATCC 33644 | − | − |
| Y. pseudotuberculosis | MD65 (III) | − (24°C) | − | Y. frederiksenii | Y225 | − | − |
| Y. pseudotuberculosis | 32 (IVA) | − (24°C) | − | Yersinia intermedia | ATCC 29909 | − | − |
| Y. pseudotuberculosis | Ikegaki (IVB) | − | − | Y. intermedia | Y229 | − | − |
| Y. pseudotuberculosis | R2 (VB) | − | − | Yersinia kristensenii | Y232 | − | − |
| Y. pseudotuberculosis | Neilson | − | − | Yersinia mollaretii | ATCC 43969 | − | − |
| Y. pseudotuberculosis | Hale | − | − | Escherichia coli | MG1655 | − | − |
| Y. pseudotuberculosis | Galligue | − | − | E. coli | C600 | − | − |
| Y. pseudotuberculosis | Parkin 413 | − | − | Shigella dysenteriae | ATCC 13313 | − | − |
| Y. pseudotuberculosis | MSU-D | − | − | Shigella flexneri | ATCC 9748 | − | − |
| Y. pseudotuberculosis | MSU-H | − | − | Salmonella enterica, sv. Typhimurium | ATCC 15277 | − | − |
| Yersinia enterocolitica | YE288 (O:3) | − | − | S. enterica, sv. Typhi | ATCC 19430 | − | − |
| Y. enterocolitica | 929-78 (O:6) | − | − | Klebsiella pneumoniae | ATCC 132 | − | − |
| Y. enterocolitica | 8081 (O:8) | − | − | K. pneumoniae | ATCC 9997 | − | − |
| Y. enterocolitica | ATCC 9610 (O:8) | − | − | Proteus vulgaris | ATCC 6896 | − | − |
| Y. enterocolitica | ATCC 23715 (O:8) | − | − | Proteus hauseri | ATCC 13315 | − | − |
| Y. enterocolitica | ATCC 27729 (O:8) | − | − | Citrobacter freundii | ATCC 6879 | − | − |
| Y. enterocolitica | ATCC 51871 (O:8) | − | − | Pantoea agglomerans | ATCC 29904 | − | − |
| Y. enterocolitica | ATCC 51872 (O:8) | − | − | Erwinia herbicola | Lot#2 (DPG) | − | − |
Notes: Phage propagation was performed at 28°C unless it is specifically indicated in the column “ϕA1122 Growth/qPCR”.
*Serovars of Y. pseudotuberculosis and Y. enterocolitica are shown in parentheses if known.
**On Y. pseudotuberculosis IB, a very weak propagation of ϕA1122 was observed at 24°C, six orders of magnitude lower than in Y. pestis CO92.
Dynamics of Y. pestis lysis by bacteriophages
The phage (ϕA1122, L-413C, or P2 vir1) was added to an early log phase BHI broth culture of Y. pestis CO92 pgm− (optical density at 600 nm, OD600 = 0.2, which corresponds to ∼1×108 CFU (colony-forming units) per ml; this was repeatedly confirmed by plating) at multiplicity of infection (MOI) of 0.1 (1 bacteriophage per 10 bacterial cells). The infected culture was incubated with shaking at 200 rpm (revolutions per minute) and 28°C (or 37°C in case of P2 vir1), and OD600 was measured every 15 minutes up to 180 min. The final concentration of live bacterial cells after 3-h incubation was determined by serial 10-fold dilutions and plating on BHI agar in triplicate.
Determination of bacteriophage burst sizes
The burst size for ϕA1122 was determined using a classic one-step growth experiment [56] modified as follows. The overnight culture of Y. pestis CO92 pgm− was diluted 1∶50 with BHI broth and incubated at 28°C with shaking at 200 rpm to get an OD600 of 0.2 (∼108 CFU/ml). 2.5×107 PFU (plaque-forming units) of phage ϕA1122 was added in a small volume of SM buffer to 5 ml of the culture to get the MOI of about 0.05 (1∶20). At 3 min post-infection, the infected culture was diluted 500-fold (60 µl was added to 30 ml of BHI in a 500-ml flask). Immediately two zero-point samples for PFU counts (0.5 ml and 1 ml) and one sample for CFU counts (0.5 ml) were taken and the flask was placed in the incubator at 28°C with shaking at 200 rpm. The PFU determinations were done by serial 10-fold dilution of the samples without chloroform (0.5 ml) and with chloroform (1 ml plus 30 µl of chloroform) in SM buffer up to 10−2 and double-layer plating in triplicates. Then 1-ml aliquots were taken at 10 minute time intervals up to 120 min postinfection, treated with chloroform, serially diluted in SM buffer and plated for PFU counts. When the burst sizes of L-413C and P2 vir1 were determined, CaCl2 was added to the bacterial culture up to 5 mM before adding the phage and samples for PFU counting were taken every 15 min. The experiment with P2 vir1 was performed at 37°C.
Phage DNA isolation
DNA of bacteriophages ϕA1122, L-413C, and P2 vir1 for qPCR calibration purpose was extracted and purified using Lambda Midi Kit (QIAGEN Inc., Valencia, California) according to the manufacturer's protocol with the following modifications: phage particles were harvested by centrifugation in a Beckman JA-17 rotor at 13,500 rpm for 3 h and treated with proteinase K at 55°C for 1 h for better disruption of phage capsids. Purity of DNA preparations was confirmed by agarose gel electrophoresis. DNA concentrations were measured by using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, Massachusetts).
Phage propagation for qPCR assays
Overnight broth bacterial cultures were diluted 1∶100 in BHI and grown with shaking until their concentration reached approximately 108 CFU/ml. Phage was added to different 10-fold dilutions of bacteria at final concentration of 105 PFU/ml to minimize the impact of the initial amount of phage on qPCR results. The infected bacterial culture was incubated with shaking for 3 h at 28°C. To prevent a non-specific low-level propagation of ϕA1122 on some strains of Y. pseudotuberculosis (see Table 1), they were incubated at 24°C. One milliliter aliquots were taken at certain time points and 30 µl of chloroform was immediately added to each sample to kill bacterial cells and release phage particles. One microliter of such a mixture (lysate) was directly used as target bacteriophage DNA for qPCR analysis without any DNA isolation step.
