Abstract
The KCNE2 gene encodes a single transmembrane domain protein that modulates a variety of K+ channel functions in various tissues. Here we show that cardiac KCNE2 transcript levels are ~10-fold upregulated at the end of pregnancy. This upregulation was mimicked by 17-β estradiol but not by 5α-dihydrotestosterone treatments in ovariectomized mice. To investigate the mechanism of KCNE2 transcriptional regulation by estrogen, we experimentally identified KCNE2 transcription start sites, delineated its gene structure and characterized its promoter region. Estrogen treatment stimulated KCNE2 promoter activity in a dose-dependent manner and ICI 182,780 blocked estrogen stimulation. A direct genomic mechanism was demonstrated by: i) the loss of estrogen responsiveness in the presence of a DNA-binding domain mutant Estrogen Receptor α or mutant KCNE2 ERE and ii) binding of ERα to the KCNE2 ERE. These findings show that a genomic mechanism of estrogen action alters KCNE2 expression, which may have important physiological implications.
Keywords: KCNE2, MiRP1, Promoter, Estrogen, ERE, Estrogen receptor alpha
1. Introduction
Ion channels are integral membrane proteins that form highly selective pores for the passage of specific ions across the plasma and internal membranes. These channels are multiprotein complexes formed by transmembrane α-subunits that coassemble with regulatory β-subunits, which can be transmembrane or cytoplasmic. These β-subunits can strongly influence ion channel properties by influencing ion fluxes across the pore of the α-subunit, thus defining the phenotype of native currents. Additionally, trafficking and subcellular localization of the α-subunits are also affected by β-subunits (Hille, 2001).
The KCNE gene family encodes a group of modulatory β-subunits: KCNE1, originally named MinK, which was cloned in 1988, and a decade later four other homologues, KCNE2-5 (MinK Related Peptides, also called MiRPs 1–4) were cloned (Takumi, Ohkubo, & Nakanishi, 1988). Among the KCNE subunits, KCNE2 is the most promiscuous one. In heterologous systems, KCNE2 can modulate a variety of K+ channel pore-forming α-subunits that may result in broad physiological effects (Abbott et al., 1999;Deschenes & Tomaselli, 2002;McCrossan & Abbott, 2004;Panaghie & Abbott, 2006;Zhang, Jiang, & Tseng, 2001). Inherited KCNE2 mutations are associated with human cardiac arrhythmogenesis and long QT syndrome (Abbott et al., 1999), emphasizing its clinical relevance. Ablation of the KCNE2 gene resulted in i) downregulation of two major components of murine cardiac action potential repolarization currents, the fast component of the transient outward K+ current Ito, f and the delayed rectifier K+ current Ik, slow in the heart (~25% and ~50%, respectively) (Roepke et al., 2008) and ii) a profound alteration of gastric secretion (Roepke et al., 2006). Furthermore, KCNE2 expression was downregulated in gastric cancer, while its overexpression suppressed cell proliferation and tumorgenesis in a gastric cancer cell line (Yanglin et al., 2007). These characteristics make KCNE2 an important ion channel regulatory subunit that may be involved in several cellular functions. Therefore, the elucidation of the regulatory mechanism(s) of KCNE2 promoter elements should provide new insights into KCNE2 gene transcription and expression.
Several K+ channel α- and/or regulatory β-subunits have been reported to be regulated by 17-β estradiol (E2) in different tissues (Drici et al., 1996;Eghbali et al., 2005;Kundu et al., 2007;O’Mahony et al., 2007;Ranki et al., 2002a;Roepke et al., 2007;Song et al., 1999;Song et al., 2001;Wong et al., 2008). Studies on the gender differences in expression levels of cardiac KATP channels as well as KCNQ1 further support the role of E2 in ion channel regulation (Moller & Netzer, 2006;O’Mahony et al., 2007;Ranki et al., 2001;Ranki et al., 2002a;Ranki et al., 2002b). Estrogen levels dramatically increase at the end of pregnancy. The maternal heart also undergoes major remodeling as a result of volume overload, mechanical stretch and hormonal changes during pregnancy. This remodeling is accompanied by an increased QT interval as well as an increased risk of cardiac arrhythmias (Gowda et al., 2003). As mutations of the KCNE2 gene are associated with cardiac arrhythmias in humans (Abbott et al., 1999) and ablation of KCNE2 gene expression has been observed to result in a reduction of two major cardiac action potential repolarizing currents in mice (Roepke et al., 2008), we explored whether cardiac KCNE2 can be regulated by pregnancy.
We found that cardiac mouse KCNE2 transcript levels are upregulated at the end of pregnancy and estrogen but not 5α-dihydrotestosterone (DHT) is the main player in this regulation. Recently, the transcription start site(s) (TSSs) of human KCNE2 has been identified. However, the 5′-flanking sequence of the TSS did not show any detectable promoter activity (Lundquist et al., 2006). Therefore, the promoter regulatory regions of the KCNE2 gene of any species remained to be characterized. Here, we have characterized the murine KCNE2 promoter region as to better understand the molecular mechanism of estrogen action.
2. Materials And Methods
2.1. Animals and hormonal treatment
Non-pregnant (NP, at diestrus-2 stage), late pregnant (LP, 20 days) and ovariectomized (OVX) mice were used (C57/BL). The stage in the animal’s estrous cycle was determined by examination of smears prepared from vaginal fluid. The mice used in all studies cycled regularly at intervals of ~four days. The cycle involved passage through each of the following stages: diestrus-2 (characterized by the presence of many leucocytes), proestrus (characterized by predominant appearance of nucleated epithelial cells and none to few leucocytes), estrus (in which a vast amount of large cornified cells are present), and finally metestrus/diestrus-1 (when many leucocytes and a few cornified cells are present). Protocols received institutional review and committee approval. For animals undergoing hormonal treatment, one-step continuous release sterile pellets were used (Innovative Research of America) to maintain a constant rate of hormonal delivery. The pellets were implanted under anesthesia subcutaneously in the lateral side of the neck using a stainless steel reusable precision trochar. OVX mice were treated with 17-β estradiol (E2) (4 μg/Kg/day for 10 days) alone, E2 (4 μg/kg/day)+ICI 182,780 (ICI) (400 μg/kg/day), or DHT, a non-metabolizable analog of testosterone (32 mg/Kg/day for 3 weeks) pellets. Placebo pellets (containing five intert compounds: cholesterol, lactose, cellulose, phosphates and cerates) were implanted in control animals (Sham). Plasma E2 levels were measured by radioimmunoassay at the end of the each treatment (Diagnostic Systems Laboratories, TX, USA).
2.2. Cell culture
HeLa (human cervical adenocarcinoma epithelial cells) and MCF7 (human breast adenocarcinoma cells) cells were purchased from American Type Culture Collection (ATCC). Cells were grown in cell culture media consisting of Dulbecco’s modified Eagle’s medium (DMEM, high glucose, with L-glutamine) (Invitrogen), supplemented with 50 U/ml penicillin, 50μg/ml streptomycin, and 10% fetal bovine serum under 5% CO2 air mixture at 37°C. Cells were subcultured according to ATCC instructions.
2.3. Real-Time PCR
Total RNA from mouse heart and myometrium tissues was isolated using Trizol (Invitrogen). Total RNA (2 μg) was reverse transcribed with gene specific primers using Omniscript RT kit (Qiagen). Gene specific primers for KCNE2 are given in Table 1 (GeneBank™ accession no. BC022699). GAPDH was used as the internal reference gene, and the sequence of the forward primer was CCTGCACCACCAACTGC TTAG, and reverse primer ATGACCTTGCCCACAGCCTTG (GeneBank™ accession no. BC083149). A single peak was detected from the first derivative of fluorescence (dF/dT) vs. temperature plots (melting curve) indicating amplification of a single product. Agarose gel electrophoresis at the end of the reaction also confirmed the amplification of a single product of the expected size. Controls consisted of: (1) the reaction cocktail without reverse transcriptase tested in a regular 30 cycle PCR; and (2) H2O instead of cDNA tested in parallel with the real-time PCR.
Table 1.
Primers used in RACE, primer extension, real time PCR and in generating deletion and mutant constructs.
| Primer name | Orientation relative to mRNA | Sequence | Position in KCNE2 gene from TSS (nt) |
|---|---|---|---|
| KCNE2R | Reverse | ttttttccatggCCATGCTTCCCCCTTGCTGTGTGGTATGTAGCAATGC | +5463 to + 5482 |
| −270/+UTR | Forward | ttttttctcgagTGTGGGAATCAACAGTACAAATAGGGTGG | −272 to −244 |
| −951/+UTR | Forward | ttttttctcgagAGACCATTCAGTAAATACGGAACGAAAATCGATTAA | −878 to −844 |
| −1461/+UTR | Forward | ttttttctcgagGGCTCAGGTTGGCTTTGGCTT | −1388 to−1368 |
| −2978/+UTR | Forward | ttttttctcgagGTCATTGCAGGGACACATCTC | −2905 to −2883 |
| mutERE | Forward | TCTGTATTATCCGAGTAAGCTAACACAGTTGTAAAGGA | −70 to −33 |
| GSP1 | Reverse | CTCTGCCCCTCTTCAGGACTGGATGT | +1343 to +1368 |
| GSP2 | Reverse | GGGCACTTTCAGCTTGAGACCTGTAAGC | +1300 to +1327 |
| GSP3 | Reverse | GATGTCACCAGAGCTTCTGCGACCC | +95 to +119 |
| GSP4 | Reverse | GGATGGCCACCACGATGAACGAGAACAT | +5650 to +5677 |
| PE | Reverse | CTTGGCTGCTCTCTTCCTGG GCATC | +40 to +64 |
| Primer a | Forward | GACCCAGTTGTAAAGGACTTT | −48 to −28 |
| Primer b | Reverse | ATGTCACCAGAGCTTCTGCGA | +100 to +120 |
| Primer c | Forward | TTAGTGTGGCCACAGATCCTA | +193 to +213 |
| Primer d | Reverse | AAACGAAGTCCTGTGAGCCTC | +739 to+759 |
| RT-F | Forward | CATCCTGTACCTCATGGTGATG | +5622 to +5643 |
| RT-R | Reverse | TGGCCTTGGAGTCTTCCAGAT | +5780 to +5800 |
TSS is at 5478 nt upstream of adenine residue of ATG translation start site. Nucleotides in uppercase are homologous to KCNE2 gene, nucleotides in lowercase are sequences introduced for cloning purposes, and restriction sites are underlined. PE1 is the primer used in primer extension. RT-F and RT-R are used in real time PCR and mutERE was used to mutate KCNE2 ERE. Mutated nucleotides are italicized and underlined.