Phage amplification in simulated clinical samples containing Y. pestis
Simulated clinical tests were done with EDTA-treated whole human blood (Biological Specialty Corp.). The blood was diluted 10-fold with BHI broth (such dilutions are routinely used in bacteriological analysis of blood for plague [8]) containing log-phase culture of Y. pestis CO92 pgm− at concentrations ranging from 108 to 103 CFU/ml. Phage ϕA1122 was added to the final concentration of 105 PFU/ml. One milliliter aliquots were taken every 60 min during 5 h, and 30 µl of chloroform was added. Each sample was diluted 20-fold with SM buffer to minimize the inhibitory effect of blood on qPCR, phage plating was performed for PFU counts and 1 µl of each dilution was used for qPCR reaction. The phage DNA extraction step was omitted.
Primer design
Primers for qPCR monitoring of phage amplification were designed by using Beacon Designer™ program (Premier Biosoft Int., http://www.premierbiosoft.com). The target for the ϕA1122-based assay was the RNA polymerase gene, and primers were selected from its variable sequences to minimize potential cross-reactions with relative T7 and other T7-like phages. The primers for L-413C were selected from the unique tail fiber gene H having a mosaic structure [50]. Several pairs of primers with lowest self- and cross-complementarity were checked in qPCR with 100 PFU of ϕA1122 or L-413C (without a DNA extraction step), and two pairs providing the highest amplification signals were selected for further work (Table 2). The primers were analyzed using BLAST (Basic Local Alignment Search Tool) engine at the NCBI web site (http://blast.ncbi.nlm.nih.gov) against the nonredundant (nr) database to make sure that at least one primer from the pair represented a unique nucleotide sequence. The primers were also tested for specificity against Y. pestis genomic DNA (suspension of strain CO92 pgm− in distilled water boiled for 5 min; about 5×108 CFU/ml) and against the closest relatives of phages ϕA1122 (T7) and L-413C (P2). Primers for L-413C DNA amplification were specific for this phage and did not amplify DNA of P2 vir1. Despite the fact that ϕA1122 and T7 RNA polymerase genes share 93% nucleotide sequence identity, no cross reaction of ϕA1122 primers was observed after 40 cycles of qPCR with 103 PFU of T7. However, a weak cross reaction (Ct = 28.37) was observed with a high dose of T7, 106 PFU, although the reaction with 106 PFU of ϕA1122 was much more robust (Ct = 7.50).
Table 2. Primers used for qPCR in this study.
| Designation | DNA Sequence | Length (bp) | Tm (°C) | GC% | Product (bp) |
| ϕA1122-F | 5′-CCAAATGGAAGCACTGCCCTGTAG-3′ | 24 | 61.8 | 54.2 | 105 |
| ϕA1122-R | 5′-ATGCGGTGAGAGCCTCAGGATTC-3′ | 23 | 62.1 | 56.5 | |
| L-413C-F | 5′-ACGTGGTCATGTCCGTCACAATC-3′ | 23 | 60.9 | 52.2 | 75 |
| L-413C-R | 5′-CAGAACCCCATTGCCTTTATCTTCAG-3′ | 26 | 60.3 | 46.2 |
Quantitative real-time PCR assay
Maxima™ SYBR Green/ROX qPCR Master Mix 2× (Fermentas Inc., Glen Burnie, Maryland) was used in all qPCR experiments according to the vendor's recommendations. Reactions were performed in a total volume of 20 µl containing 1 µl of DNA template (pure phage DNA or lysates containing live phage particles), 10 µl of the master mix, and 0.9 µM of each primer and were run on a LightCycler 2.0 (Roche Applied Science, Indianapolis, Indiana). The cycling parameters were: 95°C, 10 min; 40× (95°C, 20 s; 60°C, 60 s) with fluorescence measurement at the end of each cycle.
qPCR sensitivity test
Y. pestis CO92 pgm− was grown in BHI broth at 28°C until OD600 reached 0.2. The culture was serially diluted from 108 CFU/ml to 103 CFU/ml. Each bacterial suspension was infected with ϕA1122 or L-413C at final concentration of 105 PFU/ml (100 PFU/µl). 1-ml aliquots from each culture were collected every 30 minutes for ϕA1122 or every 60 minutes for L-413C within 3 h post-infection, treated with chloroform (30 µl), and 1µl was used for qPCR.
Evaluation of inhibitory effect of blood on qPCR
Whole human blood (Biological Specialty Corp.) and dilutions of 1∶10 and 1∶20 with SM buffer were tested. Purified DNA of ϕA1222 was added at concentrations ranging from 0.5 ng/µl to 0.5 fg/µl, and the qPCR reactions were run in duplicate. Phage DNA solutions in SM buffer at the same concentrations were used as a negative control.
Statistical analysis
Results of all qPCR tests of phage amplification are presented as the mean values of three independent experiments. Statistical significance was determined by One Way ANOVA (analysis of variance) analysis using a free online program at the web site of Vassar College (http://faculty.vassar.edu/lowry/anova1u.html). P values <0.05 were considered significant.
Results
Lytic activities against Y. pestis and propagation rates of three bacteriophages
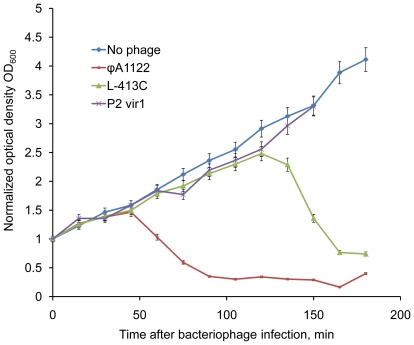
To develop a phage-based qPCR system for Y. pestis detection, we tested plague diagnostic phages ϕA1122 [7], [48], [49], [51] and L-413C [50], [52]–[54], as well as a clear plaque mutant of coliphage P2, P2 vir1 [57]. We have previously shown that P2 vir1 lyses Y. pestis at 37°C, has relatively low plaquing efficiency at 28°C, but is not active against a wild-type strain of Y. pseudotuberculosis at either temperature [50]. Since the speed of phage propagation is critical for qPCR efficiency, we first checked some parameters of amplification of ϕA1122, L-413C, and P2 vir1 on Y. pestis CO92 pgm−. Phage propagation rates can be indirectly measured by their lytic activity. The plaque sizes of the three phages after 24-h incubation at 37°C were measured. This temperature of growth was chosen to enhance lytic properties of P2 vir1 and standardize the conditions (ϕA1122 and L-413C at 28°C appeared identically). The mean diameters of plaques (measured for 20 plaques) for ϕA1122, L-413C, and P2 vir1 were 10, 3, and 1 mm, respectively. Dynamics of bacterial lysis by each of the three phages in BHI broth was then measured at 28°C, the optimal temperature for Y. pestis growth (Fig. 1). In case of ϕA1122, markedly detectable lysis occurred 45 minutes after infection. In contrast with this, clarification of culture suspension infected with L-413C occurred only in 2 h but was vigorous. No lysis of Y. pestis culture infected with P2 vir1 was observed at 28°C. It is important that ϕA1122 efficiently killed Y. pestis cells during its propagation; we were unable to detect any viable bacterial cells after 1-h incubation with ϕA1122.
Figure 1. Lytic properties of bacteriophages ϕA1122, L-413C, and P2 vir1 towards Y. pestis CO92 pgm−.
The dynamics of lysis was determined in BHI broth at multiplicity of infection of 0.1. Optical density was normalized to the start of infection (1 on the Y axis corresponds to the initial OD600 = 0.2).
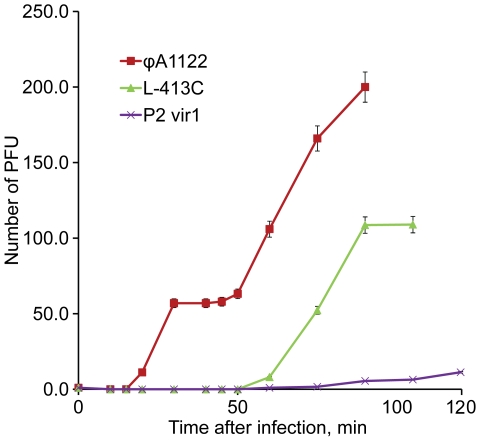
Phage propagation rates were also estimated more directly by determination of burst size (Fig. 2), which is the number of phage particles released from a single host cell. The burst sizes of ϕA1122, L-413C, and P2 vir1 for Y. pestis CO92 pgm− were about 57, 115, and 9 PFU, respectively (see the plateaus on the phage growth curves in Fig. 2). The lengths of lytic cycles of these three phages were approximately 30, 90, and 90 min. Thus, L-413C has the highest burst size, about 115 particles from one Y. pestis cell, but its lytic cycle is relatively long, approximately 90 min. Phage ϕA1122 has a lower burst size, 57 PFU, but in 90 min a single phage is supposed to make three lytic cycles and produce, under ideal conditions, 573≈1.9×105 phages. Thus, ϕA1122 showed the maximum lytic activity and the highest propagation rate; P2 vir1 had low lytic activity and was slow growing; and L-413C had intermediate growth characteristics. Due to the comparatively slow and weak amplification of P2 vir1 on Y. pestis, this phage was excluded from further experiments.
Figure 2. Determination of lysis speed and burst sizes for bacteriophages ϕA1122, L-413C, and P2 vir1 on Y. pestis CO92 pgm−.
Phage burst sizes (an average phage progeny produced by one bacterial cell) correspond to plateaus on the curves.
Performance testing of qPCR with purified DNA from Y. pestis-specific phages and with intact phage particles
The quantitative parameters of phage-based qPCR were first tested by using serial ten-fold dilutions of DNA purified from ϕA1122 and L-413C. These tests were performed in triplicate and yielded a log linear relationship between DNA concentrations and threshold cycle number (Ct), spanning an 8-log dilution series, from 5 ng down to 0.5 fg of DNA (Fig. 3, A and B). We calculated that this corresponds to 12 to 1.2×108 genome equivalents of ϕA1122 and 15 to 1.5×108 genome equivalents of L-413C based on the facts that their genome sizes are 37,555 [49] and 30,728 bp [50], respectively. To compare qPCR results obtained using purified DNA and intact phage particles, a series of 10-fold phage lysate dilutions was prepared in SM buffer, and qPCR reactions were run on different concentrations of phage particles ranging from 101 to 107 PFU per 1 µl of phage lysate (per 20 µl of qPCR sample) for ϕA1122 and from 101 to 108 PFU for L-413C. The number of viable particles per sample was confirmed by plaque assays. The results of qPCR with the particles presented in Fig. 3 (C and D) are in accordance with the data obtained with phage DNA (Fig. 3, A and B). Phage lysates treated with DNase I showed the same results as untreated phage particles (data not shown).
Figure 3. Parameters of ϕA1122- and L-413C-based qPCR tests for phage DNA and live phage particles determined by linear regression method.
A and B, standard curves plotted for DNA concentrations of ϕA1122 and L-413C, respectively. C and D, standard curves plotted for live phage particles of ϕA1122 and L-413C, respectively.
Sensitivity of phage-based qPCR detection of Y. pestis
To determine the sensitivity of phage-mediated detection of Y. pestis, we conducted a series of experiments where reporter phages ϕA1122 and L-413C were propagated on various concentrations of Y. pestis CO92 pgm− cells with subsequent PFU counts and qPCR tests. The phage titer rises determined by qPCR (Fig. 4) were calculated based on Ct values and our calibration data (Fig. 3). The starting points of infection corresponding to 100 PFU per 20-µl sample (containing 1 µl of phage lysate) were normalized to 1 (Fig. 4). Our data showed that the detection of Y. pestis using ϕA1122 is robust and there are marked differences in phage yields at different concentrations of Y. pestis. A significant rise in phage concentration and qPCR signal was observed even at the lowest possible starting concentration of Y. pestis, 103 CFU/ml, or one bacterium per 20 µl of qPCR sample (about 50-fold, see Fig. 4, A). This indicates that the limit of ϕA1122-based qPCR detection of Y. pestis was 103 CFU/ml, equivalent to one host bacterium per 20 µl of qPCR sample (or per 1 µl of phage lysate). At the same time, a rise in L-413C titer and the qPCR signal was statistically significant only at higher concentrations of host cells, 108–105 CFU/ml that corresponds to 105–102 CFU in 20 µl of qPCR sample (Fig. 4, B). Thus, the sensitivity of L-413C-mediated qPCR assay was 105 CFU/ml, or 100 CFU per sample. The entire detection test included 3-h phage propagation and 1-h qPCR reaction and took 4 h.