2.4. 5′-RACE analysis
Rapid amplification of cDNA 5′-ends (5′-RACE) analysis was performed with 2 different methods, SMART™ RACE (BD-Clontech) and GeneRacer (Invitrogen) following manufacturers’ protocols. Poly-A RNA was purified using Oligotex™ mRNA Purification System (Qiagen) from mouse heart and myometrium total RNA. The sequences of gene specific primers (GSPs) are given in Table 1. Briefly, in SMART™ RACE, purified mRNA was reverse transcribed with PowerScript Reverse Transcripatse and primed with 5′-CDS primer SMART II A oligo (SMART™ RACE KIT, Clontech). 5′-RACE PCR was performed with a tagged 5′ primer (included in the kit) and GSP1, followed by a 5′-RACE nested PCR using 5′ tagged nested oligo (included in the kit) and GSP2. As a negative control, 5′-RACE cDNA was amplified with only one primer. As RACE can produce a false TSS by premature termination of the reverse transcription reaction, another 5′-RACE PCR was performed with a forward 5′ tagged oligo (included in the kit) and a reverse primer GSP3. In GeneRacer, mouse heart and myometrium mRNA was dephosphorylated with calf intestinal phosphatase, followed by tobacco acid pyrophosphatase treatment to decap the mRNA. GeneRacer RNA oligo was then ligated to decapped mRNA using T4 RNA ligase. The ligated mRNA was reverse transcribed by SuperScript™ III with GSP4. 5′-RACE PCR was performed with GeneRacer 5′ primer (included in the kit) and GSP3, followed by a 5′-RACE nested PCR using GeneRacer 5′ nested primer and GSP2. The nested PCR products in both methods were cloned into pCR2.1-TOPO vector (Invitrogen) and were sequenced (Laragen, Inc., Los Angeles, CA).
2.5. Primer Extension analysis
Primer extension assay was performed using the oligonucleotide CTTGGCTGCTCTCTTCCTGGGCATC, which spans the nucleotides +41 and +65 downstream of the TSS identified by RACE analysis. The primer was end-labeled with [γ-32P]-ATP (GE Healthcare) using T4 Polynucleotide Kinase (New England BioLabs). The labeling reaction was heat-inactivated and purified by passage through a spin column (Roche). Mouse heart and myometrium total RNA (50 μg) was ethanol precipitated and resuspended in denaturing hybridization buffer (40 mM PIPES pH 6.4, 1 mM EDTA, 0.4M NaCl, 80% formamide). The labeled primer was hybridized to total RNA overnight at 45°C followed by ethanol precipitation. The primer was then extended with 10U AMV reverse transcriptase (Promega) at 42°C for 1 hr following the manufacturer’s protocol. The cycle sequencing was performed with the fmol DNA Cycle Sequencing System (Promega), using the same oligonucleotide primer and a corresponding fragment of the genomic clone as the template. The primer extension products were separated along with the sequencing ladder on an 8% denaturing polyacrylamide gel, and autoradiography was performed according to the standard protocol.
2.6. Plasmid constructs
All plasmids were generated using standard recombinant DNA techniques. The BAC clone RP23-312L7 (Children’s Hospital Oakland Research Institute) was used to obtain the KCNE2 genomic region of the mouse chromosome 16. Our sequencing results revealed that this BAC clone did not have 270 nt in the repetitive region of the 5′ flanking sequence as was reported in the genomic DNA sequence (NC_000082). The untranslated region (UTR) of KCNE2 (1426 nt) together with up to 2978 nt of the KCNE2 promoter were subcloned between the XhoI and NcoI restriction sites of the pGL4.10 [luc2] vector (Promega) and named −2978/+UTR construct. Promoter constructs consisting of −1461/+UTR, −951/+UTR and −270/+UTR were then generated by PCR amplification using the −2978/+UTR construct as template, and also subcloned between the XhoI and NcoI restriction sites of the pGL4.10 [luc2] vector. All ERE and ½ ERE mutations were created by PCR using the Quick-Change Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol, and they all used the −270/+UTR promoter construct as template.
The human Estrogen Receptor α (ERα) DNA-binding domain mutant (DBDM; E203A-G204A) was generated using a similar protocol with the primer GAGTCTGGTCCTGTGCAGCTTGCAAGGCCTTCTTC (mutations are italicized and underlined). Restriction mapping and subsequent sequencing (Laragen, Inc., Los Angeles, CA) confirmed the accuracy of all clones.
2.7. Cell transfection and Luciferase assay
Cells were grown in cell culture media to ~70% confluence and transfected with the promoter-luciferase constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. To account for the different sizes of the constructs used, equal molar amount of DNA was transfected and the total amount of DNA was kept constant for every transfection with the addition of pGem3Zf vector to the transfection mixture. Transfection with the empty vector served as the no promoter control. To normalize Firefly Luciferase (Luc) activity to the transfection efficiency, the vector pRL-TK, which directs Renilla Luciferase (Rluc) activity, was used as the transfection control. For E2 responsiveness assays, human ERα (subcloned in the pcDNA3 vector) was co-transfected only in HeLa cells at 0.133 times the concentration of the promoter constructs (MCF-7 endogenously express ERα, thus the ER was not co-transfected with the promoter constructs). Transfection was carried out overnight (16 h) in Opti-MEM serum-free media (Gibco). The next day, cells were induced with either estradiol (0.1–30 nM), 1 nM estradiol + ICI 182,780 (0.1–100 nM), or vehicle (ethanol <0.01%). Charcoal–dextran stripped serum (HyClone) and phenol red-free media were always used. The cells were washed with phosphate-buffered saline 24 h post treatments, lysed with passive lysis buffer and assayed using the Dual Luciferase Assay kit (Promega) and the Veritas luminometer (Turner Biosystems). All transfections were repeated at least three times and in triplicate.
2.8. Electrophoretic mobility shift assay (EMSA)
Nuclear extract of E2 treated MCF7 cell was purchased from Active Motif. Estrogen Response Element (ERE) consensus and Sp1 duplexes were from Santa Cruz Biotechnology and Promega Inc., respectively. Double stranded oligonucleotide probes (3.5 pmol) of KCNE2 ERE and mutated KCNE2 ERE were end-labeled using polynucleotide kinase and [γ-32P]ATP. Purified labeled probe (3 fmol) was used in a 10 μl binding reaction in a buffer containing 4 % glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, 10 mM Tris-HCl pH 7.5 and 500 ng poly (dI-dC)-poly(dI-dC). Before addition of labeled probe, the nuclear extract (5 μg) was pre-incubated in binding buffer for 15 min. For competition assays, competitor molecules were added during the pre-incubation stage. DNA-protein complexes were separated by electrophoresis in a 0.5 X TBE-5% PAGE (10°C, 220V) and visualized by autoradiography.
2.9. Statistics
All experiments were performed with three different RNA preparations (3 animals per each preparation of RNA). Student’s t-test was used. P values ≤ 0.05 were considered statistically significant. Values are mean ± SE.
2.10. Software
Promoter2.0 (Knudsen, 1999), Eponine (Down & Hubbard, 2002) and FirstEF (Davuluri, Grosse, & Zhang, 2001) software were used for TSS analysis. Transcription factor binding sites were identified using Transcription Element Search System (TESS) (Schug J, 2003) and MatInspector from Genomatrix Software, GmbH, München, Federal Republic of Germany. Homology search and alignments were done using NCBI BLAST and VISTA (Frazer et al., 2004) tools.
3. Results
3.1. Pregnancy and E2 alters cardiac KCNE2 mRNA level in mouse
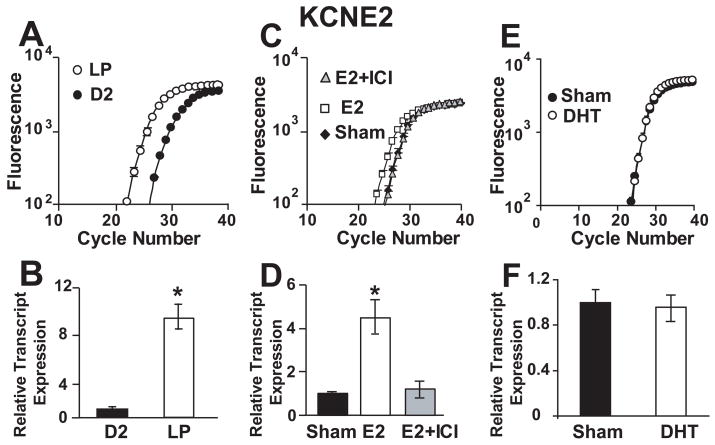
We compared cardiac KCNE2 transcript levels in non-pregnant (NP) versus late pregnant (LP) mice. Female mice have an estrous cycle of four to five days, similar to that of female rats (Lapolt). During proestrus, plasma estrogen concentrations peak for a brief period of time; the estrus stage is thus primed by the surge of estrogen occurring during proestrus, while diestrus-1 and diestrus-2 are marked by low estrogen levels. For animals in the NP group, we chose mice in their diestrus-2 stage, as it is at this stage of their cycle that the mice have been exposed to the longest periods of low estrogen levels. Fig 1A shows that the curves of the fluorescence cycle number of the cardiac KCNE2 left transcript levels were shifted in LP with respect to NP at diestrus-2 (~3 cycles), thus indicating an upregulation of KCNE2 transcripts at the end of pregnancy (~10 fold, Fig. 1A, B). GAPDH was used as a reference gene and its transcript levels were similar in NP and LP samples (not shown). As both estrogen and testosterone levels increase dramatically at the end of pregnancy, we therefore tested the possible role of estrogen and testosterone in the regulation of KCNE2 transcripts. Ovariectomized (OVX) mice were treated either with E2 alone (at a dose which produces E2 plasma concentrations similar to LP), with E2 together with the ER antagonist ICI 182, 780, or with DHT. The KCNE2 transcript levels were 5-fold upregulated by E2 treatment, while ICI 182, 780 prevented this upregulation (Fig 1C, D). Compared to E2 treatment, KCNE2 transcripts were not affected by DHT treatment (Fig. 1 E, F). Transcript levels of GAPDH were similar in OVX mice treated with Placebo (Sham), E2, E2+ICI and DHT (not shown). These data strongly suggest that estrogen but not testosterone play a key role in the upregulation of cardiac KCNE2 transcript level.
Figure 1.
Cardiac KCNE2 transcripts are upregulated by estrogen but not by DHT. (A), Cardiac KCNE2 fluorescence vs. cycle number plots in NP at diestrus-2 (●) and LP (○). (B), Bar graph showing the mean relative transcript expression normalized to NP, for KCNE2: NP 1.0±0.2 (n=4), LP 9.4±1.1 (n=4) and for GAPDH: NP 1.0±0.03 (n=4), LP 0.97±0.1 (n=4). (C). Cardiac KCNE2 fluorescence vs. cycle number plots of OVX mice treated with vehicle (sham) (◆), E2 (□), and E2+ICI (▲). (D). Bar graph showing the mean relative transcript expression normalized to sham, for KCNE2: sham 1.0±0.1 (n=4); E2-treated, 4.5±0.7 (n=4) and E2+ICI-treated, 1.2±0.3 (n=4) and for GAPDH: sham 1.0±0.08 (n=4); E2-treated, 0.99±0.02 (n=4) and E2+ICI-treated, 0.96±0.09 (n=4). (E). Cardiac KCNE2 fluorescence vs. cycle number plots of OVX mice treated with placebo (Sham, ●) and DHT (○). (F). Bar graph showing the mean relative transcript expression normalized to sham, for KCNE2: sham 1.0±0.1 (n=3) and DHT-treated, 0.96±0.1 (n=3) and for GAPDH: sham 1.0±0.1 (n=3) and DHT-treated, 1.1±0.1 (n=3). All measurements were done in triplicate, with n indicating the number of animals with independent total RNA isolations. E2, ICI 182, 780 and DHT treatment protocols are outlined in the MATERIALS AND METHODS section. *, Statistical significance P<0.01.
3.2. Identification of the mouse KCNE2 gene TSSs
The previous data strongly suggested that the transcriptional activity of the KCNE2 gene can be subject to E2 regulation. We used the reported mouse KCNE2 gene sequence (NM_134110) which contains two exons (1 and 2, Fig. 2A) and examined a 5000 nt fragment of the KCNE2 gene (NC_000082:92287769-92292769 Mus musculus chromosome 16, reference assembly, C57BL/6J) to identify possible TSS and promoter regions. Initially, the web-based software Eponine, FirstEF and Promoter2 were used for this purpose. Eponine failed to locate any promoter within this region. FirstEF correctly predicted the exon 1 end as it is found in mRNA (NM_134110). It also predicted a TSS at 141 nt upstream of exon 1 end and a non-CpG related promoter region spanning from 72 nt to 641 nt upstream of exon 1 end. Promoter2 predicted a TSS at 2750 nt upstream of exon 1 end and another within the intronic region between exon 1 and 2. Since none of the TSS predictions matched nor were precisely consistent with the 5′-end found in Unigene sequences, and since there was sparse prior evidence of the 5′-end of the KCNE2 transcript, we found it necessary to first experimentally determine the TSS of KCNE2 using RACE.
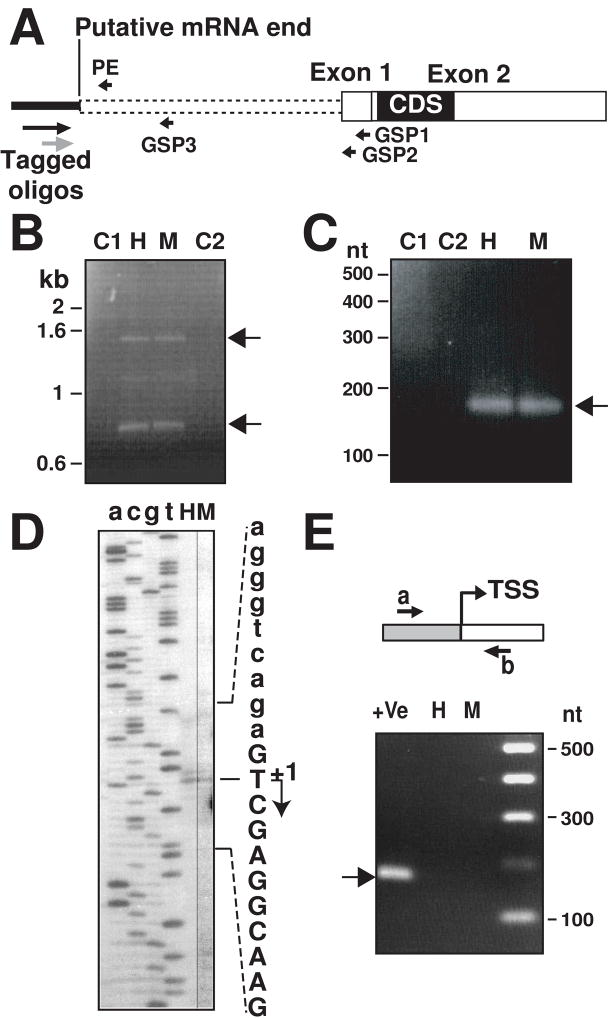
Figure 2.
Determination of KCNE2 gene TSS by 5′-RACE and primer extension analysis. (A), Scheme of putative full length KCNE2 mRNA (based on GenBank sequence NM_134110), showing relative position of primers used. Gene specific primers (GSP) 1 to 3 and tagged oligos used for 5′-RACE analysis are shown. PE primer was used for primer extension. Filled box denotes coding sequence (CDS) and white boxes are untranslated regions. Dotted lines indicate putative 5′-UTR sequence. (B), Two RACE PCR products (arrows) of ~1.5 kb and ~0.8 kb were obtained with with tagged-oligos and GSP1 primers from heart (H) and myometrium (M). C1 and C2 are negative controls with tagged oligos (C1) and GSP1 (C2). (C), RACE PCR products obtained tagged oligos and GSP3 showing a distinct ~0.16 kb (arrow); C1, C2, H and M are as in (B). (D), Autoradiogram of primer extension products using mouse total RNA from heart (H) and myometrium (M). a, c, g, t denotes corresponding bases. The sequence of the opposite strand is shown. The identified TSS is marked with an arrow (+1) and the mRNA sequence is in uppercase letters. (E), Validation of the identified TSS. Primers a and b failed to generate a product from cDNA (H, heart; M, myometrium). As a positive control, these primers amplified genomic DNA of the corresponding BAC clone (+Ve lane, arrow). Gray box, genomic sequence; white box, mRNA sequence.
For this purpose, we first used the SMART™-RACE method using KCNE2 gene specific reverse primers (GSP1 and 2, Fig. 2A and Table 1). The 5′-RACE PCR using GSP1 and 5′-primer from the tagged oligo amplified two products of ~1.5 kb and ~0.8 kb from both mouse heart and myometrium cDNAs (Fig. 2B, lanes H and M, respectively; arrows). As negative controls, PCR was performed either with only a forward or with only a reverse primer, which did not yield any product (Fig. 2B, lanes C1 and C2). Next, we did another internal PCR using the gene-specific reverse primer GSP2 and nested tagged oligo (Fig. 2A, gray arrow). Cloning and sequencing of the amplified bands revealed that they had the same TSS (G, +3 in supplemental Fig. 1 suggesting that KCNE2 mRNA can have two different 5′-UTRs of 1424 nt and 787 nt.
Since there is a possibility that reverse transcriptase could stall due to the premature termination of the reaction, we also performed another RACE reaction with a gene specific primer GSP3 (Fig. 2A), designed close to the identified TSS. The RACE PCR amplified a ~0.15 kb product in mouse heart and myometrium (Fig. 2C, arrow) as expected, ruling out a premature termination of the first RACE reaction (Fig. 2B). Cloning and sequencing of the ~0.15 kb band confirmed the determined TSS.
An alternative RACE method (GeneRacer RACE, Invitrogen), which captures only the complete full-length capped mRNA, further validated our findings by identifying the same TSS (at G, +3). Furthermore, in one of the three analyzed clones, this method also detected a contiguous TSS (G, −1 in supplemental Fig. 1). In summary, using RACE we found two vicinal TSSs for murine KCNE2.
To determine the predominant KCNE2 TSS, we performed primer extension assay using a reverse primer close to the TSSs (Fig. 2A, PE). The autoradiogram in Fig. 2D shows the presence of two TSSs, a predominant one at thymine (T) (Fig. 2D, +1 arrow and supplemental Fig. 1 arrow) and a weaker one guanine (G) (−1 in supplemental Fig. 1). A close inspection of the gel also revealed the presence of a very faint band that would correspond to the guanine residue identified by both RACE methods (G, +3 in supplemental Fig. 1). We verified this finding by performing primer extension analysis with four other total RNA preparations from heart and myometrium, and then mapped the mouse KCNE2 predominant TSS at the thymine residue 1414 nt upstream of exon 1 end (5477 nt upstream of the translation start site) and have assigned position +1 to this residue (Fig. 2D).
To confirm that transcription cannot start upstream (~ 50 bp) of the identified TSSs, we performed RT-PCR analysis using a forward primer designed in the promoter region upstream of the TSSs (Fig. 2E, primer a) and a reverse primer located downstream of the TSSs (Fig. 2E, primer b). Confirming the TSSs, these primers failed to amplify a product from cDNA of mouse heart (H) and myometrium (M) RNAs (Fig. 2E). The efficiency of these primers was validated with a positive control using genomic DNA (BAC clone), which generated the expected 170 nt product (Fig. 2E, lane +Ve, arrow).