Figure 4. Dynamics of growth of phages ϕA1122 and L-413C on different concentrations of Y. pestis cells detected by qPCR.
The starting points of phage infection correspond to 100 PFU per 1 µl sample and are normalized to 1. A. The titer rise of ϕA1122. B. L-413C amplification.
Specificity of qPCR assays
The specificity of phage-based detection was tested on 62 strains belonging to 19 bacterial species of Enterobacteriaceae including nine Yersinia species (Table 1). Both the ability of ϕA1122 and L-413C to propagate on various bacteria at 28°C and the potential rise in qPCR signal intensity were studied. L-413C did not form any plaque and did not cause any decrease in Ct value on bacterial cultures other than Y. pestis, confirming the high specificity of this phage towards Y. pestis [50], [52], [53]. Both propagation and qPCR tests of ϕA1122 incubated at 28°C were negative with nonpathogenic Yersinia, as well as with each of 17 Y. enterocolitica strains. However, bacteriophage ϕA1122 grew on 4 out of 20 different strains of Y. pseudotuberculosis (the closest phylogenetic relative of Y. pestis), and the phage propagation was easily detected by qPCR analysis. To enhance the specificity, we reduced the incubation temperature during ϕA1122 infection from 28°C to 24°C [7], [51]. This allowed the assay to get practically 100% specificity: only a low-level amplification of ϕA1122 was observed on one strain of Y. pseudotuberculosis, IB (the phage yield was 106 lower than that determined for Y. pestis CO92). The same result was obtained after propagation of ϕA1122 at 20°C.
Simulated clinical tests
Comparison of qPCR efficiencies for detection of ϕA1122 in EDTA-treated human blood diluted 1∶20 and in SM buffer showed virtually no inhibition of the reaction by blood components (Fig. 5, A). To evaluate the possibility of using phage-based qPCR for diagnostics of human plague, we performed ϕA1122 propagation and qPCR analysis in human blood experimentally contaminated with Y. pestis (Fig. 5, B). Phage ϕA1122 propagated in blood diluted with BHI broth 1∶10, although slower than on the control culture grown in BHI, and this propagation was detected by qPCR. A statistically significant rise in phage titer was observed after 5 h at the lowest concentration of Y. pestis of 105 CFU/ml in diluted blood, which corresponded to 100 CFU in a qPCR sample and 106 CFU/ml in the undiluted whole blood sample.
Figure 5. qPCR tests on simulated clinical human blood samples.
A. Linear regression of ϕA1122 DNA concentration in blood diluted 1∶20 in comparison with SM buffer data. B. ϕA1122-based detection of Y. pestis in artificially contaminated blood diluted 10-fold with BHI broth. To calculate the actual bacterial loads in the undiluted blood samples, the CFU numbers shown should be multiplied by 10. The starting points of phage infection correspond to 100 PFU per 1 µl sample and are normalized to 100 = 1.
Discussion
We report the development of a novel quantitative real-time PCR assay for indirect detection of Y. pestis based on amplification of non-modified reporter Y. pestis-specific bacteriophages in the presence of the host bacteria. This qPCR method utilizing phages ϕA1122 [7], [48], [49], [51] and L-413C [50], [52]–[54] is a reasonable alternative to a standard phage lysis test. It was shown to be easy, reliable, rapid, highly sensitive and specific. Since bacteriophages propagate only on viable and culturable cells [40], [58], our approach allows detection of live and metabolically active cells of Y. pestis, which may be specifically important in forensic studies and for characterization of activity of natural plague foci. qPCR with primers specific for phage DNA has been previously used for quantitation of bacteriophage λ instead of routine plating assay [43] and then employed for phage-based detection of several bacterial species [44]–[46]. In case of the use of gamma phage for indirect detection of B. anthracis, the assay took only about 5 h, its sensitivity reached one bacterial cell per sample, and amplification was not observed with four other species of Gram-positive and Gram-negative bacteria [44].
We analyzed three bacteriophages capable of lysing Y. pestis, ϕA1122, L-413C, and P2 vir1 [57], as potential reporters for qPCR phage-based detection of the bacterium. Since the developed assay is based on the detection and quantitation of rapidly replicating phage DNA, we first studied some parameters of lysis and propagation rates of the three phages including mean diameters of plaques, dynamics of bacterial lysis, burst size and duration of lytic cycle. Plague diagnostic bacteriophages ϕA1122 and L-413C displayed high lytic activities and propagation rates on Y. pestis, particularly ϕA1122 (Fig. 1 and 2). Phage ϕA1122 formed the largest plaques, had the shortest time of the lytic cycle and caused the fastest and most robust lysis of the bacterial culture. Importantly, ϕA1122 efficiently inactivated Y. pestis in all diagnostic samples, which is advantageous when working with this infectious agent. Of the three phages studied, L-413C showed the biggest burst size, a longer lytic cycle and delayed but vigorous lysis of bacterial cells. P2 vir1 displayed low lytic activity, slow and weak growth on Y. pestis and thus was excluded from further tests. These data were essential for designing our diagnostic assay, but also can be used for future bacteriophage plague therapy applications [59] and add some knowledge to the biology of phages ϕA1122, L-413C, and P2 vir1.