3.3. Identification of a new KCNE2 exon and an intron retained in mRNA
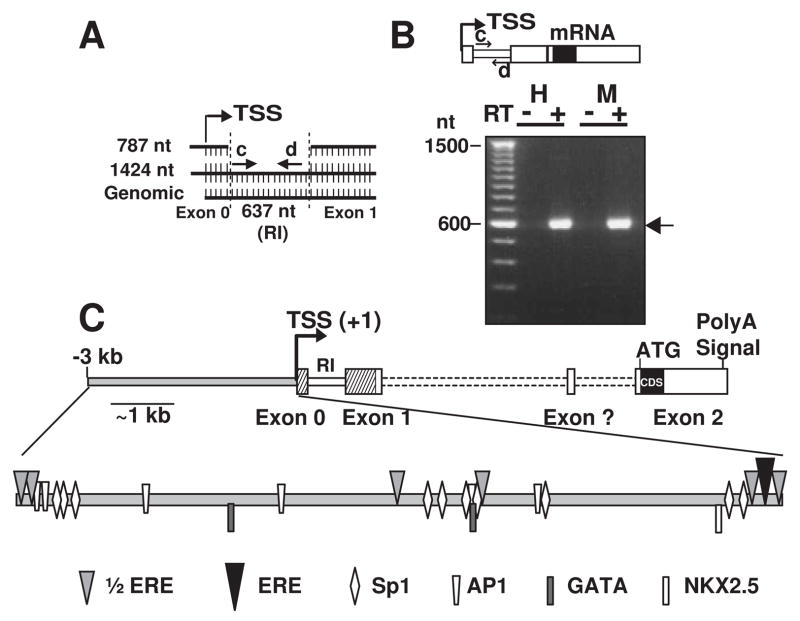
The alignment of 1424 nt and 787 nt 5′-UTRs (sharing the same TSS) with the genomic DNA sequence (NC_000082) revealed that internal 637 nt were absent in the 787 nt 5′-UTR (Fig. 3A). We predicted that these 637 nt constitute an intron, as it is flanked by the classical intron delimiting base pairs GT-AG(Breathnach & Chambon, 1981). The intronic nature of this sequence was confirmed by two different programs of splice site predictors (NetGene2 and NNSPLICE 0.9, Berkeley Drosophila Genome Project) with >99% confidence. Thus, the 787 nt 5′-UTR would predict a new exon 0 and a longer exon 1 than that predicted (NC_000082); while the 1424 nt 5′-UTR containing the 637 nt intronic sequence (supplemental Fig. 2) could be considered an alternatively spliced variant produced by intron retention (Galante et al., 2004a). To confirm intron retention in the mRNA, we performed RT-PCR using primers flanking the 5′ and 3′ ends of the retained intron (RI, Fig. 3A and B, primers c and d). We detected the expected ~600 nt product in the reactions with reverse transcriptase (Fig. 3B, arrow, RT+ lanes), while no product was detected in its absence (Fig. 3B, RT-lanes).
Figure 3.
Outline of KCNE2 gene structure. (A), Alignment of the 5′-RACE products with the mouse KCNE2 genomic sequence. Retained intron (RI) is marked between dotted lines (supplemental Fig. 2, red letters). (B), Product of retained intronic sequence in mRNA obtained with primers c and d. As control, no products were detected without reverse transcriptase. H. Heart and M, myometrium, RT, reverse transcriptase. (C), Map of KCNE2 gene showing: TSS (+1); exons 0, 1 and 2 (dashed regions were described here); coding sequence (CDS); intronic sequence (dotted line); retained intron between exons 0 and 1 and a putative exon (?) as predicted in GenBank (BG261965 and BC022699). 3 kb upstream of the TSS is shown (gray filled bar) with relative positions of transcription factor binding sites related to heart specific regulation and estrogen responsiveness.
In summary, our results show that the mouse KCNE2 mRNA has: i) a much longer 5′-UTR than previously reported, ii) a novel exon (referred to here as exon 0) encoding part of 5′-UTR, and iii) the ability to undergo alternative splicing by intron retention, thus generating mRNA with different 5′-UTR lengths. Based on these findings and the GenBank reference sequence for mRNA (NM_134110), we have outlined the mouse KCNE2 gene structure as shown in Fig. 3C.
3.4. Analysis of KCNE2 5′-flanking sequences for the presence of core promoter elements
We have analyzed the KCNE2 promoter region for classical transcriptional regulatory elements such as a TATA box, Initiator (Inr), Motif Ten Element (MTE), Downstream Promoter Element (DPE), TFIIB-Recognition Element (BRE) and Downstream Core Element (DCE) in their consensus positions (Maston, Evans, & Green, 2006). KCNE2 gene has a quasi-consensus TATA box (TATAA/TAAG/A), −29TtTAAAtG−22 (lowercase indicating the mismatches) located in the consensus position (−26 to −31) upstream of the TSS (Lue & Kornberg, 1993). We have also located two DPE (+8 to +12 and +84 to +88) in slightly different locations than their consensus positions (+28 to +32). The KCNE2 core promoter does not seem to otherwise contain any Inr, BRE, MTE or DCE elements in their typical positions (Maston, Evans, & Green, 2006;O’Lone et al., 2004). This promoter is also devoid of a CpG island. The fact that KCNE2 has a TATA box-like sequence at the typical position from the TSS classifies the KCNE2 gene as a non-CpG related TATA box containing gene.
3.5. Regulatory cis-acting elements in the promoter region
Transcription Element Search Software (TESS) and MatInspector (Genomatrix) were used to search the KCNE2 gene for cis-acting elements required for estrogen regulation (ERE and ½ERE) and for transcription factor binding sites (AP-1, Sp1, CCAAT/Enhancer binding protein α (C/EBP-α), C/EBP-β, Signal Transducers and Activators of Transcription (STAT5), NFκβ and GATA-1) known to play a role in estrogen-mediated transcription (Bjornstrom & Sjoberg, 2005;Klinge, 2001;O’Lone et al., 2004). Within 3 kb upstream of the main TSS, KCNE2 promoter has one quasi perfect ERE [−56GGgCAAGCTGACC−44] (the lowercase g instead of a T indicating one mismatch) and six ½EREs [TGACC, or GGTCA] (Fig. 3C and supplementary Fig. 1). Furthermore, this region has also other regulatory motifs involved in estrogen regulation, such as 10 Sp1, 6 AP-1, 3 C/EBP-α, and 1 C/EBP-β binding sequences (supplementary Fig. 1). We have marked only the sites detected with high level of confidence (p>0.8). STAT5 and NFκβ sites were not detected in this region.
Since KCNE2 is expressed in the heart (Jiang et al., 2004), we have also searched for transcription factor binding sites that can regulate gene expression in cardiac myocytes such as: T-box element that can bind heart-specific TBX5 (Plageman, Jr. & Yutzey, 2005), MEF2C, SRF (Serum response factor), SALL4, JAG1, GATA and NKX2.5 (NKE, TNNAGTG) (Hatcher et al., 2003;Li et al., 2007;Xin et al., 2006). Our analysis only located numerous GATA-like sequences and two consensus GATA sites (A/TGATAA/G)- −1983AGATAG−1978 and −954TGATAG−949-that could bind GATA4/6, as well as one consensus NKX2.5 site −281TCTAGTG−275 (Fig. 3C and supplementary Fig. 1). The presence of these consensus GATA and NKX2.5 sites indicates that the KCNE2 gene could be differentially regulated in the heart (Hatcher et al., 2003).
3.6. Basal and E2-induced promoter activity of KCNE2 gene
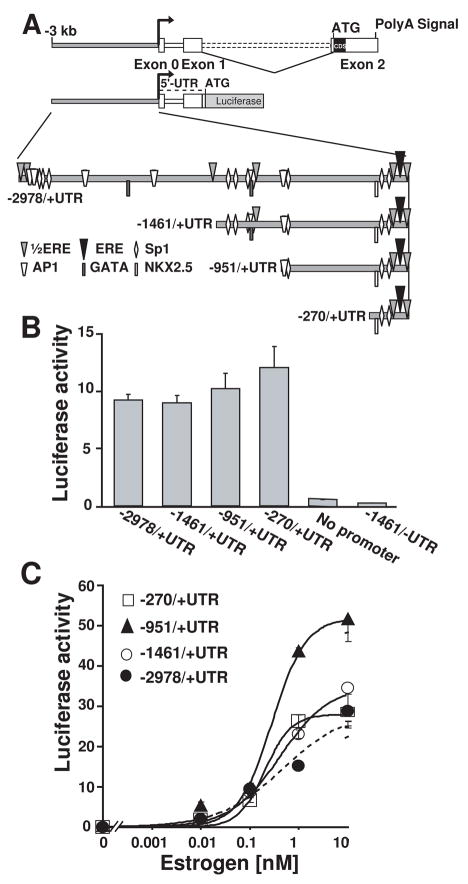
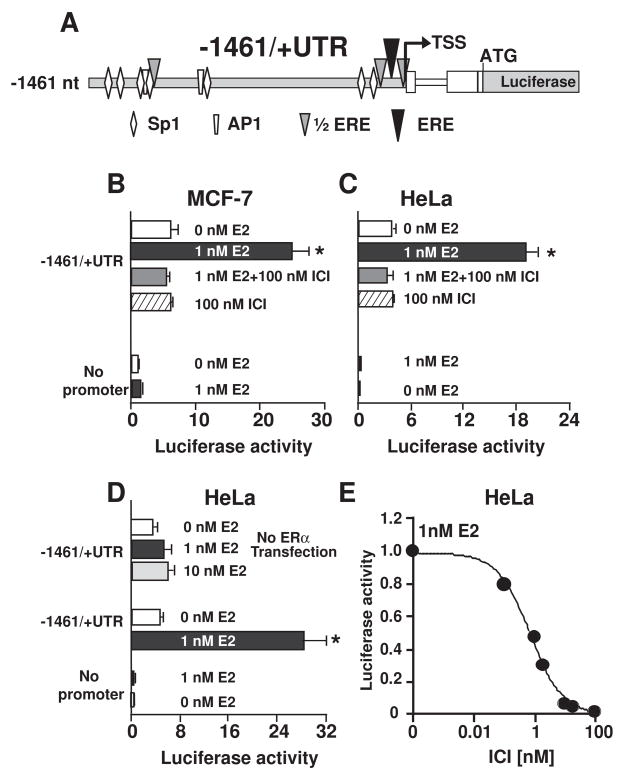
To identify mouse KCNE2 5′ sequences necessary for basal promoter activity, we made four constructs containing different lengths of the promoter region followed by KCNE2 entire 5′-UTR, including the spliced intron designed to direct luciferase expression (−2978/+UTR, −1461/+UTR, −951/+UTR and −270/+UTR, Fig. 4A).
Figure 4.
KCNE2 basal and estrogen responsive promoter activity. (A), Scheme of deletion constructs from the TSS used to measure basal (unstimulated) activity. All constructs had KCNE2 5′-UTR fused with the luciferase reporter gene. (B), Similar basal promoter activity in all constructs in HeLA cells (n=3). In the absence of 5′-UTR (−1461/-UTR), there was no promoter activity (n=3). (C). All constructs responded to estrogen in a dose dependent manner. Dose response curves in all construct could be well fitted a Hill function except for −2978/+UTR. The Hill equation used is (Luc/RLuc)-B=max/(1+(EC50/[estrogen])N, where Luc/RLuc is the promoter activity reported by luciferase (Luc) normalized by the activity of a transfection control vector reported by Luciferase (RLuc); B, basal promoter activity; EC50, concentration of 50% of maximum effect; max=maximum effect, [estrogen], estrogen concentration; and N=Hill coefficient. EC50 and max values were: −1460/+UTR, 0.41±0.05 nM and 34.58 ±1.6; −951/+UTR, 0.28 ± 0.05 nM and 51.68 ± 2.49; and −270/+UTR, 0.20 ± 0.03 nM and 28.59 ± 0.9 (n=4). N was close to 1.