Testing of the quantitative parameters of phage-based qPCR with purified DNA of phages ϕA1122 and L-413C showed reliable standard curves down to 0.5 fg (12–15 genome equivalents) that, based on burst sizes, corresponds to 0.1–0.2 cells of Y. pestis. The calculated numbers of phage genome equivalents correlated well with the standard curves based on qPCR with intact phage particles (Fig. 3). Multiple experiments on the propagation of a standard number of ϕA1122 and L-413C particles on various concentrations of Y. pestis cells with subsequent plaque counts and qPCR tests showed that the limit of ϕA1122-mediated detection of Y. pestis was as low as 103 CFU/ml, or one bacterium per qPCR sample. The sensitivity of L-413C-based test was 105 CFU/ml, or 100 CFU per sample (Fig. 4), and the duration of the whole procedure in both cases was 4 h. These results were obtained without the concentration of samples or DNA purification, which is a standard step of conventional real-time PCR assays targeting bacterial genes [60]. Several groups have reported on the use of different variants of real-time PCR targeting plasmid and chromosomal genes of Y. pestis [16]–[26]. The best detection limit observed in these studies also reached one bacterial cell per sample [18], [22], [25] and the shortest assay time described was the same as in our work, 4 h [22].
The use of simplex real-time PCR targeting a single plasmid [16], [17], [19] or chromosomal [21] gene can result in missed detection of mutant strains and the entire biovars of Y. pestis. For example, enzootic strains of Y. pestis isolated from voles in Transcaucasian Highland and Mountain Dagestan are missing the pPst plasmid [61], the most prevalent target for diagnostic PCR. The loss from laboratory strains of Y. pestis of one, two, or three plasmids [61], or the extensive chromosomal pigmentation region [62] can occur, and some strains cured of the pPst [63]–[65] or pFra [63], [66], [67] plasmid maintain full virulence. Real-time PCR in multiplex format [18], [22]–[26] broadens the spectra of detected strains but this makes the reaction more technically complicated. Based on the facts that ϕA1122 [7], [49], [51] and L-413C [50], [52], [53] can lyse almost all (99.8–99.9%) Y. pestis strains of several thousand tested, we propose a broad-range qPCR method in a simplex format.
The specificity of this phage-based detection procedure was tested on 62 strains of 19 bacterial species including a variety of isolates of pathogenic and non-pathogenic Yersinia. Both the ability of ϕA1122 and L-413C to propagate on various bacterial cultures and phage qPCR signals in the presence of the bacteria were determined (Table 1). Bacteriophage ϕA1122 has been shown to lyse practically all strains of Y. pestis and some isolates of Y. pseudotuberculosis at 26–28°C and higher temperatures [7], [49], [51]. This issue can be bypassed by using lower temperatures for growth, 20–25°C [7], [51]. In our experiments, ϕA1122 grew at 28°C on 20% (4 of 20) strains of Y. pseudotuberculosis, and the phage amplification was registered by qPCR. The reduction of growth temperature to 24°C allowed us to reach virtually 100% specificity: only one Y. pseudotuberculosis strain (IB) displayed a low degree of ϕA1122 propagation and a weak qPCR signal (the propagation rate was six orders of magnitude lower than that obtained on Y. pestis CO92). The same weak growth of ϕA1122 was observed at 20°C. All ϕA1122 propagation and qPCR tests were negative with 17 Y. enterocolitica strains and 9 isolates of nonpathogenic Yersinia. The L-413C assay did prove to be 100% specific when using the regular temperature of incubation, 28°C: the amplification of this phage was detected by using plaque counts and qPCR only on Y. pestis (Table 1). Our data confirmed the high specificity of L-413C to Y. pestis [50], [52], [53]. Inter-laboratory trials of L-413C on 6,000 global isolates of Y. pestis and 2,000 strains of Y. pseudotuberculosis have shown an absolute specificity of this phage for the plague agent [53]. Based on this extraordinary specificity of L-413C confirmed in our tests, we propose to use parallel qPCR assays with both phages for rapid and reliable detection and identification of Y. pestis.
The clinical performance of this phage-based qPCR assay was evaluated with EDTA-treated human blood artificially contaminated with Y. pestis. The appropriate dilution of blood with SM buffer (1∶20) allowed us to avoid inhibition of ϕA1122-specific qPCR by blood components (Fig. 5, A) but this decreased the assay sensitivity twenty-fold. Phage ϕA1122 was shown to propagate in the spiked blood diluted 1∶10 with BHI by the methods of plating and qPCR. Such dilutions with a nutrient broth are typically done in bacteriological tests of blood for plague [8]. Statistically significant phage amplification was detected in 5 h, and the detection limit was 100 CFU of Y. pestis in a qPCR sample, which corresponded to 105 CFU/ml in diluted blood (Fig. 5, B) and to 106 CFU/ml in undiluted blood. It has been previously shown that the efficiency of Y. pestis detection by real-time PCR from blood of infected nonhuman primates is lower compared with sera and especially with swabs from the oropharyngeal cavity [16]. The detection threshold observed by those authors was 2.1×105 copies of a gene located in the pPst plasmid [16] known to have 186 copies per genome [68]; this makes the detection limit 1,129 CFU, which is higher than in our tests. Other investigators achieved lower detection limits for real-time PCR assays with simulated human respiratory specimens, but only after multi-step sample treatment [19], [22]. For example, a detection limit of 85 CFU was observed after the pretreatment of sputum with a mucolytic agent, centrifugation, resuspending in TE buffer, artificial contamination with Y. pestis, and DNA extraction [22]. This sensitivity level of our test with spiked blood was satisfactory without pretreatment because colony counts in blood cultures of plague patients can reach 4×107 CFU/ml [69]. The sensitivity level could be enhanced using a concentration step.
Bacteriophages have been used for Y. pestis detection and plague diagnosis since the early 1930s [7], [8], [47]–[50]. ϕA1122 is recommended as an essential plague diagnostic tool by the CDC [7], [49], and L-413C is routinely used for the same purpose in many countries of Eastern Europe and Central Asia [50], [52]–[54]. Most phage plating assays require the isolation of a pure culture of Y. pestis. The culture isolation together with the lysis test usually takes three days [7], [8]. A genetically engineered phage ϕA1122 expressing luciferase reporter genes has been recently used for indirect detection of Y. pestis [39]. The method was shown to be very rapid (1 h) and allowed detection of ≥820 Y. pestis cells but fluorescent signals higher than the background were observed with two Y. pseudotuberculosis strains and even with a Y. enterocolitica isolate. An additional concern about this method is the short time frame for effective application: a gradual decline in signal strength has been found when using an incubation time of the phage with Y. pestis longer than 90 min [39].