Luciferase assays showed that the basal relative transcriptional activity of these four KCNE2 gene fragments was similar (Fig. 4B). It is also worth mentioning that there was no detectable basal promoter activity in the absence of KCNE2 5′-UTR (−1461/-UTR, Fig. 4B).
To measure E2 stimulation of the promoter activity, HeLa cells were co-transfected with the four different KCNE2 promoter constructs together with human ERα. E2 treatment stimulated KCNE2 promoter activity in a dose-dependent manner in the four constructs (Fig. 4C). The shortest (−270/+UTR) construct showed significant E2 responsiveness, indicating that this short stretch contains the elements necessary for the E2-induced activity. Increasing the promoter length to −951/+UTR resulted in a stronger promoter activity, which was then reduced with further increase of the promoter length (constructs −1461/+UTR, −2978/+UTR). This data thus suggests the presence of inhibitory sequences beyond −951 nt. The dose response curves were well fitted to a Hill equation (except for −2978/+UTR construct) having a Hill coefficient close to 1 and a similar apparent E2 affinity (EC50 ranges between 0.2 to 0.4 nM).
Interestingly, the E2 dose-response curve for the longest construct (−2978/+UTR) has complex stationary kinetics, possibly underlying the action of various types of E2 regulated elements (Fig. 4C, filled circles, dotted line).
We have also tested E2 responsiveness in the MCF7 cell line, which expresses endogenous ERα. The −1461/+UTR construct (Fig. 5A) showed about a 3-fold increase in promoter activity upon stimulation with 1 nM E2, an increase similar to that observed in HeLa cells (Fig. 5B and C, black vs. white bars). Therefore, E2 is equally effective in stimulating the KCNE2 promoter in cell lines endogenously expressing ERα, such as may be the case in native tissues. We also examined the effect of on the luciferase activity in HeLa cells in the absence of ERα transfection. Fig. 5D shows that in the absence of ERα, the KCNE2 promoter activity was not significantly modulated by 1 nM E2. An even higher concentration of E2 (10 nM) was not able to significantly stimulate promoter activity in the absence of ERα transfection. This experiment indicated that the E2-mediated upregulation of KCNE2 promoter activity mainly occurs via the binding of ERα and not through the action of membrane-bound GPR30.
Figure 5.
The specific estrogen antagonist ICI 182,780 inhibits estrogen-induced upregulation of KCNE2 transcription in different cell lines. (A). Scheme of the −1461/+UTR construct used in this study. The construct was transfected in MCF7 cells (B) or in HeLa cells together with ERa (C, D) or without ERa (D) (n=3). Transfected cells were stimulated with 1 nM estrogen (filled balck bar), 10 nM estrogen (filled light gray bar), 1 nM estrogen plus 100 nM of ICI 182,780 (gray bar) or with 100 nM ICI 182, 780 alone. As a control, the pGL4.0 promoterless vector was also transfected and stimulated with estrogen (n=3); *, statistical significance between promoter activity with 1 nM estrogen and the other conditions. (D). ICI 182,780 blocked estrogen stimulation of the −1461/+UTR construct in a dose-dependent manner (n=9). Values were fitted to a Hill function: (Luc/RLuc)-B=(max/1+([ICI]/[IC50])N) where [IC50] is the half maximal inhibitory concentration. Fitted values were IC50=0.74±0.12 nM and N=0.82±0.08. Values were normalized to the maximum value in 1 nM estrogen in the absence of ICI 182,780.
3.7. Prevention of E2-induced KCNE2 promoter activity by the specific E2 antagonist ICI 182,780
To further examine the action of E2, we investigated the effect of competitively displacing E2 from ERα by the highly competitive inhibitor ICI 182,780. We transfected the −1461/+UTR construct in HeLa cells (co-transfected with ERα) and in MCF7 cells. Cells were treated with 1 nM E2, 1 nM E2 plus 100 nM ICI 182,780, 100 nM ICI 182,780 alone, or no E2 to measure the basal promoter activity control. E2 activation was completely blocked with ICI 182,780 (100 nM) in both cell lines (Fig. 5B and C, compare black bar vs. gray bars). Treatment with only ICI 182,780 did not have any detectable effect (bar with hatched lines) on the basal activity (activity under no E2 stimulation; white bars). Furthermore, transfected HeLa cells treated with different doses of ICI 182,780 after 1 nM E2 stimulation showed that ICI 182,780 antagonized E2-mediated upregulation in a dose-dependent manner following a Hill equation with N=1 and IC50 of 0.74 nM. These results show that E2 binds to ERα to act upon the KCNE2 promoter, as E2 can be displaced in the presence of its antagonist ICI 182,780 in a dose-dependent manner.
3.8. Elucidation of the mechanism of E2 action
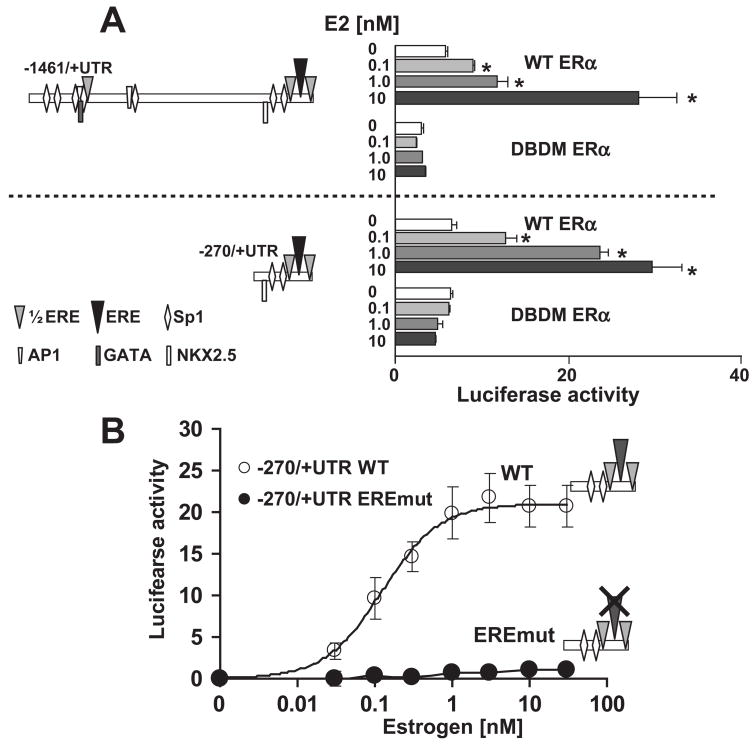
The fact that the −270/+UTR construct shows a robust E2 stimulation (Fig. 4C) suggests that the minimal E2 responsive sequences should be present within −270 nt of TSS. In this region, we have located one ERE-like sequence (−56GGgCAAGCTGACC−44), two ½ EREs and at least two Sp-factor binding sites. In addition, longer constructs, such as −1460/+UTR, had an even greater number of Sp-factor binding sites, along with AP-1 and other factors that could also mediate E2 responses. Therefore, it is possible that E2 activates these promoter regions via ERα direct binding, by indirect genomic action through another transcription factor, or both. To distinguish between the mechanisms of E2 action, we have initially used the well-characterized DNA-binding domain mutant of ERα (DBDM-ERα), which lacks the ability to bind to DNA but still can interact with other factors that eventually can up-regulate promoter activity (Jakacka et al., 2001). Thus, failure of promoter activation in the presence of this mutant would indicate that direct binding of ERα to the DNA is essential for KCNE2 promoter activation. Indeed, we observed that in the presence of DBDM-ERα, the −1461/+UTR and the −270/+UTR constructs were not stimulated by E2, whereas in the presence of wild type ERα, the KCNE2 promoter activity of both constructs is stimulated in a dose dependent manner (Fig 6A). These results strongly suggest that direct interaction of ERα is the predominant mechanism for KCNE2 promoter activation of both constructs.
Figure 6. E2 induced KCNE2 promoter activation.
(A), Comparison of estrogen responsiveness with WT and DBDM ERα in two different promoter constructs. Left, Maps of the constructs used. Right, Luciferase activities; * statistical significance between promoter activity with WT ERα and DBDM ERα. (B). Role of KCNE2-ERE element in estrogen responsiveness. Estrogen stimulated KCNE2 WT, but not KCNE2-ERE mutant, in a dose dependent manner (−270/+UTR constructs). Data of KCNE2 WT were fitted to a Hill function as in Fig. 4C with EC50=0.12±0.01 nM and N =~1.
As the −270/+UTR construct has the same robust E2 stimulation as the −1461/+UTR construct, we decided to determine whether ERα interacts with the quasi-perfect KCNE2 ERE element in the −270/+UTR construct. We took the site-directed mutagenesis approach to locate the sites within the promoter responsible for the E2-induced response. We disrupted both halves of the putative ERE (−56GGgCAAGCTGACC−44 to −56GAgTAAGCTAACA−44, the italicized and underlined nucleotides indicating the mutated bases). E2-stimulated activity of wild type (WT) and mutant constructs was compared in the presence of WT ERα. Figure 6B shows that E2 stimulation of the KCNE2 promoter was abolished upon disruption of the putative ERE. We have also mutated the other two ½ EREs present in this region, either individually or along with the quasi-perfect ERE. Individual ½ ERE mutations only had a small effect on E2 responsiveness requiring the mutation of the quasi-perfect ERE to abolish E2 responsiveness (data not shown). This data strongly argues that the quasi-perfect ERE in KCNE2 promoter mediates direct binding of ERα to the promoter.