We propose qPCR with the use of both ϕA1122 and L-413C as a reasonable alternative to routine phage lysis tests for detection and identification of Y. pestis. Our assay is simple (because it utilizes native, non-modified phages; this is also important for possible expanding the panel of phages used), rapid (4 h), highly sensitive (up to 1 cell per sample) and specific for Y. pestis. This method can be used for plague diagnostics, forensic purposes and the monitoring of plague foci. Another potential application is for pharmacokinetics studies and the evaluation of phage propagation in vivo during bacteriophage therapy. Such applications are important due to the emergence of multidrug-resistant strains of Y. pestis [5], [6].
Acknowledgments
We wish to thank Dr. Carmen Fernandez-Prada for her help with the human subjects use protocol. Dr. Patrick McGann is gratefully acknowledged for advising on qPCR. The findings and opinions expressed herein belong to the authors and do not necessarily reflect the official views of the WRAIR, the U.S. Army or the Department of Defense.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Defense Threat Reduction Agency, Joint Science and Technology Office, Medical S&T Division. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perry RD, Fetherston JD. Yersinia pestis – etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gage KL, Kosoy MY. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- 3.Butler T. Plague into the 21st Century. Clin Infect Dis. 2009;49:736–742. doi: 10.1086/604718. [DOI] [PubMed] [Google Scholar]
- 4.Inglesby TV, Dennis DT, Henderson DA, Bartlett JG, Ascher MS, et al. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 2000;283:2281–2290. doi: 10.1001/jama.283.17.2281. [DOI] [PubMed] [Google Scholar]
- 5.Galimand M, Carniel E, Courvalin P. Resistance of Yersinia pestis to antimicrobial agents. Antimicrob Agents Chemother. 2006;50:3233–3236. doi: 10.1128/AAC.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, et al. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One Mar. 2007;21; 2(3):e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu MC. Laboratory manual of plague diagnostic tests. Fort Collins: Centers for Disease Control and Prevention; 2000. 129 [Google Scholar]
- 8.Naumov AV, Samoilova LV, editors. Manual on prophylaxis of plague. Saratov: Russian Research Anti-Plague Institute “Microbe” Press.; 1992. 278 [Google Scholar]
- 9.Campbell J, Lowe J, Walz S, Ezzell J. Rapid and specific identification of Yersinia pestis by using a nested polymerase chain reaction procedure. J Clin Microbiol. 1993;31:758–759. doi: 10.1128/jcm.31.3.758-759.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch J, Schwan TG. New method for plague surveillance using polymerase chain reaction to detect Yersinia pestis in fleas. J Clin Microbiol. 1993;31:1511–1514. doi: 10.1128/jcm.31.6.1511-1514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norkina OV, Kulichenko AN, Gintsburg AL, Tuchkov IV, Popov YA, et al. Development of a diagnostic test for Yersinia pestis by the polymerase chain reaction. J Appl Bacteriol. 1994;76:240–245. doi: 10.1111/j.1365-2672.1994.tb01622.x. [DOI] [PubMed] [Google Scholar]
- 12.Trebesius K, Harmsen D, Rakin A, Schmelz J, Heesemann J. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J Clin Microbiol. 1998;36:2557–2564. doi: 10.1128/jcm.36.9.2557-2564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahalison L, Vololonirina E, Ratsitorahina M, Chanteau S. Diagnosis of bubonic plague by PCR in Madagascar under field conditions. J Clin Microbiol. 2000;38:260–263. doi: 10.1128/jcm.38.1.260-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neubauer H, Meyer H, Prior J, Aleksic S, Hensel A, et al. A combination of different polymerase chain reaction (PCR) assays for the presumptive identification of Yersinia pestis. J Vet Med B Infect Dis Vet Public Health. 2000;47:573–580. doi: 10.1046/j.1439-0450.2000.00384.x. [DOI] [PubMed] [Google Scholar]
- 15.Melo AC, Almeida AM, Leal NC. Retrospective study of a plague outbreak by multiplex-PCR. Lett Appl Microbiol. 2003;37:361–364. doi: 10.1046/j.1472-765x.2003.01377.x. [DOI] [PubMed] [Google Scholar]
- 16.Higgins JA, Ezzell J, Hinnebusch BJ, Shipley M, Henchal EA, et al. 5′ nuclease PCR assay to detect Yersinia pestis. J Clin Microbiol. 1998;36:2284–2288. doi: 10.1128/jcm.36.8.2284-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iqbal SS, Chambers JP, Goode MT, Valdes JJ, Brubaker RR. Detection of Yersinia pestis by pesticin fluorogenic probe-coupled PCR. Mol Cell Probes. 2000;14:109–114. doi: 10.1006/mcpr.2000.0295. [DOI] [PubMed] [Google Scholar]
- 18.Tomaso H, Reisinger EC, Al Dahouk S, Frangoulidis D, Rakin A, et al. Rapid detection of Yersinia pestis with multiplex real-time PCR assays using fluorescent hybridization probes. FEMS Immunol Med Microbiol. 2003;38:117–126. doi: 10.1016/S0928-8244(03)00184-6. [DOI] [PubMed] [Google Scholar]
- 19.Loïez C, Herwegh S, Wallet F, Armand S, Guinet F, et al. Detection of Yersinia pestis in sputum by real-time PCR. J Clin Microbiol. 2003;41:4873–4875. doi: 10.1128/JCM.41.10.4873-4875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAvin JC, McConathy MA, Rohrer AJ, Huff WB, Barnes WJ, et al. A real-time fluorescence polymerase chain reaction assay for the identification of Yersinia pestis using a field-deployable thermocycler. Mil Med. 2003;168:852–855. [PubMed] [Google Scholar]
- 21.Chase CJ, Ulrich MP, Wasieloski LP, Kondig JP, Garrison J, et al. Real-time PCR assays targeting a unique chromosomal sequence of Yersinia pestis. Clin Chem. 2005;51:1778–1785. doi: 10.1373/clinchem.2005.051839. [DOI] [PubMed] [Google Scholar]
- 22.Woron AM, Nazarian EJ, Egan C, McDonough KA, Cirino NM, et al. Development and evaluation of a 4-target multiplex real-time polymerase chain reaction assay for the detection and characterization of Yersinia pestis. Diagn Microbiol Infect Dis. 2006;56:261–268. doi: 10.1016/j.diagmicrobio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Stewart A, Satterfield B, Cohen M, O'Neill K, Robison R. A quadruplex real-time PCR assay for the detection of Yersinia pestis and its plasmids. J Med Microbiol. 2008;57:324–331. doi: 10.1099/jmm.0.47485-0. [DOI] [PubMed] [Google Scholar]
- 24.Tomaso H, Jacob D, Eickhoff M, Scholz HC, Al Dahouk S, et al. Preliminary validation of real-time PCR assays for the identification of Yersinia pestis. Clin Chem Lab Med. 2008;46:1239–1244. doi: 10.1515/CCLM.2008.251. [DOI] [PubMed] [Google Scholar]
- 25.Matero P, Pasanen T, Laukkanen R, Tissari P, Tarkka E, et al. Real-time multiplex PCR assay for detection of Yersinia pestis and Yersinia pseudotuberculosis. APMIS. 2009;117:34–44. doi: 10.1111/j.1600-0463.2008.00013.x. [DOI] [PubMed] [Google Scholar]
- 26.Amoako KK, Goji N, Macmillan T, Said KB, Druhan S, et al. Development of multitarget real-time PCR for the rapid, specific, and sensitive detection of Yersinia pestis in milk and ground beef. J Food Prot. 2010;73:18–25. doi: 10.4315/0362-028x-73.1.18. [DOI] [PubMed] [Google Scholar]
- 27.Craigie J. The significance and application of bacteriophage in bacteriological and virus research. Bacteriol Rev. 1946;10:73–88. [PubMed] [Google Scholar]
- 28.Aucken HM, Westwell K. Reaction difference rule for phage typing of Staphylococcus aureus at 100 times the routine test dilution. J Clin Microbiol. 2002;40:292–293. doi: 10.1128/JCM.40.1.292-293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNerney R, Kambashi BS, Kinkese J, Tembwe R, Godfrey-Faussett P. Development of a bacteriophage phage replication assay for diagnosis of pulmonary tuberculosis. J Clin Microbiol. 2004;42:2115–2120. doi: 10.1128/JCM.42.5.2115-2120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abshire TG, Brown JE, Ezzell JW. Production and validation of the use of gamma phage for identification of Bacillus anthracis. Clin Microbiol. 2005;43:4780–4788. doi: 10.1128/JCM.43.9.4780-4788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster G, Osterman BS, Godfroid J, Jacques I, Cloeckaert A. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int J Syst Evol Microbiol. 2007;57:2688–2693. doi: 10.1099/ijs.0.65269-0. [DOI] [PubMed] [Google Scholar]
- 32.De Lappe N, Connor JO, Doran G, Devane G, Cormican M. Role of subtyping in detecting Salmonella cross contamination in the laboratory. BMC Microbiol. 2009;9:155. doi: 10.1186/1471-2180-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders MF. A rapid bioluminescent technique for the detection and identification of Listeria monocytogenes in the peresence of Listeria innocua. In: Campbell AK, Kricka LJ, Stanley PE, editors. Bioluminescence and chemiluminescence: fundamental and applied aspects. Chichester: John Wiley & Sons; 1995. pp. 454–457. [Google Scholar]
- 34.Blasco R, Murphy MJ, Sanders MF, Squirrell DJ. Specific assays for bacteria using phage mediated release of adenylate kinase. Appl Microbiol. 1998;84:661–666. doi: 10.1046/j.1365-2672.1998.00393.x. [DOI] [PubMed] [Google Scholar]
- 35.Neufeld T, Schwartz-Mittelmann A, Biran D, Ron EZ, Rishpon J. Combined phage typing and amperometric detection of released enzymatic activity for the specific identification and quantification of bacteria. Anal Chem. 2003;75:580–585. doi: 10.1021/ac026083e. [DOI] [PubMed] [Google Scholar]
- 36.Loessner MJ, Rees CE, Stewart GS, Scherer S. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl Environ Microbiol. 1996;62:1133–1140. doi: 10.1128/aem.62.4.1133-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banaiee N, Bobadilla-Del-Valle M, Bardarov S, Jr, Riska PF, Small PM, et al. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. Clin Microbiol. 2001;39:3883–3888. doi: 10.1128/JCM.39.11.3883-3888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schofield DA, Westwater C. Phage-mediated bioluminescent detection of Bacillus anthracis. J Appl Microbiol. 2009;107:1468–1478. doi: 10.1111/j.1365-2672.2009.04332.x. [DOI] [PubMed] [Google Scholar]
- 39.Schofield DA, Molineux IJ, Westwater C. Diagnostic bioluminescent phage for detection of Yersinia pestis. J Clin Microbiol. 2009;47:3887–3894. doi: 10.1128/JCM.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oda M, Morita M, Unno H, Tanji Y. Rapid detection of Escherichia coli O157:H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl Environ Microbiol. 2004;70:527–534. doi: 10.1128/AEM.70.1.527-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R, McKinstry M, Hwang J, Oppenheim AB, Fekete RA, et al. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc Natl Acad Sci USA. 2006;103:4841–4845. doi: 10.1073/pnas.0601211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yim PB, Clarke ML, McKinstry M, De Paoli Lacerda SH, Pease LF, 3rd, et al. Quantitative characterization of quantum dot-labeled lambda phage for Escherichia coli detection. Biotechnol Bioeng. 2009;104:1059–1067. doi: 10.1002/bit.22488. [DOI] [PubMed] [Google Scholar]