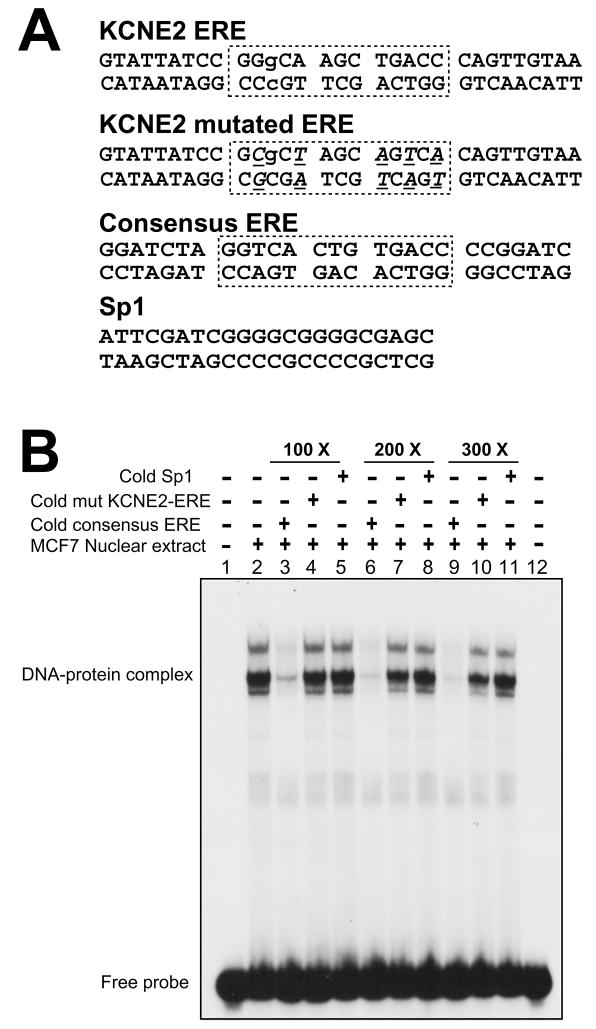
3.9. Physical interaction of ERα with the ERE
To further establish the direct interaction of ERα with the KCNE2 ERE, we examined whether ERα binds to this site using EMSA. Nuclear extract from E2-treated MCF7 cell line was used as the source of dimerized ERα. Duplexed DNA probe (KCNE2 ERE) and the competitor sequences (KCNE2 mutated ERE, Consensus ERE and Sp1) are shown in Fig. 7A. The EMSA in Fig. 7B shows that MCF7 cell nuclear extract incubated with the KCNE2-ERE probe formed three distinguishable DNA-protein complexes. This suggests that in MCF7 cells, additional proteins could interact with the DNA-ERα complex. Most importantly, all these signals were efficiently competed in a dose-dependent manner with the consensus ERE duplex (lanes 3, 6 and 9), but not with the mutant form of KCNE2-ERE (lanes 4, 7 and 10) or with the Sp1 consensus duplex (lanes 5, 8 and 11). These results conclusively demonstrate that ERα can directly bind to the KCNE2-ERE.
Figure 7.
Direct interaction of ERα with the ERE of the KCNE2 gene. (A), Duplex oligos used in EMSA. Boxes delineate the KCNE2 ERE in wild type, mutated and the perfect consensus ERE of the commercial oligo (Santa Cruz Biotechnology). In the KCNE2 mutated ERE, mutations are underlined in italics. Lowercase letters indicate the mismatch from the consensus ERE. (B), EMSA with nuclear extract of estrogen-treated MCF7 cells showing ERα binding with the KCNE2-ERE forming DNA-ERα complexes. Non-radioactive (cold) consensus ERE could compete with radiolabeled KCNE2-ERE for protein binding (lanes 3, 6 and 9). This competition is absent with mutated KCNE2-ERE (lanes 4, 7 and 10) and Sp1 duplex (lanes 5, 8 and 11). The dose dependence of the competition highlights the specificity of this reaction.
4. Discussion
4.1. E2 upregulation of cardiac KCNE2 transcripts
We hereby demonstrated for the first time the direct regulation of cardiac KCNE2 gene -an ion channel ancillary subunit- by E2. The observations that cardiac KCNE2 transcripts were upregulated in E2-primed mice and that ICI 182,780, which inhibits ER activity, reverses this upregulation is consistent with the fact that KCNE2 is an E2-responsive gene regulated by the ER. Our findings are in agreement with the view that E2 can regulate expression of the regulatory sulfonylurea receptor (SUR2A) subunit of cardiac KATP channels (Ranki et al., 2002a). Functional cardiac KATP channels have also shown to be more abundant in females than males; higher prevalence of KATP channels occurred mainly due to higher levels of the regulatory SUR2A, rather than the expression of the pore-forming inwardly rectifying K+ (Kir6.2) channels (Ranki et al., 2001). Aging was also associated with lower expression of functional cardiac KATP channels in females but not in males (Ranki et al., 2002b). These studies on the gender/aging differences in the expression levels of cardiac KATP further support the role of estrogen in heart ion channel regulation.
Among the KCNE family member genes (KCNE1 to 5), KCNE1 and KCNE2 are the 2 well-characterized cardiac ion channel β-subunit genes. These gene products are responsible for the Long QT syndrome (LQTS), an inherited cardiac arrhythmia. E2 also regulates KCNE1. Unlike KCNE2, cardiac KCNE1 transcript levels were downregulated by E2 treatment in OVX rabbits via an unknown mechanism (Drici et al., 1996). However, in contrast to the heart, in the uterus, KCNE1 transcripts are upregulated at the end of pregnancy and by E2 treatment (Boyle et al., 1987;Felipe et al., 1994;Folander et al., 1990;Pragnell et al., 1990) in rats. Therefore, regulation of KCNE1 by E2 may be tissue and/or species-dependent. In addition to E2, KCNE1 was also downregulated by DHT (Drici et al., 1996), whereas KCNE2 transcripts were not affected by DHT treatment in OVX mice. These observations suggest that different hormones in different tissues independently regulate each of the β-subunits. Since KCNE2 has a variety of functional roles in different tissues such as heart (Abbott et al., 1999) and stomach (Roepke et al., 2006) and also controls cancer cell proliferation (Yanglin et al., 2007), it would be important to determine the estrogen-induced, KCNE2-related physiological changes in these tissues.
4.2. KCNE2 TSSs and promoter
We have experimentally identified mouse KCNE2 TSSs and proposed its gene structure. KCNE2 has a TATA-containing promoter with several vicinal start sites. This is consistent with studies showing that many TATA-related genes can start at multiple sites (Suzuki et al., 2001). A single site of initiation has only been observed in genes that contain both a TATA box in combination with an initiator (Inr) sequence (Sandelin et al., 2007). Our inability to locate any Inr-like sequence around the identified KCNE2 TSSs further supports the previously described observations that transcription starts at multiple sites in the absence of a TATA-Inr combination. It has been postulated that TSS selection is not random, and that, although not a universal phenomenon, PyPu (−1+1) is usually preferred (Carninci et al., 2005). Our initial RACE (SMART) analysis identified a TSS (G, +3), which falls in this category (CG, −1+1). However, our second RACE reaction and primer extension, in addition to confirming this G, +3 site (though the signal was very faint in primer extension), showed the existence of another 2 TSSs (T, +1; G, −1). From the band intensity of the extended product of our primer extension analysis (Fig. 2D), it is clear that the TSS at T (+1) is predominant. Therefore, we have used this TSS as our reference point. It is worth mentioning that analysis of the gene sequence using the web-based software Eponine, FirstEF and Promoter2.0 failed to detect a promoter region immediately upstream of our experimental TSSs. Eponine and FirstEF rely on the presence of a CpG island for promoter prediction, but since KCNE2 promoter is not GC rich, these software tools could not detect the promoter precisely. Alternatively, Promoter2.0 relies on the presence of a TATA box for its promoter prediction algorithm. Since KCNE2 contains a TATA-like sequence that is not in full consensus with the TATA box sequence, Promoter2.0 was also unable to predict the actual promoter region of KCNE2, only predicting a promoter distal to our TSSs. This in-silico analysis highlights the relevance of experimentally defining the KCNE2 TSSs for the purpose of accurately locating the promoter region.
It is important to note that the promoter activity required the presence of the untranslated region (Fig. 4B). Many mammalian promoters are known to have this property (Smale & Kadonaga, 2003), which is attributed to the involvement of downstream cis-acting elements like DCE (downstream core element), MED-1 (multiple start element downstream) and Downstream Promoter Elements (DPE) (Ince & Scotto, 1995). Analysis of KCNE2 revealed the presence of two DPE at +8 to +12 and +84 to +88 that may play a significant role in promoter activity, although they are not located in their consensus positions (+28 to +32).
4.3. Mouse KCNE2 gene structure revisited: a novel exon and retention of intron generates mRNA diversity
Our analysis detected a 5′-UTR 1279 nt longer than previously reported, including a novel exon and a retained intron (Fig. 3C). In some mRNAs this intron was properly spliced out (Fig. 2B). Intron retention was observed in about 50% of KCNE2 mRNAs, both in heart and myometrium. Intron retention is a common type of alternative splicing, and in mammals it has been observed in many genes. In humans, intron retention has been reported to occur in up to 15% of the genes, with the majority of these occurrences taking place in the 5′-UTR (Galante et al., 2004b).
Based on our findings and the GenBank reference sequence (NM_134110), we have determined that the mouse KCNE2 gene contains at least 3 exons. Interestingly, sequences from mouse testis and retina point to the existence of an alternative 5′-UTR (GenBank BG261965 and BC022699), which predicts another exon in the intronic region between exons 1 and 2 (Fig. 3C, Exon ?). The potential presence of another promoter within this region remains to be studied. Nevertheless, our results and this data indicate that the KCNE2 gene can generate multiple mRNAs. This mRNA diversity could define translation efficiency and transport to the cytoplasm (Mansilla et al., 2005;Sakabe & de Souza, 2007).
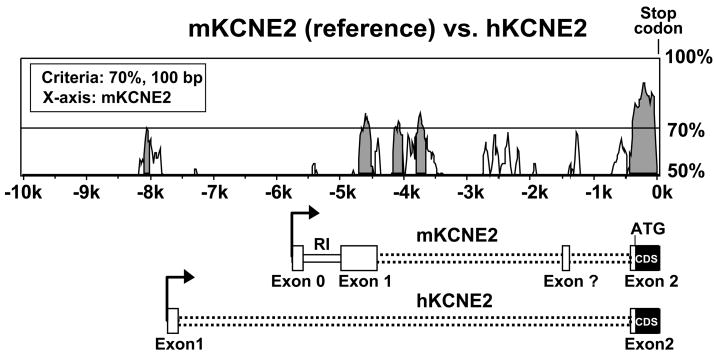
A close comparison of 20000 nt upstream of the KCNE2 coding sequences (CDS) of mouse (GeneID: 246133) and human (GeneID: 9992) genes revealed some homology, particularly between the TSSs and the stop codon, but displayed virtually no homology upstream of the TSS up to 20 kb, except around −8 kb (Fig. 8). As expected, the CDS contained in exon 2 are evolutionarily conserved, while the exon 1 sequences, which contribute to the 5′-UTR, are different. Furthermore, part of the mouse exon 1 is homologous to the human intronic sequences between exon 1 and 2. Thus, the newly identified exon 0 and promoter region is not well conserved between mouse and human, indicating that this gene may be regulated differentially between species.