- 43.Edelman DC, Barletta J. Real-time PCR provides improved detection and titer determination of bacteriophage. Biotechniques. 2003;35:368–375. doi: 10.2144/03352rr02. [DOI] [PubMed] [Google Scholar]
- 44.Reiman RW, Atchley DH, Voorhees KJ. Indirect detection of Bacillus anthracis using real-time PCR to detect amplified gamma phage DNA. J Microbiol Methods. 2007;68:651–653. doi: 10.1016/j.mimet.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Kutin RK, Alvarez A, Jenkins DM. Detection of Ralstonia solanacearum in natural substrates using phage amplification integrated with real-time PCR assay. J Microbiol Methods. 2009;76:241–246. doi: 10.1016/j.mimet.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Ogorzaly L, Gantzer C. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: application to urban raw wastewater. J Virol Methods. 2006;138:131–139. doi: 10.1016/j.jviromet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Pokrovskaya MP. A plague bacteriophage in dead susliks. Gigiena Epidemiol. 1929;12:31–34. [Google Scholar]
- 48.Advier M. Etude d'un bactériophage antipesteux. Bull Soc Pathol Exotiques. 1933;26:94–99. [Google Scholar]
- 49.Garcia E, Elliott JM, Ramanculov E, Chain PS, Chu MC, et al. The genome sequence of Yersinia pestis bacteriophage ϕA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J Bacteriol. 2003;185:5248–5262. doi: 10.1128/JB.185.17.5248-5262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia E, Chain P, Elliott JM, Bobrov AG, Motin VL, et al. Molecular characterization of L-413C, a P2-related plague diagnostic bacteriophage. Virology. 2008;372:85–96. doi: 10.1016/j.virol.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 51.Gunnison JB, Larson A, Lazarus AS. Rapid differentiation between Pasteurella pestis and Pasteurella pseudotuberculosis by action of bacteriophage. J Infect Dis. 1951;88:254–255. doi: 10.1093/infdis/88.3.254. [DOI] [PubMed] [Google Scholar]
- 52.Larina VS, Anisimov PI, Adamov AK. A novel strain of plague bacteriophage for identification of Pasteurella pestis. Probl Particularly Dangerous Infect. 1970;11:132–136. [Google Scholar]
- 53.Imamaliev OG, Serebryakova VG, Anisimova TI, Plotnikov OP, Sergeeva GM, et al. Comparative estimation of activity and specificity of diagnostic plague bacteriophages, L-413C, and the Pokrovskaya phage. In: Bektemirov TA, Zhouravleva YZ, Litvinova MY, editors. Standards, strains, and methods of control of bacterial and viral preparations. Moscow: Mechnikov Institute Press; 1986. pp. 102–106. [Google Scholar]
- 54.Bobrov AG, Kirillina OA, Filippov AA, Kutyrev VV. Restriction mapping of DNA of plague diagnostic phages of Pokrovskaya and L-413C. Probl Particularly Dangerous Infect. 1999;79:138–144. [Google Scholar]
- 55.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56.Ellis EL, Delbrück M. The growth of bacteriophage. J Gen Physiol. 1939;22:365–384. doi: 10.1085/jgp.22.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bertani LE. The effect of the inhibition of protein synthesis on the establishment of lysogeny. Virology. 1957;4:53–71. doi: 10.1016/0042-6822(57)90043-0. [DOI] [PubMed] [Google Scholar]
- 58.Awais R, Fukudomi H, Miyanaga K, Unno H, Tanji Y. A recombinant bacteriophage-based assay for the discriminative detection of culturable and viable but nonculturable Escherichia coli O157:H7. Biotechnol Prog. 2008;22:853–859. doi: 10.1021/bp060020q. [DOI] [PubMed] [Google Scholar]
- 59.Anisimov AP, Amoako KK. Treatment of plague: promising alternatives to antibiotics. J Med Microbiol. 2006;55:1461–1475. doi: 10.1099/jmm.0.46697-0. [DOI] [PubMed] [Google Scholar]
- 60.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Filippov AA, Solodovnikov NS, Kookleva LM, Protsenko OA. Plasmid content in Yersinia pestis strains of different origin. FEMS Microbiol Lett. 1990;55:45–48. doi: 10.1016/0378-1097(90)90165-m. [DOI] [PubMed] [Google Scholar]
- 62.Fetherston JD, Schuetze P, Perry RD. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 63.Kutyrev VV, Filippov AA, Shavina NI, Protsenko OA. Genetic analysis and simulation of the virulence of Yersinia pestis. Mol Gen Mikrobiol Virusol. 1989;8:42–47. [PubMed] [Google Scholar]
- 64.Samoilova SV, Samoilova LV, Yezhov IN, Drozdov IG, Anisimov AP. Virulence of pPst+ and pPst− strains of Yersinia pestis for guinea-pigs. J Med Microbiol. 1996;45:440–444. doi: 10.1099/00222615-45-6-440. [DOI] [PubMed] [Google Scholar]
- 65.Welkos SL, Friedlander AM, Davis KJ. Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain CO92. Microbial Pathogenesis. 1997;23:211–223. doi: 10.1006/mpat.1997.0154. [DOI] [PubMed] [Google Scholar]
- 66.Drozdov IG, Anisimov AP, Samoilova SV, Yezhov IN, Yeremin SA, et al. Virulent non-capsulate Yersinia pestis variants constructed by insertion mutagenesis. J Med Microbiol. 1995;42:264–268. doi: 10.1099/00222615-42-4-264. [DOI] [PubMed] [Google Scholar]
- 67.Welkos SL, Davis KM, Pitt LM, Worsham PL, Friedlander AM. Studies on the contribution of the F1 capsule-associated plasmid pFra to the virulence of Yersinia pestis. Contrib Microbiol Immunol. 1995;13:299–305. [PubMed] [Google Scholar]
- 68.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 69.Butler T, Levin J, Linh NN, Chau DM, Adickman M, et al. Yersinia pestis infection in Vietnam. II. Quantiative blood cultures and detection of endotoxin in the cerebrospinal fluid of patients with meningitis. J Infect Dis. 1976;133:493–499. doi: 10.1093/infdis/133.5.493. [DOI] [PubMed] [Google Scholar]