Figure 8.
Homology plot of the mouse and human KCNE2 genes upstream sequences (−10000 of the stop codon). Analysis was performed using Vista browser and LAGAN algorithm. Gray regions indicate conserved sequences with at least 70% homology in 100 bp segments, mKCNE2 considered as reference. Corresponding regions in the mouse KCNE2 gene can be compared with the KCNE2 mRNA scheme. The KCNE2 mRNA scheme description is similar to Fig. 3C.
4.4. Mechanisms of KCNE2 gene genomic regulation by E2
Our transcript measurements revealed that KCNE2 is highly upregulated by E2. E2 can regulate gene expression via a direct genomic pathway, in which E2 binding to the ER α or β leads to the formation of homo/hetero dimers, which then may translocate to the nucleus and bind directly to a gene’s EREs (Klinge, 2001). Dimerized ER can also bind to multiple ½ EREs, sometimes concomitantly with binding to Sp1 sequences (O’Lone et al., 2004;Sanchez et al., 2002;Watanabe et al., 1998). Alternatively, E2 stimulates gene transcription via an indirect pathway in which E2 receptors can interact with transcription factors such as Sp1 and AP-1 (Bjornstrom & Sjoberg, 2005;Dong, Tsai-Morris, & Dufau, 2006;Nilsson et al., 2001). To investigate the molecular mechanisms involved in the E2-mediated upregulation of KCNE2 transcripts, promoter activity assays were performed using constructs of different fragments of the KCNE2 gene fused with a luciferase reporter gene. Our data revealed that the KCNE2 promoter was highly stimulated by E2. Subsequent sequence analysis of the KCNE2 promoter revealed the presence of an imperfect ERE in close proximity [−56GGgCAAGCTGACC−44] to the TSSs, along with 6 ½EREs within 3 kb upstream of the TSSs. This region also contains Sp1, AP-1, GATA and cEBP binding sites, which are known to be involved in E2 regulation either via a direct or indirect pathway. Because ICI 182,780 is known to activate E2-responsive promoters via Sp1 (Fleming et al., 2006), the results showing that ICI 182,780 alone has no effect (Fig. 5B and C) eliminate this potential pathway of E2 action. To distinguish the predominant mode of E2 action on KCNE2, we initially used the well-characterized DBDM-ERα (Jakacka et al., 2001), which cannot bind to DNA but still can interact with other factors. Our findings demonstrated that the E2 stimulation of KCNE2 occurred through a direct mode of action.
Our E2 responsiveness assays with promoter constructs of various lengths showed that the even the shortest −270/+UTR construct was significantly stimulated and that maximum activity was obtained with the −951/+UTR construct. However, the level of stimulation did not increase with increased promoter length up to −2978 nt (−2978/+UTR), which indicates that the minimal region required for the E2 stimulation resides within the −270 nt, which contains an imperfect ERE. It is known that ERE-mediated E2-responsiveness of a promoter also depends on the content of the promoter (Barkhem et al., 2002;Fleming et al., 2006). This could explain the increased E2-induced stimulation of the −951/+UTR construct when compared to the minimal responsive −270/+UTR construct. This occurrence may be explained by one or more of the following: 1) the presence of other ERE-like sequences (we have marked only consensus ½ EREs and the ERE with at least one consensus half site) that can bind the ER (Klinge, 2001), 2) the binding of other factors in the promoter region between −270 nt and −951 nt, which may aid in ERα-mediated stimulation, 3) favorable chromatin assembly in −951 nt construct after stimulation. In −1461/+UTR and −2978/+UTR constructs, the presence of inhibitory sequences further upstream may be responsible for lower stimulation efficiency compared to the −951/+UTR construct. Nevertheless, as our results indicated that −270/+UTR construct contained the minimal E2-responsive region, and as this region of the promoter also contains an imperfect ERE, we proceeded to test the role of this ERE in the E2 stimulation of KCNE2 transcripts. To do so, we have introduced a mutation which disrupted the ERE site and observed that this mutation completely abolished the E2-induced transcript upregulation of KCNE2. These results thus confirm that E2 stimulates the KCNE2 gene via a direct genomic mechanism, in which the ER binds directly to the ERE sequence. Our EMSA results further substantiated these findings, as binding of the ER to the KCNE2 ERE was efficiently competed with a consensus ERE. The mutant form of the KCNE2 ERE could partially compete at a very high concentration (Fig. 7, lines 7,9); this could be due to the fact that few nucleotides were mutated, and the remaining sequence has some residual binding capacity to ERα; alternatively, the surrounding sequences may also play a role in binding of ERα to the KCNE2 ERE (Hall, McDonnell, & Korach, 2002;Klinge et al., 2001).
4.5. Functional implications of E2-induced KCNE2 regulation of cardiac excitability
Here we have shown for the first time that cardiac KCNE2 transcripts are upregulated by E2 in mice. In expression systems, KCNE2 can modulate current kinetics of several Kv channel α-subunits like Kv4.2, Kv4.3, ERG1 (KCNH2) and KVLQT1(Abbott et al., 1999;McCrossan & Abbott, 2004). In particular, KCNE2 can potentiate current amplitude of Kv4.2/Kv4.3 (molecular correlates of Ito,f) channels (Deschenes & Tomaselli, 2002;Zhang, Jiang, & Tseng, 2001). We also found that KCNE2 targeting to the t-tubules is regulated by E2 and it favors KCNE2 association with Kv4.2/Kv4.3 (unpublished data).
E2 is known to have a cardioprotective effect, as it activates several protective signaling pathways(Kim et al., 2006;Patten et al., 2004;Patten & Karas, 2006;Pedram et al., 2008). On the other hand, QT interval is prolonged in high estrogenic conditions, such as in late pregnancy (Eghbali et al., 2005) and in E2-treated OVX mice via a downregulation of Ito,f and Ik,slow (Saito et al., 2008), which may favor arrhythmogenesis. It is important to address here the functional implication of the E2-induced regulation of KCNE2 (both of the transcript levels and of subcellular localization) on cardiac excitability. Upregulation of the KCNE2 transcripts, together with KCNE2 targeting to the t-tubules under high E2 conditions, would result in a higher interaction of KCNE2 with Kv4.3/Kv4.2 and therefore a potentiation of Ito,f currents. Thus, we speculate that E2-induced KCNE2 modulation will result in an increase in Ito,f currents, which may play a compensatory role by mitigating estrogen-induced prolongation of the QT interval. Further studies are however necessary to characterize the functional impact of KCNE2 association with other putative Kv α-subunits on cardiac excitability (Panaghie & Abbott, 2006).
In summary, we have delineated the mouse KCNE2 gene structure and showed that the KCNE2 gene belongs to the novel class of membrane proteins that are regulated by direct genomic action of estrogen. A better understanding of the mechanisms by which estrogen regulates expression of KCNE2 will provide novel means of controlling cell function under healthy and diseased conditions.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- Barkhem T, Haldosen LA, Gustafsson JA, Nilsson S. Transcriptional synergism on the pS2 gene promoter between a p160 coactivator and estrogen receptor-alpha depends on the coactivator subtype, the type of estrogen response element, and the promoter context. Mol Endocrinol. 2002;16:2571–2581. doi: 10.1210/me.2002-0051. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- Boyle MB, Azhderian EM, Maclusky NJ, Naftolin F, Kaczmarek LK. Xenopus oocytes injected with rat uterine RNA express very slowly activating potassium currents. Science. 1987;235:1221–1224. doi: 10.1126/science.2434999. [DOI] [PubMed] [Google Scholar]
- Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Grosse I, Zhang MQ. Computational identification of promoters and first exons in the human genome. Nat Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- Deschenes I, Tomaselli GF. Modulation of Kv4.3 current by accessory subunits. FEBS Lett. 2002;528:183–188. doi: 10.1016/s0014-5793(02)03296-9. [DOI] [PubMed] [Google Scholar]
- Dong J, Tsai-Morris CH, Dufau ML. A novel estradiol/estrogen receptor alpha-dependent transcriptional mechanism controls expression of the human prolactin receptor. Journal of Biological Chemistry. 2006;281:18825–18836. doi: 10.1074/jbc.M512826200. [DOI] [PubMed] [Google Scholar]
- Down TA, Hubbard TJ. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drici MD, Burklow TR, Haridasse V, Glazer RI, Woosley RL. Sex hormones prolong the QT interval and downregulate potassium channel expression in the rabbit heart. Circulation. 1996;94:1471–1474. doi: 10.1161/01.cir.94.6.1471. [DOI] [PubMed] [Google Scholar]
- Eghbali M, Deva R, Alioua A, Minosyan TY, Ruan H, Wang Y, Toro L, Stefani E. Molecular and functional signature of heart hypertrophy during pregnancy. Circ Res. 2005;96:1208–1216. doi: 10.1161/01.RES.0000170652.71414.16. [DOI] [PubMed] [Google Scholar]
- Felipe A, Knittle TJ, Doyle KL, Snyders DJ, Tamkun MM. Differential expression of Isk mRNAs in mouse tissue during development and pregnancy. Am J Physiol. 1994;267:C700–705. doi: 10.1152/ajpcell.1994.267.3.C700. [DOI] [PubMed] [Google Scholar]
- Fleming JG, Spencer TE, Safe SH, Bazer FW. Estrogen regulates transcription of the ovine oxytocin receptor gene through GC-rich SP1 promoter elements. Endocrinology. 2006;147:899–911. doi: 10.1210/en.2005-1120. [DOI] [PubMed] [Google Scholar]
- Folander K, Smith JS, Antanavage J, Bennett C, Stein RB, Swanson R. Cloning and expression of the delayed-rectifier ISK channel from neonatal rat heart and diethylstilbestrol-primed rat uterus. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:2975–2979. doi: 10.1073/pnas.87.8.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004a;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004b;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda RM, Khan IA, Mehta NJ, Vasavada BC, Sacchi TJ. Cardiac arrhythmias in pregnancy: clinical and therapeutic considerations. International Journal of Cardiology. 2003;88:129–133. doi: 10.1016/s0167-5273(02)00601-0. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Diman NY, McDermott DA, Basson CT. Transcription factor cascades in congenital heart malformation. Trends Mol Med. 2003;9:512–515. doi: 10.1016/j.molmed.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. 3. Sinauer; 2001. [Google Scholar]
- Ince TA, Scotto KW. A conserved downstream element defines a new class of RNA polymerase II promoters. Journal of Biological Chemistry. 1995;270:30249–30252. doi: 10.1074/jbc.270.51.30249. [DOI] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. Journal of Biological Chemistry. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- Jiang M, Zhang M, Tang DG, Clemo HF, Liu J, Holwitt D, Kasirajan V, Pond AL, Wettwer E, Tseng GN. KCNE2 protein is expressed in ventricles of different species, and changes in its expression contribute to electrical remodeling in diseased hearts. Circulation. 2004;109:1783–1788. doi: 10.1161/01.CIR.0000124225.43852.50. [DOI] [PubMed] [Google Scholar]
- Kim JK, Pedram A, Razandi M, Levin ER. Estrogen prevents cardiomyocyte apoptosis through inhibition of reactive oxygen species and differential regulation of p38 kinase isoforms. Journal of Biological Chemistry. 2006;281:6760–6767. doi: 10.1074/jbc.M511024200. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM, Jernigan SC, Smith SL, Tyulmenkov VV, Kulakosky PC. Estrogen response element sequence impacts the conformation and transcriptional activity of estrogen receptor alpha. Mol Cell Endocrinol. 2001;174:151–166. doi: 10.1016/s0303-7207(01)00382-3. [DOI] [PubMed] [Google Scholar]
- Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- Kundu P, Alioua A, Stefani E, Toro L. Regulation of mouse slo gene expression: Multiple promoters, transcription start sites and genomic action of estrogen. Journal of Biological Chemistry. 2007 doi: 10.1074/jbc.M704777200. [DOI] [PubMed] [Google Scholar]
- Li T, Li YM, Jia ZQ, Chen P, Ma KT, Zhou CY. Carboxyl terminus of Nkx2.5 impairs its interaction with p300. Journal of Molecular Biology. 2007;370:976–992. doi: 10.1016/j.jmb.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Lue NF, Kornberg RD. A possible role for the yeast TATA-element-binding protein in DNA replication. Proc Natl Acad Sci USA. 1993;90:8018–8022. doi: 10.1073/pnas.90.17.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist AL, Turner CL, Ballester LY, George AL., Jr Expression and transcriptional control of human KCNE genes. Genomics. 2006;87:119–128. doi: 10.1016/j.ygeno.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Mansilla A, Lopez-Sanchez C, de la Rosa EJ, Garcia-Martinez V, Martinez-Salas E, de PF, Hernandez-Sanchez C. Developmental regulation of a proinsulin messenger RNA generated by intron retention. EMBO Rep. 2005;6:1182–1187. doi: 10.1038/sj.embor.7400539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. Transcriptional Regulatory Elements in the Human Genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- McCrossan ZA, Abbott GW. The MinK-related peptides. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Moller C, Netzer R. Effects of estradiol on cardiac ion channel currents. Eur J Pharmacol. 2006;532:44–49. doi: 10.1016/j.ejphar.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- O’Lone R, Frith MC, Karlsson EK, Hansen U. Genomic Targets of Nuclear Estrogen Receptors. Mol Endocrinol. 2004 doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- O’Mahony F, Alzamora R, Betts V, LaPaix F, Carter D, Irnaten M, Harvey BJ. Female gender-specific inhibition of KCNQ1 channels and chloride secretion by 17beta-estradiol in rat distal colonic crypts. Journal of Biological Chemistry. 2007;282:24563–24573. doi: 10.1074/jbc.M611682200. [DOI] [PubMed] [Google Scholar]
- Panaghie G, Abbott GW. The impact of ancillary subunits on small-molecule interactions with voltage-gated potassium channels. Curr Pharm Des. 2006;12:2285–2302. doi: 10.2174/138161206777585175. [DOI] [PubMed] [Google Scholar]
- Patten RD, Karas RH. Estrogen replacement and cardiomyocyte protection. Trends Cardiovasc Med. 2006;16:69–75. doi: 10.1016/j.tcm.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Patten RD, Pourati I, Aronovitz MJ, Baur J, Celestin F, Chen X, Michael A, Haq S, Nuedling S, Grohe C, Force T, Mendelsohn ME, Karas RH. 17beta-estradiol reduces cardiomyocyte apoptosis in vivo and in vitro via activation of phospho-inositide-3 kinase/Akt signaling. Circ Res. 2004;95:692–699. doi: 10.1161/01.RES.0000144126.57786.89. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Lubahn D, Liu J, Vannan M, Levin ER. Estrogen Inhibits Cardiac Hypertrophy: Role of Estrogen Receptor Beta to Inhibit Calcineurin. Endocrinology. 2008 doi: 10.1210/en.2008-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman TF, Jr, Yutzey KE. T-box genes and heart development: putting the “T” in heart. Dev Dyn. 2005;232:11–20. doi: 10.1002/dvdy.20201. [DOI] [PubMed] [Google Scholar]
- Pragnell M, Snay KJ, Trimmer JS, Maclusky NJ, Naftolin F, Kaczmarek LK, Boyle MB. Estrogen induction of a small, putative K+ channel mRNA in rat uterus. Neuron. 1990;4:807–812. doi: 10.1016/0896-6273(90)90207-v. [DOI] [PubMed] [Google Scholar]
- Ranki HJ, Budas GR, Crawford RM, Davies AM, Jovanovic A. 17Beta-estradiol regulates expression of K(ATP) channels in heart-derived H9c2 cells. J Am Coll Cardiol. 2002a;40:367–374. doi: 10.1016/s0735-1097(02)01947-2. [DOI] [PubMed] [Google Scholar]
- Ranki HJ, Budas GR, Crawford RM, Jovanovic A. Gender-specific difference in cardiac ATP-sensitive K(+) channels. J Am Coll Cardiol. 2001;38:906–915. doi: 10.1016/s0735-1097(01)01428-0. [DOI] [PubMed] [Google Scholar]
- Ranki HJ, Crawford RM, Budas GR, Jovanovic A. Ageing is associated with a decrease in the number of sarcolemmal ATP-sensitive K+ channels in a gender-dependent manner. Mech Ageing Dev. 2002b;123:695–705. doi: 10.1016/s0047-6374(01)00415-8. [DOI] [PubMed] [Google Scholar]
- Roepke TK, Kontogeorgis A, Xu X, Young J, Purtell K, Gutstein D, Lerner DJ, Abbott GW. The role of MiRP1 (KCNE2) in murine ventricular repolarization. Biophysical Journal 1751-Platform 2008 [Google Scholar]
- Roepke TA, Malyala A, Bosch MA, Kelly MJ, Ronnekleiv OK. Estrogen regulation of genes important for K+ channel signaling in the arcuate nucleus. Endocrinology. 2007;148:4937–4951. doi: 10.1210/en.2007-0605. [DOI] [PubMed] [Google Scholar]
- Roepke TK, Anantharam A, Kirchhoff P, Busque SM, Young JB, Geibel JP, Lerner DJ, Abbott GW. The KCNE2 potassium channel ancillary subunit is essential for gastric acid secretion. Journal of Biological Chemistry. 2006;281:23740–23747. doi: 10.1074/jbc.M604155200. [DOI] [PubMed] [Google Scholar]
- Saito T, Ciobotaru A, Toro L, Stefani E, Eghbali M. Gender and Estrogen Determines Outward Potassium Current Densities in Mouse Ventricular Myocytes. Biophysical Journal. 2008 2988-Pos. [Google Scholar]
- Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Nguyen D, Rocha W, White JH, Mader S. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays. 2002;24:244–254. doi: 10.1002/bies.10066. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Schug J. Current protocols in Bioinformatics. Wiley Interscience; 2003. Using TESS to Predict Transcription Factor Binding Sites in DNA Sequence; pp. 1–16. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Song M, Helguera G, Eghbali M, Zhu N, Zarei MM, Olcese R, Toro L, Stefani E. Remodeling of Kv4.3 Potassium Channel Gene Expression under the Control of Sex Hormones. Journal of Biological Chemistry. 2001;276:31883–31890. doi: 10.1074/jbc.M101058200. [DOI] [PubMed] [Google Scholar]
- Song M, Zhu N, Olcese R, Barila B, Toro L, Stefani E. Hormonal control of protein expression and mRNA levels of the MaxiK channel alpha subunit in myometrium. FEBS Lett. 1999;460:427–432. doi: 10.1016/s0014-5793(99)01394-0. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Taira H, Tsunoda T, Mizushima-Sugano J, Sese J, Hata H, Ota T, Isogai T, Tanaka T, Morishita S, Okubo K, Sakaki Y, Nakamura Y, Suyama A, Sugano S. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2001;2:388–393. doi: 10.1093/embo-reports/kve085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi T, Ohkubo H, Nakanishi S. Cloning of a membrane protein that induces a slow voltage-gated potassium current. Science. 1988;242:1042–1045. doi: 10.1126/science.3194754. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Inoue S, Hiroi H, Orimo A, Kawashima H, Muramatsu M. Isolation of estrogen-responsive genes with a CpG island library. Mol Cell Biol. 1998;18:442–449. doi: 10.1128/mcb.18.1.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Tsang SY, Yao X, Chan FL, Huang Y. Differential effects of estrogen and progesterone on potassium channels expressed in Xenopus oocytes. Steroids. 2008;73:272–279. doi: 10.1016/j.steroids.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Xin M, Davis CA, Molkentin JD, Lien CL, Duncan SA, Richardson JA, Olson EN. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc Natl Acad Sci USA. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanglin P, Lina Z, Zhiguo L, Na L, Haifeng J, Guoyun Z, Jie L, Jun W, Tao L, Li S, Taidong Q, Jianhong W, Daiming F. KCNE2, a down-regulated gene identified by in silico analysis, suppressed proliferation of gastric cancer cells. Cancer Lett. 2007;246:129–138. doi: 10.1016/j.canlet.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Zhang M, Jiang M, Tseng GN. minK-related peptide 1 associates with Kv4.2 and modulates its gating function: potential role as beta subunit of cardiac transient outward channel? Circ Res. 2001;88:1012–1019. doi: 10.1161/hh1001.090839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.